Abstract
The evolution of gene expression regulation has contributed to species differentiation. The 3′ untranslated regions (3′UTRs) of mRNAs include regulatory elements that modulate gene expression; however, our knowledge of their implications in the divergence of bacterial species is currently limited. In this study, we performed genome-wide comparative analyses of mRNAs encoding orthologous proteins from the genus Staphylococcus and found that mRNA conservation was lost mostly downstream of the coding sequence (CDS), indicating the presence of high sequence diversity in the 3′UTRs of orthologous genes. Transcriptomic mapping of different staphylococcal species confirmed that 3′UTRs were also variable in length. We constructed chimeric mRNAs carrying the 3′UTR of orthologous genes and demonstrated that 3′UTR sequence variations affect protein production. This suggested that species-specific functional 3′UTRs might be specifically selected during evolution. 3′UTR variations may occur through different processes, including gene rearrangements, local nucleotide changes, and the transposition of insertion sequences. By extending the conservation analyses to specific 3′UTRs, as well as the entire set of Escherichia coli and Bacillus subtilis mRNAs, we showed that 3′UTR variability is widespread in bacteria. In summary, our work unveils an evolutionary bias within 3′UTRs that results in species-specific non-coding sequences that may contribute to bacterial diversity.
INTRODUCTION
A prototypical bacterial mRNA comprises the protein coding sequence (CDS) and the 5′ and 3′ untranslated regions (5′UTR and 3′UTR), which flank the CDS upstream and downstream, respectively. In addition to the Shine-Dalgarno (SD) sequence, large bacterial 5′UTRs often contain riboswitches, thermosensors and RNA structures that are targeted by sRNAs and RNA-binding proteins. These regulatory elements control premature transcription termination, mRNA translation, and/or mRNA processing (1–3). Recent discoveries have revealed that 3′UTRs contain regulatory elements that modulate central metabolism, virulence and biofilm formation through different mechanisms. Several of these regulatory elements control the expression of the protein encoded in the same mRNA (cis-acting mechanism), while others behave as regulatory RNAs that base pair with other mRNAs (trans-acting mechanism) (4–19).
About cis-acting 3′UTRs, we previously showed that the long 3′UTR of the icaR mRNA, which encodes the main repressor of the PIA-PNAG exopolysaccharide synthesis in Staphylococcus aureus, interacts with the 5′UTR of the same mRNA to inhibit IcaR translation. This occurs through base-pairing of the anti-SD motif (UCCCC sequence located in the 3′UTR) at the ribosome binding site (RBS), impairing ribosome accessibility by creating a double stranded RNA substrate that is cleaved by the endoribonuclease III (RNase III) (5). Similar 5′-3′UTR interactions were recently described in the hbs mRNA of Bacillus subtilis. The cleavage of the hbs 5′UTR by RNase Y is countered by a 14-nt perfect base-pairing interaction between the 5′ and the 3′UTR, which partially overlaps with the cleavage site (8). Other bacterial 3′UTRs are often used as direct entry points for ribonucleases to initiate mRNA degradation. This is the case for the hmsT mRNA, which encodes a protein that modulates c-di-GMP synthesis, required for allosteric activation of polysaccharide production in Yersinia pestis biofilm formation. Polynucleotide phosphorylase (PNPase)-dependent cleavage of the hmsT 3′UTR affects the turnover of the mRNA and, ultimately, HsmT expression (6,9). Additionally, several AU-rich 3′UTRs of Y. pestis are targeted by PNPase when the transcriptional terminator (TT) is Rho-dependent (9). In Salmonella, HilD, one of the main transcriptional activator factors of the pathogenicity island 1 (SPI-1) (20), is processed through its 3′UTR by RNase E and PNPase (21). Similarly, the aceA mRNA in Corynebacterium glutamicum, which encodes the isocitrate lyase protein that is part of the glyoxylate cycle (22), is processed by endoribonucleases E/G (RNase E/G) at the 3′UTR (4). On the contrary, there are examples of 3′UTRs that prevent mRNA degradation. When Escherichia coli grows under iron-limiting conditions, the apo-AcnB protein interacts with a stem-loop structure located at the 3′UTR of its own mRNA, and prevents RyhB-induced degradation. This occurs because the apo-AcnB binds close to the RNase E cleavage site, which is essential for acnB mRNA degradation (7).
Regarding trans-acting regulatory RNAs, S. aureus RNAIII was for decades the sole example of a prokaryotic mRNA with a regulatory 3′UTR. RNAIII consists of a 5′UTR of 84-nt, a short CDS encoding the δ-toxin (hld), and a long structured 3′UTR (352-nt) that controls the temporal expression of several virulence factors and the transcriptional repression of Rot exotoxins by targeting their mRNAs (23,24). More recently, several small RNAs (sRNAs) derived from 3′UTRs have been described, some of which are expressed from alternative mRNA-internal promoters (Type I 3′UTR-derived sRNAs), while others are the consequences of specific mRNA processing events (Type II 3′UTR-derived sRNAs) (18).
Previous studies have shown the sequence conservation of some 3′UTRs to be variable depending on the bacterial species. For example, the 3′UTR of dapB mRNA, which produces the type I dapZ 3′UTR-derived sRNA, is not conserved among enterobacterial species (except for the R1 seed and the Rho-independent terminator) (11). Similarly, the sdhX sRNA, which is generated by processing the 3′ end of the sdh-suc operon by RNase E in E. coli and Salmonella, also shows nucleotide variations between different enterobacterial species (16,17). Interestingly, sequence differences in the sdhX sRNA could establish an mRNA target divergence between Salmonella and E. coli (16).
Although, these and other pioneering studies have indicated the importance of 3′UTRs as a new class of post-transcriptional regulatory elements controlling relevant physiological processes in bacteria, several questions remain unanswered. Are 3′UTR sequences preserved within and between bacterial species? How often do conserved genes from closely related bacteria contain different 3′ UTRs and how is 3′UTR variability originated? Do differences in 3′UTRs between orthologous genes have consequences in their expression at the protein level?
In this study, we aimed to evaluate the evolutionary relationship between the CDSs and their corresponding 3′UTR sequences in bacteria. To this end, we performed genome-wide comparative analyses of orthologous mRNAs among closely related species of the genus Staphylococcus. As a result, we discovered that most of the 3′UTRs from these mRNAs were rather variable at the sequence level. In most cases, the inter-species nucleotide conservation of mRNAs was lost downstream of the coding sequence, suggesting a large variability within the 3′UTRs. In vivo mapping using RNA-seq of the transcript boundaries in three species of the genus Staphylococcus confirmed that 3′UTRs vary in length and sequence when compared with the S. aureus transcriptomic data. We demonstrated that such differences may have an impact on the expression of orthologous genes and, thus, the functionality of the 3′UTRs could differ depending on the analysed species. Finally, by extending the 3′UTR conservation analysis to other bacteria genera, we found similar sequence variations. In summary, this study proposes that 3′UTRs from orthologous genes may be selectively targeted by evolution to create sequence differences that contribute to diversity in bacteria.
MATERIALS AND METHODS
Strains, plasmids, oligonucleotides and growth conditions
The bacterial strains, plasmids and oligonucleotides used in this study are listed in Supplementary Tables S1, S2, and S3, respectively. Staphylococcus sp. strains were grown in Tryptic Soy Broth (Pronadisa) supplemented with 0.25% glucose (TSBg) or, when indicated, in Brain Heart Infusion (BHI). Escherichia coli was grown in Luria-Bertani (LB) broth (Pronadisa). The B2 (casein hydrolysate, 10 g l−1; yeast extract, 25 g l−1; NaCl, 25 g l−1; K2HPO4, 1 g l−1; glucose, 5 g l−1; pH 7.5) and SuperBroth (tryptone, 30 g l−1; yeast extract, 20 g l−1; MOPS, 10 g l−1; pH 7) media were used to prepare S. aureus and E. coli competent cells, respectively. For selective growth, media were supplemented with the appropriated antibiotics at the following concentrations: Erythromycin (Erm), 10 μg ml−1; Ampicillin (Amp), 100 μg ml−1.
Nucleotide conservation analysis
The conservation of UTRs among orthologous monocistronic mRNAs from phylogenetically-related bacterial species was determined by performing blastn comparisons using the Microbial Nucleotide BLAST tool (https://blast.ncbi.nlm.nih.gov/). Monocistronic mRNA boundaries were manually annotated by visualizing the S. aureus transcriptomic maps from previous studies (5,25–26) in the Jbrowse application (27). For the batch comparison of S. aureus 3′UTRs, a file including the query sequences in FASTA format was used. Each sequence included the annotated 3′UTR and the last 200 nt of the CDS of a monocistronic operon (Supplementary Figure S1). This restriction was applied to normalize the starting point for each of the UTRs from the blastn results, facilitating further analysis and plotting. We considered that 200 nt from the CDS were enough to properly identify their corresponding orthologous genes. Similarly, for batch comparison of S. aureus 5′UTRs, a FASTA file including the annotated 5′UTRs and the first 200 nt of the CDSs were used. In order to normalize the starting point of the CDS, we used reverse-complement query sequences for 5′UTR blastn comparisons (Supplementary Figure S1). The UTR conservation length was registered as the last nucleotide from the query sequence showing a positive alignment with the region of the genome under comparison. For this purpose, the information contained in the blastn result TXT files was transformed into tables using a custom R script. Nucleotide alignments that started at values higher than 100, finished at values lower than 100, or were shorter than 80 nucleotides were not considered. The UTR conservation lengths of each Staphylococcus species were plotted against the reference UTR lengths of S. aureus (Supplementary Figure S1). The square correlation coefficient (R2) was calculated in R from the resulting fitted linear model. This analysis was also applied to compare the mapped 3′UTRs of B. subtilis (28) and E. coli (29) with phylogenetically-related species of the Bacillus genus and Enterobacteriaceae family, respectively. Note that in the case of B. subtilis, 3′UTRs longer than 150 nt were not annotated (28).
RNA sequencing and data analysis
In order to perform the transcriptomic maps of the S. simiae CCM 7213T, S. capitis SK14 and S. epidermidis RP62A, strains were grown in TSBg at 37°C and 200 rpm until exponential phase was reached. Total RNAs were extracted as previously described (30). RNA sequencing and preliminary data analysis was carried out by the Stab Vida company. The RNA-seq reads from the S. simiae CCM 7213T, S. capitis SK14 and S. epidermidis RP62A samples were aligned using the Rockhopper program (31) and the complete genome sequences from S. simiae NCTC13838 (NZ_LT906460.1), S. capitis AYP1020 (NZ_CP007601.1) and S. epidermidis RP62A (NC_002976.3) as references, respectively. The obtained read coverage files (.wig) from the plus and minus strands were converted to BigWig (.bw) files with the wig2BigWig program. The generated .bw files and the gene annotation files, including the positions of transcriptional terminators (TTs) predicted by TransTerm HP program (32), were loaded into a public web server based on Jbrowse (27) (http://rnamaps.unavarra.es/). The 3′UTR lengths of the orthologous monocistronic mRNAs were manually annotated and plotted against the lengths of previously described S. aureus 3′UTRs (5).
Simultaneous mapping of 5′ and 3′ ends from RNA molecules
The mapping of 5′ and 3′ ends of mRNAs was performed using a modified version of the Rapid Amplification of cDNA Ends (mRACE) method (33). Briefly, total RNA samples were treated with the Cap-Clip Acid Pyrophosphatase (Tebu-Bio) following the manufacturer's recommendations. After 1 h of incubation, RNAs were extracted by phenol–chloroform and precipitated with sodium acetate and cold ethanol. Serial dilutions of Cap-Clip and non-Cap-Clip treated RNA were ligated using the T4 RNA Ligase I (New England Biolabs) at 16°C ON. RT-PCRs for all the ligated RNA dilutions were carried out using the SuperScript™ III One-Step RT-PCR System with the Platinum Taq DNA polymerase (Invitrogen) and the outward primers A and B (Supplementary Table S3). For the mapping of the 3′UTR of rpiRc from S. epidermidis and S. capitis primers RACE-3XFLAG-A and RACE-RpiRc-sau-B were used. PCR products were run in 2.5% agarose gels and bands of the expected size were purified and ligated into the pGEM-T Easy vector (Promega). The resulting reactions were used to transform E. coli XL1-Blue cells (Stratagene). Ten isolated white colonies were then analyzed by Sanger sequencing. Transcript boundaries were determined by blastn analysis and nucleotide frequencies representing the 5′ and 3′ ends, respectively, were registered as percentages (%).
Chromosomal mutagenesis
The mutants generated in this study (Supplementary Table S1) were obtained by a two-step homologous recombination that exchanges a specific chromosomic region by the mutant allele present in the pMAD plasmid (34), as previously described (35). The marker-less mutants were verified by PCR using oligonucleotides E and F (Supplementary Table S3) and Sanger sequencing.
