Abstract
Development of curative therapies for chronic hepatitis B virus (HBV) infection will likely require new animal models. Here, we evaluate HBV infection in squirrel monkeys based on the high‐sequence homology of the HBV receptor, Na+/taurocholate co‐transporting peptide (NTCP), between humans and squirrel monkeys. HBV PreS1 peptide was examined for binding human and squirrel monkey NTCP. Immunodeficient Fah −/−, NOD, Rag1 −/−, Il2Rg null (FNRG) mice engrafted with human or squirrel monkey hepatocytes were challenged with HBV or Woolly Monkey HBV (WMHBV). In addition, adult squirrel monkeys were inoculated with HBV, WMHBV, adeno‐associated virus containing an infectious genome of HBV (AAV‐HBV), and AAV‐WMHBV. Finally, neonate squirrel monkeys were assessed for the potential of chronic infection with WMHBV. PreS1 peptide efficiently bound to human and squirrel monkey NTCP but not to mouse or capuchin NTCP. FNRG mice engrafted with squirrel monkey hepatocytes were susceptible to infection by WMHBV but not human HBV. Similarly, adult squirrel monkeys could be infected with WMHBV but not human HBV, whereas chimeric mice engrafted with human hepatocytes were susceptible to HBV but not WMHBV. Infection of squirrel monkeys with AAV‐WMHBV yielded maximum viremia of 108 genomes/mL with detectable virus for up to 8 months. Notably, covalently closed circular DNA was detected in the liver of these animals. Infection of neonates with WMHBV led to detectable viremia for up to 6 months. Conclusions: Adult and neonate squirrel monkeys exhibited prolonged WMHBV viremia lasting 6‐8 months. This is greater than twice the duration of viremia achieved in other nonhuman primates and suggests that squirrel monkeys may be a suitable model for testing HBV therapeutics.

Abbreviations
- AAV
adeno‐associated virus
- ALT
alanine aminotransferase
- anti‐HBc
antibodies to hepatitis B core antigen
- cccDNA
covalently closed circular deoxyribonucleic acid
- CXCL
chemokine (C‐X‐C motif) ligand
- ELISA
enzyme‐linked immunosorbent assay
- FNRG
Fah −/−, NOD, Rag1 −/−, Il2rg null
- GE
genome equivalents
- HBeAg
hepatitis B e antigen
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HDV
hepatitis delta virus
- HDVpsHBV
hepatitis delta virus pseudotyped with hepatitis B virus envelope proteins
- HDVpsWMHBV
hepatitis delta virus pseudotyped with woolly monkey hepatitis B virus envelope proteins
- hNTCP
human sodium taurocholate cotransporting polypeptide
- IFN
interferon
- IgG
immunoglobulin G
- NA
nucleoside/nucleotide analog
- NTBC
2‐(2‐nitro‐4‐trifluoro‐methylbenzoyl)‐1,3‐cyclohexanedione
- NTCP
sodium taurocholate cotransporting polypeptide
- PCR
polymerase chain reaction
- pgRNA
pregenomic ribonucleic acid
- qPCR
quantitative polymerase chain reaction
- RT‐PCR
real‐time polymerase chain reaction
- VP
viral particles
- WMHBV
woolly monkey HBV
Despite the availability of a vaccine for hepatitis B virus (HBV) infection, more than 257 million people worldwide are chronically infected with the virus.1 Viral persistence causes liver fibrosis that can progress to cirrhosis and liver cancer. The World Health Organization estimates that 880,000 deaths per year can be attributed to chronic HBV infection.
There are two approved treatments for chronic HBV infection, interferons (IFNs), and nucleoside/nucleotide analogs (NAs). Although IFNs and NAs can be effective at reducing HBV viral titers, they do not prevent viral entry or formation of the stable HBV genome, covalently closed circular DNA (cccDNA), in hepatocytes. Therefore, continuous treatment is necessary to maintain HBV suppression in patients. After removal of interferon or NA treatment, HBV will rebound. The goal of most future therapies is to achieve a functional cure, a combination of low viral titer with the loss of hepatitis B surface antigen (HBsAg) from peripheral blood. Loss of HBsAg would indicate that an immune response was achieved that can suppress HBV such that low or undetectable viral titers are maintained after the removal of antivirals. More challenging would be to achieve virologic cure, in which cccDNA is eliminated from infected cells.
The development of new therapies targeting HBV will likely be dependent on animal models. HBV has a narrow host range, causing chronic infection only in humans and chimpanzees. Because chimpanzees are no longer available for biomedical research, new animal models are urgently needed. Mice, even those expressing human Na+/taurocholate co‐transporting peptide (hNTCP) are not susceptible to HBV infection.2 Immunocompetent mouse models have been generated by delivering HBV genomes as transgenes in adeno‐associated virus (AAV) vectors.3, 4 Viral persistence was mouse strain–specific and variable among animals, but some animals maintained HBsAg and hepatitis B core antigen for more than 1 year after injection.5 However, it has been shown that some cccDNA is formed in AAV8‐HBV infected C57BL/6 mice, but these levels are significantly lower than what is observed in hNTCP‐expressing HepG2 cells or in HBV patient liver samples,6 thus limiting the utility of these models for the development of virologic cures.
An alternative strategy is to infect chimeric mice engrafted with human hepatocytes with HBV. Once infected, the human hepatocytes in these chimeric mice support all stages of the HBV viral life cycle, including the formation of cccDNA. One recently developed chimeric mouse model is the FNRG mouse (Fah − / −, NOD, Rag1 − / −, Il2rg null).7 In this immunodeficient model, oral administration of 2‐(2‐nitro‐4‐trifluoro‐methylbenzoyl)‐1,3‐cyclohexanedione (NTBC) prevents the buildup of toxic metabolites. Withdrawal of NTBC results in hepatocellular injury. The inducible liver injury enables the mice to be transplanted at an older age than other chimeric mouse models, allowing for greater flexibility for timing of hepatocyte transplantation. Although these murine models are useful for studying viral infection, their immunodeficient backgrounds render them unsuitable for testing therapeutic vaccines or immune‐modulatory agents.
Tree shrews are not a natural host for HBV infection, but they are the only nonprimate known to be susceptible to infection. Infection of adult animals results in weak, short‐lived viremia with limited viral replication.8 There are a number of HBV‐related viruses that infect other animal species. Duck HBV is an Avihepadnavirus that causes chronic liver infection in those birds.9 Woodchuck hepatitis virus (WHV) is another related virus belonging to the family Orthohepadnavirus. Some of the major drawbacks of these models include poor availability of animals, lack of reagents, limited annotated genomic data, and genetic variation from HBV. For example, WHV has only 70% identity to human HBV.10
Since the ban on biomedical research in chimpanzees in 2015, there has been increased effort to generate a new nonhuman primate model of chronic HBV infection. Some wild cynomolgus macaques from Mauritius Islands were reported to have low titer chronic HBV infection.11 This strain of HBV was able to infect Barbary macaques. However, a recent attempt to replicate infection of Mauritian cynomolgus macaques with that strain of HBV was unsuccessful.12 In 1998, a hepatitis B–like virus was isolated from zoo‐housed woolly monkeys suffering from fulminant hepatitis (WMHBV),13 and an infectious clone of the virus was reported in 2003.14 WMHBV is a hepadnavirus with the same genetic organization as human HBV. Woolly monkeys are endangered, but it was shown that this virus could also be used to infect closely related spider monkeys. In spider monkeys, WMHBV reached maximum titers of 105 genome equivalents (GE)/mL and infection was detectable for 6 weeks. Addition of the immunosuppressant FK506 extended the duration of infection to 16 weeks.14 Several species of spider monkeys are threatened or endangered in the wild, and therefore are not routinely available for research.
