Abstract
The transcription factor promyelocytic leukemia zinc finger protein (PLZF) is involved in the development of natural killer (NK) cells and innate lymphoid cells, including liver‐resident NK cells in mice. In human NK cells, the role of PLZF in liver residency is still unknown. Expression of PLZF in matched human peripheral blood‐ and liver‐derived NK cells and the association of PLZF expression with surface molecules and transcription factors relevant for tissue residency were investigated using multiparameter flow cytometry and assessing single‐cell messenger RNA (mRNA) levels. Intrahepatic cluster of differentiation (CD)56bright NK cells expressed significantly higher levels of PLZF than peripheral blood CD56bright NK cells, which were predominantly PLZFlo. Expression of PLZF was highest within C‐X‐C motif chemokine receptor 6 (CXCR6)+CD69+ liver‐resident NK cells among intrahepatic CD56bright NK cell populations. Association of PLZF with liver‐residency markers was also reflected at mRNA levels. A small PLZFhiCD56bright NK cell population was identified in peripheral blood that also expressed the liver‐residency markers CXCR6 and CD69 and shared functional characteristics with liver‐resident NK cells. Conclusion: PLZF is implicated as part of a transcriptional network that promotes liver residency of human NK cells. Expression of liver‐homing markers on peripheral blood PLZFhiCD56bright NK cells identifies an intermediate population potentially contributing to the maintenance of liver‐resident NK cells.
High expression of PLZF was associated with co‐expression of the liver‐residency markers CXCR6 and CD69 on human CD56bright NK cells not only in livers but also in a small NK cell subset in peripheral blood. These CXCR6+CD69+PLZFhi CD56bright NK cells in peripheral blood furthermore displayed functional characteristics similar to liver‐resident NK cells, suggesting that these cells might represent an intermediate stage and potentially contribute to the maintenance of liver‐resident NK cells.

Abbreviations
- APC
Allophycocyanin
- BUV
Brilliant Ultraviolet
- BV
Brilliant Violet
- CCR6
C‐C chemokine receptor type 6
- CD
cluster of differentiation
- Ct
cycle threshold
- CXCR6
C‐X‐C motif chemokine receptor 6
- Eomes
eomesdermin
- FDR
false discovery rate
- FITC
Fluorescein Isothiocyanate
- hi
high
- IFN‐γ
interferon‐gamma
- IHL
intrahepatic leukocyte
- ihNK
intrahepatic natural killer
- IL
interleukin
- ILC
innate lymphoid cell
- int
intermediate
- IQR
interquartile range
- LOD
limit of detection
- lrNK
liver‐resident natural killer
- lo
low
- MdFI
median fluorescence intensity
- mRNA
messenger RNA
- NK
natural killer
- NKG2C
killer cell lectin like receptor C2
- NKT
natural killer T
- PBMC
peripheral blood mononuclear cell
- pbNK
peripheral blood natural killer
- PBS
phosphate‐buffered saline
- PCR
polymerase chain reaction
- PE
Phycoerythrin
- PLZF
promyelocytic leukemia zinc finger protein
- t‐SNE
t‐Distributed Stochastic Neighbor Embedding
- T‐bet
T‐box transcription factor 21
- TNF‐α
tumor necrosis factor alpha
- v
volume
Natural killer (NK) cells are part of the innate immune system and provide a first line of defense against viral infections and malignancies.1, 2 NK cells have furthermore been suggested to regulate tissue homeostasis and tissue regeneration.2, 3 There is mounting evidence that the interplay between tissue‐resident immune cells and stromal cells contributes to an immunologic environment that is highly adapted to the requirements of a particular organ.4 Despite the emerging role of NK cells in tissue immunity and development,5, 6 factors regulating tissue residency of NK cells in humans remain largely unknown. In healthy humans, peripheral blood NK (pbNK) cells exhibit a broad variety of cellular subsets.7 Conventionally, human NK cells have been characterized by their expression of cluster of differentiation (CD)56 and CD16 into CD56bright and CD56dim NK cells, the latter representing the majority of pbNK cells.8, 9 Tissue‐resident NK cells exhibit phenotypic and functional differences compared to pbNK cells.10, 11, 12, 13, 14, 15, 16, 17 In human liver, about 40% of all lymphocytes are NK cells,18 which include a population of CD56bright liver‐resident NK (lrNK) cells.6, 10, 11, 12 The largest human lrNK cell subset co‐expresses C‐X‐C motif chemokine receptor 6 (CXCR6) and CD6910, 16 and exhibits an eomesdermin (Eomes)hiT‐box transcription factor 21 (T‐bet)lo transcription factor profile.10, 11, 17 In addition, a small distinct CD49a+ lrNK cell population has been described.12, 13, 16 Accumulation of these cells in liver tissue indicates their residing state, yet a subset of NK cells in peripheral blood also expresses CXCR6.10, 14, 19
The transcription factor promyelocytic leukemia zinc finger protein (PLZF; synonym ZBTB16) has been sufficient to induce retention of PLZF‐transgenic CD4+ T cells in murine liver.20 PLZF is furthermore highly expressed in murine innate lymphoid cell (ILC) precursors, which give rise to several ILC classes and NK cells.21, 22, 23, 24 These PLZFhi ILC precursors were able to generate high numbers of lrNK cells,21, 22, 23 which are considered to be ILC1s in mice.25 In human NK cells, PLZF expression has also been detected26, 27 but varies between different NK cell subsets.28, 29 The contribution of PLZF to the mechanisms regulating tissue homing and tissue residency of human NK cells remains unknown. In this study, we show that among CD56bright intrahepatic NK (ihNK) cells, PLZF is highly associated with markers of liver residency. We report the existence of a small population of PLZFhiCD56bright NK cells in peripheral blood, expressing the liver‐homing receptors CXCR6 and CD69 and sharing functional characteristics with lrNK cells.
Materials and Methods
Study Design and Participants
Liver tissue and matched peripheral blood samples were collected at the University Medical Center Hamburg‐Eppendorf (UKE), Hamburg, Germany, and the Asklepios Clinic Barmbek (AKB), Hamburg, Germany. Peripheral blood (60‐80 mL) was drawn immediately before or at the beginning of surgery. At the UKE, samples were collected from adult individuals who underwent liver transplantation. At the AKB, noncirrhotic nontumorous liver tissue was taken from resected livers of adult individuals who suffered from hepatic metastases. Clinical characteristics of study participants are summarized in Table 1. For in vitro experiments, peripheral blood of 7 healthy individuals was collected at the Heinrich Pette Institute in Hamburg. Written informed consent was obtained from all study participants. The study protocols were approved by the ethics committee of the medical association of Hamburg (Ärztekammer Hamburg; PV4898, PV4081, and PV4780). No donor organs were obtained from executed prisoners or other institutionalized persons. Randomization or blinding was not applied in this study.
Table 1.