Plasmid constructions
Most of the plasmids used in this study were engineered as previously described (36). Briefly, PCR fragments were amplified from chromosomic or plasmidic DNA with the DreamTaq DNA polymerase or Phusion High-Fidelity DNA Polymerase (Thermo Scientific), using the oligonucleotides listed in Supplementary Table S3. The PCR products were run and purified from agarose gels using the NucleoSpin® Gel and PCR Clean-up Macherey-Nagel kit and ligated into the pJET 1.2 vector (Thermo Scientific). The resulting plasmids were used to transform E. coli XL1-Blue cells. These were then purified from overnight (ON) cultures with the NucleoSpin® Plasmid Macherey-Nagel kit and verified by Sanger sequencing. When required, DNA fragments were excised using FastDigest restriction enzymes (Thermo Scientific) and ligated into the appropriate vector with the Rapid DNA ligation kit (Thermo Scientific). The final plasmids were introduced into S. aureus strains by electroporation, as previously described (37).
The pMAD plasmids used for chromosomal mutations were constructed by amplifying the flanking sequences (AB and CD) of the target regions using primers A/B and C/D (Supplementary Table S3). After cloning them into pJET they were digested and ligated into pMAD in a two-insert ligation process using BamHI, NheI and EcoRI (Supplementary Table S2 and S3).
The plasmids expressing chimeric icaR mRNAs were constructed taking advantage of conserved natural restriction sites located at the beginning of the icaR 3′UTRs. Specifically, the icaR mRNA regions were amplified by PCR using chromosomic DNA from the corresponding staphylococcal species and the specific oligonucleotides IcaR+1 and IcaR-Term (Supplementary Table S3). The resulting PCR products were inserted into pJET and digested with SpeI and EcoRI. In the case of S. simiae, restriction enzymes HincII and EcoRI were used. The digested fragments were ligated into the p3xFIcaRm or pIcaRm (5), producing the plasmids carrying the chimeric icaR mRNAs listed in Supplementary Table S2.
The ftnA and rpiRc genes were labelled with the 3xFLAG tag sequence by overlapping PCRs. Specifically, the tagged ftnA gene was generated by linking two partially overlapping PCR fragments that were amplified using oligonucleotides Ftn-A, 3XFLAG-ftn-izqda, 3XFLAG-ftn-drcha and Term-ftn (Supplementary Table S3). A second PCR with oligonucleotides +1-ftn and Term-ftn (Supplementary Table S3) was performed to amplify the 3xFftnA mRNA region. The PCR fragment was ligated into pJET, digested with restriction enzymes BamHI and KpnI and inserted into pEW, generating the p3xFFtnA plasmid (Supplementary Table S2). The pEW plasmid was constructed by transferring the EcoRI-NarI fragment (which includes the transcriptional terminator region) from pCN47 to the pCN40 plasmid (38). Similarly, the rpiRc gene was tagged by introducing the 3xFLAG sequence through overlapping PCRs using oligonucleotides +1-RpiRc, 3XFLAG-RpiRc-izqda, 3XFLAG-RpiRc-drcha and Term-RpiRc (Supplementary Table S3). This PCR fragment was digested with BamHI and EcoRI and inserted into pEW, generating the p3xFRpiRc plasmid (Supplementary Table S2).
Plasmids p3xFFtnAΔ3′UTR and p3xFRpiRcΔ3′UTR, which express the ftnA and rpiRc mRNAs lacking the 3′UTRs, were constructed by amplifying PCR fragments from the p3xFFtnA and p3xFRpiRc plasmids with oligonucleotide pairs +1-ftn/Δ3UTR-ftn-term and +1-RpiRc/Δ3UTR-RpiRc-term (Supplementary Table S3), respectively, and ligated them into pEW (Supplementary Table S2).
To generate the plasmids expressing the chimeric tagged ftnA and rpiRc mRNAs (Supplementary Table S2), the corresponding 3′UTRs from different staphylococcal species were fused to S. aureus FtnA or RpiRc CDSs by overlapping PCRs. First, PCR fragments including the 5′UTR and the 3xFLAG CDS from the ftnA or rpiRc genes were generated by using the p3xFFtnA and p3xFRpiRc plasmids as templates and oligonucleotide pairs +1-ftn/CDS-stop-ftn and +1-RpiRc / CDS-stop-RpiRc as primers, respectively. Second, the 3′UTR regions from different staphylococcal species were amplified using their corresponding fw and rvs oligonucleotides (Supplementary Table S3). Finally, the PCR fragments including the 3xFLAG tagged CDS and the 3′UTR were fused by PCR using oligonucleotides +1 and rvs (Supplementary Table S3). Next, they were ligated into pJET and digested with BamHI/ KpnI and BamHI/EcoRI in the case of 3xFFtnA and 3xFRpiRc, respectively, and inserted into pEW.
The p3xFRpiRc+3′UTRIS256 plasmid was generated by PCR simulating the 3′UTR of rpiRc from the S. aureus strain 2010-60-6511-5 (GenBank: JJCE00000000.1), which carries an IS256 insertion. Briefly, IS256 was amplified from the S. aureus strain 15981, using primers IS256-fw and IS256-3′UTR-RpiRc-rvs and the 3′UTR fragment was amplified from p3xFRpiRc, using the RpiRc-CDS-SpeI-fw and 3UTR-RpiRc-42-IS256-rvs primers (Supplementary Table S3). Then, an overlapping PCR with RpiRc-CDS-SpeI-fw/IS256-3′UTR-RpiRc-rvs primers was used to generate the 3′UTR carrying the IS256 insertion. The amplified product was ligated into pJET. Natural restriction sites were used to digest p3xFRpiRc with SpeI/EcoRI, pJET 3xFRpiRc with NspI/EcoRI and pJET-3′UTR+IS256 with SpeI/NspI, followed by a double-fragment ligation into the pEW plasmid in order to recreate an rpiRc gene with the IS256 insertion in its 3′UTR.
The plasmid carrying 3xFRpiRc with the IS1181 insertion in the 3′UTR, p3xFRpiRc+3′UTRIS1181, was generated simulating the 3′UTR of rpiRc from the S. aureus strain DAR1183 (GenBank: KK099086.1) by PCR. This was achieved by amplifying the IS1181 from the S. aureus strain N315 and the 3'UTR from p3xFRpiRc using primers 3′UTR-RpiRc-IS1181-fw / IS1181-rvs and IS1181-3UTR-RpiRc-fw / pCN-univ-AT, respectively (Supplementary Table S3). An overlapping PCR with the primer pair 3′UTR-RpiRc-IS1181-fw/pCN-univ-rv-AT was used to generate the 3′UTR carrying IS1181, which was then cloned into pJET. Natural restriction sites (SpeI/KasI or SpeI/HindIII) were used to digest p3xFRpiRc while HindIII/KasI were used to digest pJET-3′UTR+IS1181 with HindIII/KasI. Then, a double-fragment ligation into the pEW plasmid was performed to recreate an rpiRc gene carrying the IS1181 insertion in its 3'UTR.
The GFP reporter plasmids were constructed using the Listeria monocytogenes pAD-cGFP plasmid as a template (39). The hly 5′UTR and GFP sequences were amplified with primers Sal-GFP-fw and BcuI-TT-BamHI-GFP-rvs (Supplementary Table S3). The resulting PCR fragment was cloned into the pEW plasmid using SalI and BamHI. The 3′UTR of rpiRc was amplified using primers BamHI-EcoRI-3UTR-RpiRc-fw and SmaI-3UTR-RpiRc-rvs (Supplementary Table S3) and inserted downstream of the gfp gene using restriction sites BamHI and SmaI. The 3′UTRs carrying the IS were amplified from p3xFRpiRc+3′UTRIS256 and p3xFRpiRc+3′UTRIS1181 using BamHI-EcoRI-3UTR-RpiRc-fw and KpnI-term-RpiRc and introduced into the pEW-GFP plasmid using BamHI and KpnI. Lastly, the pGFP-Δ3′UTR-rpiRc was constructed using the pGFP-3′UTR-rpiRc as a template and primers SalI-GFP-fw and KpnI-term-RpiRc. The amplification product was then ligated into the pEW-GFP using SalI and KpnI.
Total protein extraction and Western blotting
Preinocula were grown in 5 ml of TSBg supplemented with Erm (TSBg+Erm) and incubated ON at 37°C and 200 rpm. Bacterial concentrations were estimated by measuring the optical density (OD600) of the preinocula and then normalized to an OD600 of 0.02 in Erlenmeyer flasks containing TSBg+Erm. Cultures were grown at 37°C and 200 rpm until an OD600 of 0.5 (exponential phase) was reached for strains carrying IcaR and FtnA constructs and around 5–6 (stationary phase) for those carrying the RpiRc plasmids. Culture samples were harvested by centrifugation (10 min at 4,400 g and 4°C) and bacterial pellets stored at −20°C until needed. Pellets were thawed, washed and resuspended in 1 ml of phosphate-buffered saline (PBS). Next, bacterial suspensions were transferred to Fast Prep tubes containing acid-washed 100 μm glass beads (Sigma) and cells lysed using a FastPrep-24 instrument (MP Biomedicals) at speed 6 for 45 s twice. Total cell extracts were obtained by centrifuging the Fast Prep tubes for 10 min at 21,000 g and 4°C. The total protein concentration was quantified using the Bio-Rad protein assay kit. Samples were prepared at the desired concentration in Laemmli buffer and stored at −20°C until needed.
Western blotting was performed as previously described (36). The 3xFLAG tagged IcaR, RpiRc and FtnA samples were incubated with mouse monoclonal anti-FLAG M2-Peroxidase (HRP) antibodies (Sigma) diluted 1:1,000 whereas the GFP samples were incubated with mouse monoclonal anti-GFP antibodies 1:5,000 (Living Colors, Clontech) and peroxidase-conjugated goat anti-mouse immunoglobulin G and M antibodies 1:2,500 (Pierce-Thermo Scientific). Membranes were developed using the SuperSignal West Pico Chemiluminiscent Substrate kit (Thermo Scientific). Protein bands were quantified by densitometry of Western blot images using ImageJ (http://rsbweb.nih.gov/ij/). Each of the protein levels was normalized to either the levels of S. aureus (Sau) or the WT sample.
RNA extraction and Northern blotting
Bacteria were grown as described in the previous section. Cultures were centrifuged for 3 min and 4,400 g at 4°C and pellets stored at −80°C. Total RNA extraction was performed as previously described (30). Northern blotting was performed as described by Caballero et al (36) with the following modifications. Radiolabelled-RNA probes were synthesized from PCR products carrying the T7 promoter (Supplementary Table S3) using the MAXIscript T7 transcription kit (Ambion) and 32P-α-UTP, following the manufacturer's recommendations. Probes were then purified with Illustra MicroSpin G-50 columns (GE Healthcare) and membranes were hybridized with the corresponding RNA probes at 68°C ON. mRNA levels were quantified by densitometry of Northern blot autoradiographies using ImageJ (http://rsbweb.nih.gov/ij/). Each of the mRNA levels was normalized either to the levels of S. aureus (Sau) or the WT sample.
RNA stability assay
Bacteria were grown as indicated in the previous sections. Once the exponential phase (OD600 = 0.5) was reached, six aliquots of 20 ml of the culture were transferred to 50 ml Falcon tubes containing 300 μg ml−1 of Rifampicin and incubated at 37°C for 0, 2, 4, 8, 15 and 30 min. Then, 5 ml of the Stop solution (95% ethanol, 5% phenol) were added to the samples and centrifuged for 2 min at 4,400 g. Pellets were frozen in liquid nitrogen and stored at −80°C. RNA extraction and Northern blot analysis were conducted as described above.
Identification of elements disrupting the rpiRc mRNA sequence
All available S. aureus genome sequences were retrieved from ftp://ftp.ncbi.nlm.nih.gov/genomes/ through the wget command-line utility. The Fasta files were loaded into a local nucleotide database using the Geneious software package. Next, a Blastn algorithm was locally run using the rpiRc mRNA as a query. Genomes presenting pairwise alignment disruption located at the rpiRc 3′UTR were registered. When possible, sequences surrounding the position of alignment disruptions were used to look for insertion sequences in the IS database (https://isfinder.biotoul.fr/) (40). Note that in some cases, due to the assembly protocol, repeated sequences were eliminated from contig ends avoiding the identification of the IS responsible for the alignment disruption.