Because hepadnaviruses have been found in humans and great apes (chimpanzees) but not in Old World monkeys (macaques and baboons), it was interesting to find them in New World monkeys (woolly monkeys). This observation could be explained by sequence analysis of the receptor for HBV, Na+/taurocholate co‐transporting peptide (NTCP).15, 16 NTCP is a bile acid transporter protein. NTCP sequences from 11 primate species spanning Old World monkeys and New World monkeys were recently compared. In that study, HEK293 and HepG2 cell lines were stably transfected to express the different NTCPs, and these cells were tested for binding to preS1 peptides from HBV and WMHBV. NTCPs from the Old World monkeys did not bind to the peptides from HBV or WMHBV, but the peptides did bind to NTCPs from two New World monkeys: squirrel monkeys and marmosets.17 Through some elegant mutational analysis, it was determined that the amino acid position G158 plays a significant role in binding of NTCP to HBV and WMHBV. Furthermore, this work suggested that squirrel monkeys may be a good model for HBV or WMHBV infection. In this study, using multiple different approaches, we demonstrate that WMHBV infects squirrel monkeys and can induce prolonged viremia in both adult and neonate animals.
Materials and Methods
Animal Studies
Squirrel monkeys were housed and cared for at the Southwest National Primate Research Center at Texas Biomedical Research Institute (adult and AAV studies) or Michale E. Keeling Center for Comparative Medicine and Research (neonate studies), and chimeric mouse studies were performed at Princeton University. Animals were cared for in accordance with the Guide for the Care and Use of Laboratory Animals, and all protocols were approved by the Institutional Animal Care and Use Committees. All facilities are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and operate in accordance with the National Institutes of Health and U.S. Department of Agriculture guidelines and the Animal Welfare Act. Details are presented in the Supporting Information.
Myrcludex B In Vitro Binding Assay
HEK293 cells were transfected with plasmids encoding human, squirrel monkey, or cynomolgus macaque NTCP genes. Two days after transfection, the cells were incubated with fluorescently labeled Myrcludex B for 1 hour at room temperature. Controls cells were pre‐incubated with unlabeled Myrcludex B as a blocking reagent for 30 minutes. These cells were washed then incubated with fluorescently labeled Myrcludex B. Cells were then either fixed with paraformaldehyde and incubated with Hoechst‐33342 (Thermo Fisher, Waltham, MA) and imaged by fluorescent microscopy or lysed for western blot.
Western Blot of NTCP
Detailed western blot procedures are described in the Supporting Information. Briefly, cells were lysed in Laemmli Buffer (Bio‐Rad, Hercules, CA). Cell lysate samples were prepared for deglycosylation with PNGaseF (New England BioLabs, Ipswich, MA). NTCP from the cell lysate was compared with recombinant deglycosylated NTCP protein (Abnova, Taipei, Taiwan). The primary antibodies were rabbit polyclonal SLC10A1 (MilliporeSigma, Burlington, MA) diluted 1:1,000 and mouse monoclonal beta actin (Li‐Cor) diluted 1:5,000. The secondary antibodies, IRDye 800CW goat anti‐rabbit immunoglobulin G (IgG) (Licor) and IRDye 680RD goat anti‐mouse IgG (Licor), were used at 1:10,000.
Production of Primary Hepatocytes
The procedures for production of primary hepatocytes are described in the Supporting Information and have been described previously.18
AAV Viruses
AAV‐HBV and AAV‐WMHBV were produced from greater than genome‐length clones of HBV and WMHBV (SignaGen, Rockville, MD). The HBV genome sequence used was subtype AYW accession number J02203.1. The WMHBV sequence used was GenBank accession number AY226578.1.14
Quantification of Viral DNA and RNA in Squirrel Monkeys
The TaqMan (Applied Biosystems, Foster City, CA) polymerase chain reaction (PCR) and real‐time PCR (RT‐PCR) assays used for detection of viral DNA and RNA have been described previously for HBV19 and WMHBV14 and are described in detail in the Supporting Information for serum‐derived and liver‐derived materials. The HBV assay is based on the X region and would detect all transcripts, as all transcripts span this region before the polyA site. The WMHBV RT‐PCR assay is based on primers in the surface region. This assay detects all transcripts except X. Because X is a low abundance transcript, this assay is similar to the total transcript assay for HBV.
Engraftment of Adult Squirrel Monkey and Human Hepatocytes Into FNRG Mice
FNRG mice were generated and transplanted as previously described.7, 20 Female mice between 6‐10 weeks of age were injected with 1.0 × 106 cryopreserved adult human or squirrel monkey hepatocytes. Primary human hepatocytes were obtained from BioIVT (Westbury, NY). FNRG mice were cycled on NTBC (Yecuris Inc., Tualatin, OR) supplemented in their water. FNRG mice were maintained on amoxicillin chow in standard filter top rodent cages on autoclaved bedding. All interventions were performed during the light cycle. Hepatocyte engraftment was monitored by enzyme‐linked immunosorbent assay (ELISA) for albumin as described in Supporting Information. HBV and WMHBV inocula and assays to detect viral DNA and RNA are described in the Supporting Information.
Quantification Viral DNA From Liver Chimeric Mouse Livers
HBV DNA isolated from lysed cells was PCR‐amplified using CCGTCTGTGCCTTCTCATCTG (forward primer), AGTCCAAGAGTCCTCTTATGTAAGACCTT (reverse primer), and probe FAM‐CCGTGTGCACTTCGCTTCACCTCTGC‐TAMRA.21
Quantification of Viral Pregenomic Ribonucleic Acid From Liver Chimeric Mouse Livers
Pregenomic ribonucleic acid (pgRNA) in the liver of chimeric mice was quantified with the Luna Universal One‐Step RT‐qPCR Kit (New England BioLabs). The primers for WMHBV were forward primer ACCCAATGCCCCTATCTTATC and reverse primer CAGGAAGATGCTGGAGATTG, and the primers for HBV were forward primer GAGTGTGGATTCGCACTCC and reverse primer GAGGCGAGGGAGTTCTTCT.21
Adult Squirrel Monkey Infection
Adult male squirrel monkeys (ages 5‐8) were infected by intravenous injection of 4.6 × 108 GE of HBV (animal number 34959), 5.0 × 108 GE of WMHBV (animal number 34957), 5.0 × 1012 viral particles (VP) of AAV‐HBV (animal numbers 36242 and 36243), or 5.0 × 1012 VP AAV‐WMHBV (animal numbers 36244 and 36245). Animals were bled weekly for the first month, then biweekly up to week 32. Serum was assayed for viral DNA by PCR and antigens by ELISA, as well as assayed for alanine aminotransferase (ALT) by standard serum glutamic‐pyruvic transaminase testing. Liver biopsies were taken at weeks 4 and 14, and tissue was preserved in formalin for histology, RNALater for RNA isolation (MilliporeSigma), or snap‐frozen on dry ice for DNA extraction.