Clinical Data of Individuals From the Transplant and Resection Cohort Included in This Study
| Data Set | (1) | (2) | (3) | (4) | All |
|---|---|---|---|---|---|
| Figures Corresponding to Data Set | 1‐3, S1, S2 | 4 | 5A,B; S4A,B; | 5C‐E; S4C | |
| Median (Range) or n | |||||
| Available samples | |||||
| Liver samples | 12 | 4 | 7 | 6 | 24* |
| Matched PBMC samples | 8 | 4 | 7 | 6 | 20* |
| Demographics | |||||
| Sex (F/M) | 3/9 | 2/2 | 2/5 | 3/3 | 9/16 |
| Median age (years) | 58 (48‐69) | 57 (43‐68) | 63 (51‐68) | 66 (50‐79) | 61 (43‐79) |
| Primary liver disease (transplant cohort) | |||||
| HCV | 4† | 0 | 0 | 0 | 4† |
| ALD | 0 | 0 | 1 | 0 | 1 |
| HCC | 3 | 0 | 2‡ | 0 | 4* , ‡ |
| NAFLD | 1 | 0 | 0 | 0 | 1 |
| AIH | 0 | 0 | 0 | 1 | 1 |
| Retransplantation | 1 | 0 | 0 | 1 | 2 |
| Primary cancer (resection cohort) | |||||
| Colorectal cancer | 2 | 3 | 4 | 4 | 9* |
| Uveal melanoma | 1 | 0 | 0 | 0 | 1 |
| Hepatic adenomatosis | 0 | 1 | 0 | 0 | 1 |
| CMV | |||||
| Positive/negative/unknown | 9/3/0 | 4/0/0 | 3/1/3 | 6/0/0 | 17/4/3 |
Samples of 4 donors with either HCC or colorectal cancer were used in multiple experiments.
Two individuals with HCV had also developed HCC.
One individual had HCC based on ALD.
Abbreviations: AIH, autoimmune hepatitis; ALD, alcoholic liver disease; CMV, cytomegalovirus serological status; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; NAFLD, nonalcoholic fatty liver disease.
Cell Isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood using standard density gradient centrifugation with Ficoll (Biochrom GmbH, Berlin, Germany). Liver‐derived cells were mechanically segregated from structural components without enzymatic treatment, as described.12 For functional experiments, intrahepatic leukocytes (IHLs) were purified through a density gradient centrifugation with OptiPrep (Sigma, Munich, Germany).30 PBMCs and hepatic cells from the liver transplantation and resection cohort were cryopreserved in liquid nitrogen and thawed for analysis.
Surface and Intracellular Staining of Immune Cells
PBMCs and liver‐derived cells were washed with phosphate‐buffered saline (PBS) before being incubated at room temperature in the dark for 20 minutes with surface antibodies and Zombie NIR or Zombie Aqua (BioLegend, San Diego, CA). Antibodies used in this study are listed within the Supporting Data. Afterwards, cells were washed with PBS and either fixed with 4% (mass/volume [v]) paraformaldehyde or stained intracellularly. For permeabilization and fixation during intracellular staining, the Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific, Waltham, MA) was used according to the manufacturer's protocol. Cells were analyzed using an LSR Fortessa flow cytometer (BD Bioscience, San Jose, CA). Generated flow cytometry standard data were evaluated with FlowJo version 10.7 (FlowJo, LLC, Ashland, OR).
Degranulation Assay
IHL and matched PBMC samples were thawed and cells rested in Roswell Park Memorial Institute medium (Thermo Fisher Scientific) supplemented with 10% v/v fetal bovine serum (Biochrom GmbH, Berlin, Germany) and 0.5 ng/mL interleukin (IL)‐15 (PeproTech, Hamburg, Germany) at 37°C and 5% CO2 for 6 hours. Cells were stained with anti‐CD69 (allophycocyanin [APC]) and washed with PBS before being co‐incubated with K562 cells at an effector to target cell (E:T) ratio of 5:1 in the presence of an anti‐CD107a antibody (Brilliant Violet 421 [BV421]) for 6 hours at 37°C and 5% CO2. After 1 hour, monensin (GolgiStop; BD Bioscience, San Jose, CA) and brefeldin A (GolgiPlug) were added.31 Finally, cells were intracellularly stained for cytokines as described above.
Single Cell Messenger RNA Analysis
A Fluidigm platform consisting of C1, Juno, and Biomark HD (Fluidigm, San Francisco, CA) was used to quantify messenger RNA (mRNA) expression of single cells in a targeted approach. Intrahepatic and matched peripheral blood, single, live, CD3−CD14−CD19−CD56brightCD16− NK cells of 4 individuals were sorted with a BD FACS Aria Fusion (BD Bioscience) and individually loaded onto a C1 single‐cell preamp integrated fluidic circuit (IFC), 5‐10 µm. Only microscopically validated capture sites containing one single cell were included in downstream analyses. Complementary DNA from individual cells was generated and pre‐amplified using the C1 with 96 primers, and quantitative polymerase chain reaction (PCR) was performed using the 96.96 Dynamic Array IFC in the Biomark HD according to the manufacturer's protocol. Data were analyzed with Real‐Time PCR Analysis V4.3.1 (Fluidigm). Because normalization of single‐cell expression data has been argued against, it was not applied.32 Cells without detectable mRNA for two out of three housekeeping genes (beta‐2 microglobulin [B2M], ribosomal protein L13a [RPL13A], glyceraldehyde 3‐phosphate dehydrogenase [GAPDH]) were excluded. A threshold cycle (Ct) value of 24 was defined as the limit of detection (LOD), as described,33, 34 therefore all Ct values above the LOD were set to 24. Levels of mRNA are displayed as 2(LOD–Ct).33, 34 The peak detection ranges of the melting curves in the real‐time PCR analysis software were defined as the median temperature peak ±1.2°C of all nonfailed reactions for the respective gene of all cells analyzed. Other settings were kept as factory settings (peak sensitivity, 7; peak ratio threshold, 0.8). The Ct values of all reactions that were marked as failed under these conditions were set to the LOD. Primers used in this study are listed within the Supporting Data.
t‐Distributed Stochastic Neighbor Embedding Analysis
Two‐dimensional display of high‐dimensional data was calculated using viSNE (Cytobank, Santa Clara, CA), based on the Barnes‐Hut implementation of the t‐distributed stochastic neighbor embedding (t‐SNE) algorithm.35
Statistics
Statistical analysis was performed using Prism 8 (GraphPad Software Inc., La Jolla, CA) and R (R‐3.5.1; RStudio 1.1.463, packages ggplot2 3.1.0). Due to the sample sizes, flow cytometry data were not assumed to be distributed normally. Differences between paired samples were analyzed using Wilcoxon signed‐rank tests for matched pairs. Correlations between expression levels of PLZF and liver‐residency markers were assessed using Spearman's rank correlation. Mixed‐effects regression models with a random intercept to account for intrasubject correlation were used for the statistical comparisons of single‐cell mRNA expression data. All P values presented in this study were collectively adjusted for test multiplicity using a false discovery rate (FDR) of 5% by applying the original method of Benjamini and Hochberg.36 FDR‐adjusted P values <0.05 were considered statistically significant.
Results
A Small Subset of CXCR6+CD69+ NK Cells Is Present in Peripheral Blood
NK cells derived from hepatic tissues and matched peripheral blood samples were analyzed using multiparameter flow cytometry. NK cells were defined as single live CD45+CD3−CD14−CD19− lymphocytes that expressed CD56; these were further separated into CD56bright and CD56dim NK cells (Supporting Fig. S1A,B). In line with previous studies,10, 16 viSNE analysis revealed clusters of CD56bright ihNK cells expressing CXCR6 and CD69; these were largely absent in peripheral blood (Fig. 1A). We furthermore observed a significantly increased proportion of CXCR6‐expressing cells among CD56bright pbNK cells compared to CD56dim pbNK cells (Fig. 1B). Expression patterns of CD57, CD62L, and killer cell lectin like receptor C2 (NKG2C) in livers were in line with previous results,10, 11, 19 validating our characterization of ihNK cells (Fig. 1B). Comparing co‐expression, single expression, and no expression of CXCR6 and CD69, ihNK cells harbored a large fraction of CD69+CXCR6+ cells (Fig. 1C). Interestingly, a small but clearly distinguishable and consistent NK cell subset with a CXCR6+CD69+ phenotype was also detected in peripheral blood (Fig. 1C,D). This NK cell subset was also present in peripheral blood of healthy individuals (data not shown). Proportions of CXCR6+CD69+ and CXCR6−CD69− but not of single positive NK cells significantly differed between peripheral blood and liver samples (Fig. 1D). In summary, these results demonstrate the existence of a small NK cell population in peripheral blood co‐expressing the liver‐residency markers CXCR6 and CD69.
