Highlights
-
•
The regional pattern of cerebral alterations in PLS includes the area III of the CC.
-
•
Callosal alterations of the texture parameters entropy and homogeneity were shown in PLS.
-
•
Texture and macrostructure of the callosal area III is targeted as a neuroimaging marker in PLS.
Keywords: Amyotrophic lateral sclerosis, Corpus callosum, Magnetic resonance imaging, Texture analysis, Motor neuron disease, Primary lateral sclerosis
Abstract
Background
The regional distribution of cerebral morphological alterations in primary lateral sclerosis (PLS) is considered to include the area III of the corpus callosum (CC).
Objective
The study was designed to investigate regional white matter (WM) alterations in the callosal area III by T1 weighted magnetic resonance imaging (T1w-MRI) data in PLS patients compared with healthy controls, in order to identify atrophy and texture changes in vivo.
Methods
T1w-MRI-based white matter mapping was used to perform an operator-independent CC-segmentation for the different areas of the CC in 67 PLS patients vs 82 matched healthy controls and vs 85 ALS patients. The segmentation was followed by texture analysis of the separated CC areas for the PLS patients vs controls and vs ALS patients.
Results
PLS was associated with significant atrophy in the area III of the CC (but not in the other callosal segments), while the alterations in the ALS patients were much more variable and were not significant at the group level. Furthermore, significant regional alterations of the texture parameters entropy and homogeneity in this area were shown in PLS patients and in ALS patients.
Conclusions
This T1w-MRI study demonstrated focused regional CC atrophy and texture alterations limited to the callosal area III (which comprises fibers projecting into the primary motor cortices) in PLS, in comparison to a higher variability in CC size in ALS.
1. Introduction
Involvement of the corpus callosum (CC) can be regarded as a consistent feature of motor neuron disease (MND), independent of clinical upper motor neuron involvement, and may reflect bilateral cortical involvement or interhemispheric spread of pathology (Filippini et al., 2010). Callosal involvement during the course of amyotrophic lateral sclerosis (ALS) has been demonstrated in several studies (Filippini et al., 2010; Müller et al., 2012, 2016). This finding is primarily based on diffusion tensor imaging (DTI) studies with fractional anisotropy (FA) reduction or radial diffusivity (RD) increase in the corpus callosum, extending rostrally and bilaterally to the region of the primary motor cortices (Kassubek et al., 2012). The decrease in CC white matter integrity in patients with MND has been localized to motor-related areas (Chapman et al., 2014), corresponding to segment III according to Hofer and Frahm who parcellated the corpus callosum by introducing vertical subdivisions with respect to the outcome of the fiber tractography as a modification of the Witelson scheme (Hofer and Frahm, 2006; Witelson 1989). Beyond these results by advanced neuroimaging sensitive to microstructural changes, also routine structural MRI in MND can demonstrate high T2 signal or callosal atrophy at individual level, especially in motor related areas (Riad et al., 2011; Osborn et al., 2016). Although visual inspection of the motor segment in individual patients in routine MRI suggests local atrophy, macro-structural imaging studies showed conflicting results with studies reporting local atrophy of this region on the one hand (Yamauchi et al., 1995) and studies reporting no gross size differences of the CC compared to controls on the other hand (Chapman et al., 2012). Therefore, in the current study, the focus is set to an MND with a comparatively long disease duration where hypothetically structural alterations could take place during the long lasting course of the disease: Primary lateral sclerosis (PLS) is considered to be a motor neuron disease (MND) which almost exclusively affects upper motor neurons (UMN) (Singer et al., 2007; Wais et al., 2017) as one of the restricted phenotypes of ALS (Ludolph et al., 2015). ALS and PLS share advanced neuroimaging features including WM alterations as assessed by DTI (Agosta et al., 2014), and PLS patients were also investigated by tract-of-interest-based DTI, demonstrating significant alterations in the ALS-related tract systems (Müller et al., 2018A; Müller et al., 2018B). ALS and PLS patients exhibit also further brain alterations, such as considerable brainstem atrophy (Bede et al., 2019). Given that PLS mainly affects the UMN and that patients are generally more prone to be investigated by MRI in later disease due to the longer disease durations compared to ´classical´ ALS, changes in the corpus callosum by DTI measures have been addressed in several studies (Kolind et al., 2013; Agosta et al., 2014), but regional atrophy (of the segment III) as assessed by volumetric techniques has not been systematically investigated yet.
In the current study, involvement of the CC in PLS was investigated with the aim to identify a potential imaging marker, based on standard T1-weighted (T1w) MRI recordings, in order to challenge previous findings by an objective parametrization.
2. Methods
2.1. Subjects and patient characteristics
Sixty-seven PLS patients were included who met the proposed diagnostic criteria for PLS (Pringle et al., 1992; Gordon et al., 2006; Singer et al., 2007). All PLS patients underwent standardized clinical and neurological examinations: to be eligible, subjects had to meet the following criteria: no family history of MND, no clinical diagnosis of frontotemporal dementia (FTD), age at onset > 40 years, no mutations of major genes related to hereditary spastic paraparesis if investigated, no other major systemic, psychiatric or neurological illnesses, no history of substance abuse; further mandatory criteria for inclusion were negative tests for multiple sclerosis and for central nervous system infections, and routine MRI scans excluded any brain or cervical cord abnormalities suggesting a different etiology of the clinical symptoms. All subjects gave written informed consent for the study protocol according to institutional guidelines which had been approved by the Ethics Committee of Ulm University, Germany (No. 19/12).