Haemolysis assay
Bacterial cultures were grown in 15 ml of BHI ON at 37°C and 200 rpm. Cultures were centrifuged for 10 min and 4,400 g at 4°C to collect the supernatants containing the haemolysins. Supernatants were equalized using the wet weight of the pellets and the remaining cells removed from by filtration employing 0.2 μm pore filters. Supernatants were concentrated 10 times using a SpeedVac Concentrator. 30 μl of the concentrated supernatants were transferred into 5-mm holes within Columbia Sheep blood (5%) agar plates (Biomerieux). Plates were incubated at 37°C ON and then kept at 4°C for several days.
In vitro transcription
PCR fragments containing the T7 promoter were used as templates for in vitro transcription. The T7 promoter was included in the corresponding Fw oligonucleotides (Supplementary Table S3). In vitro transcription was carried out using the T7 polymerase at 37°C ON followed by the removal of the PCR templates with a DNase I treatment. Transcripts were run on a denaturing 6% polyacrylamide gel and visualized using UV shadowing. RNAs were excised from the gels and eluted by incubating the gel fractions with elution buffer (0.5 M ammonium acetate, 1 mM EDTA, 0,1% SDS) and phenol (pH 4.5) at 4°C ON. RNAs were then purified using a phenol-chloroform extraction and precipitated with ethanol. Pellets were washed with 70% ethanol and resuspended in water. The RNA integrity was checked by polyacrylamide gel electrophoresis and their concentration measured with a NanoDrop Instrument (Agilent Technologies).
5′-End labelling of synthetic RNAs
Prior to labelling, the RNA was dephosphorylated with FastAP (Thermo Scientific) at 37°C for 15 min followed by a phenol-chloroform extraction. 7 μg of dephosphorylated RNA were incubated with 32P-γ-ATP and PNK (Thermo Scientific) at 37°C for 1 h. Labelled nucleic acids were purified on a denaturing 6% polyacrylamide gel as described inthe previous section. The efficiency of the labelling was measured using a scintillation counter.
Electrophoretic mobility shift assays
Labelled and non-labelled RNAs were separately prepared at the desired concentrations by adding sterile MilliQ water and 5× renaturing buffer (100 mM Tris–HCl pH 7.5, 300 mM KCl, 200 mM NH4Cl, 15 mM DTT). Samples were denatured at 90°C for 1 min and chilled on ice for an additional minute. Next, 10 mM of MgCl2 were added and samples were incubated for 10 min at 22°C. Subsequently, the labelled RNA was mixed with increasing concentrations of non-labelled samples in 1X reaction buffer (20 mM Tris–HCl pH 7.5, 60 mM KCl, 40 mM NH4Cl, 3 mM DTT, 10 mM MgCl2) in a total volume of 20 μl. The mixtures were then incubated at 37°C for 15 min before running them in a non-denaturing 6% polyacrylamide gel and 1X Tris-borate buffer containing 1 mM MgCl2 at 300 V and 4°C. The gel was developed by autoradiography.
PIA-PNAG quantification
PIA/PNAG exopolysaccharide was extracted and quantified as previously described (5,41). Cell surface extracts (5 μl), which were obtained from bacteria grown ON in TSBg, were spotted onto a nitrocellulose membrane using a Bio-Dot microfiltration apparatus (Bio-Rad). The membranes were blocked ON with 5% skimmed milk in phosphate-buffered saline with 0.1% Tween 20 (PBS-Tw20). After washing several times with PBS-Tw20, the membranes were incubated for 2 h with specific anti-PNAG antibodies diluted 1:20,000 (42). Bound antibodies were detected by incubating the membranes for 1 h with peroxidase-conjugated goat anti-rabbit immunoglobulin G antibodies (Jackson ImmunoResearch Laboratories, Inc., Westgrove, PA) diluted 1:10,000. Finally, membranes were washed with PBS–Tw20 and developed using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and Amersham Hyperfilm ECL films (GE Healthcare Life Sciences).
RESULTS
3′UTRs are highly variable among staphylococcal species
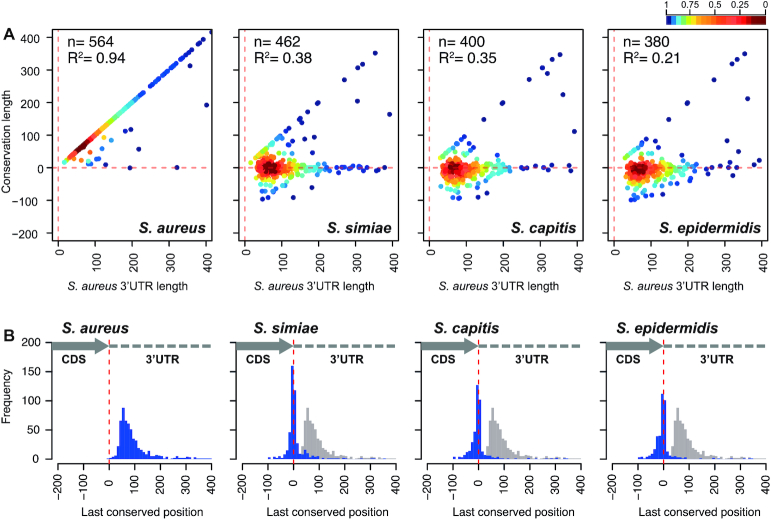
Next-generation RNA sequencing (NGS) technologies allow to accurately determine mRNA boundaries and, therefore, determine the lengths of the untranslated regions (UTRs). To evaluate the evolutionary relationship between coding sequences (CDSs) and their corresponding 3′UTRs, we performed genome-wide comparative analyses. Using high-resolution transcriptome maps of S. aureus as a reference, we analysed the 3′UTR sequence conservation of close phylogenetic members of the genus Staphylococcus (Supplementary Figure S1). In order to simplify the study, we focused on monocistronic mRNAs. This ensured that each analysed CDS was flanked by a 5′ and a 3′UTR. First, we generated a query database including the 3′UTR sequences plus the last 200 nucleotides (nt) of each monocistronic CDS (∼590 sequences). This stretch of nucleotides was intended to serve as an indicator for the correct identification of orthologous genes among staphylococcal species. Using blastn (43), we compared the selected sequences against 8 representative genomes of the most closely-related staphylococcal species according to the TimeTree knowledge-base (44) (Supplementary Figure S2). The last conserved nucleotide position of each of the orthologous mRNAs was registered (Supplementary Table S4) and plotted against the length of their corresponding S. aureus mRNA (Figure 1A and Supplementary Figure S3). The comparison of S. aureus NCTC 8325 and MW2 strains revealed that 97% of the analysed mRNAs fell on the diagonal axis of the plot, indicating 3′UTRs conservation. Only 17 (3%) out of 589 monocistronic mRNAs showed 3′UTR sequence variations, which could be explained by the presence of unique insertion sequences (ISs) or Staphylococcus aureus repeat (STAR) elements (45).
Figure 1.
High-throughput conservation analysis of 3′UTRs from mRNAs encoding orthologous proteins among phylogenetically-related staphylococcal species. (A) Scatter plots representing the conservation of 3′ end regions of S. aureus mRNAs compared to different closely-related staphylococcal species. The S. aureus NCTC 8325 3′UTR sequence query database was compared by blastn to S. aureus MW2, S. simiae NCTC13838, S. capitis AYP1020 and S. epidermidis RP62A genome sequences. Each dot represents the last conserved nucleotide (y axis) of a specific species in function of the S. aureus 3′UTR length (x axis). Based on the transcriptomic maps previously described (5), each query sequence included the last 200 nucleotides of the corresponding CDS plus the whole 3′UTR of monocistronic mRNAs. The plot was coloured by applying the Kernel density estimation, which indicates the proximity of the dots: blue, isolated dots; red, overlapping dots. The number (n) of plotted mRNAs encoding orthologous proteins and the square correlation coefficient (R2) are indicated. (B) Histogram plots showing the distribution of the last conserved nucleotide position (blue bars) among the S. aureus monocistronic mRNAs compared to the indicated staphylococcal species. Each blue bar represents the number of conserved 3′UTRs at a given position in windows of 10 nt (width of the bar). Grey bars represent the S. aureus distribution, which is included as a reference. The dashed red line indicates the position of the stop codon.
However, 3′UTRs conservation among the analysed mRNAs decreased as the phylogenetic distance between S. aureus and the other staphylococcal species increased. This phenomenon is reflected by the dots being positioned along the horizontal line that represents the stop codon in Figure 1A and Supplementary Figure S3A. The lack of conservation was already noticeable for several 3′UTRs of S. argenteus, which is the closest species to S. aureus. This increased considerably in S. simiae and left the remaining species of the genus with just a few conserved 3′UTRs (Figure 1A and Supplementary Figure S3A). As one would expect, among the conserved 3′UTRs (diagonal line) for all the staphylococcal species, we identified RNAIII and seven putative riboswitch-dependent 3′UTRs, meaning that the mRNA transcription ended at the TT of the downstream riboswitch when in the OFF configuration (5,23–24,30). We also found two mRNAs, encoding hypothetical proteins SAOUHSC_02781 and SAOUHSC_02702, which produced the 3′UTR-derived sRNAs RsaOT (i.e. srr43 or SAOUHSCs080) and Teg130 (i.e. SAOUHSCs100), respectively (46–48). The remaining mRNAs with conserved 3′UTRs encoded enolase (eno), ribosomal protein L32 (rpmF) and a small membrane protein (SAOUHSC_01055a).
It is worth noting that, in most of the mRNAs, nucleotide conservation was lost downstream of the orthologous CDS (Figure 1A and Supplementary Figure S3A). This is illustrated in Figure 1B and Supplementary Figure S3B, in which the distribution of the last conserved nucleotide position (quantified in windows of 10 nt) is shown. The left panel of this figure shows the 3′UTR conserved length distribution in S. aureus, where the maximum of the peak is about 60 nt downstream of the stop codon. Meanwhile, in most of the other staphylococcal species, the maximum is located within the region comprised between −10/+10 nt from the stop codon (Figure 1B and Supplementary Figure S3B).
It is expected that non-coding sequences accumulate more changes throughout the course of evolution than their corresponding CDSs, which cannot significantly change without dramatically affecting protein functionality. In agreement with this, we reasoned that 5′UTRs (excluding the ribosome binding region) should suffer a similar degree of nucleotide variation than the 3′UTRs. Therefore, we performed the same blastn comparison, using a query database that included the 5′UTR sequences plus the first 200 nt of the CDS (Supplementary Table S5). As shown by the scatter plots in Supplementary Figure S4, evident variations among the 5′UTRs of orthologous genes existed. However, the number of conserved 5′UTRs was higher than their 3′UTR counterparts. This was significant when comparing the 5′UTR and 3′UTR scatter plots of a specific staphylococcal species, as the number of dots closer to the diagonal line is always greater for the 5′UTRs (Supplementary Figure S4). Although this analysis may have been biased by the presence of the ribosome binding site, it supported the idea of 3′UTRs being more prone to evolutionary changes than 5′UTRs.
Most of the mRNAs encoding orthologous proteins contain 3′UTRs with different lengths and sequences in Staphylococcaceae
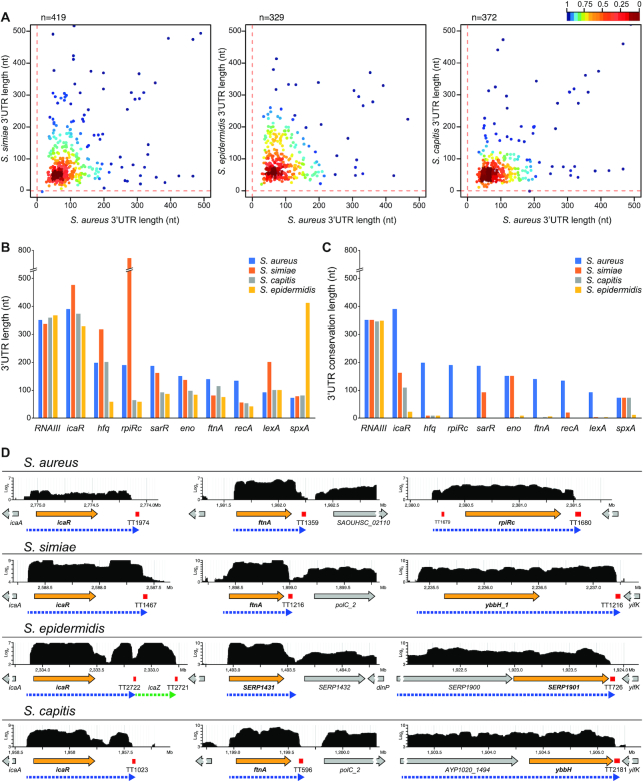
The fact that the conservation of several 3′UTRs was almost fully lost after the CDSs indicated that the mRNAs encoding orthologous proteins had different 3′UTRs. To determine and compare the real length of 3′UTRs among closely-related staphylococcal species, we performed RNA sequencing of whole transcripts from the S. simiae, S. epidermidis and S. capitis strains. The obtained transcriptomic maps were loaded into a Jbrowse-based server (27), which is publicly available at http://rnamaps.unavarra.es. Then, the lengths of the 3′UTRs from each S. aureus orthologous monocistronic gene were manually annotated by combining the RNA-seq data and the Rho-independent transcriptional terminator predictions (using TransTermHP algorithm) (32). Subsequently, we plotted the real 3′UTR lengths from each staphylococcal species against the 3′UTR lengths of their corresponding S. aureus orthologous genes. Figure 2A shows that most of the 3′UTR lengths from S. aureus did not correlate with those of the three other analysed staphylococcal species (Figure 2A). This lack of correspondence was exemplified when selecting some biologically relevant genes (Figures 2B–D). As reflected in Figures 2B and D, in most cases, the length of the 3′UTR showed variability among all the analysed species, while a similar 3′UTR length for a given gene was only preserved in a few examples. Overall, these data confirmed that most of the mRNAs encoding orthologous genes in staphylococcal species had 3′UTRs with different lengths in addition to sequence variation.