Figure 1.
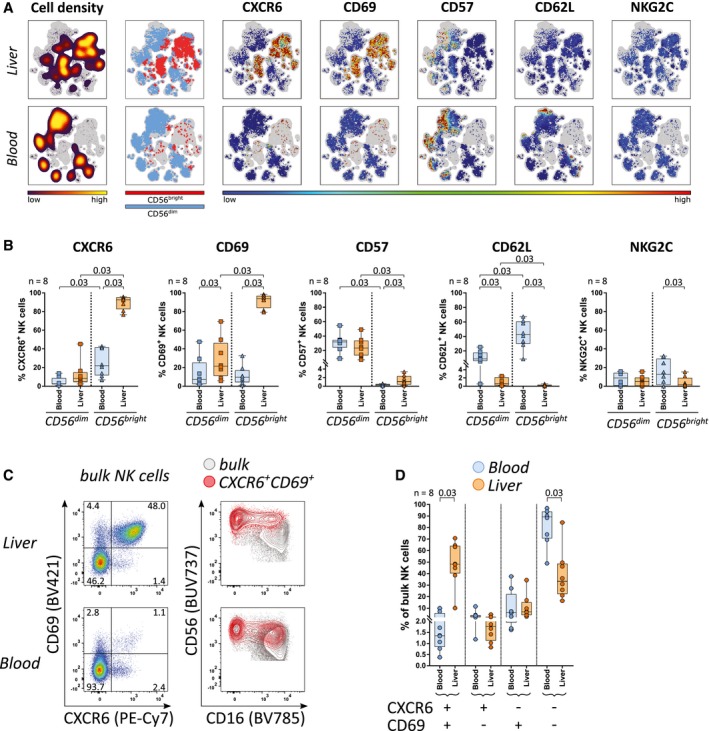
A small subset of CXCR6+CD69+ NK cells is present in peripheral blood. (A) Intrahepatic and peripheral blood NK cells of 1 representative donor were collectively mapped using viSNE. From left to rights, plots display cell density, distribution of CD56bright and CD56dim NK cells, and expression levels of CXCR6, CD69, CD57, CD62L, and NKG2C, as indicated by the color codes shown underneath. (B) Proportions of CXCR6+, CD69+, CD57+, CD62L+, and NKG2C+ NK cells within peripheral blood (blue) and intrahepatic (orange) CD56bright and CD56dim NK cells (n = 8). (C) Representative CXCR6 and CD69 expression among intrahepatic and peripheral blood NK cells (left column) and an overlay of CD69+CXCR6+ NK cells (red) and bulk NK cells (gray) (right column). (D) Proportions of CXCR6/CD69 double‐positive, single‐positive, and double‐negative NK cells in peripheral blood (blue) and liver samples (orange) (n = 8). Boxes indicate median and interquartile range (IQR), whiskers indicate total range. Wilcoxon signed‐rank tests for matched pairs followed by FDR adjustment (B,D) were used for statistical testing (significance value shown at the top of each subpanel). Abbreviation: NKG2C, killer cell lectin like receptor C2.
CD56bright NK Cells in Peripheral Blood and Liver Differentially Express the Transcription Factor PLZF
Next, we assessed the transcription factor profiles of NK cells in peripheral blood and liver samples. NK cells were categorized based on their expression of the transcription factors Eomes (low [lo], intermediate [int], high [hi]), T‐bet (lo, hi), and PLZF (lo, hi) (Fig. 2A). As expected,10, 11 high expression of Eomes and low expression of T‐bet were predominantly observed among CD56bright ihNK cells (Fig. 2B,C). High PLZF expression was detected within the majority of NK cells derived from peripheral blood and liver samples with the exception of CD56bright pbNK cells, which, unlike their intrahepatic counterparts, displayed significantly lower expression of PLZF (Fig. 2B,D). CD56dim and CD56bright ihNK cells further exhibited significantly higher median fluorescence intensity (MdFI) values of PLZF compared to pbNK cells (Fig. 2D). Overall, these data show that CD56bright NK cells significantly differ in their transcription factor profiles between peripheral blood and liver. In addition to the previously reported EomeshiT‐betlo pattern,10, 11 CD56bright ihNK cells also expressed significantly higher levels of PLZF compared to CD56bright pbNK cells.
Figure 2.
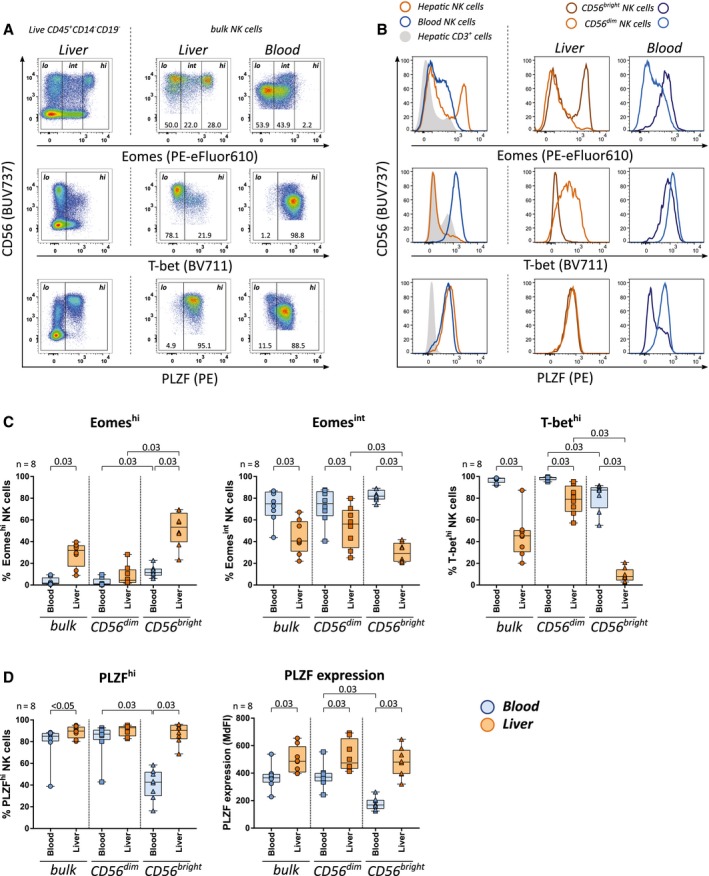
CD56bright NK cells in peripheral blood and liver differentially express the transcription factor PLZF. (A) Gating of low, intermediate, and high expression for Eomes and of low and high expression for T‐bet and PLZF, using CD45+CD3+CD14−CD19− cells as internal control for low expression. (B) Expression of Eomes, T‐bet and PLZF: intrahepatic CD3+ cells and bulk NK cells in peripheral blood and liver (left column); CD56bright and CD56dim NK cells in peripheral blood and liver (middle and right column). (C) Proportions of Eomeshi, Eomesint, and T‐bethi cells and (D) proportions of PLZFhi and MdFI of PLZF in peripheral blood (blue) and intrahepatic (orange) bulk, CD56dim, and CD56bright NK cells (n = 8). Boxes indicate median and IQR; whiskers indicate total range. Wilcoxon signed‐rank tests for matched pairs followed by FDR adjustment (C,D) were used for statistical testing (significance value shown at the top of each subpanel).