Neonatal Squirrel Monkey Infection
Six neonatal squirrel monkeys (mixed gender) were infected by intravenous injection of 1.0 × 107 GE of WMHBV. Animals were bled biweekly for 2 months, then monthly until week 24 (animal 6604 was not bled at the final timepoint due to health concerns). Serum was used for ELISA and viral quantitation.
ELISA Detection of Serum HBsAg and HBeAg Levels
HBsAg and hepatitis B e antigen (HBeAg) levels, including WMHBV antigens, in the serum of squirrel monkeys were determined by ELISA (DiaSorin, Saluggia, Italy) following the manufacturer’s protocol. Purified HBsAg or HBeAg were used to generate standard curves.
Quantification of HBV and WMHBV cccDNA From Liver Tissue by Southern Blot
A detailed protocol is included in the Supporting Information. Briefly, DNA from squirrel monkey livers was either digested with T5 exonuclease (New England BioLabs) or left undigested. After electrophoresis and transfer, the membrane was hybridized with a WMHBV probe set (Affymetrix, St. Clara, CA). After hybridization, the membrane was washed followed by pre‐amplification, amplification, and probe labeling for 60 minutes each. The membrane was incubated with CDP‐Star detection reagent (GE Healthcare, Chicago, IL). The membrane was washed and rehybridized with a HBV probe set (Affymetrix) following this same procedure.
Detection of WMHBV DNA in Tissue Sections by In Situ Hybridization
WMHBV was detected in liver sections from the squirrel monkeys using the BaseScope (ACDBio, Newark, CA) technology. Liver tissue was fixed in formalin and embedded in paraffin. Sections were prepared using standard methodologies. The details of the BaseScope detection of WMHBV DNA is described in the Supporting Information.
Quantification of IFN gamma, CXCL9, and CXCL10 Transcripts in Squirrel Monkey Liver
The methods are described in detail in the Supporting Information. Total liver RNA was prepared in the same manner as for viral RNA, and the TaqMan assay conditions are the same as those used for viral RNA. All primers and probes were from Integrated DNA Technologies (Coralville, IA) and were dual‐labeled with FAM/TAMRA. The primers and probes are listed with the accession numbers in the Supporting Information: IFN gamma (Accession No. XM_003926543), chemokine (C‐X‐C motif) ligand (CXCL) 9 (Accession No. XM_003931992), and CXCL10 (Accession No. XM_003931996).
Immunohistochemical Staining of Ki67 and Activated Caspase‐3 in Squirrel Monkey Liver
The methods are described in detail in the Supporting Information. Liver sections were deparaffinized in EZ‐DeWax (HK 585‐5K; BioGenex, San Ramon, CA) and antigen retrieval was performed in antigen retrieval solution (HK 086‐9K; BioGenex). Slides were treated sequentially with peroxidase suppressor, universal block, and avidin (36000; Thermo Fisher Scientific, Waltham, MA). Slides were incubated sequentially with primary antibody diluted in universal block containing a biotin, biotinylated goat antimouse IgG, and avidin‐biotin complex. Slides were developed with Immpact Nova Red peroxidase substrate (SK‐4805; Vector Labs, Burlingame, CA), counterstained with Mayer’s Hematoxylin (S3309; Agilent Dako, Santa Clara, CA), dehydrated, and mounted in nonaqueous mounting media (H‐5000; VectorLabs). Ki67 was detected with a mouse monoclonal antibody (M7240, MIB‐1; Agilent Dako) at 1:100 dilution and activated caspase 3 was detected with a rabbit antibody (9664S, 5A1E; Cell Signaling Technology, Danvers, MA) at 1:100 dilution.
Results
Squirrel monkey NTCP has a conserved glycine at position 15817 that was shown to be critical in HBV binding. The conservation of this residue in multiple HBV‐permissive species as well as several amino acid 158 variants that are not permissive is illustrated in Fig. 1A. Interestingly, the phylogenetic relationship between the NTCPs from these species does not predict permissiveness (Fig. 1B). To confirm that squirrel monkey NTCP binds to HBV, we performed a binding assay with Myrcludex B (Mycludex B) using HEK293 cells transfected with human, squirrel monkey, or cynomolgus NTCP. Myrcludex B is a peptide drug that mimics the amino terminus of N‐acylated preS1 (part of the large HBV envelope protein).22 NTCP expression was confirmed in transfected cells by western blot (Fig. 2A). In agreement with previous reports, fluorescently labeled Myrcludex B bound to human and squirrel monkey NTCP expressing cells but not cynomolgus macaque NTCP expressing cells.17, 23 This binding could be blocked by pre‐incubation with non‐fluorescently labeled Myrcludex B (Fig. 2B).
Figure 1.
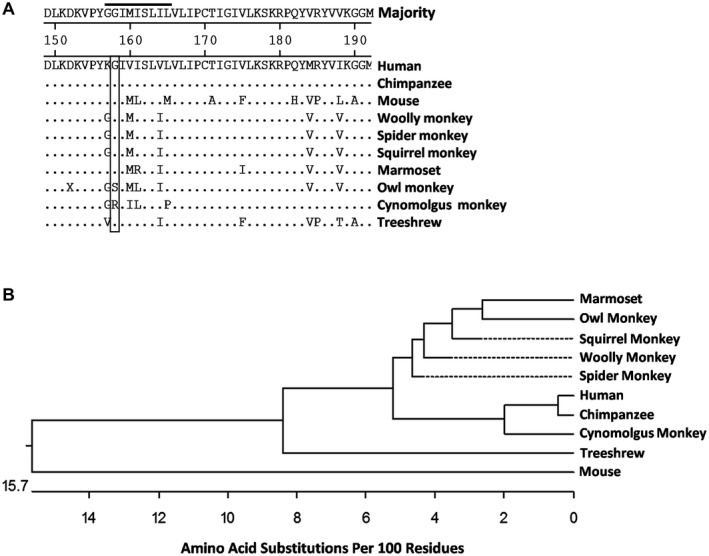
(A) Amino acid sequence alignment of NTCP from putative HBV animal models. The gray bar highlights the amino acid at position 158, which is critical for mediating binding to HBV. (B) Phylogenetic tree depicting evolutionary distance between species from (1A) based on difference in amino acid sequence of NTCP.
Figure 2.