Figure 2.
The lengths of the 3′UTRs do not correlate among staphylococcal species. (A) Scatter plots representing the real 3′UTR lengths of each staphylococcal species in function of the 3′UTR lengths of the corresponding orthologous S. aureus mRNAs. The length of the 3′UTRs from S. aureus orthologous monocistronic mRNAs were annotated by combining the transcriptomic data and the prediction of Rho-independent transcriptional terminators by TransTermHP (32). Only 3′UTRs shorter than 500 nt are represented. The plot was coloured by applying the Kernel density estimation, which indicates the proximity of the dots: blue, isolated dots; red, overlapping dots. (B) Plot representing the 3′UTR length of relevant orthologous genes in the indicated staphylococcal species. (C) Plot representing the 3′UTR conservation length of the orthologous genes analysed in B, which was determined by the blastn algorithm. RNAIII is included as an example of an mRNA with a highly conserved 3′UTR. (D) Browser images showing the RNA transcribed from the icaR, ftnA and rpiRc chromosomic regions of the S. aureus, S. simiae, S. epidermidis and S. capitis strains. The RNA-seq data was mapped to the corresponding genomes, and the transcriptomic maps were loaded onto a server based on Jbrowse (27). The complete transcriptomic maps are available at http://rnamaps.unavarra.es/.
3′UTR sequence differences are originated by local nucleotide changes and gene rearrangements
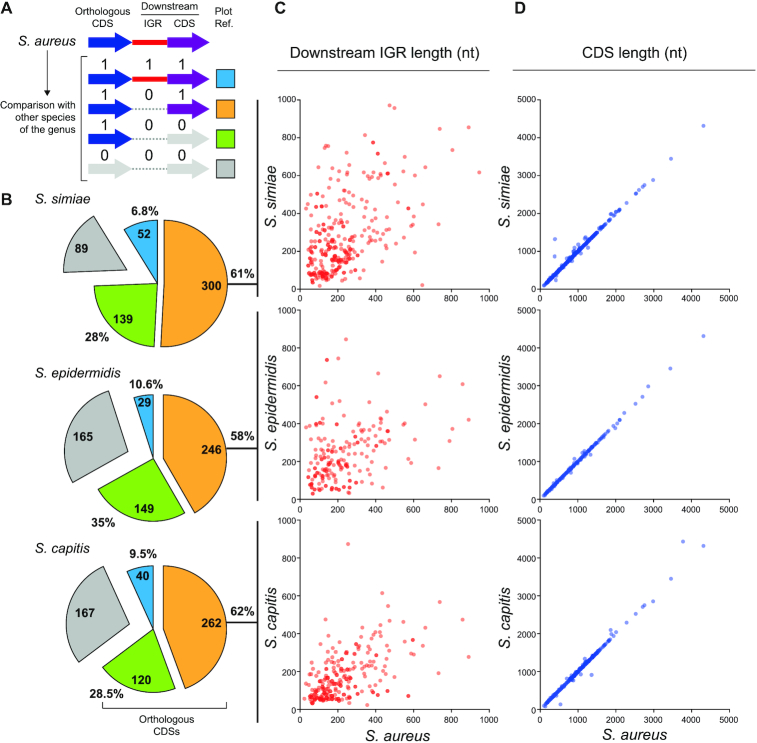
In order to understand how 3′UTR differences originated among mRNAs encoding orthologous proteins, we compared the whole genomic sequence of S. aureus (Sau) against that of S. simiae (Ssim), S. epidermidis (Sepi) and S. capitis (Scap) using Mauve (49). Note that we only focused on the monocistronic mRNAs, as stated above. We expected 3′UTR sequence variations to occur due to different genomic rearrangements, as previously described for some sRNAs located at intergenic regions (IGRs) (50). Therefore, we checked the sequence conservation in genomic regions downstream of the orthologous CDSs, including IGRs and adjacent CDSs, for each genomic pair (Sau versus Ssim, Sau versus Sepi and Sau versus Scap). We assigned a value of 1 or 0 depending on whether the downstream IGR/CDS was conserved or not, respectively (Figure 3A). Confirming previous blastn analysis (Figure 1), we found that only ∼6.8–10.6% of the S. aureus CDSs had a conserved downstream IGR/CDS when compared to the abovementioned species (Figure 3B). On the other hand, 28–35% of the orthologous CDSs lacked a conserved downstream IGR/CDS, indicating that gene rearrangements in these loci led to 3′UTR variations. The rest of CDSs showed a non-conserved downstream IGR but a preserved downstream CDS (between 58% and 62% depending on the species analysed) (Figure 3B). When performing this analysis, we realized that such variations substantially altered the IGR lengths (Figure 3C). Considering that the 3′ end conservation was lost around the protein stop codon (Figure 1), IGR length variations may be attributed to shifts in the position of the stop codon, which may ultimately affect the CDS length. To address such a possibility, we compared the lengths of S. aureus CDSs to those of their corresponding orthologous genes for the three abovementioned staphylococcal species. As Figure 3D shows, most of the orthologous CDSs had similar lengths, indicating that variations in the lengths of the IGRs were not due to differences in the analysed CDS lengths (Figure 3D). Therefore, we concluded that the differences on the 3′UTR sequences occurred partly due to gene rearrangements and mostly because of sequence variations in the IGR sequences. The causes for local variations were unknown.
Figure 3.
Nucleotide sequence variation occurring downstream of orthologous CDSs may explain 3′UTR diversity. (A) Schematic representation of the conservation analysis performed on IGRs and CDSs located downstream of an orthologous CDS. Whole-genome comparisons between S. aureus and its phylogenetically-related species were performed using Mauve (49). Values 1 and 0 were assigned to conserved and non-conserved IGRs/CDSs, respectively, and a different colour was attributed depending on the conservation configuration. Blue: the IGR and CDS downstream of the orthologous CDS are conserved; orange: the downstream CDS is conserved but not the IGR; green: both downstream regions are not conserved, and grey: no orthologous CDS was found in the analysed species. (B) Pie chart quantifying the different categories represented in A. (C) Plot showing the length of the IGR downstream of each orthologous CDS in the different species compared to that of S. aureus. (D) Plot showing the length of the orthologous CDS in the different species compared to that of S. aureus. Note that only the IGRs and CDSs that fall under the orange category are plotted in C and D.
Species-specific 3′UTRs variations affect the expression of orthologous genes
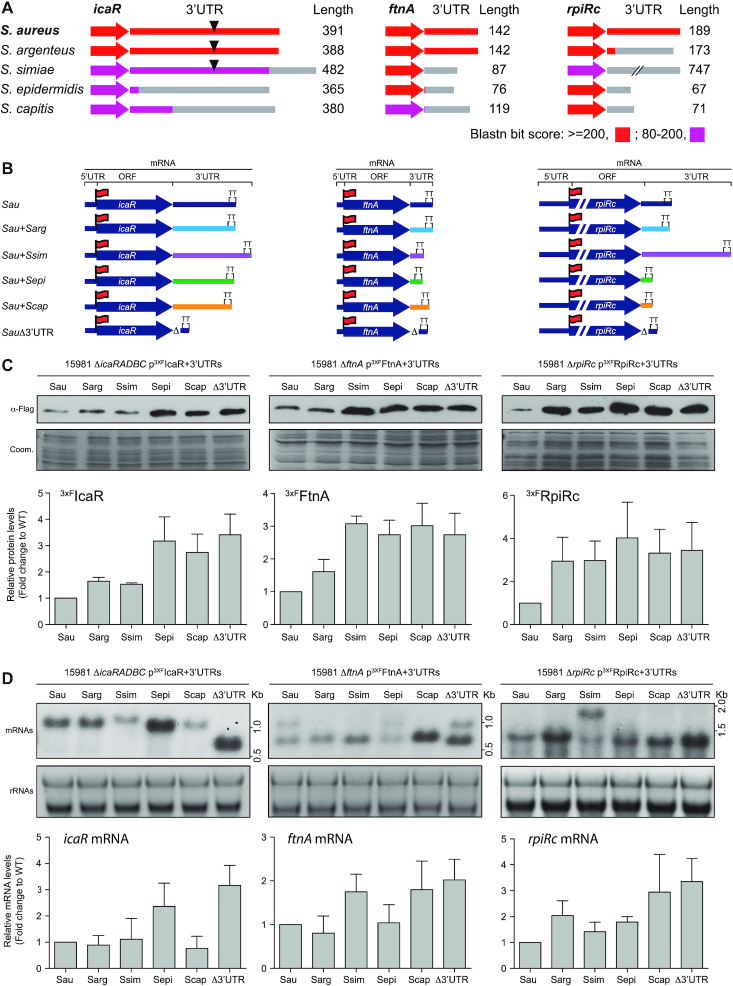
3′UTR variations would be physiologically relevant if they led to differences in the expression of the orthologous genes of a particular bacterial species. To study this in detail, we selected the icaR (5), ftnA and rpiRc genes as examples since we had previously found that their expression was affected by the deletion of their corresponding 3′UTR. In addition, according to the single nucleotide polymorphism (SNP) analysis carried out by Joseph and colleagues (51), the 3′UTRs sequences of these mRNAs are highly conserved among different S. aureus strains (Supplementary Figure S5). At the same time, these 3′UTRs present different levels of conservation when compared to their closest phylogenetically-related staphylococcal species (Figure 4A). In addition, transcript mapping by RNA-seq revealed variations in the 3′UTR lengths (Figure 2B and D). According to synteny analysis, the icaR 3′UTR showed significant variations in length and sequence due to gene rearrangements (Supplementary Figure S6A), while the differences in the ftnA and rpiRc 3′UTRs were most likely caused by local nucleotide variations in the IGRs, as no gene rearrangements were found by synteny analysis (Supplementary Figure S6B and C). Although a CDS insertion was found downstream of the ftnA in S. argenteus, it did not affect the mRNA sequence (Supplementary Figure S6B).
Figure 4.
Changes in 3′UTRs sequences affect protein expression. (A) Schematic representation of the conservation analysis of the icaR, ftnA and rpiRc mRNAs among phylogenetically-related species. The colour code indicates the blastn bit score. The 3′UTR lengths in the corresponding species are indicated and represented as grey lines. Black triangles indicate the presence of the UCCCC motif. (B) Schematic representation of the constructed chimeric mRNAs, which comprises the S. aureus CDS and a 3′UTR from a different staphylococcal species (for strains and plasmids details see supplementary data). Red flags indicate the insertion of the 3xFLAG tag in the N-terminus. (C) Western blot showing the levels of an orthologous protein when it is expressed from different chimeric mRNAs. The Western blot was developed using peroxidase conjugated anti-FLAG antibodies. A Coomassie stained gel portion is shown as a loading control. (D) Northern blot showing the mRNA levels expressed from the constructs represented in B. These were detected using CDS-specific antisense radiolabelled RNA probes. Ribosomal RNAs stained with Midori Green are shown as loading controls. Western and Northern blots images show the representative results from at least three independent replicates. Protein and mRNA levels were quantified by densitometry of Western blot images and Northern blot autoradiographies using ImageJ (http://rsbweb.nih.gov/ij/). Each of the protein or mRNA levels was normalized to the levels of S. aureus. Sau, S. aureus; Sarg, S.argenteus; Ssim,S. simiae; Sepi, S. epidermidis; Scap,S. capitis.
To confirm the transcript boundaries of the icaR, ftnA and rpiRc mRNAs from S. aureus, S. simiae, S. epidermidis and S. capitis, we performed a simultaneous mapping of their 5′ and 3′ mRNA ends using a modified version of the rapid amplification of cDNA ends technique (mRACE). Supplementary Figures S7 to S9 show the mRACE results that confirmed the mRNA mappings from the RNA-seq data in combination with the transcriptional terminator predictions. Note that rpiRc in S. epidermidis and S. capitis is the last gene of a polycistronic transcript. Therefore, we were unable to obtain mRACE mapping results from the native gene. We used Mfold to predict the putative transcriptional terminator structures of the transcripts (52). Interestingly, these stem-loops were different for the majority of the cases (Supplementary Figures S7B, S8B and S9B).
