Abstract
Exquisite control of the DnaA initiator is critical to ensure that bacteria initiate chromosome replication in a cell cycle-coordinated manner. In many bacteria, the DnaA-related and replisome-associated Hda/HdaA protein interacts with DnaA to trigger the Regulatory Inactivation of DnaA (RIDA) and prevent over-initiation events. In the Caulobacter crescentus Alphaproteobacterium, the RIDA process also targets DnaA for its rapid proteolysis by Lon. The impact of the RIDA process on adaptation of bacteria to changing environments remains unexplored. Here, we identify a novel and conserved DnaA-related protein, named HdaB, and show that homologs from three different Alphaproteobacteria can inhibit the RIDA process, leading to over-initiation and cell death when expressed in actively growing C. crescentus cells. We further show that HdaB interacts with HdaA in vivo, most likely titrating HdaA away from DnaA. Strikingly, we find that HdaB accumulates mainly during stationary phase and that it shortens the lag phase upon exit from stationary phase. Altogether, these findings suggest that expression of hdaB during stationary phase prepares cells to restart the replication of their chromosome as soon as conditions improve, a situation often met by free-living or facultative intracellular Alphaproteobacteria.
INTRODUCTION
All cells must be able to integrate environmental and internal cues to decide when to start the duplication of their genome. The key players, called ORC in eukaryotic and archaeal cells, and DnaA in bacteria, are conserved AAA+ (ATPases Associated with diverse cellular activities) proteins. In bacteria, DnaA typically binds to several so-called DnaA boxes located on a chromosomal origin and promotes the opening of the DNA double helix at a nearby AT-rich region, initiating the loading of the DNA polymerase onto the DNA (1,2). The correct timing of chromosome replication is directly influenced by the availability and the activity of DnaA, even in bacteria that also have additional mechanisms regulating this process (3–5). In most bacteria, DnaA must be bound to ATP to initiate replication and the ATPase activity of DnaA modulates its capacity to act as an initiator (1).
In Escherichia coli, the Regulatory Inactivation of DnaA (RIDA) process ensures the timely inactivation of DnaA during the replication cycle of the chromosome (1,2). This process involves a conserved protein homologous to the AAA+ domain III of DnaA, named Hda, which interacts with the β-sliding clamp through a conserved QLSLF motif located at its N-terminus (6–8). The most recent model suggests that two β-clamps associate with four ADP-associated Hda molecules to form inactive octameric complexes prior to replication initiation by DnaA-ATP (9). Then, once replication starts, the octameric complex dissociates when β-clamps are loaded onto the DNA, inducing the formation of active trimeric or tetrameric complexes that can interact with DnaA (9). Detailed analysis also demonstrated that the interaction between the replisome-associated Hda protein and DnaA requires both an interaction between Hda and the DnaA domain IV and an interaction between the AAA+ domains of each protein, leading to the conversion of DnaA-ATP into DnaA-ADP (10). Altogether, this process ensures that E. coli cells start the replication of their chromosome once and only once per cell cycle. Consistently, RIDA-deficient cells over-initiate DNA replication, which leads to an accumulation of DNA strand breaks inhibiting growth and colony forming ability under aerobic conditions (11). Remarkably, Hda is a very conserved protein, suggesting that the RIDA process is a common strategy used by many bacteria to regulate the timing of replication initiation, although this has been tested in an only limited number of species so far. Still, it was found that the HdaA homolog of Hda in Caulobacter crescentus plays a similar role (4,5). In this oligotrophic and aerobic environmental Alphaproteobacterium, the hdaA gene is essential (12) and HdaA was shown to interact with the β-clamp through a QFKLPL motif located at its N-terminus and to co-localize with the replisome throughout the whole S phase of the cell cycle (13). Notably, this motif, together with a conserved R-finger in its AAA+ domain, were both found to be critical for the essential activity of HdaA, suggesting that interactions between HdaA, the β-clamp and DnaA are also important steps during the RIDA process in C. crescentus (13). Moreover, characterization of the RIDA process in this bacterium uncovered a second layer in DnaA regulation that is not found in E. coli: once DnaA-ATP is converted into DnaA-ADP in an HdaA-dependent manner, it appears to be more efficiently degraded by the Lon ATP-dependent protease (14–16). Thus, the HdaA protein of C. crescentus plays a dual role in the RIDA process, by simultaneously provoking the inactivation and the degradation of DnaA, providing a remarkably robust control system (4). In addition to this RIDA mechanism, C. crescentus encodes a conserved response regulator named CtrA, which binds to the chromosomal origin to inhibit replication initiation in G1 phase cells and during late stages of the S phase (4,17,18).