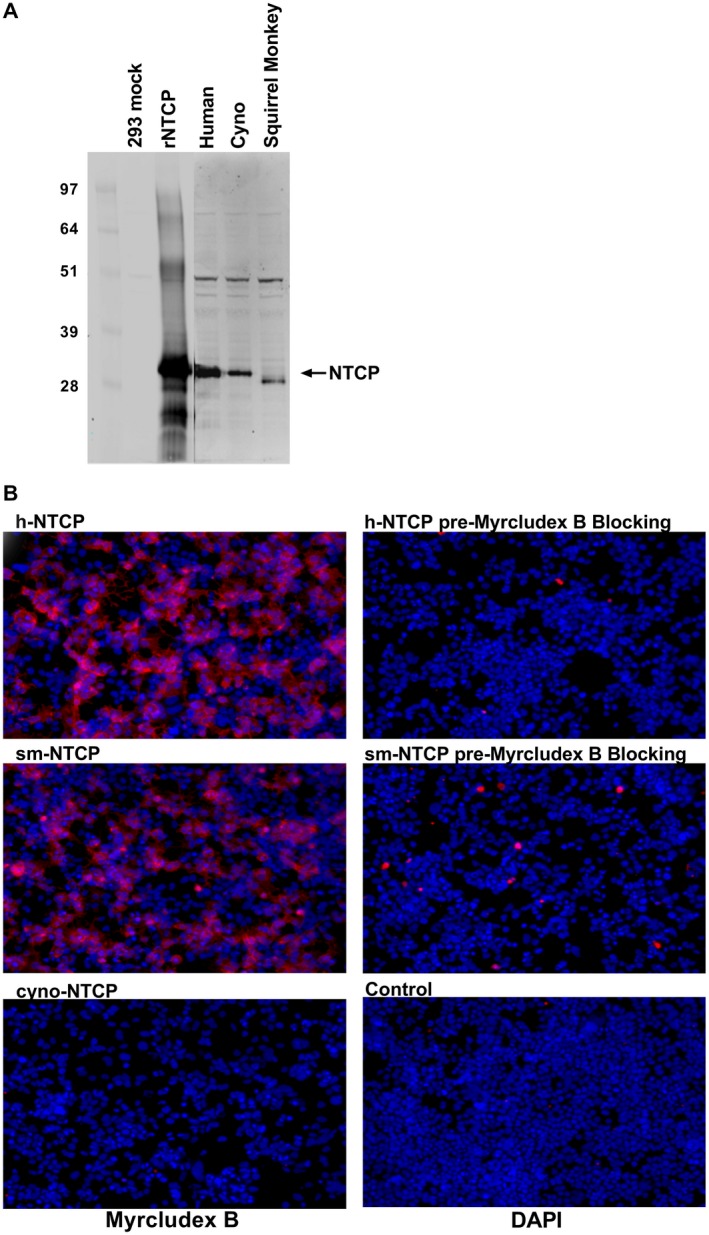
Myrcludex B binds squirrel monkey NTCP. HEK293 cells were transfected with protein expression vectors that express species‐specific NTCPs. (A) Two days after transfection, the cells were either lysed for western blot analysis of NTCP expression. (B) NTCP‐expressing HEK293 cells were incubated with fluorescently labeled Myrcludex B (left column). Fluorescent Myrcludex B binding could be blocked by pre‐incubation with unlabeled Myrcludex B (right column). Abbreviations: DAPI, 4´,6‐diamidino‐2‐phenylindole (a type of stain).
To examine the ability of squirrel monkey hepatocytes to support the replication of HBV and WMBHV, studies were conducted in FNRG mice that were engrafted with either human or squirrel monkey hepatocytes isolated by collagenase perfusion. Engraftment was estimated using a circulating plasma albumin assay, which demonstrated that both human and squirrel monkey hepatocyte engraftment could be maintained over 150 days (Fig. 3A,B). There was a decrease in the albumin levels of squirrel monkey hepatocyte engrafted FNRG mice 3 to 4 weeks following infection. This is most likely due to the prolonged cycling of the mice on NTBC, as this chemical inhibits the buildup of toxic metabolites that would lead to the endogenous murine hepatocytes death, thus reducing the growth advantaged for the engrafted squirrel monkey hepatocytes. Squirrel monkey or human hepatocyte engrafted FNRG mice were given intraperitoneal injection of serum containing 108 genomes/animal of WMHBV or HBV. Viral genomes and pgRNA copies were determined by PCR from liver tissue (Fig. 3C,D). For the HBV‐infected animals with human hepatocytes, each milligram of tissue contained approximately 108 copies of viral DNA and 107 copies of pregenomic RNA. HBV‐infected mice with squirrel monkey hepatocytes had approximately 103 HBV genome copies and 103 copies of pgRNA per milligram of tissue, which was at or below the limit of detection of the assay. For WMHBV‐infected animals with human hepatocytes, viral DNA and pgRNA were undetectable. WMHBV‐infected mice with squirrel monkey hepatocytes had 105 viral DNA copies/mg and more than 107 copies of WMHBV pgRNA/mg. Taken together, these data suggest that squirrel monkey hepatocyte engrafted FNRG mice can be infected with WMHBV but not HBV.
Figure 3.
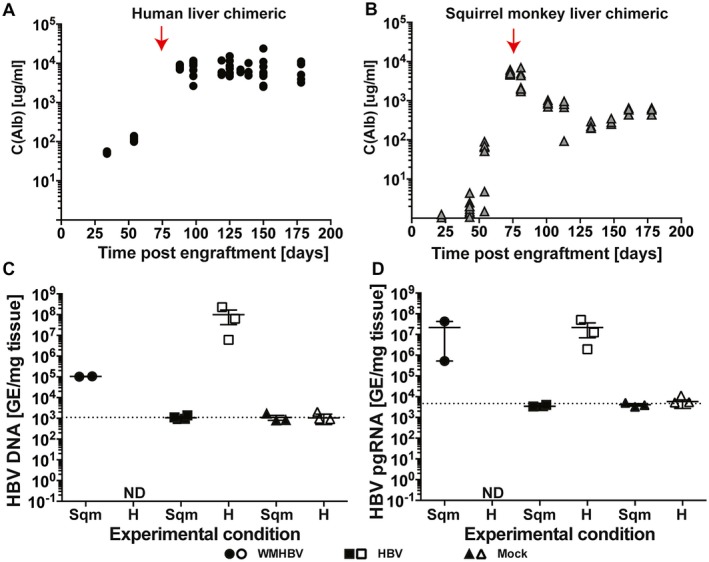
Squirrel monkey liver chimeric FNRG mice support WMHBV infection and transcription. FNRG mice were engrafted with either primary human or squirrel monkey hepatocytes and were subsequently challenged with HBV or WMHBV. Primary hepatocyte engraftment was determined by ELISA for circulating albumin of the respective species. (A) Albumin levels for FNRG mice engrafted with primary human hepatocytes. (B) Albumin levels for FNRG mice engrafted with squirrel monkey hepatocytes. The red arrow depicts when mice were challenged with either HBV or WMHBV. The dotted line indicates the limit of detection for the assay. Quantification of total HBV and WMHBV DNA (C) and pgRNA (D) in liver tissues from human and squirrel monkey liver chimeric mice that had been challenged with either HBV or WMHBV. Abbreviation: Alb, albumin.
Although our previous in vivo and in vitro data14, 24 suggested that spider monkeys might be a good model for WMHBV infection, several species of spider monkeys are endangered and spider monkeys are not normally bred for biomedical research. In contrast, squirrel monkeys are bred for biomedical research, so WMHBV infection was tested in these animals. Similar to the mouse studies, we found that squirrel monkeys inoculated intravenously with 109 GE of HBV did not show signs of infection or viral replication (Fig. 4A). However, inoculation with WMHBV resulted in acute infection with up to 1.1 × 105 GE/mL serum 1 week after infection (Fig. 4B). Viremia was variable and was cleared by 10 weeks following infection. No rise in ALT was detected during the infection.
Figure 4.