Based on the information collected, we constructed chimeric mRNAs that combined the CDSs of the three selected S. aureus genes with the 3′UTR of their corresponding orthologous genes (from the same staphylococcal species analysed in Figure 4A). We fused such 3′UTRs to their corresponding S. aureus CDS, which carried the 3xFLAG sequence at the N-terminal, to monitor their protein expression (Figure 4B). These plasmids were then electroporated into the mutant strains that lacked the original genes to facilitate the detection of the mRNAs that were expressed from the plasmids. To confirm the mapping of the 3′ end of the chimeric rpiRc mRNA carrying the 3′UTRs from S. epidermidis and S. capitis, we performed mRACE. The results showed that the transcripts ended at the expected sites, which were downstream of the predicted stem-loops (Supplementary Figure S9).
Expression analysis of the 3xFLAG-tagged proteins by Western blotting revealed that the deletion of the 3′UTRs produced an increase of the IcaR, Ftn and RpiRc proteins. Interestingly, the chimeric icaR mRNAs harbouring the UCCCC motif (3′UTRs from S. simiae and S. argenteus) (Figure 4A), which was required for modulation of IcaR translation in S. aureus (5), presented similar IcaR protein levels to those shown by the S. aureus wild-type (WT) mRNA. In contrast, the chimeric icaR mRNAs lacking the UCCCC motif (S. epidermidis and S. capitis icaR 3′UTRs) expressed a comparable amount of the IcaR protein to that of a S. aureus mRNA carrying the 3′UTR deletion (Figure 4C). Chimeric mRNAs expressing FtnA and RpiRc showed protein yields similar to those of the same mRNAs lacking the 3′UTR. The exception to such behaviour was the chimeric construct carrying the ftnA mRNA in combination with the 3′UTR of S. argenteus, whose sequence was conserved when compared to S. aureus (Figure 4A). The same samples were then subjected to Northern blot analyses. The results showed that, in most cases, the steady-state mRNA levels correlated with the protein levels (Figure 4D). This suggested that the increase in protein levels, which occurred when the original 3′UTR sequence was substituted by the one present in its orthologous mRNA, was most likely due to an increase in mRNA stability, as previously described for the icaRΔ3′UTR mRNA (5). To confirm this, we measured the half-life of the chimeric icaR+3′UTRSepi mRNA as an example. Supplementary Figure S10 shows that this chimeric mRNA presented a higher half-life in comparison with the one from the icaR WT mRNA (Supplementary Figure S10).
Note that the icaR-3′UTRScap and ftnA-3′UTRSepi chimeras did not show increased mRNA levels, implying that protein expression could be affected in other ways. Altogether, these results indicated that species-specific variations in the 3′UTR sequences could not reproduce the expression levels found in S. aureus, suggesting putative distinct functional roles for 3′UTRs in the different species.
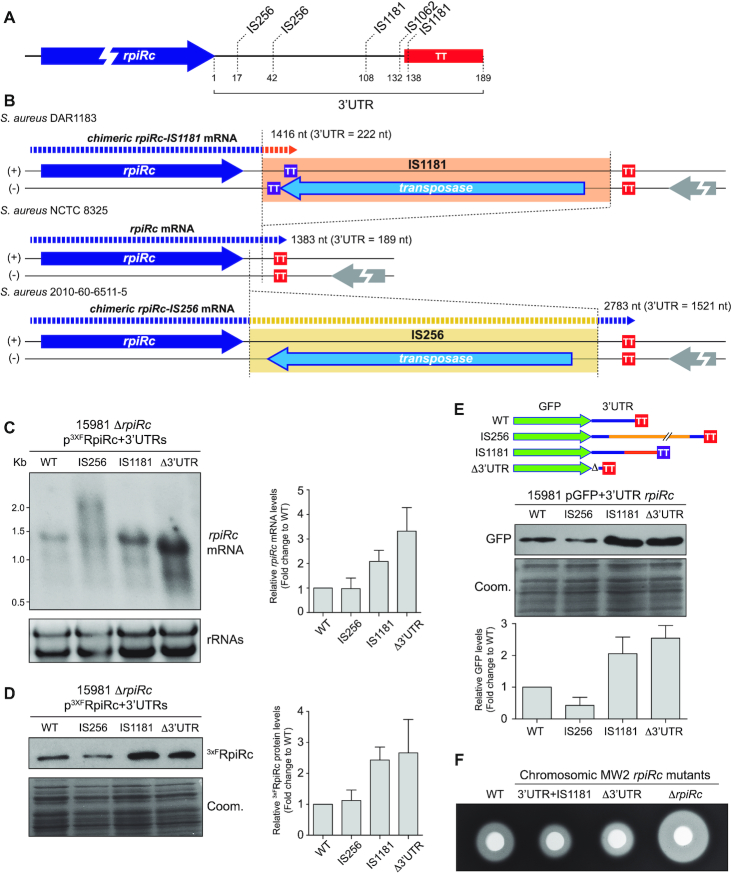
IS1181 transposition to rpiRc 3′UTR modifies RpiRc expression and haemolysis production
The rpiRc 3′UTR in the S. simiae strain is considerably longer than the 3′UTRs from other staphylococcal species (Figure 4A). A deep sequence analysis revealed that the S. simiae rpiRc 3′UTR contains three copies of STAR elements (45), which are distributed among staphylococcal species (53). Since the number and genomic locations of the STAR element copies are variable among species and strains (53), we investigated whether insertions of STAR elements may also occur in the rpiRc 3′UTR of S. aureus and, therefore, alter protein expression as observed in the chimeric rpiRc construct carrying the 3′UTR of S. simiae (Figure 4C). Since SNP analysis were not indicative of the presence of insertion/deletion sequences, we performed rpiRc mRNA sequence pairwise alignments to look for alignment disruptions within the rpiRc 3′UTR. We reasoned that alignment disruptions could help us identify repeated regions. Following this idea, we used the whole rpiRc mRNA sequence as the query to perform blastn against all draft and complete S. aureus genomes available on the NCBI database. The results showed that 33 S. aureus genomes presented alignment disruptions within the rpiRc 3′UTR (Supplementary Table S6). Since many of these genomes were not fully closed, some alignment disruptions occurred due to the query sequence aligning to different contigs. By analysing the extremes of the aligned contigs, we identified that 16 out of the 33 genomes contained IS insertions in different locations of the rpiRc 3′UTR sequence. Specifically, we were able to map nine IS1181 (54), five IS256 (55) and two ISs of the IS30 transposase family (Figure 5A and Supplementary Table S6). The remaining genomes presented sequence deletions or duplications. Interestingly, we identified duplications of 8–12 nt in some of the strains, which were indicative of the presence/excision of ISs. The fact that no IS sequences were found in the related contigs may be explained by either the use of assembly algorithms that eliminated repeated sequences from the extremes of the contigs or the ISs being excised. However, the latter would not explain why these contigs were not assembled together, suggesting that an unknown IS may be inserted in the rpiRc 3′UTR of these genomes.
Figure 5.
Disruption of the rpiRc 3′UTR sequence by IS transposition events affects the expression of RpiRc. (A) Schematic representation showing the main insertion sites of ISs in the rpiRc 3′UTR of S. aureus. (B) Schematic representation showing the putative chimeric mRNAs that are generated when IS1181 and IS256 are inserted in the rpiRc 3′UTR of the S. aureus DAR1183 and 2010-60-6511-5 strains, respectively. Since IS1181 harbours a stem loop that serves as a transcriptional terminator (TT) in the same strand as the rpiRc gene, which is opposite to the transposase gene, the insertion generates a chimeric 3′UTR that is 222 nt-long. In contrast, the IS256 lacks TTs, generating a chimeric mRNA that includes the whole IS256 sequence. (C) Northern blots showing the mRNA levels expressed from the constructs harbouring the WT rpiRc gene and the IS insertions that mimic the configurations found in the S. aureus DAR1183 and 2010-60-6511-5 strains. The plasmid expressing the rpiRc mRNA Δ3′UTR was included as a control. The mRNA levels of rpiRc were detected with a CDS-specific antisense radiolabelled RNA probe. Ribosomal RNAs stained with Midori Green are shown as loading controls. (D) Western blot showing the 3xFRpiRc protein levels expressed from the constructions used in C. The Western blot was developed using peroxidase conjugated anti-FLAG antibodies. A Coomassie stained gel portion is shown as a loading control. The Western and Northern blot images show the representative results from at least three independent experiments. The protein and mRNA levels were quantified by densitometry of Western blot and Northern blot autoradiographies using ImageJ (http://rsbweb.nih.gov/ij/). Each of the protein or mRNA levels was normalized to the levels of the WT rpiRc mRNA. (E) Western blot showing the GFP protein levels of the constructs harbouring the gfp fused to the 3′UTR, 3′UTR+IS256, 3′UTR+IS1181 of rpiRc and the Δ3′UTR as a control. Western blot was developed using monoclonal anti-GFP antibodies and quantified as described in D. (F) Haemolytic halos produced by the haemolysins contained in the supernatant of the cultures from S. aureus MW2 wild-type (WT) and their isogenic chromosomic mutants: Δ3′UTR, ΔrpiRc and 3′UTR+IS1181. The supernatants were concentrated 10 times and loaded into 5-mm holes in Columbia Sheep blood (5%) agar plates.
In order to analyse the consequences of such insertions, we constructed two plasmids containing the IS256 and IS1181 sequences 42 and 108 nt downstream of the RpiRc stop codon. These mimicked the chromosomic configurations found in the rpiRc region of the S. aureus 2010-60-6511-5 and DAR1183 strains, respectively (Figure 5B). In both cases, the insertion orientation of the IS placed the transposase gene in the DNA strand opposite to that of rpiRc. Northern blot analysis revealed different chimeric mRNA lengths (Figure 5C). On the one hand, IS1181 carried a stem-loop downstream of the transposase gene, in the same DNA strand as the rpiRc gene, that acted as a transcriptional terminator (Figure 5B). Therefore, the IS1181 insertion produced a chimeric rpiRc-IS1181 mRNA of ∼1.4 kb (3′UTR-IS1181 of 222 nt) that included a few nucleotides of the IS1181 sequence. This band migrated in a similar manner as the original rpiRc mRNA. The impact of this insertion was an increase in the mRNA levels (Figure 5B and C), which correlated with an increase in the 3xFRpiRc protein levels (Figure 5D). On the other hand, since the IS256 lacked stem-loops able to prevent transcription from adjacent genes, a chimeric rpiRc-IS256 mRNA of ∼2.8 kb (3′UTR-IS256 ∼1.5 kb) was produced. In contrast to the IS1181 insertion, the chimeric rpiRc-IS256 mRNA included the whole IS256 sequence (Figure 5B and C). In this case, there were no considerable changes neither in the mRNA nor 3xFRpiRc protein levels (Figure 5C and D). Overall, these examples indicated that IS insertions may produce different RpiRc protein expression outcomes depending on the type of IS, the insertion site and the orientation of the IS.
To further confirm that the rpiRc 3′UTR had a functional role that could be affected by IS insertions, we analysed the effect of the WT 3′UTR and the chimeric 3′UTR-ISs on the expression of a heterologous gene, such as the green fluorescent protein (gfp). To this end, we fused the rpiRc 3′UTR WT, the 3′UTR-IS1181 and 3′UTR-IS256 sequences downstream of the gfp gene. As a control, we included the Δ3′UTR plasmid that carried the rpiRc TT downstream of gfp (Figure 5E). Western blot analyses showed that the rpiRc 3′UTR lead to a lower GFP expression when compared to the Δ3′UTR version (Figure 5E). This indicated that the rpiRc 3′UTR could reduce the expression of a heterologous gene and, therefore, act as a functional module. However, the presence of IS1181 in the rpiRc 3′UTR increased GFP expression while IS256 only slightly decreased it. This confirmed that the transposition of ISs in the rpiRc 3′UTR could have different consequences on protein expression depending on the insertion events (Figure 5E).