The PLS patient group (31 m/36f, age 60 ± 10 years) was compared both with a group of 82 age- and gender-matched controls (40 m/42f, age 62 ± 14 years) and with a group of 85 patients with ALS (38 m/47f, age 60 ± 10 years (Table 1). Out of the 82 healthy controls, 21 controls obtained a second T1w scan within a follow-up period of ΔT = 10 months in average.
Table 1.
Subjects' characteristics. ALS-FRS-R – revised ALS functional rating scale.
| PLS | ALS | controls | p | |
|---|---|---|---|---|
| male/female | 31/36 | 47/38 | 40/42 | Kruskal-Wallis: 0.60 |
| age/years (mean ± std. dev.) | 60 ± 10, range (38–81) | 60 ± 10, range (35–76) | 62 ± 14, range (20–82) | Kruskal-Wallis: 0.54 |
| ALS-FRS-R (mean ± std. dev.) | 38 ± 7, range (16–45) | 40 ± 7, range (15–46) | – | t-test: 0.15 |
| disease duration / years (mean ± std. dev.) | 5 ± 3 range (3–21) median 3 years | 1 ± 1 range (0–3) median 1 year | – | t-test: <0.0001 |
2.2. MRI acquisition
MRI scanning was performed on a 1.5 Tesla Magnetom Symphony (Siemens Medical, Erlangen, Germany); the study protocol consisted of a T1w scan (magnetization-prepared rapid gradient-echo – MPRAGE) with 144 slices, 256 × 256 pixels, slice thickness 1.2 mm, pixel size 1.0 mm × 1.0 mm); the echo time (TE) and repetition time (TR) were 4.2 ms and 1640 ms, respectively.
Out of the 67 patients with PLS, 40 patients additionally obtained a diffusion tensor imaging (DTI) scan (Müller et al., 2018B). DTI study protocol A (13 PLS patients) consisted of 13 volumes (45 slices, 128 × 128 pixels, slice thickness 2.2 mm, pixel size 1.5 mm × 1.5 mm) representing 12 gradient directions (b = 800 s/mm2), and one scan with gradient 0 (b = 0). The echo time (TE) and repetition time (TR) were 93 ms and 8000 ms, respectively. Five scans were averaged online by the scanner software in image space. DTI study protocol B (27 PLS patients) consisted of 52 volumes (64 slices, 128 × 128 pixels, slice thickness 2.8 mm, pixel size 2.0 mm × 2.0 mm), representing 48 gradient directions (b = 1000 s/mm2), and four scans with b = 0. TE and TR were 95 ms and 8000 ms. For details of the DTI analysis cascade, please refer to (Müller et al., 2018B).
2.3. Data analysis
The pre- and postprocessing was performed by use of the analysis software Tensor Imaging and Fiber Tracking (TIFT – Müller et al., 2007) which was initially developed for DTI analysis and now has been extended by a new package for CC texture analysis. The automated analysis procedure included the following main processing steps:
(i) isometric and affine alignment to the anterior commissure/posterior commissure line, (ii) adjustment of intensity threshold to automatically segment the CC, (iii) subdivision of the CC into areas I–V according to the Hofer and Frahm scheme (Hofer and Frahm, 2006), (iv) calculation of area sizes and texture parameters entropy, homogeneity, kurtosis, and skewness, (v) statistical comparison at the group level.
2.3.1. Isometric and affine alignment of T1w data to the AC/PC line
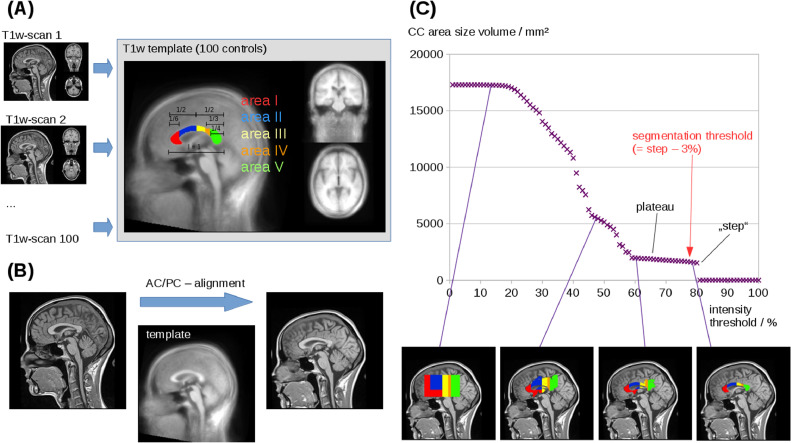
Alignment of T1w data was performed iteratively: in a first step, AC and PC were manually defined in 100 data sets of healthy controls and a T1w template was created by arithmetic averaging of all data sets (Fig. 1A). In a second step, T1w data of all study participants were fit to the T1w template by a conjugated simplex fitting procedure (Fig. 1B).
Fig. 1.
Procedure for the fully automated CC segmentation and texture analysis. (A) Alignment of T1w data: AC and PC were manually defined in 100 data sets of healthy controls so that a T1w template could be created by arithmetic averaging of all data sets. (B) T1w data of all study participants were fit to the T1w template. (C) The intensity threshold was varied from 100% to 0% of maximum intensity and the area size of the midsagittal CC plane was calculated. Beginning with an area size of 0 mm2 at threshold 100%, the area size of the CC plane remains stable at 0 mm2 during the initial decrease of the intensity threshold until a “step” occurs between about 80% and 60% of maximum intensity; the intensity threshold for CC segmentation was then defined as this “step”-threshold minus 3% (red arrow). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3.2. Adjustment of intensity threshold to automatically segment the midsagittal CC plane
In order to automatically segment the CC, the following procedure was applied: the intensity threshold was varied from 100% to 0% of maximum intensity and the area size of the midsagittal CC plane was calculated. Starting with an area size of 0 mm2 at threshold 100%, the size of the midsagittal plane remains at 0 mm2 during the initial decrease of the intensity threshold until a “step” occurs (see Fig. 1C) and the CC plane size shows a plateau; the intensity threshold for CC segmentation was then defined as this “step”-threshold minus 3% in order to obtain stability of the results (Fig. 1C). Supplementary Figure 1 shows the reproducibility of this procedure in two T1w scans of the same control subject, recorded at a time interval of 6 months.