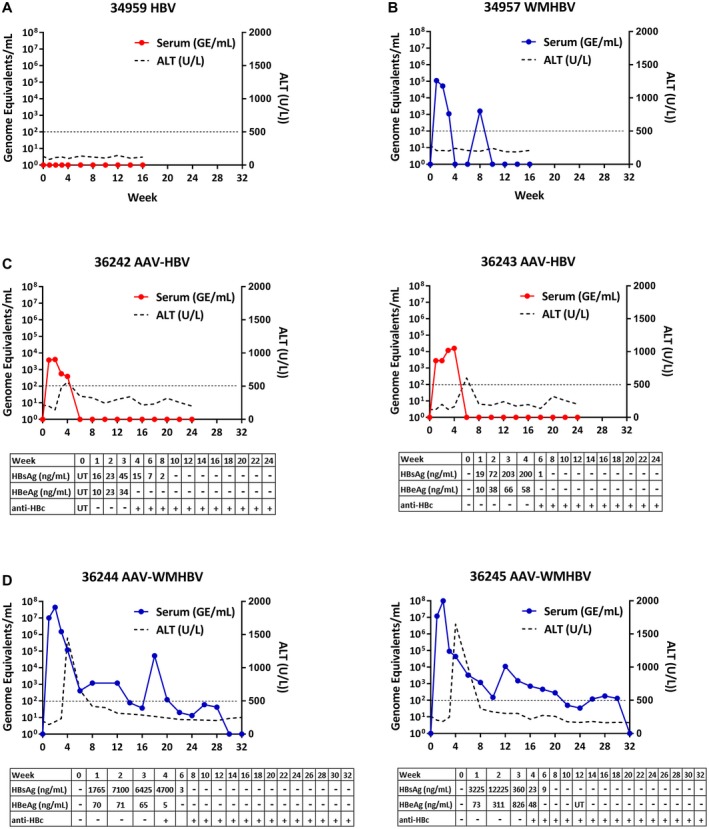
AAV‐WMHBV elicits early high‐titer viremia that remains detectable for up to 8 months. Serum viral titers and ALT levels from adult squirrel monkeys infected with HBV (A), WMHBV (B), AAV‐HBV (C), or AAV‐WMHBV (D). Accompanying tables show ELISA measurements of HBsAg (ng/mL) and HBeAg (ng/mL) as well as the presence of anticore antibodies. The line indicates the limit of detection for the PCR assays, 102 GE/mL of serum.
AAV has been used to deliver the HBV genomes to mice as a mechanism to infect murine hepatocytes normally refractory to infection.4, 25 We hypothesized that the weak acute infection we observed with WMHBV infection of squirrel monkeys might be due to modest efficiency of infection of squirrel monkey hepatocytes. To improve infection, we inoculated two squirrel monkeys with AAV‐HBV and two squirrel monkeys with AAV‐WMHBV. AAV8 is the serotype of AAV, used because of its strong liver tropism. Squirrel monkeys inoculated with 5.0 × 1012 AAV‐HBV viral particles experienced an acute infection of short duration (Fig. 4C). Viral titers peaked between weeks 2 and 4 after infection, reaching 104 GE/mL serum. In contrast, squirrel monkeys inoculated with AAV‐WMHBV exhibited peak viral titers of 9.9 × 107 GE/mL serum at week 2, and the virus was detectable for up to 32 weeks (Fig. 4D). Unlike the viral infection with serum containing WMHBV, ALT flared to 1.5 × 103 U/L at week 4 following infection, after which viremia decreased to between 102 and 103 GE/mL serum. HBsAg was detectable for 6 weeks following AAV‐WMHBV infection (peak 7.0 × 103 ng/mL to 1.2 × 104 ng/mL). This corresponds well with the levels observed in patients with HBV during the immune tolerant phase having 104 ng/mL to 105 ng/mL. HBeAg was detectable for 4 weeks, and antibodies to hepatitis B core antigen (anti‐HBc) were detected starting at 4 weeks.
Liver tissue was collected for nucleic acid characterization. Liver DNA was assayed for both genomic viral DNA by PCR and for cccDNA by southern blot. Total RNA was used to determine HBV RNA copies. In contrast to the circulating viral genomes, liver HBV genomes were relatively stable at 4 and 14 weeks (Fig. 5A). Copy numbers ranged from 3.1 × 103 to 8.2 × 104 per microgram of liver DNA. In contrast, viral transcripts decreased by 2‐4 logs between weeks 4 and 14, with WMHBV decreasing from 4.5 × 107 to 1.2 × 103 GE/µg of liver RNA (Fig. 5B). These data suggest that AAV vector may have contributed to the liver viral DNA, while replication dramatically decreased after week 4.
Figure 5.
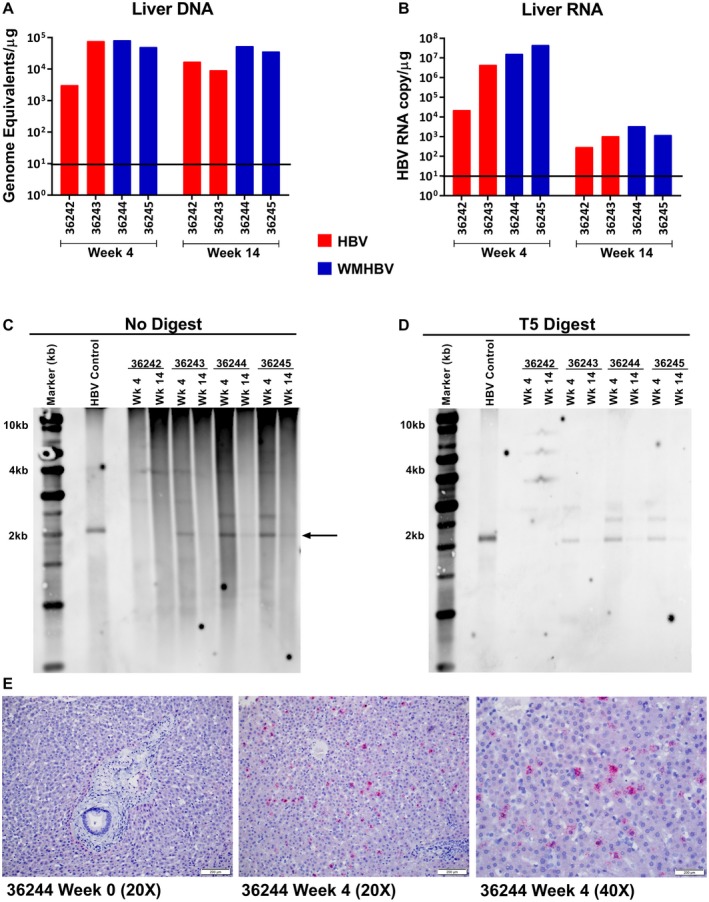
HBV cccDNA and viral transcripts are detectable in the liver of AAV‐WMHBV‐infected squirrel monkeys. Squirrel monkeys that had been infected with AAV‐HBV or AAV‐WMHBV had viral DNA and RNA isolated from the liver. (A) HBV and WMHBV genome copies were measured from the total liver DNA of the indicated adult squirrel monkeys at weeks 4 and 14. The line indicates the limit of detection for the PCR assays, 101 GE/g of DNA. (B) RNA transcripts from liver tissue were measured by RT‐PCR. The line indicates the limit of detection for the PCR assay, 101 GE/g of RNA. (C) DNA from the liver DNA were analyzed by southern blot for viral DNA. Arrow highlighting the 2kB band indicates the predicted band size for cccDNA. (D) DNA from liver biopsies was treated with T5 nuclease to digest non‐supercoiled DNA and then analyzed by southern blot. cccDNA is resistant to T5 digestion. (E) In situ hybridization (RNAscope) for woolly monkey RNA in formalin‐fixed liver sections from squirrel monkey 36244, before and after infection with AAV‐WMHBV.