Next, we decided to analyse the relevance of the rpiRc 3′UTR on the S. aureus physiology and the implications of the IS insertions in its role. Previous studies showed that RpiRc repressed the expression of RNAIII. RNAIII is the master regulator of S. aureus virulence, which activates many virulence factors, including haemolysins. A mutant lacking the rpiRc gene exhibited an increased haemolysis capacity due to the lack of RNAIII repression (56–58). To test whether alterations in the rpiRc 3′UTR could modify the haemolytic activity of S. aureus, we constructed chromosomal mutants in S. aureus MW2, a community-acquired agr-positive methicillin-resistant (MRSA) strain. On the one hand, we performed a 115-bp chromosomal deletion that removed most of the 3′UTR of rpiRc while preserving its intrinsic TT (MW2 Δ3′UTR), as we did for the aforementioned construct. On the other hand, we inserted IS1181 in the same rpiRc region as it is naturally found in S. aureus DAR1183 (MW2 3′UTR+IS1181). As a control, we deleted the whole rpiRc gene (MW2 ΔrpiRc). These strains were grown overnight and the production of haemolysins in the culture supernatants was tested in sheep-blood agar plates. As expected, the ΔrpiRc mutant showed an increase in haemolysis when compared to the WT strain (Figure 5F). In contrast, the deletion of the rpiRc 3′UTR and the IS1181 insertion in the 3′UTR, which produced higher protein levels of the RpiRc repressor (Figure 5D), resulted in lower levels of haemolysis (Figure 5F). Overall, these results highlighted the relevance of the rpiRc 3′UTR in the physiology of S. aureus while demonstrating that the RpiRc function could be altered by the insertion of IS1181 in the rpiRc 3′UTR.
Species-specific icaR 3′UTRs produce different PNAG levels in S. aureus
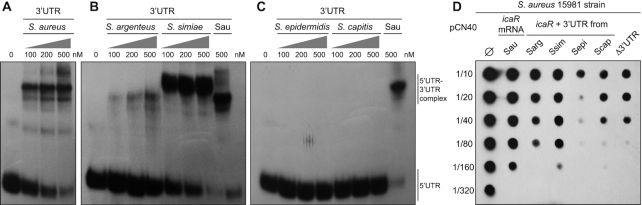
The expression analysis of chimeric icaR mRNAs indicated that the icaR 3′UTRs from S. epidermidis and S. capitis were unable to reproduce the 3′UTR-mediated regulation observed in S. aureus (Figure 4C). We previously showed that the icaR 3′UTR interacted with the 5′UTR of the same mRNA molecule through a UCCCC motif to repress IcaR translation and, in return, modulate PIA-PNAG biosynthesis and biofilm formation in S. aureus (5). The absence of the UCCCC motif in the S. epidermidis and S. capitis icaR 3′UTRs may account for the differences in IcaR expression. This may be attributed to the lack of interaction between their 5′ and 3′UTRs. To test this hypothesis, we carried out electrophoretic mobility shift assays (EMSAs). The S. aureus icaR 5′UTR was synthesized in vitro, radioactively labelled and then incubated with increasing concentrations of cold icaR 3′UTRs from the five staphylococcal species analysed in Figure 4. Figure 6A shows that the S. aureus icaR 3′UTR produced the expected gel-shifts when interacting with the 5′UTR, as previously described (5). Consistently, a similar behaviour was observed when using the 3′UTR of S. simiae (Figure 6B). In the case of the S. argenteus icaR 3′UTR, despite sharing a relatively high sequence identity with its S. aureus counterpart (82% of identity), a much lower binding affinity was found (Figure 6B). This indicated that although S. argenteus preserved the UCCCC motif, the nucleotide differences might affect the binding affinity in vitro. S. epidermidis and S. capitis 3′UTRs, as one would expect, did not produce gel shifts when incubated with the 5′UTR due to the absence of the UCCCC motif in their sequences (Figure 6C). Moreover, recent results showed that the 5′ and 3′UTRs from S. epidermidis, unlike what was observed for S. aureus, did not interact between them, excluding the existence of an orthologous mechanism (59). In contrast, it was found that the S. epidermidis-specific sRNA IcaZ, downstream of the icaR 3′UTR, targeted the icaR 5′UTR and prevented IcaR translation (59). Altogether, these results indicated that sequence variations around the 3′UTR determined the presence or absence of specific motifs and, ultimately, the generation of functional distinctions.
Figure 6.
Species-specific variations in icaR 3′UTRs resulted in different PIA-PNAG levels. Electrophoretic mobility shift assays (EMSAs) between the synthetic S. aureus icaR 5′UTR and the synthetic icaR 3′UTR from: (A) S. aureus as a positive control; (B) S. argenteus and S. simiae, which carry the UCCCC motif necessary for 5′’UTR interaction, as previously described (5); (C) S. epidermidis and S. capitis, which lack the UCCCC motif. The autoradiographies of the band-shifts result from incubating a 32P-labelled synthetic S. aureus 5′UTR RNA fragment (40,000 cpm) with increasing amounts of the different synthetic 3′UTR RNAs (100–500 nM). The UTR complexes are indicated on the right side of the autoradiography. (D) Quantification of PIA-PNAG exopolysaccharide production in the S. aureus 15981 strains expressing the different chimeric icaR mRNAs. Serial dilutions of the samples were spotted onto nitrocellulose membranes and PIA-PNAG expression was developed with specific anti-PIA-PNAG antibodies. ∅ indicates the presence of an empty pCN40 plasmid. Sau, S. aureus; Sarg, S. argenteus; Ssim, S. simiae; Sepi, S. epidermidis; Scap, S. capitis. Images show representative results from at least two independent experiments.
In order to demonstrate that the differences observed in terms of IcaR expression by chimeric mRNAs (Figure 4D) were enough to alter PIA-PNAG biosynthesis and, thus, be of biological relevance, we introduced the plasmids expressing these chimeric mRNAs into the S. aureus 15981 strain, which produced high levels of PIA-PNAG exopolysaccharide. The quantification of PIA-PNAG levels by dot-blot using specific antibodies against the exopolysaccharide showed that the strains expressing higher levels of IcaR (the chimeric Scap/Sepi and Δ3′UTRs strains, which lacked the UCCCC motif) presented lower levels of PIA-PNAG in comparison with the strains producing IcaR from mRNAs containing the UCCCC motif (the WT and chimeric Sarg/Ssim strains) (Figure 6D). These results confirmed that evolutionary variations in 3′UTRs, which occur among phylogenetically-related bacteria, are enough to create gene expression differences that affect important biological processes, such as exopolysaccharide production. This suggested that the synthesis of orthologous proteins might be differentially affected by 3′UTR sequence variations.
3′UTR variability among mRNAs encoding orthologous proteins is a common trait in bacteria
Having proved that the inter-species 3′UTR variability had consequences on S. aureus gene expression, we decided to extend the 3′UTR conservation analysis to other bacterial genera. First, we focused on several previously described functional 3′UTRs (4,6–8). We compared the last 200 nt of the CDSs plus the entire 3′UTR sequences from the E. coli acnB, Y. pestis hmsT, C. glutamicum aceA and B. subtilis hbs mRNAs with those of their corresponding phylogenetically-related species (Supplementary Figures S11A–S14A). Although the CDSs were highly conserved across all closely-related species, there were 3′UTR sequence variations in all four gene examples (Figure 7). The E. coli acnB 3′UTR was conserved in the Citrobacter, Enterobacter and Klebsiella species with only a few nucleotide variations. In contrast, and according to synteny analysis by SyntTax (60), the Salmonella, Cronobacter and Serratia acnB 3′UTRs sequences varied due to gene rearrangements (Supplementary Figure S11B). For example, in S. enterica serovar Typhimurium SL1344, an insertion of the SL1344_0160 gene (which encodes a putative HNH restriction endonuclease) deleted the sequence downstream of the acnB CDS and included a different TT. As a result, the Salmonella acnB 3′UTR was only 34 nt long and it lacked the stem-loop recognized by the apo-AcnB protein (7).
Figure 7.
Schematic representation of the conservation analysis of regulatory 3′UTRs among phylogenetically-related species. Blastn analyses were performed using the E. coli acnB (A), Y. pestis hmsT (B), C. glutamicum aceA (C) and B. subtilis hbs (D) mRNAs as queries, which carried 3′UTRs with proven regulatory capacities (4,6–8). The colour code indicates the blastn bit score. The estimated 3′UTR lengths are indicated and represented by grey lines.
Sequence variations were also present in the C. glutamicum aceA, B. subtilis hbs and Yersinia hmsT 3′UTRs. Although the hbs and hmsT CDSs are widely distributed across the Bacillus and Yersinia genera, their 3′UTRs were only conserved in the closest species: B. amyloliquefaciens, B. licheniformis and B. pumilus in the former and Y. pestis biovars and Y. pseudotuberculosis in the latter (Figure 7). Finally, none of the 10 compared species of the genus Corynebacterium that carried the aceA CDS presented a similar sequence to that of the C. glutamicum aceA 3′UTR (Figure 7). We carried out the prediction of Rho-independent transcriptional terminator structures in the non-conserved 3′UTRs to determine the length variability (Figure 7 and Supplementary Figures S11–S14). Most of these 3′UTRs variations were explained by the gene rearrangements occurring downstream of CDSs, with the exception of the hbs 3′UTR, in which variations may have occurred locally (Supplementary Figures S11–S14).
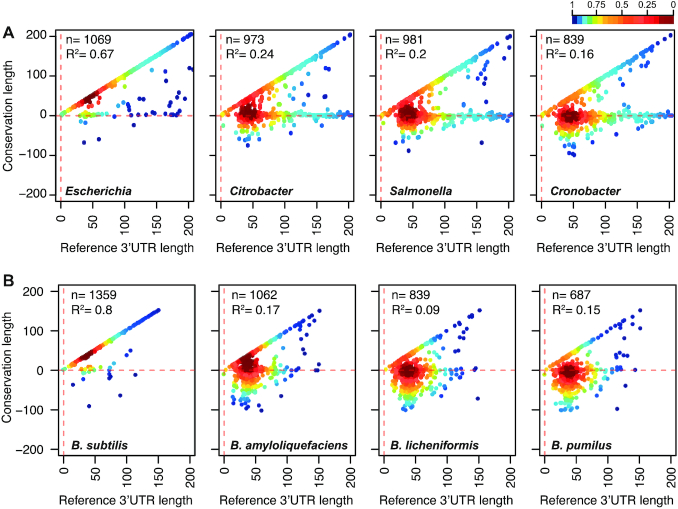
Finally, to further investigate whether 3′UTR variations could also be distributed among other genes of E. coli and B. subtilis, as observed in S. aureus, we performed similar genome-wide 3′UTR conservation analyses. Since the 3′-end transcript boundaries were recently released for E. coli and B. subtilis (28,29), we generated a query database including the sequences of all mapped 3′UTRs plus the last 200 nt of their corresponding CDSs. Using blastn, we compared the E. coli and B. subtilis databases against representative genomes of species from the Enterobacteriaceae family and the genus Bacillus, respectively. We found a similar outcome to the one observed for the genus Staphylococcus, meaning that 3′UTR sequences were also variable among mRNAs encoding orthologous proteins (Figure 8, Supplementary Figure S15, Supplementary Tables S7 and S8). The bacterial species of the Enterobacteriaceae family showed a higher number of conserved 3′UTRs in comparison with the Bacillus species. This was probably due to the phylogenetic distances between the species in the former being shorter than the ones in the latter. However, both bacterial groups presented a very high variation rate of their 3′UTR sequences, as illustrated in Figure 8 and Supplementary Figure S15, where most of the dots fell onto the horizontal line, which represented the protein stop codon. This indicated that nucleotide conservation was also lost downstream of most CDSs (Supplementary Figure S16). On the one hand, these comparative analyses confirmed that the nucleotide conservation analysis might only be useful for detecting a small percentage of inter-species conserved 3′UTRs. On the other hand, the results indicated that the 3′UTRs from orthologous genes underwent different evolutionary events, a phenomenon that seems to be widespread in bacteria.
Figure 8.
3′UTR sequence variations are widely distributed in bacteria. Scatter plots representing the conservation of the 3′ end regions of E. coli (A) and B. subtilis (B) mRNAs compared to their corresponding 3′ end regions in phylogenetically-related species. The E. coli BW25113 3′UTR sequence query database was compared to the E. coli O157-H7 str EC4115, Citrobacter koseri ATCC BAA-895, Salmonella Typhimurium SL1344 and Enterobacter aerogenes KCTC 2190 strains. The B. subtilis 168 3′UTR sequence query database was compared to the B. subtilis OH 131.1, B. amyloliquefaciens DSM7, B. licheniformis ATCC 14580, and B. pumilus SH-B9 strains. Each dot represents the last conserved nucleotide, according to blastn, of an indicated species (y axis) in function of the E. coli and B. subtilis 3′UTR lengths (x axis). The plot was coloured by applying the Kernel density estimation, which indicates the proximity of the dots. Blue: isolated dots; red: overlapping dots. The number (n) of plotted mRNAs encoding orthologous proteins and the square correlation coefficient (R2) are indicated.