2.3.3. Subdivision of the CC into areas I–V
According to the Hofer and Frahm scheme (Hofer and Frahm, 2006) based on the scheme by Witelson (Witelson, 1989), the segmented CC was subdivided into areas I – V. To this end, the CC was split into areas I, II and areas III-V at the half of the total length of the CC. One third of the anterior half is defined as area 1, the remaining 2/3 of the anterior half is area II; 1/3 of the posterior half is defined as area III, the remaining posterior part is divided by 1/3 for area IV and 2/3 to area V.
2.3.4. Calculation of area sizes and texture parameters
Area sizes were calculated by counting the voxels of the segmented area. In order to obtain increased stability of the results, the areas were calculated in the three central sagittal slices and the arithmetic average was calculated. In order to account for individual CC sizes, CC areas I – V were then calculated as percent of total CC size. The intensity values for the segmented CC were z-normalized to µ = 0 and σ = 1. Texture alterations in PLS and ALS patients have already been detected by DTI metrics (especially fractional anisotropy – FA) for the corticospinal tract, the CC, and further brain areas. The loss of fiber integrity as indicated by FA reduction has been shown in a DTI-based study in ALS patients (Chapman et al., 2014). These microstructural alterations could hypothetically also be reflected by textural changes in morphological T1w scans. A loss of fiber integrity could lead to a more inhomogeneous distribution of voxel intensity values in the respective brain areas. To assess these features, texture parameters (Stockman and Shapiro, 2001) that measure the intensity distribution could be used; entropy, skewness, and kurtosis measure the global distribution of intensity values in a certain area, whereas the homogeneity measures the intensity alterations in the direct vicinity of a voxel. Thus, a loss of fiber tract integrity could lead to a wider range of intensity distribution, and also to a more inhomogeneous pattern in the respective brain areas. These features could be assessed by the following texture parameters:
The entropy M(x) is defined as the average amount of information produced by a stochastic source of data. It is a measure of randomness of gray level distribution; structures with larger number of grey levels have larger entropy
| (1) |
where I is the pixel intensity and P the probability of a pixel with intensity I.
The skewness S(x) is a measure of asymmetry of data around sample mean:
| (2) |
The kurtosis K(x) is a measure of sharpness of peak of the frequency distribution curve:
| (3) |
The (in)homogeneity H(x) is a measure of the differences of voxel intensities of neighbored voxels:
| (4) |
Nd(i,j) is the intensity difference of a voxel i to its neighbors j.
Note that all texture parameters were calculated independently for all areas, i.e. for each area µ = 0 and σ = 1 is not valid (in contrast to µ = 0 and σ = 1 for the whole CC).
2.3.5. CC segmentation and planimetry by atlas-based volumetry (ABV)
As a reference analysis for segmentation and planimetry of the CC, standardized Atlas-Based Volumetry (ABV) (Huppertz et al., 2010) was used. The method is fully automated and based on algorithms of SPM (Wellcome Department of Imaging Neuroscience, London, UK; http://www.fil.ion.ucl.ac.uk/spm) for image segmentation into grey matter and white matter (WM) compartments and for normalization to Montreal Neurological Institute (MNI) stereotaxic space. For the purpose of the current study, the area sizes of the midsagittal CC plane and its segments I–V according to the definition by Hofer and Frahm (Hofer and Frahm, 2006) were measured. To this end, the midsagittal CC plane was determined in MNI space by multiplication of the segmented cerebral WM compartment with a midsagittal plane. The CC segments within this plane were delineated by an automated parcellation procedure coded in MATLAB® script and following the description of the CC parcellation by Hofer and Frahm (cf. chapter 2.3.3). Due to modulation of the WM images, the effect of normalization (i.e., extension or shrinkage of the investigated structures) was compensated for so that the area sizes were determined for the midsagittal CC plane and its segments in native space.
2.3.6. Statistical comparison at the group level
Since the subject groups were large enough to show a Gaussian distribution of parameters, Student's t-test was used for comparison at the group level – a non-parametric Mann-Whitney-U test revealed similar results. To investigate associations between different parameters, scatter plots were used and Pearson correlation coefficient was calculated for the areas sizes and for the texture parameters of the different areas I – V. All statistical tests were performed at a unified significance level of α = 0.01 (2-tailed), p-values were considered as significant (< 0.01) or highly significant (< 0.001). Due to the exploratory nature of the trial, all outcomes of statistical tests must be interpreted as hypotheses generating.
3. Results
3.1. Reproducibility of the results from two independent scans of the same subject
Twenty-one healthy controls obtained a follow-up T1w scan which was used to test the analysis results for reproducibility. In summary, absolute averaged differences between the two scans, arithmetically averaged for the areas I – V, were 1.6% for area size (values were calculated as portion of the total midsagittal CC area size), 1.0% for entropy, 1.7% for homogeneity, 4.2% for kurtosis, and 3.5% for skewness. Details of the reproducibility analysis are summarized in Supplementary Table 1.