Southern blot for supercoiled viral DNA revealed bands co‐migrating with the positive control circular HBV DNA in the two AAV‐WMHBV‐infected animals as well as one of the AAV‐HBV‐infected animals (Fig. 5C). These bands were not affected by treatment with T5 nuclease, which degrades relaxed circular and other non‐supercoiled forms of the viral DNA (Fig. 5D). Band intensity decreased in all cases between weeks 4 and 14 following infection. In addition to the cccDNA co‐migrating with the control circular HBV DNA, slower migrating bands were observed that may represent AAV‐HBV and AAV‐WMHBV concatamers.26
To further confirm the presence of WMHBV RNA transcripts in squirrel monkey hepatocytes and to estimate the percentage of infected cells, in situ hybridization was performed on liver sections from the week‐4 postinfection biopsies. RNAscope probes were designed against the WMHBV × region to detect all viral transcripts (Fig. 5E). Based on this assay, approximately 13% of hepatocytes were positive at week 4.
Next, we examined the liver tissue for gene‐expression markers consistent with an adaptive immune response. Quantitative RT‐PCR was used to determine the levels of the transcripts for IFN gamma, and the IFN gamma–induced genes CXCL9 (i.e., MIG) and CXCL10 (i.e., IP‐10) (Fig. 6A). The data are expressed as the fold difference in transcript levels in comparison to four uninfected animals. The two AAV‐WMHBV‐infected animals (36244, 36245) had 4.3‐fold and 3.7‐fold elevations for IFN gamma at week 4, but no increase at week 14. AAV‐HBV animals exhibited no increase in IFN gamma at either time point (data not shown). Consistent with the increase in IFN gamma expression in AAV‐WMHBV animals at week 4 following infection, CXCL9 was elevated 3.8‐fold and 3.1‐fold, and CXCL10 was elevated 2.9‐fold and 2.4‐fold. In addition, to determine whether the ALT flare was associated with significant death and replacement of hepatocytes, as would be expected if an adaptive immune response had eliminated infected cells, liver sections were stained for the cell death marker activated caspase‐3 and the proliferation marker Ki67. Numerous activated caspase‐3‐positive cells were observed in the liver of AAV‐WMHBV‐infected animals at week 4 (Fig. 6B, upper left panel), but not at week 14 (Fig. 6B, lower left panel), or either time point for AAV‐HBV‐infected animals (data not shown). In AAV‐WMBHV‐infected animals at week 4, up to 13% of hepatocytes stained for Ki67, indicating replacement of a large number of hepatocytes (Fig. 6B upper right panel). At week 14, the number of cells staining for Ki67 was minimal (Fig. 6B lower right panel). Very few Ki67 positive cells were observed in the liver of AAV‐HBV‐infected animals at week 4 or 14 (data not shown). Collectively, these data are consistent with an adaptive immune response that eliminated most of the cells expressing WMHBV antigens, as AAV8 itself does not induce liver injury.
Figure 6.
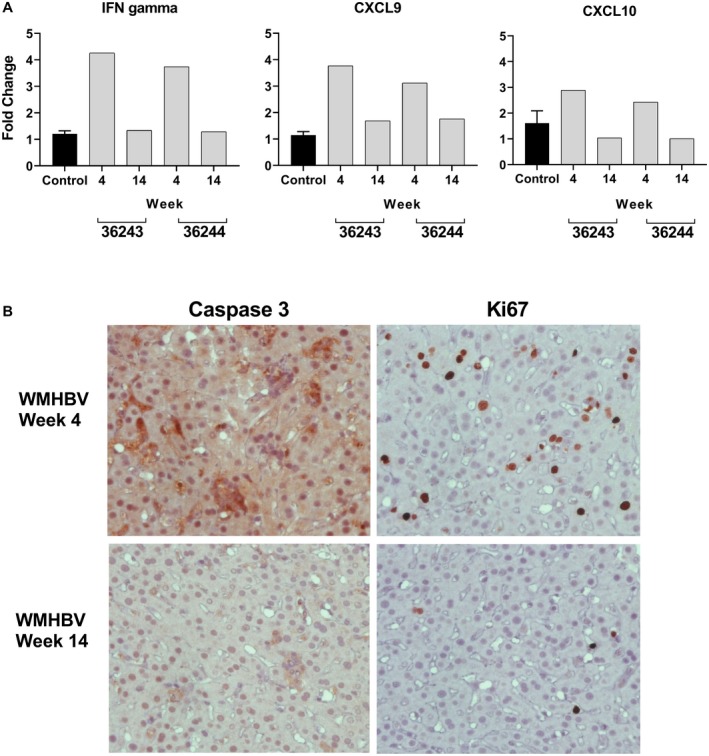
Changes in IFN gene expression and markers of cell death and proliferation in the liver of AAV‐WMHBV‐infected monkeys. (A) TaqMan RT‐PCR was performed on total liver RNA from four control animals (C) and two animals infected with AAV‐WMHBV (36244, 36245) at 4 and 14 weeks following infection. Primers and probes for squirrel monkey IFN gamma, CXCL9 (i.e., Mig), and CXCL10 (i.e., IP‐10) were designed based on the genomic sequence of squirrel monkeys as described in the Materials and Methods section. The values for infected animals are expressed as a fold change in comparison to the controls. C is the average of four animals and is expressed as a fold change of 1, no fold change, with the range shown. (B) Immunohistochemical staining of liver sections from an AAV‐WMHBV‐infected monkey at week 4 and 14 for the cell death marker–activated caspase‐3 (left, upper, and lower panels) and for the cell proliferation maker Ki67 (right, upper, and lower panels).
Most adults infected with HBV do not become chronically infected. However, up to 90% of people born from HBV‐positive mothers become chronically infected through vertical transmission. In an attempt to replicate vertical transmission in squirrel monkeys, 6‐day‐old squirrel monkey neonates were infected with 108 GE of WMHBV. Serum viral DNA was quantified by PCR. All six of the inoculated animals had detectable viremia for 3 months, and four of the animals had detectable viremia greater than 102 GE/mL at 6 months (Fig. 7). A 4‐log increase in viral DNA occurred in one animal at 3 months and coincided with an increase in HBsAg (animal number 6560 with maximum viral titer of 2.7 × 105 ng/mL and 3.6 × 102 ng/mL HBsAg) (data not shown). However, viral clearance occurred in the following month.
Figure 7.
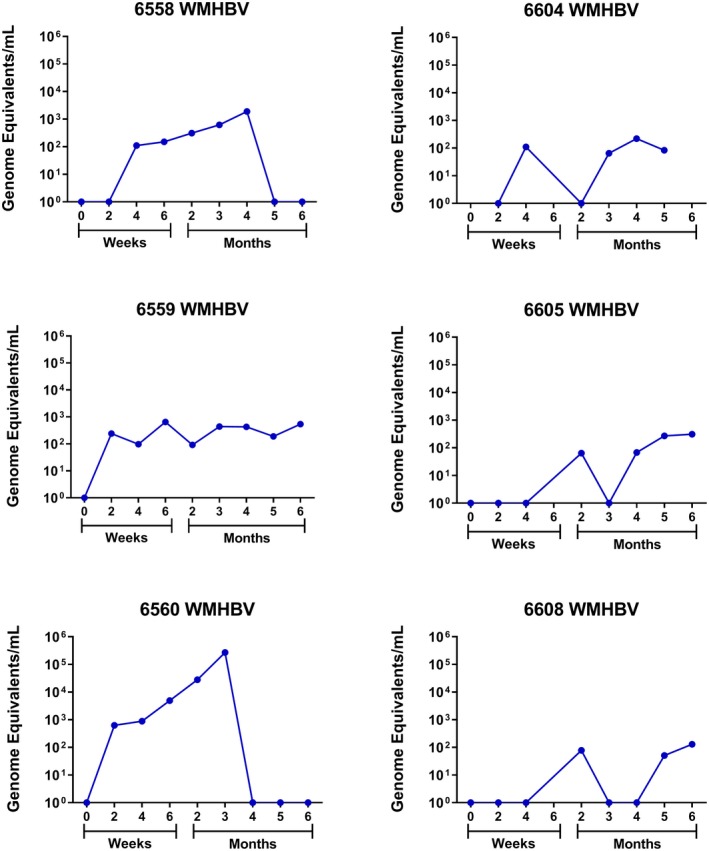
Squirrel monkeys infected with WMHBV as neonates remain viremic for up to 6 months. Quantitation of WMHBV genomes in serum from six squirrel monkey neonates infected with WMHBV.