DISCUSSION
Bacterial IGRs, which have been traditionally defined as the genomic regions between two CDSs, are known for being poorly conserved among phylogenetically-related bacteria (61–64). For this reason, they have often been regarded as ‘junk’ genetic material lacking relevant biological functions. However, recent RNA-seq technologies have revealed that IGRs may be more complex than initially anticipated, accommodating a plethora of regulatory elements that are either transcribed independently as sRNAs or fully functional from within the UTRs of mRNAs. In recent years, several studies have shown the biological relevance of 3′UTRs both in bacteria and eukaryotic organisms (for recent reviews see (18–19,65)). Since the 3′UTR-regulatory elements are physically- and functionally-related to the adjacent CDSs through mRNAs, an evolutionary connection between them might be expected in phylogenetically-related species. This indicates that the 3′UTR sequences would be preserved to the same degree as their corresponding CDSs. However, in this study, we observed this to be true for just a few examples, since most of the 3′UTRs from mRNAs encoding orthologous proteins, in closely-related bacterial species, display different lengths and sequences. The idea of synthetizing hundreds of nucleotides at the 3′ ends without a relevant function seems contradictory as bacteria tend to efficiently manage energy and clones with a lower fitness are rapidly eliminated from populations. Therefore, an explanation for this relatively high 3′UTR variation rate between related species should be sought. First, it is important to highlight that 3′UTR sequences are more highly conserved in comparison with non-transcribed regions of strains from the same bacterial species (5). This implies that an evolutionary pressure to preserve 3′UTR sequences in a particular species exists. In addition, our data indicates that specific inter-species variations in 3′UTR sequences often occur. Therefore, these variations could differentially affect the expression of orthologous proteins from different species. This could be a strategy used by bacteria to create diversity without changing the CDS, an idea supported by previous studies that show how bacterial genes can shift rapidly between multiple regulatory nodes. Oren et al demonstrated the existence of promoter-regulatory regions that changed among orthologous genes, contributing to expression divergence and conferring distinct fitness advantages (66). Analogously, the finding that 3′UTR sequence variations can affect protein expression depending on the species may be an evolutionarily-selected trait to create species-specific post-transcriptional regulatory processes (16,18,67). Since the 5′UTRs are closely related with the initiation of translation, any significant sequence variations could impair protein synthesis and, thus, explain the bias in variability within 3′UTRs. This hypothesis is in agreement with the role of 3′UTRs in eukaryotes, which have been specifically targeted through the course of evolution to contribute to the divergence of species by accumulating regulatory elements in their sequences (65,68–69). It is worth noting that longer 3′UTRs (including multiple miRNA target sites) are preferred in functionally critical eukaryotic genes that are spatially or temporally expressed. In contrast, housekeeping genes are selected to have shorter 3′UTRs (70). Strikingly, the 3′UTRs of Salmonella hilD and S. aureus icaR mRNAs (which encode the main transcriptional regulators of the SP-1 pathogenicity island and biofilm formation, respectively) are among the longest bona fide 3′UTRs found in bacteria. Resembling the eukaryotic process in which mRNAs are targeted by miRNAs, it has been recently shown that hilD and icaR 3′UTRs are targeted by sRNAs (71,72). In the former, the Spot 42 sRNA (transcriptionally repressed by CRP-cAMP) targets the hilD 3′UTR, exerting a positive effect on HilD expression (71,73). In the latter, the RsaI sRNA modulates the PIA-PNAG synthesis by interacting with the 3′UTR of the icaR mRNA (72). The RsaI interacting region is located between the UCCCC motif, which is required for the icaR 3′-5′UTR interaction, and the transcriptional terminator of the icaR mRNA (72). Interestingly, while the UCCCC motif is conserved across the S. aureus, S. argenteus and S. simiae icaR mRNAs, the RsaI paring region is only present in the S. aureus icaR mRNA. Note that the RsaI sRNA is conserved in most species of the genus Staphylococcus. The Spot 42 sRNA is present in several bacterial genera, while the hilD gene is exclusive to Salmonella (74). Overall, this shows that the 3′UTRs can accumulate more than one regulatory motif, which may be differentially selected during evolution.
The acquisition of icaR 3′UTR variations may be explained by gene rearrangements that created mRNA chimeras among staphylococcal species. The S. aureus icaR CDS is only present in 5 out of 9 staphylococcal species that encode the icaADBC operon. It is interesting to note that in the four species lacking the icaR CDS either DspB (hexosaminidase), a TetR-like regulator, or proteins of a two-component system are encoded in place of the icaR gene. These different genomic organizations indicate that the regulatory locus evolved independently from the icaADBC operon and may also explain the appearance of 3′UTR variations in the icaR mRNA when such a locus was acquired or mobilized. Similarly, gene rearrangements could also be responsible for the disparity of sRNA content in phylogenetically-related bacterial species and may also justify some of the 3′UTR variations (50).
In addition, there may be other phenomena beyond gene rearrangements behind the 3′UTR variations, since only a minor percentage of them (∼28–35%) fall under this category (Figure 3). It is likely that mobile insertion sequences and/or repeated sequences, such as those found in the IGRs of S. aureus, change the functionality of the 3′UTRs of orthologous mRNAs (53,75–76). Interestingly, we showed that the presence of STAR elements and ISs that interrupt 3′UTR sequences affect the expression of RpiRc in S. aureus (Figure 5). This suggests that alternative IS transposition/excision processes may generate local 3′UTR variations that can be evolutionarily selected if they produce an advantage for a specific clone in comparison to the rest of the bacterial population. Alternatively, ISs might be also responsible for phase variation mechanisms (77,78). Here, we showed that the presence of IS1181 in the rpiRc 3′UTR of several S. aureus strains reduced haemolysin production by increasing the levels of the RNAIII-repressor RpiRc (Figure 5). Further studies would be required to determine whether the transposition of ISs into the rpiRc 3′UTR provide an additional way to regulate S. aureus virulence through the agr locus, as seen for the cases of agr-phase-variation (79).
Some of the genomic repeats include stem-loops, which serve as transcriptional terminators (Figure 5) or provide specific targets for RNase III (80). Moreover, changes in the 3′UTR sequence could also affect mRNA turnover by modifying the 3′UTR accessibility to exoribonucleases. This indicates that the accumulation/elimination of ribonuclease target sites could be an alternative way to specifically modulate the expression of a protein at the post-transcriptional level in a particular species. In agreement with this, the deletion of 3′UTRs (e.g. hilD, aceA, hmsT and icaR), and with them, the putative ribonuclease target sites, increases protein expression (4–6,9,21). Similarly, the presence of specific target sites for RNA-binding proteins in 3′UTRs could also contribute to regulatory differences among bacterial species (7,11).
In addition to a differential effect on orthologous genes, it should be also considered that variations in 3′UTR sequences may affect the expression of downstream genes. For example, if variations in the 3′UTR sequence alter the strength of the corresponding TT or even eliminate it (e.g. Figure 4 shows that transcriptional read-through occurs in ftnA of some staphylococcal species), transcription would continue to increase the expression of downstream genes or create antisense RNAs if the downstream gene is encoded in the opposite direction (81). Moreover, if a downstream gene is close enough, variations in the 3′UTR sequence could also alter promoter regulatory elements of the next transcription unit, thereby affecting their expression.
Further studies are needed to improve our knowledge of how sequence variations in 3′UTRs may specifically affect gene expression in different bacterial species. The development of RNA sequencing techniques, mapping the precise 3′-ends of transcripts, as well as ribonuclease cleavage regions and RNA-protein binding sites will considerably contribute to this (28,82–84). Combining such information with studies like the one presented here will help improving our understanding of the evolutionary features responsible for the generation of species-specific sequences that may, ultimately, lead to bacterial species differentiation.
DATA AVAILABILITY
The transcriptomic maps of S. simiae CCM 7213T, S. capitis SK14 and S. epidermidis RP62A strains are available at http://rnamaps.unavarra.es/. The transcriptomic data has been also deposited in GEO under accession GSE143600. Other data or material that support the findings of this study can be made available by the corresponding author upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jaione Valle, Prof. Cristina Solano, Prof. Iñigo Lasa, Prof. Josep Casadesús and Prof. Pascale Romby for their excellent advice and their critical reading of the manuscript; Prof. Tomas Maira-Litrán for the anti-PIA-PNAG antibodies; B. Garcia for her helpful assistance with RNA extraction. We also thank the following organizations for providing strains and plasmids: Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for providing pCN40 (NR-46131) and pCN47 (Ref. NR-46141) plasmids constructed by Prof. Emmanuelle Charpentier and Staphylococcus aureus strains N315 (NR-45898) and Staphylococcus epidermidis strain RP62A (Ref. ATCC 35984) through BEI Resources, NIAID, NIH and the Human Microbiome Project for providing Staphylococcus capitis strain SK14 (Ref HM-117) also through BEI Resources, NIAID, NIH.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A.T.-A. was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program [ERC-CoG-2014-646869] and the Spanish Ministry of Economy and Competitiveness [BFU2014-56698-P] grants; C.J.C. was supported by a predoctoral contract from the Public University of Navarre; I.C. was supported by recurrent funding from the Centre National de la Recherche Scientifique (CNRS) and by a PICS program (‘Projet International de Coopération Scientifique’, No. PICS07507, CNRS) between France and Spain. Funding for open access charge: European Research Council (ERC), European Union's Horizon 2020 research and innovation program [ERC-CoG-2014-646869].
Conflict of interest statement. None declared.