3.2. Atrophy of CC area III in patients with PLS in comparison with ALS and controls
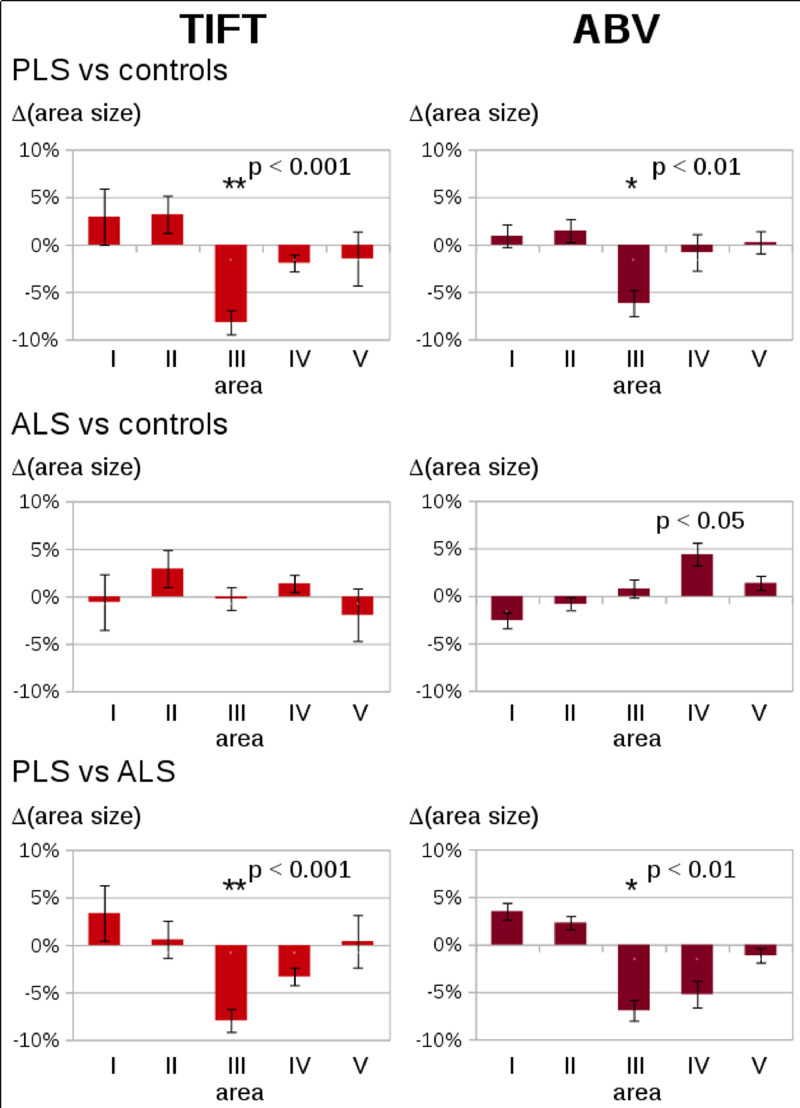
Planimetry results for the global CC showed no significant size alterations between the subject groups (PLS, ALS, and controls – Supplementary Figure 2). Planimetry results for the different CC areas in 67 patients with PLS, 85 patients with ALS, and 82 healthy controls at the group level are summarized in Supplementary Figure 3. A significant reduction in size (p < 0.01, corrected for multiple comparisons) in PLS patients vs controls and in PLS patients vs ALS patients was observed for area III, while no significant differences for PLS patients vs controls and for PLS patients vs ALS patients were observed in the other areas I, II, IV, V (Fig. 2, left). The deviation of about 8% (atrophy of area III in PLS patients compared to healthy controls and to ALS patients, respectively) is markedly larger than the deviation calculated from the reproducibility scans (see Section 3.1.). These results could be confirmed by the ABV analysis which also demonstrated a significant reduction in area size for area III (p < 0.01, corrected for multiple comparisons), while no significant differences between PLS patients and controls and for PLS patients vs ALS patients were observed in the other areas I, II, IV, V (Fig. 2, right). Correlation analysis of two different methodological approaches, i.e., TIFT results (area sizes in% of total midsagittal CC area size) and ABV results revealed a high significance of p < 0.001 for all areas.
Fig. 2.
Differences in area sizes for CC areas I-V (in% of the total CC area size) in patients with PLS and ALS as compared to healthy controls, determined by two different methods (i.e., TIFT and ABV). The figure displays the average difference with the standard error of the mean as error bars – left: Analysis by TIFT, right: analysis by ABV. * p < 0.01; ** p < 0.001.
In the PLS patients, a correlation (p = 0.02) was observed for ALS-FRS-R with area size of area III (in percent of total midsagittal CC area size); no association was found for disease duration. For PLS and ALS patients together, a correlation (p = 0.03) was found for ALS-FRS-R to area size of area III; area size of area III was highly significant correlated to disease duration (p < 0.001).