PLZF Is Highly Expressed by CXCR6+CD69+CD56bright NK Cells in Liver and Peripheral Blood
We subsequently investigated whether PLZF expression in NK cells was associated with phenotypic characteristics of liver residency. In liver, PLZFhiCD56bright NK cells displayed significantly higher proportions of cells expressing CXCR6, CD69, high levels of Eomes, and low levels of T‐bet compared to PLZFloCD56bright NK cells (Fig. 3A,B). Furthermore, expression levels of PLZF in CD56bright ihNK cells positively correlated with expression levels of CXCR6 and CD69 (r = 0.76, P = 0.03 and r = 0.71, P = 0.03, respectively; Supporting Fig. S2A). Similar to ihNK cells, the fraction of pbNK cells expressing CXCR6 and CD69 was significantly increased within PLZFhiCD56bright pbNK cells compared to PLZFloCD56bright pbNK cells (Fig. 3A,B). These observations indicate a consistent association of high PLZF expression with expression of CXCR6 and CD69 among CD56bright NK cells not only in liver but also in peripheral blood.
Figure 3.
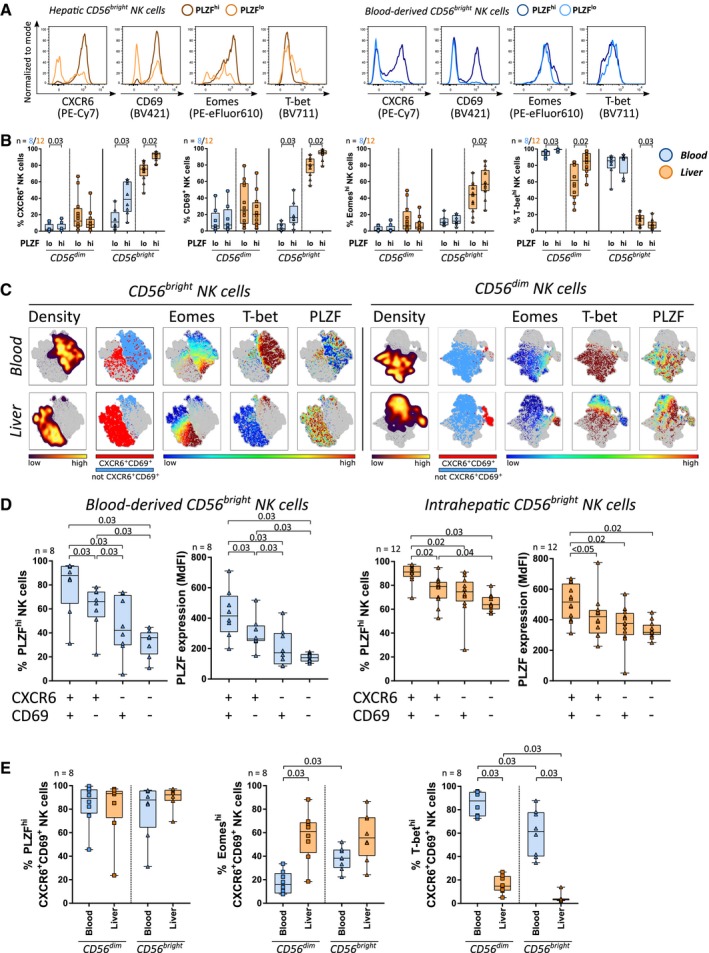
PLZF is highly expressed by CXCR6+CD69+CD56bright NK cells in liver and peripheral blood. (A) Expression of CXCR6, CD69, Eomes, and T‐bet in intrahepatic (orange, left panel) and peripheral blood (blue, right panel) CD56bright NK cells with high (dark) or low (light) PLZF expression. (B) Proportions of CXCR6+, CD69+, Eomeshi, and T‐bethi cells among peripheral blood (blue, n = 8) or intrahepatic (orange, n = 12) CD56bright and CD56dim NK cells with a PLZFhi or PLZFlo profile. (C) Representative viSNE plots showing cell density and clustering of CXCR6+CD69+ NK cells as well as expression of Eomes, T‐bet, and PLZF among CD56bright and CD56dim NK cells in peripheral blood and liver. (D) Proportion of PLZFhi cells and PLZF MdFI among peripheral blood‐derived (n = 8, blue) and intrahepatic (n = 12, orange) CXCR6/CD69 double‐positive, single‐positive, and double‐negative CD56bright NK cells. (E) Proportions of PLZFhi, Eomeshi, and T‐bethi NK cells within CD56dim or CD56bright CXCR6+CD69+ NK cells in peripheral blood (blue) and liver (orange, n = 8). Boxes indicate median and IQR; whiskers indicate total range. Wilcoxon signed‐rank tests for matched pairs followed by FDR adjustment (B,D,E) were used for statistical testing (significance value shown at the top of each subpanel).
Taking into account that lrNK cells are characterized by co‐expression of CXCR6 and CD69, we examined CXCR6+CD69+ NK cells for their PLZF expression. In a viSNE analysis, clusters of CXCR6+CD69+CD56bright pbNK cells overlapped with their liver‐resident counterpart and expressed high levels of PLZF (Fig. 3C). In line with these observations, CXCR6+CD69+CD56bright NK cells in peripheral blood and liver contained significantly higher proportions of PLZFhi cells and expressed significantly higher levels of PLZF compared to CD56bright NK cells with single or no expression of CXCR6 and CD69 (Fig. 3D). Notably, proportions of PLZFhi cells within CXCR6+CD69+CD56bright pbNK cells were comparable to CXCR6+CD69+CD56bright lrNK cells (Fig. 3E). Like most CD56dim pbNK cells, the population of CXCR6+CD69+CD56dim pbNK cells also expressed high levels of PLZF (Fig. 3C,E). In contrast to lrNK cells, the majority of CXCR6+CD69+CD56bright pbNK cells exhibited intermediate or low expression of Eomes and high expression of T‐bet (Fig. 3E). In vitro culture of PBMCs from healthy donors with IL‐12 led to a significant enrichment of PLZFhiCXCR6+CD69+ NK cells (Supporting Fig. S3A‐C), in line with published information.16 In summary, CXCR6+CD69+ NK cells exhibited the highest expression of PLZF among CD56bright NK cells in liver and peripheral blood. CXCR6+CD69+CD56bright NK cells thereby constituted a specific subpopulation of NK cells in peripheral blood that differed from the majority of CD56bright pbNK cells expressing low PLZF levels. Among the investigated transcription factors, only high expression of PLZF was a shared characteristic between CXCR6+CD69+CD56bright pbNK cells and lrNK cells while expression of Eomes and T‐bet differed between those two subsets.