REFERENCES
- 1. Waters L.S., Storz G.. Regulatory RNAs in bacteria. Cell. 2009; 136:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gripenland J., Netterling S., Loh E., Tiensuu T., Toledo-Arana A., Johansson J.. RNAs: regulators of bacterial virulence. Nat. Rev. Micro. 2010; 8:857–866. [DOI] [PubMed] [Google Scholar]
- 3. Bouloc P., Repoila F.. Fresh layers of RNA-mediated regulation in Gram-positive bacteria. Curr. Opin. Microbiol. 2016; 30:30–35. [DOI] [PubMed] [Google Scholar]
- 4. Maeda T., Wachi M.. 3′ untranslated region-dependent degradation of the aceA mRNA, encoding the glyoxylate cycle enzyme isocitrate lyase, by RNase E/G in Corynebacterium glutamicum. Appl. Environ Microbiol. 2012; 78:8753–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruiz de Los M,I., Vergara-Irigaray M., Segura V., Villanueva M., Bitarte N., Saramago M., Domingues S., Arraiano C.M., Fechter P., Romby P. et al.. Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 2013; 9:e1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu H., Mao X.-J., Guo X.-P., Sun Y.-C.. The hmsT 3′ untranslated region mediates c-di-GMP metabolism and biofilm formation in Yersinia pestis. Mol. Microbiol. 2015; 99:1167–78. [DOI] [PubMed] [Google Scholar]
- 7. Benjamin J.-A.M., Massé E.. The iron-sensing aconitase B binds its own mRNA to prevent sRNA-induced mRNA cleavage. Nucleic Acids Res. 2014; 42:10023–10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braun F., Durand S., Condon C.. Initiating ribosomes and a 5′/3′-UTR interaction control ribonuclease action to tightly couple B. subtilis hbs mRNA stability with translation. Nucleic Acids Res. 2017; 45:11386–11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao J.-P., Zhu H., Guo X.-P., Sun Y.-C.. AU-Rich Long 3′ untranslated region regulates gene expression in bacteria. Front Microbiol. 2018; 9:633–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawano M., Reynolds A.A., Miranda-Rios J., Storz G.. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005; 33:1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chao Y., Papenfort K., Reinhardt R., Sharma C.M., Vogel J.. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012; 31:4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng T., Berghoff B.A., Oh J.-I., Weber L., Schirmer J., Schwarz J., Glaeser J., Klug G.. Regulation of a polyamine transporter by the conserved 3′ UTR-derived sRNA SorX confers resistance to singlet oxygen and organic hydroperoxides in Rhodobacter sphaeroides. RNA Biol. 2016; 13:988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chao Y., Vogel J.. A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell. 2016; 61:352–363. [DOI] [PubMed] [Google Scholar]
- 14. Guo M.S., Updegrove T.B., Gogol E.B., Shabalina S.A., Gross C.A., Storz G.. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Devel. 2014; 28:1620–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Updegrove T.B., Kouse A.B., Bandyra K.J., Storz G.. Stem-loops direct precise processing of 3′ UTR-derived small RNA MicL. Nucleic Acids Res. 2018; 47:1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyakoshi M., Matera G., Maki K., Sone Y., Vogel J.. Functional expansion of a TCA cycle operon mRNA by a 3′ end-derived small RNA. Nucleic Acids Res. 2018; 250:1727–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Mets F., van Melderen L., Gottesman S.. Regulation of acetate metabolism and coordination with the TCA cycle via a processed small RNA. Proc. Natl. Acad Sci. U.S.A. 2019; 116:1043–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miyakoshi M., Chao Y., Vogel J.. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr. Opin. Microbiol. 2015; 24:132–139. [DOI] [PubMed] [Google Scholar]
- 19. Ren G.-X., Guo X.-P., Sun Y.-C.. Regulatory 3′ untranslated regions of bacterial mRNAs. Front. Microbiol. 2017; 8:633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellermeier J.R., Slauch J.M.. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 2007; 10:24–29. [DOI] [PubMed] [Google Scholar]
- 21. López-Garrido J., Puerta-Fernández E., Casadesús J.. A eukaryotic-like 3′ untranslated region in Salmonella enterica hilD mRNA. Nucleic Acids Res. 2014; 42:5894–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gerstmeir R., Wendisch V.F., Schnicke S., Ruan H., Farwick M., Reinscheid D., Eikmanns B.J.. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 2003; 104:99–122. [DOI] [PubMed] [Google Scholar]
- 23. Novick R.P., Ross H.F., Projan S.J., Kornblum J., Kreiswirth B., Moghazeh S.. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993; 12:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bronesky D., Wu Z., Marzi S., Walter P., Geissmann T., Moreau K., Vandenesch F., Caldelari I., Romby P.. Staphylococcus aureus RNAIII and Its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. 2016; 70:299–316. [DOI] [PubMed] [Google Scholar]
- 25. Lasa I., Toledo-Arana A., Dobin A., Villanueva M., de los Mozos I.R., Vergara-Irigaray M., Segura V., Fagegaltier D., Penadés J.R., Valle J. et al.. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20172–20177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koch G., Yepes A., Förstner K.U., Wermser C., Stengel S.T., Modamio J., Ohlsen K., Foster K.R., Lopez D.. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell. 2014; 158:1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dar D., Shamir M., Mellin J.R., Koutero M., Stern-Ginossar N., Cossart P., Sorek R.. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science. 2016; 352:aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dar D., Sorek R.. High-resolution RNA 3′-ends mapping of bacterial Rho-dependent transcripts. Nucleic Acids Res. 2018; 46:6797–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., Loh E., Gripenland J., Tiensuu T., Vaitkevicius K. et al.. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009; 459:950–956. [DOI] [PubMed] [Google Scholar]
- 31. Tjaden B. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol. 2015; 16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kingsford C.L., Ayanbule K., Salzberg S.L.. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007; 8:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Britton R.A., Wen T., Schaefer L., Pellegrini O., Uicker W.C., Mathy N., Tobin C., Daou R., Szyk J., Condon C.. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 2007; 63:127–138. [DOI] [PubMed] [Google Scholar]
- 34. Arnaud M., Chastanet A., Debarbouille M.. New vector for efficient allelic replacement in naturally nontransformable, low-GC-Content, Gram-positive bacteria. Appl. Environ. Microbiol. 2004; 70:6887–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Valle J., Toledo-Arana A., Berasain C., Ghigo J.M., Amorena B., Penadés J.R., Lasa I.. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 2003; 48:1075–1087. [DOI] [PubMed] [Google Scholar]
- 36. Caballero C.J., Menendez-Gil P., Catalan-Moreno A., Vergara-Irigaray M., García B., Segura V., Irurzun N., Villanueva M., Ruiz de Los Mozos I., Solano C. et al.. The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic Acids Res. 2018; 46:1345–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J.C. Electrotransformation of staphylococci. Methods Mol. Biol. 1995; 47:209–216. [DOI] [PubMed] [Google Scholar]
- 38. Charpentier E., Anton A.I., Barry P., Alfonso B., Fang Y., Novick R.P.. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 2004; 70:6076–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balestrino D., Hamon M.A., Dortet L., Nahori M.-A., Pizarro-Cerda J., Alignani D., Dussurget O., Cossart P., Toledo-Arana A.. Single-cell techniques using chromosomally tagged fluorescent bacteria to study Listeria monocytogenes infection processes. Appl. Environ. Microbiol. 2010; 76:3625–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M.. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006; 34:D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cramton S.E., Gerke C., Schnell N.F., Nichols W.W., Götz F.. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999; 67:5427–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maira-Litrán T., Kropec A., Goldmann D.A., Pier G.B.. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated staphylococcal poly-N-acetyl-β-(1-6)-glucosamine. Infect. Immun. 2005; 73:6752–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L.. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008; 36:W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hedges S.B., Marin J., Suleski M., Paymer M., Kumar S.. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015; 32:835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cramton S.E., Schnell N.F., Götz F., Brückner R.. Identification of a new repetitive element in Staphylococcus aureus. Infect. Immun. 2000; 68:2344–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bohn C., Rigoulay C., Chabelskaya S., Sharma C.M., Marchais A., Skorski P., Borezée-Durant E., Barbet R., Jacquet E., Jacq A. et al.. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010; 38:6620–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beaume M., Hernandez D., Farinelli L., Deluen C., Linder P., Gaspin C., Romby P., Schrenzel J., Francois P.. Cartography of Methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS ONE. 2010; 5:e10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carroll R.K., Weiss A., Broach W.H., Wiemels R.E., Mogen A.B., Rice K.C., Shaw L.N.. Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in Staphylococcus aureus. mBio. 2016; 7:e01990-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Darling A.C.E., Mau B., Blattner F.R., Perna N.T.. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004; 14:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Raghavan R., Kacharia F.R., Millar J.A., Sislak C.D., Ochman H.. Genome rearrangements can make and break small RNA genes. Genome Biol. Evolut. 2015; 7:557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Joseph S.J., Li B., Petit R.A., Qin Z.S., Darrow L., Read T.D.. The single-species metagenome: subtyping Staphylococcus aureus core genome sequences from shotgun metagenomic data. PeerJ. 2016; 2016:e2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Purves J., Blades M., Arafat Y., Malik S.A., Bayliss C.D., Morrissey J.A.. Variation in the genomic locations and sequence conservation of STAR elements among staphylococcal species provides insight into DNA repeat evolution. BMC Genomics. 2012; 13:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chesneau O., Lailler R., Derbise A., Solh El N.. Transposition of IS1181 in the genomes of Staphylococcus and Listeria. FEMS Microbiol. Lett. 1999; 177:93–100. [DOI] [PubMed] [Google Scholar]
- 55. Lyon B.R., Gillespie M.T., Skurray R.A.. Detection and characterization of IS256, and insertion sequence in Staphylococcus aureus. J. Gen. Microbiol. 1987; 133:3031–3038. [DOI] [PubMed] [Google Scholar]
- 56. Zhu Y., Nandakumar R., Sadykov M.R., Madayiputhiya N., Luong T.T., Gaupp R., Lee C.Y., Somerville G.A.. RpiR homologues may link Staphylococcus aureus RNAIII synthesis and pentose phosphate pathway regulation. J. Bacteriol. 2011; 193:6187–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gaupp R., Wirf J., Wonnenberg B., Biegel T., Eisenbeis J., Graham J., Herrmann M., Lee C.Y., Beisswenger C., Wolz C. et al.. RpiRc Is a pleiotropic effector of virulence determinant synthesis and attenuates pathogenicity in Staphylococcus aureus. Infect. Immun. 2016; 84:2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Balasubramanian D., Ohneck E.A., Chapman J., Weiss A., Kim M.K., Reyes-Robles T., Zhong J., Shaw L.N., Lun D.S., Ueberheide B. et al.. Staphylococcus aureus coordinates Leukocidin expression and pathogenesis by sensing metabolic fluxes via RpiRc. mBio. 2016; 7:e00818-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lerch M.F., Schoenfelder S.M.K., Marincola G., Wencker F.D.R., Eckart M., Förstner K.U., Sharma C.M., Thormann K.M., Kucklick M., Engelmann S. et al.. A non-coding RNA from the intercellular adhesion (ica) locus of Staphylococcus epidermidis controls polysaccharide intercellular adhesion (PIA)-mediated biofilm formation. Mol. Microbiol. 2019; 111:1571–1591. [DOI] [PubMed] [Google Scholar]
- 60. Oberto J. SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics. 2013; 14:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Molina N., van Nimwegen E.. Universal patterns of purifying selection at noncoding positions in bacteria. Genome Res. 2008; 18:148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Luo H., Tang J., Friedman R., Hughes A.L.. Ongoing purifying selection on intergenic spacers in group A streptococcus. Infect. Genet. Evol. 2011; 11:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sridhar J., Sabarinathan R., Balan S.S., Rafi Z.A., Gunasekaran P., Sekar K.. Junker: An intergenic explorer for bacterial genomes. Genomics Proteomics Bioinformatics. 2011; 9:179–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thorpe H.A., Bayliss S.C., Hurst L.D., Feil E.J.. Comparative analyses of selection operating on nontranslated intergenic regions of diverse bacterial species. Genetics. 2017; 206:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mayr C. Regulation by 3′-Untranslated regions. Annu. Rev. Genet. 2017; 51:171–194. [DOI] [PubMed] [Google Scholar]
- 66. Oren Y., Smith M.B., Johns N.I., Kaplan Zeevi M., Biran D., Ron E.Z., Corander J., Wang H.H., Alm E.J., Pupko T.. Transfer of noncoding DNA drives regulatory rewiring in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:16112–16117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Updegrove T.B., Shabalina S.A., Storz G.. How do base-pairing small RNAs evolve. FEMS Microbiol. Rev. 2015; 39:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mazumder B., Seshadri V., Fox P.L.. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 2003; 28:91–98. [DOI] [PubMed] [Google Scholar]
- 69. Mayr C., Bartel D.P.. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009; 138:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cheng C., Bhardwaj N., Gerstein M.. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics. 2009; 10:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mouali El Y., Gaviria-Cantin T., Sánchez-Romero M.A., Gibert M., Westermann A.J., Vogel J., Balsalobre C.. CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet. 2018; 14:e1007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bronesky D., Desgranges E., Corvaglia A., Francois P., Caballero C.J., Prado L., Toledo-Arana A., Lasa I., Moreau K., Vandenesch F. et al.. A multifaceted small RNA modulates gene expression upon glucose limitation in Staphylococcus aureus. EMBO J. 2019; 38:e99363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mouali El Y., Balsalobre C.. 3′ untranslated regions: regulation at the end of the road. Curr. Genet. 2019; 65:127–131. [DOI] [PubMed] [Google Scholar]
- 74. Bækkedal C., Haugen P.. The Spot 42 RNA: A regulatory small RNA with roles in the central metabolism. RNA Biol. 2015; 12:1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hardy K.J., Ussery D.W., Oppenheim B.A., Hawkey P.M.. Distribution and characterization of staphylococcal interspersed repeat units (SIRUs) and potential use for strain differentiation. Microbiology. 2004; 150:4045–4052. [DOI] [PubMed] [Google Scholar]
- 76. Geissmann T., Chevalier C., Cros M.-J., Boisset S., Fechter P., Noirot C., Schrenzel J., Francois P., Vandenesch F., Gaspin C. et al.. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009; 37:7239–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ziebuhr W., Krimmer V., Rachid S., Lößner I., Götz F., Hacker J.. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: Evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 1999; 32:345–356. [DOI] [PubMed] [Google Scholar]
- 78. Vandecraen J., Chandler M., Aertsen A., Van Houdt R.. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017; 43:709–730. [DOI] [PubMed] [Google Scholar]
- 79. Gor V., Takemura A.J., Nishitani M., Higashide M., Medrano Romero V., Ohniwa R.L., Morikawa K.. Finding of Agr phase variants in Staphylococcus aureus. mBio. 2019; 10:e00796-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. De Gregorio E., Abrescia C., Carlomagno M.S., Di Nocera P.P.. Ribonuclease III-mediated processing of specific Neisseria meningitidis mRNAs. Biochem. J. 2003; 374:799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lasa I., Toledo-Arana A., Gingeras T.R.. An effort to make sense of antisense transcription in bacteria. RNA Biol. 2012; 9:1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chao Y., Li L., Girodat D., Förstner K.U., Said N., Corcoran C., Śmiga M., Papenfort K., Reinhardt R., Wieden H.-J. et al.. In vivo cleavage map illuminates the central role of rnase E in coding and non-coding RNA pathways. Mol. Cell. 2017; 65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Altuvia Y., Bar A., Reiss N., Karavani E., Argaman L., Margalit H.. In vivo cleavage rules and target repertoire of RNase III in Escherichia coli. Nucleic Acids Res. 2018; 17:138–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Holmqvist E., Vogel J.X.R.. RNA-binding proteins in bacteria. Nat. Rev. Micro. 2018; 16:601–615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic maps of S. simiae CCM 7213T, S. capitis SK14 and S. epidermidis RP62A strains are available at http://rnamaps.unavarra.es/. The transcriptomic data has been also deposited in GEO under accession GSE143600. Other data or material that support the findings of this study can be made available by the corresponding author upon request.