3.3. Alterations of texture of the CC in patients with PLS in comparison with ALS and controls
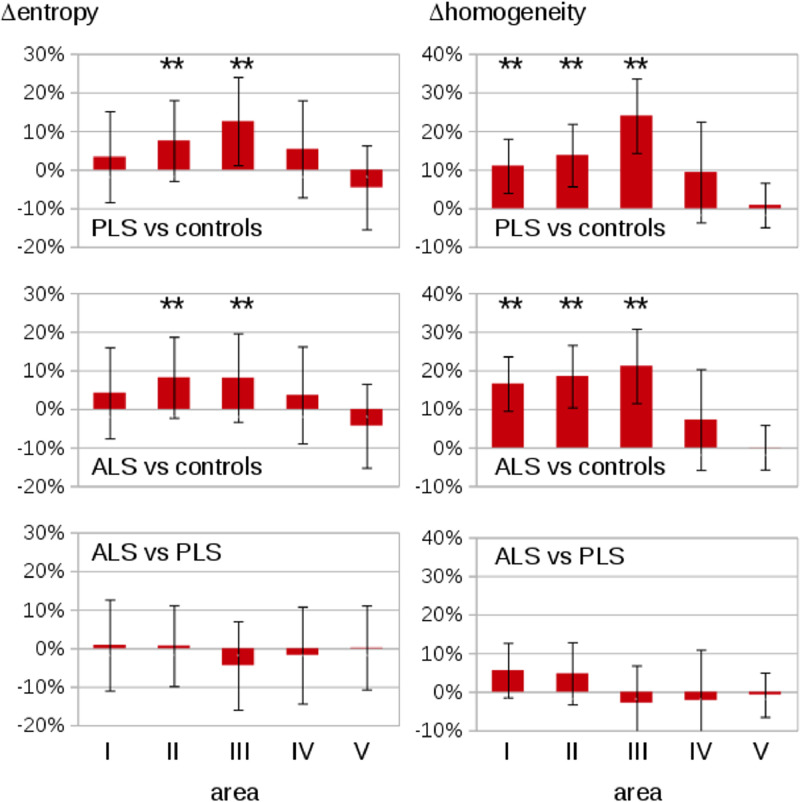
Texture analysis for the global CC showed no significant alteration between the subject groups (PLS, ALS, and controls – Supplementary Figure 2). The texture parameters for 67 patients with PLS, 85 patients with ALS, and 82 healthy controls at the group level are summarized in Supplementary Figure 3. PLS patients and ALS patients showed highly significantly increased entropy in the texture of CC area II (p < 0.001) and area III (p < 0.001) compared to controls, while areas I, IV, and V show no significant differences (Fig. 3 left panel). PLS patients and ALS patients demonstrated increased inhomogeneity in CC area I (p < 0.001), area II (p < 0.001), and area III (p < 0.001) compared to age-matched controls, while areas IV and V showed no significant differences (Fig. 3 right panel). Correlations for PLS patients were observed between texture parameters (entropy p = 0.01; homogeneity p = 0.02) of CC area III and ALS-FRS-R; for the whole group of PLS and ALS patients together, significant correlations were found for entropy in the texture of CC area III and ALS-FRS-R (p < 0.01) and homogeneity and ALS-FRS-R (p < 0.01). No association was found for disease duration.
Fig. 3.
Texture results for CC areas I-V in the patients with PLS and ALS as compared to healthy controls. PLS and ALS patients show an increased entropy in texture of CC area II (**) and area III (**) compared to matched controls. Furthermore, PLS and ALS patients show an increased inhomogeneity in texture of CC area I (**), area II (**), and area III (**) compared to matched controls. error bars are the standard error of the mean, * p < 0.01; ** p < 0.001.
No significant differences between PLS patients and controls and between ALS patients and controls were found for the texture parameters skewness and kurtosis. The texture parameters are independent from the number of analyzed voxels, i.e., the area III atrophy was not associated with texture parameters.
3.4. Receiver operating characteristics (ROC) curves of texture parameters
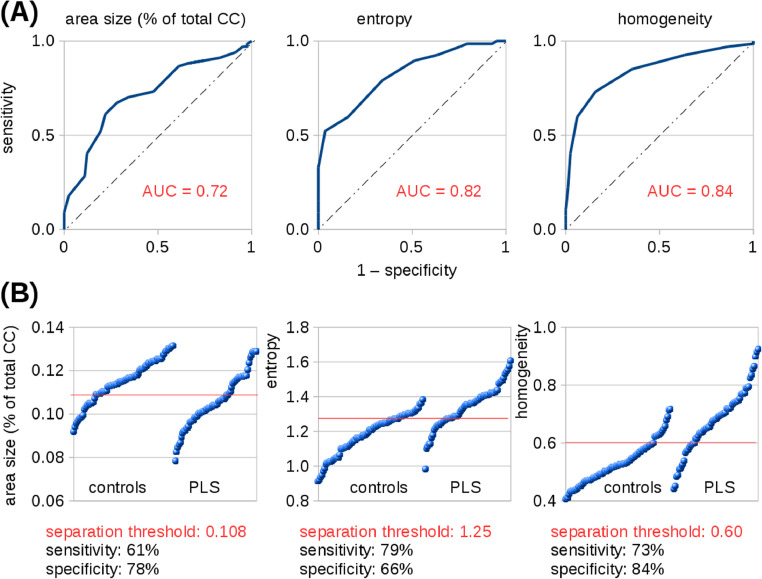
Despite significant differences, there was an overlap between control and patient groups both in CC atrophy and also in CC texture parameters of area III. Since an objective of this study was to provide an insight into texture and its performance as a diagnostic biomarker, classification accuracy was estimated by use of ROC curves which were calculated for PLS patients vs controls for area III atrophy (area under curve (AUC) = 0.72 (“fair”)), entropy (AUC = 0.82 (“good”)), and homogeneity (AUC = 0.84 (“good”)) (Fig. 4A). Optimum separation thresholds of area III revealed a sensitivity of 61% and a specificity of 78% for area III atrophy (threshold 10.8%), a sensitivity of 79% and a specificity of 66% for entropy (threshold 1.25), and a sensitivity of 73% and a specificity of 84% for homogeneity (threshold 0.60) (Fig. 4B).
Fig. 4.