Discussion
The development of new therapies for curing chronic HBV infection would be greatly accelerated by the availability of a robust animal model of chronic infection, preferably a nonhuman primate due to the similarities to humans. Chimpanzees were the only research animal that faithfully recapitulated chronic human infection, but because chimpanzees are no longer available for biomedical research, a new model is needed.
Due to the conserved glycine at position 158 of the HBV receptor NTCP, squirrel monkeys were hypothesized to be a suitable NHP host for both HBV and WMHBV (Fig. 1A). This was supported by Myrcludex B binding data from both Muller et al.17 and our study. In addition, in accordance with that report, we independently confirmed that cynomolgus macaques are not permissive to Myrcludex B binding.
We then generated FNRG liver chimeric mice with both human and squirrel monkey hepatocytes. Mice engrafted with human hepatocytes supported very high levels of HBV replication, but not WMHBV. No replication of HBV was detected in mice with squirrel monkey hepatocytes, whereas these animals were able to support infection and replication of WMHBV. The same was noted in vivo in squirrel monkeys themselves. Squirrel monkeys were not permissive to HBV infection but supported WMHBV infection. In addition, delivery of the WMHBV genome to the liver using AAV vectors in adult squirrel monkeys yielded high‐level viremia, which remained detectable for up to 8 months. We also confirmed persistent WMHBV infection in neonate squirrel monkeys without the need to use AAV vector delivery.
In vitro studies have suggested that hNTCP expression in primary hepatocytes from cynomolgus macaques, rhesus macaques, and pigs render these cells susceptible to hepatitis delta virus (HDV) and presumably HBV infection and replication.23 This concept was recently tested in vivo for macaques. Burwitz et al.12 infected rhesus macaques with HBV after first infecting these animals with AAV or helper‐dependent adenovirus‐expressing hNTCP. Three of four animals showed some evidence of HBV replication. The maximum viral load obtained was 103 GE/mL serum, and viremia was only present for 8 weeks. cccDNA was not detected in these macaques. Improvement upon this model may require a much higher percentage of AAV or helper‐dependent adenovirus transduction in the liver or creation of an hNTCP transgenic monkey. It is also possible that the in vitro data from macaque or pig hepatocytes expressing hNTCP may not predict in vivo replication due to the need for numerous host factors that support HBV replication. HDV infection only models the entry step for HBV replication.
Two other nonhuman primate models have also had limited success in supporting chronic HBV or WMHBV infection. A genotype D subtype ayw3 HBV variant isolated from Mauritius Island cynomolgus macaques was used to infect Barbary macaques with maximum viral titers of 1.5 × 103 GE/mL.11 However, attempts to replicate those findings were unsuccessful.12 When we initially isolated WMHBV, we examined spider monkeys as surrogate hosts, since woolly monkeys are endangered. In that study, maximum viral titers of 105 GE/mL were obtained with viremia that lasted only six weeks. The addition of the T‐cell inhibitor FK506 (tacrolimus) extended viremia through 16 weeks but had no effect on maximum viral titer.14 Infecting spider monkeys with AAV‐WMHBV may be worthwhile to test; however, the poor availability of spider monkeys for biomedical research as well as the lack of robust reagents for studying host immune response in these animals will continue to make it a challenging model to develop further.
The squirrel monkey WMHBV infection data presented in this study are a step forward toward modeling human HBV infection. The viral titers approaching 108 GE/mL serum are several logs higher than observed in any other nonhuman primate model. Similarly, the 8 months of viremia obtained in adult squirrel monkeys infected with AAV‐WMHBV and the 6 months of viremia observed in neonates infected with WMHBV are essentially double the duration of previous nonhuman primate infections. The ability to obtain these results in an immunocompetent animal that supports the formation of cccDNA using a virus that is genetically close to HBV is also a major strength of this model. Despite the progress made in the squirrel monkey model, it is still limited in the low viral titers that are sustained, the clearing of HBsAg, the formation of anti‐HBc antibodies, and the eventual clearance of the virus. Several approaches for improvement of this model will be pursued in the future. Adaptation of the virus to the squirrel monkey by serial passage of the virus may improve replication in both adults and neonates without the use of an AAV vector. This approach has been successful for a number of viruses in adaptation to a different species, presumably due to selection of variants that improve interactions with host proteins needed for replication or evasion of the host innate and adaptive immune responses. Improved replication and longer duration of viremia will provide opportunities for induction of chronic infection, more detailed characterization of the immune response, and potential evaluation of novel therapeutics. Numerous immunological reagents are available for these studies that are cross‐reactive to squirrel monkey, as described in Nehete et al.28 and the National Institutes of Health Nonhuman Primate Resource (http://www.nhpreagents.org), but newer technologies may prove to be more efficient, such as the RNAscope technology used in Fig. 5. This technology allows synthesis of probes for any gene of interest and is not limited by cross‐reactive antibodies that may or may not be fully cross‐reactive.
A benefit of the squirrel monkey–WMHBV infection model described here over AAV‐HBV‐infected mice is 2‐fold. First, squirrel monkeys are not immune‐tolerant to viral antigens, which is the case for AAV‐HBV mice.4, 27 Immune tolerance could only be broken in these mice by vaccination with HBV DNA or HBsAg/cytosine‐guanine dinucleotide. Second, squirrel monkeys are more genetically similar to humans than mice; thus, the host response of squirrel monkeys to WMHBV antigens may better mimic what is observed in human patients chronically infected with HBV. There are numerous antibodies that can be used to immune phenotype squirrel monkey cells,28 and newer technologies such as RNA flow29 and RNA scope30 can be used to investigate the squirrel monkey host response to infection.
It is likely that future studies combining several of these methods will lead to a better model of chronic HBV infection. Launching WMHBV infection in neonate squirrel monkeys using an AAV‐WMHBV would combine two of the methods that were shown to improve infection and duration of viremia. Infecting the adult squirrel monkeys with AAV‐WMHBV in the presence of immune suppression (as done previously with the spider monkeys) could also lead to immune tolerance of WMHBV proteins and enable the selection of a squirrel monkey–adapted virus or a T‐cell escape mutant. NTCP sequences from additional nonhuman primate species suggest alternatives to the squirrel monkey. G158 is also conserved in marmosets. Replication of both WMHBV and HBV in marmosets should be explored, perhaps initially using AAV vectors to initiate high titer infection. The high fecundity of marmosets makes them amenable to prenatal infection studies, large‐scale preclinical trials, and perhaps generation of an hNTCP transgenic animal.