Single‐Cell mRNA Analysis Shows Association of PLZF and Markers of Liver Residency in CD56bright ihNK Cells
To support the results obtained at the protein level, we quantified respective mRNA expression levels in unstimulated single CD56bright NK cells from peripheral blood and liver, using the Fluidigm technology. Although mRNA levels do not necessarily correspond to protein levels,37 we still observed a significant difference in PLZF mRNA expression between CD56bright pbNK and ihNK cells, with more cells carrying PLZF mRNA in liver (Fig. 4A). The proportion of cells expressing mRNAs for CXCR6 and Eomes was increased in liver similar to corresponding protein expression patterns, whereas mRNAs for CD69 and T‐bet were detectable in similar proportions of CD56bright pbNK and ihNK cells, despite significant differences at the protein level (Fig. 4B). Consequently, we defined cells with a PLZF mRNA signal as PLZFhi and cells without a PLZF mRNA signal as PLZFlo and analyzed both groups for expression of CXCR6, CD69, Eomes, and T‐bet mRNA (Fig. 4A,B). CD56brightPLZFhi ihNK cells contained significantly higher mRNA levels of CXCR6 and CD69 (Fig. 4B). Moreover, CD56brightPLZFhi ihNK cells had a tendency to express higher mRNA levels of Eomes (Fig. 4B). Taken together, high PLZF mRNA expression was associated with increased mRNA levels of CXCR6, CD69, and Eomes in CD56bright ihNK cells, corresponding to the results obtained from flow cytometric protein analysis. The overall consistent association between PLZF and liver‐residency markers implicates PLZF in the regulation of liver homing of human CD56bright NK cells.
Figure 4.
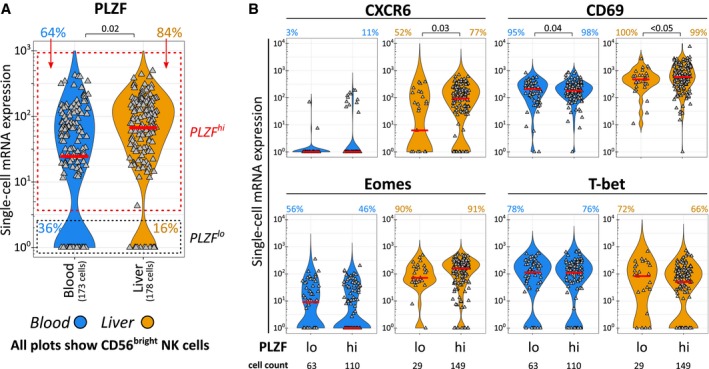
Single‐cell mRNA analysis shows association of PLZF and markers of liver residency in CD56bright ihNK cells. The analysis included 173 single peripheral blood CD56bright(CD16−) NK cells (blue) and 178 single intrahepatic CD56bright(CD16–) NK cells (orange) derived from 4 individuals. (A) Single‐cell PLZF mRNA levels in peripheral blood and intrahepatic CD56bright NK cells and classification of PLZFhi and PLZFlo cells. Proportions of cells in the respective PLZF gates are displayed as colored numbers. (B) Single‐cell mRNA levels of CXCR6, CD69, Eomes, and T‐bet in peripheral blood or intrahepatic CD56bright NK cells with a PLZFhi or PLZFlo mRNA profile. Colored percentages represent the proportion of cells with an mRNA signal. Gray triangles represent individual cells. The median expression is indicated by a red bar. Mixed‐effects regression models followed by FDR adjustment (A,B) were used for statistical testing (significance value shown at the top of each subpanel).
CXCR6+CD69+CD56bright NK Cells in Peripheral Blood and lrNK Cells Share Functional Characteristics
To gain deeper insights into similarities and differences between CXCR6+CD69+ pbNK cells and lrNK cells, we investigated their functional properties. To this end, granzyme and perforin expression were measured in unstimulated NK cells (Supporting Fig. S4A). In peripheral blood and liver, CD56bright NK cells most commonly expressed granzyme K but not granzyme B and perforin, whereas CD56dim NK cells showed the opposite expression pattern (Supporting Fig. S4B). CXCR6+CD69+CD56bright pbNK cells harbored significantly higher proportions of granzyme K+ cells and significantly lower proportions of granzyme B+ and perforin+ cells compared to CXCR6+CD69+CD56dim pbNK cells (Fig. 5A,B) and were thereby more similar to CXCR6+CD69+CD56bright lrNK cells, which likewise expressed high levels of granzyme K and low levels of granzyme B and perforin (Fig. 5B).
Figure 5.
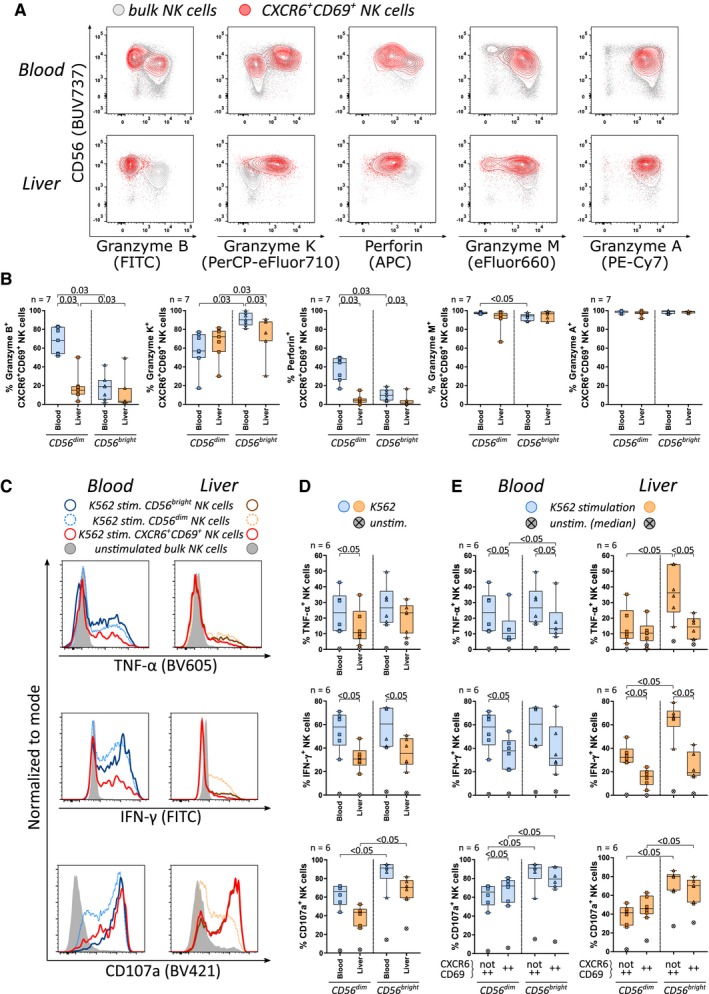
CXCR6+CD69+CD56bright NK cells in peripheral blood and lrNK cells share functional characteristics. (A) Overlay of bulk (gray) and CXCR6+CD69+ NK cells (red) showing the expression of granzyme B, granzyme K, perforin, granzyme M, and granzyme A in peripheral blood and liver. (B) Proportions of granzyme B+, granzyme K+, perforin+, granzyme M+, and granzyme A+ cells within blood‐derived (blue) and intrahepatic (orange) CD56dim and CD56bright CXCR6+CD69+ NK cells (n = 7). (C) Histograms showing TNF‐α, IFN‐γ, and CD107a expression among K562‐stimulated CD56bright (dark), CD56dim (light, dashed), and CXCR6+CD69+ (red) as well as unstimulated NK cells (gray) in peripheral blood (left) and liver (right). Proportions of TNF‐α+, IFN‐γ+, and CD107a+ cells within K562‐stimulated peripheral blood‐derived (blue) and intrahepatic (orange) (D) CD56dim and CD56bright NK cells or (E) CXCR6+CD69+ and not CXCR6+CD69+ CD56dim and CD56bright NK cells (n = 6). Crossed gray dots represent the median positive proportion in the respective unstimulated NK cell subset (D,E). Boxes indicate median and IQR; whiskers indicate total range. Wilcoxon signed‐rank tests for matched pairs followed by FDR adjustment (B,D,E) were used for statistical testing (significance value shown at the top of each subpanel). Abbreviations: APC, allophycocyanin; FITC, fluorescein isothiocyanate; stim., stimulated; unstim., unstimulated.