Overlap between PLS patient group and controls in CC area III atrophy and texture parameters. (A) ROC curves for area III atrophy (area under curve (AUC) = 0.72 (“fair”)), entropy (AUC = 0.82 (“good”)), and homogeneity (AUC = 0.84 (“good”)) for PLS patients vs controls. (B) Optimum separation thresholds for atrophy (threshold 10.8%), for entropy (threshold 1.25), and for homogeneity (threshold 0.60), respectively, of area III.
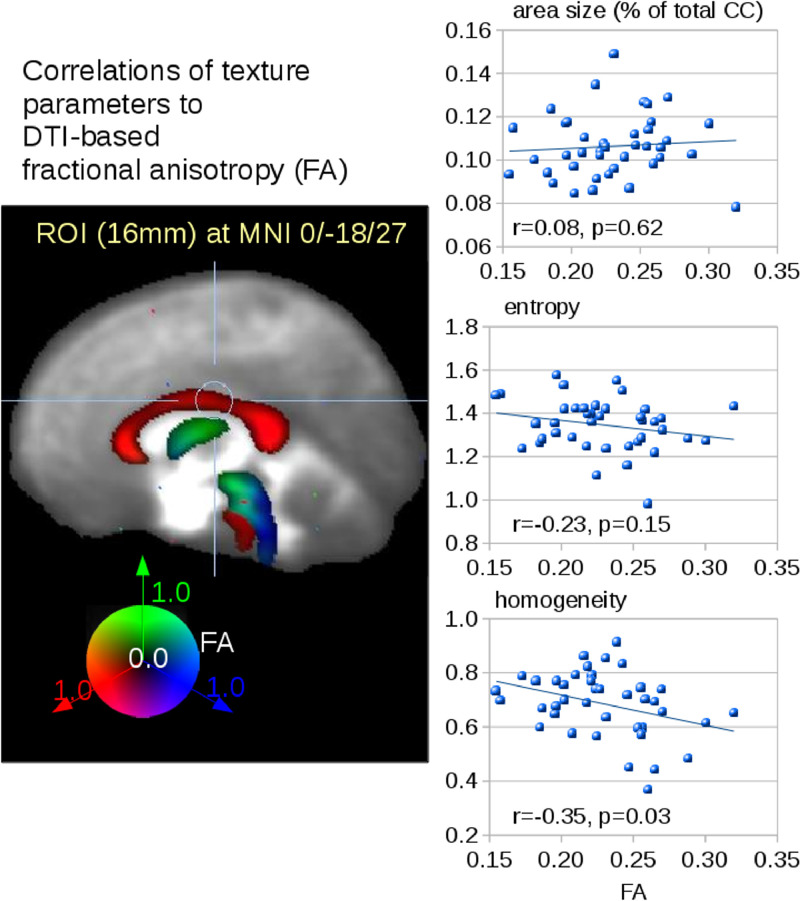
3.5. Correlation of texture parameters to DTI analysis
Microstructure analysis of the CC was assessed by fractional anisotropy (FA) mapping of DTI scans. Correlation analysis of microstructure (FA) and the texture parameters entropy and homogeneity could be performed for 40 PLS patients. Fig. 5 shows the correlation of ROI-based FA values in CC area III (size 16 mm; center MNI 0/−18/27). No correlation was found for atrophy of area III and FA values, a trend of decreased entropy with increasing FA values could be observed. A correlation (r=−0.35, p = 0.03) was detected for the texture parameter homogeneity and FA values in area III.
Fig. 5.
Correlation analysis between texture and FA values. Correlations of atrophy and texture parameters of area III of the CC to FA values in a ROI (16 mm) in area III of the CC (MNI 0/−18/27).
4. Discussion
First, this systematic unbiased MRI study demonstrated the generally accepted “neuroimaging textbook knowledge” (Osborn et al., 2016) that PLS patients often show atrophy in the area III of the corpus callosum. The unbiased automated planimetry could demonstrate, beyond visual assessment, that the PLS group exhibited a highly significant reduction of the callosal area III, as confirmed by two independent techniques (TIFT implementation and ABV, respectively). However, although highly significant at group level, a full separation of the groups could not be achieved so that atrophy of the area III of the CC does not seem to be a consistent finding in all PLS patients. In addition, the size of the CC area III was even more variable in ALS so that group results were not significant. Thus, atrophy of the area III of the CC can be regarded as a feature in PLS (although not observed in each and every patient), whereas – although its texture is significantly altered - the variability in size in ALS does not allow to consider it a marker at group level and has to be investigated in a future study with higher numbers of participants with ALS. As a consequence, the use as a diagnostic marker in a clinical setting which requires the identification of pathology at an individual patient´s level cannot be provided.
Furthermore, the results expand the knowledge of CC alterations in association with MND. Beyond regional atrophy, loss of fiber integrity as indicated by FA reduction in areas II and III has already been shown in a DTI-based study in ALS patients (e.g. Chapman et al., 2014). Beyond regional atrophy, loss of fiber integrity as indicated by FA reduction in areas II and III has already been shown in a DTI-based study in ALS patients (e.g. Chapman et al., 2014). Three-dimensional texture maps from T1w images have been shown to correlate with the DTI metrics within the CST (Ishaque et al., 2019), in accordance with the findings of the current study that FA values correlate with texture measures. Thus, one may conclude that texture changes may be related to white matter microstructural abnormalities. This loss might hypothetically also result in a more inhomogeneous distribution of grey levels in T1w scans. As a consequence, the entropy and the (in)homogeneity have also been analyzed in the current study. Texture analysis has already been applied to Alzheimer`s disease (Maani et al., 2015) and to a group of 19 ALS patients (Maani et al., 2016). In the latter study, the CC was not reported to show a significantly different texture in ALS compared to controls. However, the difference to the current study might result from the different texture parameters studied or the different number of subjects in these two studies; further investigations are required to resolve these discrepancies. The entropy in a sample increases when gray values in a sample distribution show a more inhomogeneous pattern, while the inhomogeneity rises when gray level differences between neighboring voxels increase. Average intensity and variance were not considered for the CC subareas given that the global CC values were z-normalized. The additional parameters kurtosis and skewness, however, were candidates for detecting increased inhomogeneity, but the results showed no significant difference between the PLS patient group and the control group. Finally, differences in texture could be detected by increased entropy in areas II and III and by increased (in)homogeneity in areas I, II, and III – the texture alterations in areas II and III at the group level were in agreement with fractional anisotropy reductions (loss of fiber integrity), as reported previously (Chapman et al., 2014). Texture analysis of the CC may provide further evidence for a consistent involvement of the CC across patients with PLS; moreover, in addition to DTI-based analysis, an association between texture and FA in area III could be shown. While CC involvement might relate to interhemispheric spread, it might equally reflect secondary damage due to independent bilateral cortical processes.