Supporting information
Supported by: Gilead Sciences; The Southwest National Primate Research Center resources are supported by NIH grant P51‐OD011133 from the Office of Research Infrastructure Programs/Office of the Director; The Michale E. Keeling Center for Comparative Medicine and Research, Squirrel Monkey Breeding and Research Resource is supported by NIH Grant P41‐OD010938‐38 from the Office of Research Infrastructure Programs/Office of the Director. In addition, A.P. is supported by a Burroughs Wellcome Fund Award for Investigators in Pathogenesis and B.Y.W. is supported by and an NIH/NRSA Ruth L. Kirschstein Predoctoral award.
Potential conflict of interest: Drs. Chen and Lanford received grants from Gilead. Dr. Eng owns stock in and is employed by Gilead. Dr. Salas owns stock in and is employed by Gilead. Dr. Delaney owns stock in, is employed by, and has intellectual property rights in Gilead. Dr. Ploss received grants from Gilead. Dr. Voitenleitner owns stock in Gilead.
References
Authors names in bold designate shared‐first authorship.
- 1. Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 2015;386:1546‐1555. [DOI] [PubMed] [Google Scholar]
- 2. Winer BY, Shirvani‐Dastgerdi E, Bram Y, Sellau J, Low BE, Johnson H, et al. Preclinical assessment of antiviral combination therapy in a genetically humanized mouse model for hepatitis delta virus infection. Sci Transl Med 2018;10:eaap9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deng Q, Mancini‐Bourgine M, Zhang X, Cumont MC, Zhu R, Lone YC, et al. Hepatitis B virus as a gene delivery vector activating foreign antigenic T cell response that abrogates viral expression in mouse models. Hepatology 2009;50:1380‐1391. [DOI] [PubMed] [Google Scholar]
- 4. Dion S, Bourgine M, Godon O, Levillayer F, Michel ML. Adeno‐associated virus‐mediated gene transfer leads to persistent hepatitis B virus replication in mice expressing HLA‐A2 and HLA‐DR1 molecules. J Virol 2013;87:5554‐5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang LR, Wu HL, Chen PJ, Chen DS. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc Natl Acad Sci U S A 2006;103:17862‐17867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lucifora J, Salvetti A, Marniquet X, Mailly L, Testoni B, Fusil F, et al. Detection of the hepatitis B virus (HBV) covalently‐closed‐circular DNA (cccDNA) in mice transduced with a recombinant AAV‐HBV vector. Antiviral Res 2017;145:14‐19. [DOI] [PubMed] [Google Scholar]
- 7. de Jong YP, Dorner M, Mommersteeg MC, Xiao JW, Balazs AB, Robbins JB, et al. Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci Transl Med 2014;6:254ra129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walter E, Keist R, Niederost B, Pult I, Blum HE. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology 1996;24:1‐5. [DOI] [PubMed] [Google Scholar]
- 9. Mason WS, Seal G, Summers J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol 1980;36:829‐836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winer BY, Ploss A. Determinants of hepatitis B and delta virus host tropism. Curr Opin Virol 2015;13:109‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dupinay T, Gheit T, Roques P, Cova L, Chevallier‐Queyron P, Tasahsu SI, et al. Discovery of naturally occurring transmissible chronic hepatitis B virus infection among Macaca fascicularis from Mauritius Island. Hepatology 2013;58:1610‐1620. [DOI] [PubMed] [Google Scholar]
- 12. Burwitz BJ, Wettengel JM, Muck‐Hausl MA, Ringelhan M, Ko C, Festag MM, et al. Hepatocytic expression of human sodium‐taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques. Nat Commun 2017;8:2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lanford RE, Chavez D, Brasky KM, Burns RB 3rd, Rico‐Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci U S A 1998;95:5757‐5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lanford RE, Chavez D, Barrera A, Brasky KM. An infectious clone of woolly monkey hepatitis B virus. J Virol 2003;77:7814‐7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 2012;1:e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, et al. Hepatitis B and D viruses exploit sodium taurocholate co‐transporting polypeptide for species‐specific entry into hepatocytes. Gastroenterology 2014;146:1070‐1083. [DOI] [PubMed] [Google Scholar]
- 17. Muller SF, Konig A, Doring B, Glebe D, Geyer J. Characterisation of the hepatitis B virus cross‐species transmission pattern via Na+/taurocholate co‐transporting polypeptides from 11 New World and Old World primate species. PLoS ONE 2018;13:e0199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lanford RE, Estlack L. A cultivation method for highly differentiated primary chimpanzee hepatocytes permissive for hepatitis C virus replication. Methods Mol Med 1999;19:501‐515. [DOI] [PubMed] [Google Scholar]
- 19. Wooddell CI, Yuen MF, Chan HL, Gish RG, Locarnini SA, Chavez D, et al. RNAi‐based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 2017;9:eaan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winer BY, Huang T, Low BE, Avery C, Pais MA, Hrebikova G, et al. Recapitulation of treatment response patterns in a novel humanized mouse model for chronic hepatitis B virus infection. Virology 2017;502:63‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winer BY, Huang TS, Pludwinski E, Heller B, Wojcik F, Lipkowitz GE, et al. Long‐term hepatitis B infection in a scalable hepatic co‐culture system. Nat Commun 2017;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barrera A, Guerra B, Notvall L, Lanford RE. Mapping of the hepatitis B virus pre‐S1 domain involved in receptor recognition. J Virol 2005;79:9786‐9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lempp FA, Wiedtke E, Qu B, Roques P, Chemin I, Vondran FWR, et al. Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes. Hepatology 2017;66:703‐716. [DOI] [PubMed] [Google Scholar]
- 24. Barrera A, Guerra B, Lee H, Lanford RE. Analysis of host range phenotypes of primate hepadnaviruses by in vitro infections of hepatitis D virus pseudotypes. J Virol 2004;78:5233‐5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang YH, Fang CC, Tsuneyama K, Chou HY, Pan WY, Shih YM, et al. A murine model of hepatitis B‐associated hepatocellular carcinoma generated by adeno‐associated virus‐mediated gene delivery. Int J Oncol 2011;39:1511‐1519. [DOI] [PubMed] [Google Scholar]
- 26. Penaud‐Budloo M, Le Guiner C, Nowrouzi A, Toromanoff A, Cherel Y, Chenuaud P, et al. Adeno‐associated virus vector genomes persist as episomal chromatin in primate muscle. J Virol 2008;82:7875‐7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang D, Liu L, Zhu D, Peng H, Su L, Fu YX, et al. A mouse model for HBV immunotolerance and immunotherapy. Cell Mol Immunol 2014;11:71‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nehete PN, Hanley PW, Nehete BP, Yang G, Ruiz JC, Williams L, et al. Phenotypic and functional characterization of lymphocytes from different age groups of Bolivian squirrel monkeys (Saimiri boliviensis boliviensis). PLoS ONE 2013;8:e79836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Douam F, Hrebikova G, Albrecht YE, Sellau J, Sharon Y, Ding Q, et al. Single‐cell tracking of flavivirus RNA uncovers species‐specific interactions with the immune system dictating disease outcome. Nat Commun 2017;8:14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin‐fixed, paraffin‐embedded tissues. J Mol Diagn 2012;14:22‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