As a next step, tumor necrosis factor alpha (TNF‐α) and interferon‐gamma (IFN‐γ) production as well as degranulation (CD107a expression) of NK cells were determined by stimulating PBMCs and IHLs with major histocompatibility complex class I‐devoid K562 cells (Supporting Fig. S4C). The majority of CXCR6+CD69+ pbNK and ihNK cells contained significantly lower proportions of TNF‐α+ and IFN‐γ+ cells than NK cells without co‐expression (Fig. 5C,E). Significantly higher frequencies of CD56bright NK cells in peripheral blood and liver expressed CD107a compared to CD56dim NK cells (Fig. 5D). While CD107a responses to K562 stimulation were comparable among NK cell subsets, CD56bright NK cells in peripheral blood and liver had substantially increased baseline degranulation, even in the absence of K562 cells (Fig. 5E). The highest baseline expression of CD107a was detected among CXCR6+CD69+CD56bright lrNK cells (Fig. 5E). In summary, lrNK cells can be identified by a functional phenotype that combines high expression of granzyme K, low expression of granzyme B and perforin with poor TNF‐α and IFN‐γ response, and an increased baseline expression of CD107a. CXCR6+CD69+CD56bright pbNK cells displayed specific functional characteristics similar to lrNK cells, suggesting that both NK cell populations are not only phenotypically but potentially also functionally related (Fig. 6).
Figure 6.
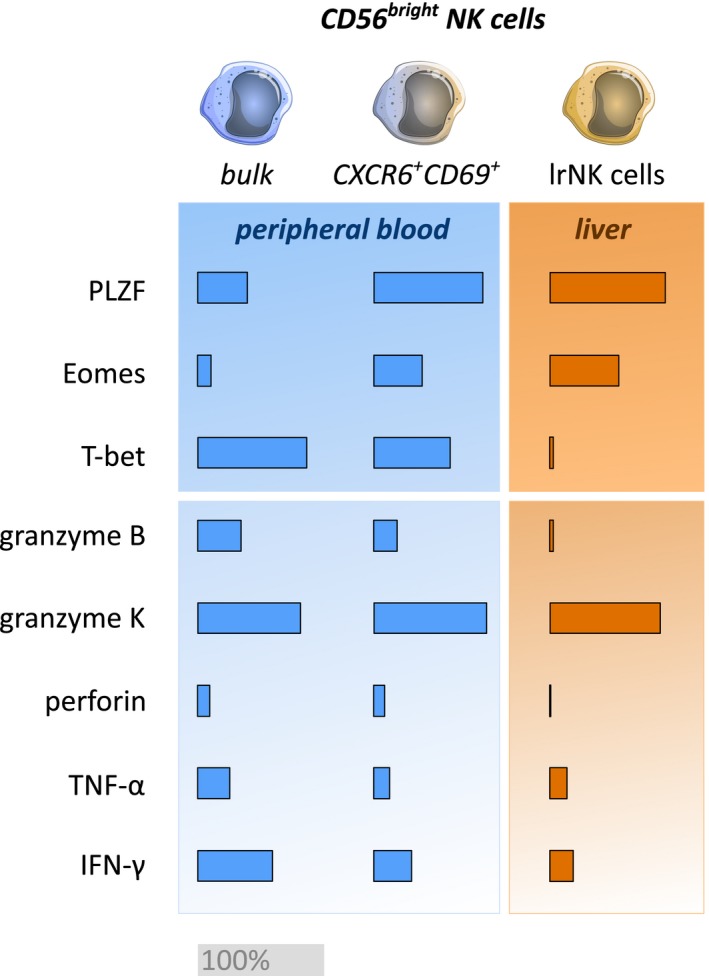
Summary of the transcription factor profiles and functional characteristics of CXCR6+CD69+CD56bright pbNK cells and lrNK cells. Comparison of proportions of PLZFhi, Eomeshi, T‐bethi, granzyme B+ (gzmB), granzyme K+ (gzmK), perforin+, TNF‐α+, and IFN‐γ+ cells among CD56bright pbNK cells, CXCR6+CD69+CD56bright pbNK cells, and CXCR6+CD69+CD56bright lrNK cells. An equivalent proportion of 100% positive cells is indicated by a gray bar.
Discussion
Studies in mice and humans have revealed the existence of lrNK cells that express characteristic surface markers10, 11, 12, 13, 25, 38 and are regulated by39, 40 or associated with10, 11, 13, 17 specific transcription factors. Specifically, human lrNK cells were described to display an EomeshiT‐betlo profile.10, 11, 17 Unraveling the network of factors that determine liver homing and liver residency of human NK cells is important to gain better insights into liver‐specific immunity. In this study, we identified a subset of CXCR6+CD69+CD56bright NK cells expressing high levels of PLZF not only as the major liver‐resident NK cell population in human liver but also as a small population in peripheral blood. CXCR6+CD69+CD56bright pbNK cells exhibited functional properties similar to lrNK cells. These PLZFhiEomesintT‐bethiCXCR6+CD69+CD56bright NK cells in peripheral blood phenotypically might represent an intermediate stage of NK cells related to lrNK cells, suggesting a potential transition from classical circulating PLZFloEomesintT‐bethiCXCR6−CD69−CD56bright to liver‐resident PLZFhiEomeshiT‐betloCXCR6+CD69+CD56bright NK cells.
PLZF has been described to define several immune cell linages in mice and to be involved in the development of natural killer T (NKT) cells,41 NK cells, and ILCs, including lrNK cells (lrILC1s).22, 23, 24 In humans, PLZF has been reported to affect function and development of NKT cells, γδ T cells, CD8+ T cells, mucosa‐associated invariant T cells, and NK cells.26 In contrast to results observed in mice,21, 22, 42 the majority of human NK cells express PLZF.26, 28, 29 Consistent with these findings, we observed high PLZF expression in CD56dim NK cells derived from liver and peripheral blood. While CD56bright ihNK cells also exhibited high PLZF expression, most CD56bright pbNK cells expressed PLZF at low levels. Overall, the differences of PLZF expression between human and murine NK cells suggest that PLZF has additional functions in human NK cells. This is supported by the observation that PLZF transcription generates more splice variants in human cells compared to murine cells.43 Furthermore, significant differences in PLZF expression between human CD56bright NK cells derived from liver and peripheral blood observed here indicate a potential role of PLZF in regulating the ability of CD56bright NK cells to persist in or home to liver tissues.
Available data on PLZF in the context of tissue residency of lymphocytes are limited. Accumulation of PLZF‐transgenic CD4+ T cells in murine livers has been described,20 and PLZF can regulate CXCR6 expression on immune cells in mice.21, 44, 45 In humans, PLZF has been shown to regulate C‐C chemokine receptor type 6 expression in T helper 17 cells46; this is critical for trafficking into Peyer's patches in the small intestine.47 We detected significantly increased proportions of CXCR6+ and CD69+ NK cells among PLZFhiCD56bright NK cells not only in human liver but also in peripheral blood. These data demonstrate that high expression of PLZF is characteristic of CD56bright NK cells expressing liver‐residency markers and suggest that PLZF is a transcription factor influencing liver homing of CD56bright NK cells. The observation that CD56dim NK cells uniformly expressed high levels of PLZF in liver and peripheral blood indicates that PLZF expression alone is not sufficient to determine liver residency of human NK cells but rather the requirement of a transcription factor network regulating liver homing of NK cells. In this line, studies in mice did not detect direct binding of PLZF to the CXCR6 gene region despite up‐regulation of CXCR6 gene expression in CD4+ thymocytes of mice with transgenic PLZF expression.45 This indicated an indirect modulation of CXCR6 expression by PLZF that might depend on other factors that potentially explain variations between different cell subsets. While an EomeshiT‐betlo NK cell phenotype is characteristic of lrNK cells, most CXCR6+CD69+CD56bright pbNK cells displayed an EomesintT‐bethi profile but expressed high levels of PLZF comparable to lrNK cells. Hence, acquisition of high PLZF expression in CD56bright pbNK cells might represent an initial step in recruiting CD56bright pbNK cells to the liver.