The study was not without limitations. The design was cross-sectional, and thus results will have to be re-evaluated in a longitudinal design in future studies. In addition, clinical evidence of central CC involvement such as the observation of mirror movements in patients, reflecting impaired interhemispheric inhibition (Karandreas et al., 2007) were not assessed, neither were the MRI data combined with electrophysiological evidence for altered callosal function by transcranial magnetic stimulation (Wittstock et al., 2007).
In summary, the current MRI data demonstrate PLS-associated morphometric changes in the central nervous system in vivo, but they do not provide the clinician with one imaging marker that contributes to an accurate clinical diagnosis with sufficient sensitivity and specificity or discriminative power. However, given the significant difference in CC topography with respect to size of area III together with entropy and homogeneity changes, the (unbiased) recognition of CC area III alterations might help to develop neuroimaging criteria in support of the diagnosis MND in general or PLS in particular. Such an MRI-based diagnostic classification has to await future confirmation, but might be composed by a combination of multiparametric imaging metrics with different measures of planimetric (or volumetric) changes, changes in microstructure, and other alterations in MND-associated patterns. For ALS, potential algorithms have been proposed recently (Ferraro et al., 2017; Bede et al., 2017). Based on such a multiparametric protocol which is still to be developed but might include CC planimetry as a candidate, a composite score from different MRI-based approaches might be defined in the diagnostic pathway to MND. With this aim in mind, the current study addressed PLS as the ALS variant with prominent UMN involvement for the analysis of the CC, but future studies will expand such MRI-based CC substructure analyses to ´classical´ ALS in order to assess their value as a biological marker for the neuroimaging signature of MND.
Author disclosures
All authors: All authors report no conflicts of interest.
Statement
All human studies have been approved by the appropriate ethics committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
CRediT authorship contribution statement
Hans-Peter Müller: Conceptualization, Software, Validation, Formal analysis, Project administration, Writing - original draft. Jens Dreyhaupt: Formal analysis, Writing - review & editing. Francesco Roselli: Investigation, Writing - review & editing. Magdalena Schlecht: Investigation, Formal analysis, Writing - review & editing. Albert C. Ludolph: Funding acquisition, Writing - review & editing. Hans-Jürgen Huppertz: Formal analysis, Writing - review & editing. Jan Kassubek: Conceptualization, Investigation, Supervision, Project administration, Writing - original draft.
Acknowledgments
Acknowledgements
Sonja Fuchs is thankfully acknowledged for her great help in the acquisition of MRI data. The authors would like to thank the Ulm University Center for Translational Imaging MoMAN for its support.
Funding
This study was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG Grant Number LU 336/15-1), the German Network for Motor Neuron Diseases (BMBF 01GM1103A), and Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.nicl.2020.102223.
Appendix. Supplementary materials
References
- Agosta F., Galantucci S., Riva N. Intrahemispheric and interhemispheric structural network abnormalities in PLS and ALS. Hum. Brain Mapp. 2014;35:1710–1722. doi: 10.1002/hbm.22286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Iyer P.M., Finegan E. Virtual brain biopsies in amyotrophic lateral sclerosis: diagnostic classification based on in vivo pathological patterns. Neuroimage Clin. 2017;15:653–658. doi: 10.1016/j.nicl.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Chipika R.H., Finegan E. Brainstem pathology in amyotrophic lateral sclerosis and primary lateral sclerosis: a longitudinal neuroimaging study. Neuroimage Clin. 2019;24 doi: 10.1016/j.nicl.2019.102054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M.C., Jelsone-Swain L., Fling B.W. Corpus callosum area in amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2012;13:589–591. doi: 10.3109/17482968.2012.708935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M.C., Jelsone-Swain L., Johnson T.D. Diffusion tensor MRI of the corpus callosum in amyotrophic lateral sclerosis. J. Magn. Reson. Imaging. 2014;39:641–647. doi: 10.1002/jmri.24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro P.M., Agosta F., Riva N. Multimodal structural MRI in the diagnosis of motor neuron diseases. Neuroimage Clin. 2017;16:240–247. doi: 10.1016/j.nicl.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N., Douaud G., Ce M. Corpus callosum involvement is a consistent feature of amyotrophic lateral sclerosis. Neurology. 2010;75:1645–1652. doi: 10.1212/WNL.0b013e3181fb84d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon P.H., Cheng B., Katz I.B. The natural history of primary lateral sclerosis. Neurology. 2006;66:647–653. doi: 10.1212/01.wnl.0000200962.94777.71. [DOI] [PubMed] [Google Scholar]