It has been suggested that lrNK cells as a large fraction of total intrahepatic lymphocytes play a central role in regulating the immunologic balance of the liver. In our studies, lrNK cells displayed a specific functional profile with low expression of granzyme B and perforin in combination with high expression of granzyme K at baseline as well as low expression of TNF‐α and IFN‐γ after K562 stimulation, similar to other reports.10, 11, 17 Although the functional profile of lrNK cells was different compared to the majority of pbNK cells, CXCR6+CD69+CD56bright pbNK cells showed very similar functional characteristics, supporting a potential relationship. The mechanisms underlying the maintenance of lrNK cells in humans are incompletely understood. A recent study reported that Eomeshi lrNK cells from the original organ donor were still detectable 13 years after liver transplantation even though most Eomeshi lrNK cells originated from the recipient.17 These data suggest that lrNK cells can be replenished both from local hepatic precursor cells and through retention of circulating NK cells in the liver. CXCR6 enables retention of CD56bright NK cells in hepatic sinusoids where sinusoidal endothelial cells highly express the corresponding C‐X‐C motif chemokine ligand 16 (CXCL16).48 Consequently, the subpopulation of CXCR6+CD69+PLZFhiCD56bright NK cells we detected in peripheral blood might have the potential to repopulate liver tissues. In line with previous findings,16 we were able to increase proportions of CXCR6+CD69+PLZFhi pbNK cells in vitro by culturing them in the presence of IL‐12, a cytokine produced by antigen‐presenting cells during inflammatory processes.49 Most individuals participating in this study suffered from diseases accompanied by severe liver inflammation and damage. However, the presence of CXCR6+CD69+PLZFhiCD56bright NK cells in peripheral blood of healthy individuals indicates that this population exists under physiologic conditions. We furthermore observed that CXCR6+CD69+PLZFhiCD56bright pbNK cells can be distinguished from lrNK cells by their expression of T‐bet and Eomes. Processes leading to the retention of pbNK cells including adaptations in the expression of transcription factors are likely to involve factors within the intrahepatic microenvironment (e.g., IL‐15, transforming growth factor β [TGF‐β]), as recently suggested for the expression of T‐bet and Eomes.17, 50 Taken together, cytokine‐dependent acquisition of liver‐residency markers and changes within transcription factor profiles might represent a mechanism by which recruitment of NK cells into the liver can be regulated.
In conclusion, our study identified a subset of PLZFhiCD56bright NK cells in peripheral blood that shares phenotypic and functional characteristics with liver‐resident NK cells while lacking a classic liver residency‐associated EomeshiT‐betlo transcription factor profile. Our results suggest a model in which PLZF regulates the expression of liver‐residency markers on human CD56bright NK cells and that PLZFhiEomesintT‐bethiCXCR6+CD69+CD56bright NK cells represent an intermediate state of NK cells in peripheral blood that can acquire the ability to be retained in hepatic tissue (Fig. 7). Unraveling the processes that determine human NK cells to become liver resident will not only grant better insight into tissue‐specific immunity but also help to improve upcoming cell‐based therapeutic approaches that target hepatic diseases.
Figure 7.
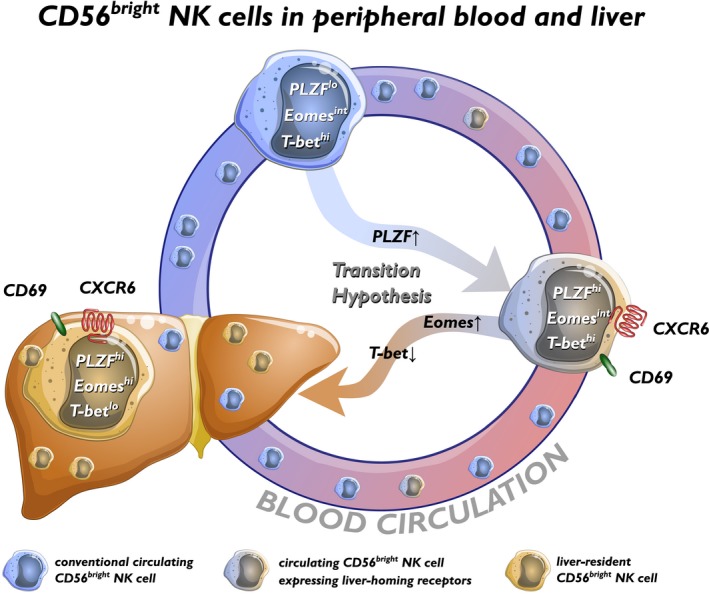
Model for transition from circulating to liver‐resident NK cells. CXCR6+CD69+CD56bright NK cells in peripheral blood express high levels of PLZF while largely exhibiting an EomesintT‐bethi profile. Based on the data of this study and considering evidence for the replenishment of lrNK cells from peripheral blood,17 we propose a model for human NK cell recruitment into liver tissue by the up‐regulation of liver‐homing markers CXCR6 and CD69 controlled by sequential changes in the transcription factor profile. We suggest that CXCR6+CD69+CD56bright NK cells in peripheral blood represent an intermediate stage that has the potential to be retained in liver tissues when exposed to factors of the intrahepatic microenvironment (e.g., IL‐15, TGF‐β). Abbreviation: TGF‐β, transforming growth factor β.
Supporting information
Acknowledgment
We thank all donors participating in this study at the University Medical Center Hamburg‐Eppendorf and the Asklepios Hospital Barmbek in Hamburg as well as all nurses and surgeons of the Department of Hepatobiliary and Transplant Surgery and Department of General and Abdominal Surgery.
Supported by the German Research Foundation (SFB841 to M.A., C.S., G.T., and S.L.; SFB841 graduate school to A.Z., H.G., and L.H.; and KFO306 to M.A., C.S., A.L., T.P., G.R., and G.T.) and the Helmut and Hannelore Greve Foundation (to C.S.).
The funders had no influence on study design, data collection and analysis, the decision to publish, or contents of the manuscript.
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Jost S, Altfeld M. Control of human viral infections by natural killer cells. Annu Rev Immunol 2013;31:163‐194. [DOI] [PubMed] [Google Scholar]
- 2. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol 2008;9:503‐510. [DOI] [PubMed] [Google Scholar]
- 3. Tosello‐Trampont A, Surette FA, Ewald SE, Hahn YS. Immunoregulatory role of NK cells in tissue inflammation and regeneration. Front Immunol 2017;8:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fan X, Rudensky AY. Hallmarks of tissue‐resident lymphocytes. Cell 2016;164:1198‐1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Björkström NK, Ljunggren H‐G, Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol 2016;16:310‐320. [DOI] [PubMed] [Google Scholar]
- 6. Melsen JE, Lugthart G, Lankester AC, Schilham MW. Human circulating and tissue‐resident CD56(bright) natural killer cell populations. Front Immunol 2016;7:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horowitz A, Strauss‐Albee DM, Leipold M, Kubo J, Nemat‐Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 2013;5:208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Michel T, Poli A, Cuapio A, Briquemont B, Iserentant G, Ollert M, et al. Human CD56bright NK cells: an update. J Immunol 2016;196:2923‐2931. [DOI] [PubMed] [Google Scholar]
- 9. Poli A, Michel T, Thérésine M, Andrès E, Hentges F, Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology 2009;126:458‐465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stegmann KA, Robertson F, Hansi N, Gill U, Pallant C, Christophides T, et al. CXCR6 marks a novel subset of T‐bet(lo) Eomes(hi) natural killer cells residing in human liver. Sci Rep 2016;6:26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harmon C, Robinson MW, Fahey R, Whelan S, Houlihan DD, Geoghegan J, et al. Tissue‐resident Eomes(hi) T‐bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol 2016;46:2111‐2120. [DOI] [PubMed] [Google Scholar]
- 12. Martrus G, Kautz T, Lunemann S, Richert L, Glau L, Salzberger W, et al. Proliferative capacity exhibited by human liver‐resident CD49a+CD25+ NK cells. PLoS One 2017;12:e0182532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marquardt N, Béziat V, Nyström S, Hengst J, Ivarsson MA, Kekäläinen E, et al. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol 2015;194:2467‐2471. [DOI] [PubMed] [Google Scholar]
- 14. Lugthart G, Melsen JE, Vervat C, van Ostaijen‐ten Dam MM, Corver WE, Roelen DL, et al. Human lymphoid tissues harbor a distinct CD69+ CXCR6+ NK cell population. J Immunol 2016;197:78‐84. [DOI] [PubMed] [Google Scholar]
- 15. Salzberger W, Martrus G, Bachmann K, Goebels H, Heß L, Koch M, et al. Tissue‐resident NK cells differ in their expression profile of the nutrient transporters Glut1, CD98 and CD71. PLoS One 2018;13:e0201170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hydes T, Noll A, Salinas‐Riester G, Abuhilal M, Armstrong T, Hamady Z, et al. IL‐12 and IL‐15 induce the expression of CXCR6 and CD49a on peripheral natural killer cells. Immunity Inflamm Dis 2018;6:34‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cuff AO, Robertson FP, Stegmann KA, Pallett LJ, Maini MK, Davidson BR, et al. Eomeshi NK cells in human liver are long‐lived and do not recirculate but can be replenished from the circulation. J Immunol 2016;197:4283‐4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao B, Jeong WI, Tian Z. Liver: an organ with predominant innate immunity. Hepatology 2008;47:729‐736. [DOI] [PubMed] [Google Scholar]
- 19. Angelo LS, Bimler LH, Nikzad R, Aviles‐Padilla K, Paust S. CXCR6+ NK cells in human fetal liver and spleen possess unique phenotypic and functional capabilities. Front Immunol 2019;10:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomas SY, Scanlon ST, Griewank KG, Constantinides MG, Savage AK, Barr KA, et al. PLZF induces an intravascular surveillance program mediated by long‐lived LFA‐1‐ICAM‐1 interactions. J Exp Med 2011;208:1179‐1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, Dinner AR, et al. PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci U S A 2015;112:5123‐5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature 2014;508:397‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, et al. An Id2RFP‐reporter mouse redefines innate lymphoid cell precursor potentials. Immunity 2019;50:1054‐1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walker JA, Clark PA, Crisp A, Barlow JL, Szeto A, Ferreira ACF, et al. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity 2019;51:104‐118.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang L, Peng H, Zhou J, Chen Y, Wei H, Sun R, et al. Differential phenotypic and functional properties of liver‐resident NK cells and mucosal ILC1s. J Autoimmun 2016;67:29‐35. [DOI] [PubMed] [Google Scholar]
- 26. Eidson M, Wahlstrom J, Beaulieu AM, Zaidi B, Carsons SE, Crow PK, et al. Altered development of NKT cells, γδ T cells, CD8 T cells and NK cells in a PLZF deficient patient. PLoS One 2011;6:e24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Björklund ÅK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single‐cell RNA sequencing. Nat Immunol 2016;17:451‐460. [DOI] [PubMed] [Google Scholar]
- 28. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 2015;42:443‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, et al. Epigenetic modification and antibody‐dependent expansion of memory‐like NK cells in human cytomegalovirus‐infected individuals. Immunity 2015;42:431‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langeneckert AE, Lunemann S, Martrus G, Salzberger W, Hess LU, Ziegler AE, et al. CCL21‐expression and accumulation of CCR7+ NK cells in livers of patients with primary sclerosing cholangitis. Eur J Immunol 2019;49:758‐769. [DOI] [PubMed] [Google Scholar]
- 31. Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods 2004;294:15‐22. [DOI] [PubMed] [Google Scholar]
- 32. McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, et al. Data exploration, quality control and testing in single‐cell qPCR‐based gene expression experiments. Bioinformatics. 2013;29:461‐467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hipp N, Symington H, Pastoret C, Caron G, Monvoisin C, Tarte K, et al. IL‐2 imprints human naive B cell fate towards plasma cell through ERK/ELK1‐mediated BACH2 repression. Nat Commun 2017;8:1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livak KJ, Wills QF, Tipping AJ, Datta K, Mittal R, Goldson AJ, et al. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods 2013;59:71‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van der Maaten L. Accelerating t‐SNE using tree‐based algorithms. J Mach Learn Res 2014;15:3221‐3245. [Google Scholar]
- 36. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289‐300. [Google Scholar]
- 37. Gong H, Wang X, Liu B, Boutet S, Holcomb I, Dakshinamoorthy G, et al. Single‐cell protein‐mRNA correlation analysis enabled by multiplexed dual‐analyte co‐detection. Sci Rep 2017;7:2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sojka DK, Plougastel‐Douglas B, Yang L, Pak‐Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue‐resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, et al. T‐bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med 2014;211:563‐577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T‐bet and eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao J, Weng X, Bagchi S, Wang CR. Polyclonal type II natural killer T cells require PLZF and SAP for their development and contribute to CpG‐mediated antitumor response. Proc Natl Acad Sci U S A 2014;111:2674‐2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, et al. The BTB‐zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol 2008;9:1055‐1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suliman BA, Xu D, Williams BRG. The promyelocytic leukemia zinc finger protein: two decades of molecular oncology. Front Oncol 2012;2:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gleimer M, von Boehmer H, Kreslavsky T. PLZF controls the expression of a limited number of genes essential for NKT cell function. Front Immunol 2012;3:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, et al. Multiple layers of transcriptional regulation by PLZF in NKT‐cell development. Proc Natl Acad Sci U S A 2016;113:7602‐7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh SP, Zhang HH, Tsang H, Gardina PJ, Myers TG, Nagarajan V, et al. PLZF regulates CCR6 and is critical for the acquisition and maintenance of the Th17 phenotype in human cells. J Immunol 2015;194:4350‐4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang C, Kang SG, Lee J, Sun Z, Kim CH. The roles of CCR6 in migration of Th17 cells and regulation of effector T‐cell balance in the gut. Mucosal Immunol 2009;2:173‐183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hudspeth K, Donadon M, Cimino M, Pontarini E, Tentorio P, Preti M, et al. Human liver‐resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun 2016;66:40‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. El‐emshaty HM, Nasif WA, Mohamed IE. Serum cytokine of IL‐10 and IL‐12 in chronic liver disease: the immune and inflammatory response. Dis Markers 2015;2015:707254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harmon C, Jameson G, Almuaili D, Houlihan DD, Hoti E, Geoghegan J, et al. Liver‐derived TGF‐β maintains the eomeshitbetlo phenotype of liver resident natural killer cells. Front Immunol 2019;10:1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