- Hofer S., Frahm J. Topography of the human corpus callosum revisited–comprehensive fiber tractography using diffusion tensor magnetic resonance imaging. Neuroimage. 2006;32:989–994. doi: 10.1016/j.neuroimage.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Huppertz H.J., Kröll-Seger J., Klöppel S. Intra- and interscanner variability of automated voxel-based volumetry based on a 3D probabilistic atlas of human cerebral structures. Neuroimage. 2010;49:2216–2224. doi: 10.1016/j.neuroimage.2009.10.066. [DOI] [PubMed] [Google Scholar]
- Ishaque A., Mah D., Seres P. Corticospinal tract degeneration in ALS unmasked in T1-weighted images using texture analysis. Hum. Brain Mapp. 2019;40:1174–1183. doi: 10.1002/hbm.24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandreas N., Papadopoulou M., Kokotis P. Impaired interhemispheric inhibition in amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2007;8:112–118. doi: 10.1080/17482960601030113. [DOI] [PubMed] [Google Scholar]
- Kassubek J., Ludolph A.C., Müller H.P. Neuroimaging of motor neuron diseases. Ther. Adv. Neurol. Dis. 2012;5:119–127. doi: 10.1177/1756285612437562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolind S., Sharma R., Knight S. Myelin imaging in amyotrophic and primary lateral sclerosis. Amyotroph. Lateral Scler. Frontotemporal. Degener. 2013;14:562–573. doi: 10.3109/21678421.2013.794843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludolph A., Drory V., Hardiman O. A revision of the el escorial criteria - 2015. Amyotroph. Lateral Scler. Frontotemporal. Degener. 2015;29:1–2. doi: 10.3109/21678421.2015.1049183. [DOI] [PubMed] [Google Scholar]
- Maani R., Yang Y.H., Kalra S. Voxel-based texture analysis of the brain. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0117759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maani R., Yang Y.H., Emery D., Kalra S. Cerebral degeneration in amyotrophic lateral sclerosis revealed by 3-Dimensional texture analysis. Front Neurosci. 2016;10:120. doi: 10.3389/fnins.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.P., Unrath A., Ludolph A.C. Preservation of diffusion tensor properties during spatial normalization by use of tensor imaging and fibre tracking on a normal brain database. Phys. Med. Biol. 2007;52:N99–109. doi: 10.1088/0031-9155/52/6/N01. [DOI] [PubMed] [Google Scholar]
- Müller H.-.P., Unrath A., Huppertz H.-.J. Neuroanatomical patterns of cerebral white matter involvement in different motor neuron diseases as studied by diffusion tensor imaging analysis. Amyotroph. Lateral Scler. 2012;13:254–264. doi: 10.3109/17482968.2011.653571. [DOI] [PubMed] [Google Scholar]
- Müller H.P., Turner M.R., Grosskreutz J. A large-scale multicentre cerebral diffusion tensor imaging study in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry. 2016;87:570–579. doi: 10.1136/jnnp-2015-311952. [DOI] [PubMed] [Google Scholar]
- Müller H.P., Agosta F., Gorges M. Cortico-efferent tract involvement in primary lateral sclerosis and amyotrophic lateral sclerosis: a two-centre tract of interest-based DTI analysis. Neuroimage Clin. 2018;20:1062–1069. doi: 10.1016/j.nicl.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.P., Gorges M., Kassubek R. Identical patterns of cortico-efferent tract involvement in primary lateral sclerosis and amyotrophic lateral sclerosis: a tract of interest-based MRI study. Neuroimage Clin. 2018;18:762–769. doi: 10.1016/j.nicl.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A.G., Salzman K.L., Jhaveri M.D. 3rd Edition. Elsevier LTD; Oxford: 2016. Diagnostic Imaging: Brain. [Google Scholar]
- Pringle C.E., Hudson A.J., Munoz D.G. Primary lateral sclerosis. Clinical features, neuropathology and diagnostic criteria. Brain. 1992;115:495–520. doi: 10.1093/brain/115.2.495. [DOI] [PubMed] [Google Scholar]
- Riad S.M., Hathout H., Huang J.C. High T2 signal in primary lateral sclerosis supports the topographic distribution of fibers in the corpus callosum: assessing disease in the primary motor segment. AJNR Am. J. Neuroradiol. 2011;32:E61–E64. doi: 10.3174/ajnr.A2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M.A., Statland J.M., Wolfe G.I. Primary lateral sclerosis. Muscle Nerve. 2007;35:291–302. doi: 10.1002/mus.20728. [DOI] [PubMed] [Google Scholar]
- Stockman G., Shapiro L.G. Prentice Hall PTR; Upper Saddle River, NJ, USA: 2001. Computer Vision, Chapter 7. [Google Scholar]
- Wais V., Rosenbohm A., Petri S. The concept and diagnostic criteria of primary lateral sclerosis. Acta Neurol. Scand. 2017;136:204–211. doi: 10.1111/ane.12713. [DOI] [PubMed] [Google Scholar]
- Witelson S.F. Hand and sex differences in the isthmus and genu of the human corpus callosum. A Postmortem morphological study. Brain. 1989;112:799–835. doi: 10.1093/brain/112.3.799. [DOI] [PubMed] [Google Scholar]
- Wittstock M., Wolters A., Benecke R. Transcallosal inhibition in amyotrophic lateral sclerosis. Clin. Neurophysiol. 2007;118:301–307. doi: 10.1016/j.clinph.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Yamauchi H., Fukuyama H., Ouchi Y. Corpus callosum atrophy in amyotrophic lateral sclerosis. J. Neurol. Sci. 1995;134:189–196. doi: 10.1016/0022-510x(95)00220-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.