When bacteria face adverse growth conditions, they must adjust their cell cycle accordingly to ensure survival. These adaptation mechanisms are still poorly understood, although most bacteria are believed to inhibit the replication of their chromosome under such conditions. In C. crescentus, translation of the dnaA transcript is strongly inhibited in response to nutrient limitations, leading to a rapid clearance of DnaA by Lon (19,20). Thus, C. crescentus cells that enter stationary phase are mostly arrested at the pre-divisional stage of their cycle (G2 phase) or as small new-born G1 cells (21,22). How such cells can then exit from stationary phase, re-start the replication of their genome and proliferate again when conditions get better remains unclear, although this must be a frequent need in natural environments. In this study, we uncovered the existence of a novel and conserved DnaA-related protein that is specifically expressed during stationary phase, most likely preparing cells to initiate DNA replication during exit from stationary phase.
MATERIALS AND METHODS
Plasmids and strains
Oligonucleotides and plasmids used in this study are described in Supplementary Table S1 and Table S2, respectively. Bacterial strains used in this study are described in Supplementary Table S3. Material and methods used to construct novel plasmids and strains are described in Supplementary Material and Methods.
Growth conditions and synchronization
E. coli strains were cultivated at 37°C in LB medium or on LB + 1.5% agar (LBA). C. crescentus was cultivated at 30°C in peptone yeast extract (PYE) complex medium or on PYE + 1.5% agar (PYEA). When required for selections or to maintain plasmids or genetic constructs, antibiotics were added at the following concentrations for solid/liquid media (μg/ml), respectively: tetracycline (Tet; PYE: 2/1, LB: 10/10), kanamycin (Km; PYE: 25/5, LB: 50/25), gentamycin (Gent; PYE: 5/1, LB: 10/10), spectinomycin (Spec; PYE: 100/25, LB: 50/50), streptomycin (Strep; PYE: 5/5, LB: 30/30) and nalidixic acid (Nal; PYEA: 20). When mentioned, glucose and/or xylose were added at a final concentration of 0.2% and/or 0.3%, respectively. When indicated, vanillate was added at a final concentration of 1 mM. Synchronized cultures of C. crescentus were obtained by centrifugation in a Percoll (Sigma, USA) density gradient followed by isolation of swarmer cells using a protocol adapted from (23). Swarmer cells were then released into PYE medium for cell cycle studies.
Immunoblot analysis
Proteins were resolved on 10% (for DnaA) or 12.5% (for HdaA, M2-HdaB, CcrM or GFP-HdaA) SDS-PAGE gels. Proteins were electro-transferred to PVDF membranes (Merck Millipore, MA, USA). Immuno-detection was performed with polyclonal antibodies. Anti-DnaA (24), anti-HdaA (12), anti-CcrM (25) (all three produced by immunizing rabbits at Josman LLC) and anti-rabbit conjugated to horse-radish peroxidase (HRP) (A0545 from Sigma, USA) sera were diluted 1:10 000. Anti-GFP (MA5–15256-HRP from Thermo Fischer Scientific, USA) and anti-FLAG (F3165 from Sigma, USA) sera were diluted 1:10 000. Anti-mouse conjugated to HRP (W4021 from Promega, USA) was diluted 1:5000. Immobilon Western Blotting Chemo-luminescence HRP substrate (Merck Millipore, MA, USA) was used and membranes were scanned with a Fusion FX (Vilber Lourmat, FR) instrument to detect the chemiluminescent signal. Relative band intensities on images were quantified using the ImageJ software.
β-Galactosidase assays
β-Galactosidase assays were done using a standard procedure as described previously (26).
Microscopy and image analysis
Cells were immobilized on a slide using a thin layer of 1% agarose dissolved in dH2O and imaged immediately. Phase contrast (Ph3) and fluorescence images were taken using the microscope system described in (27). The MicrobeJ software (28) was then used: (i) to estimate the proportion of dead cells on fluorescence images of LIVE/DEAD-stained cells using default parameters of the Bacteria mode of detection option; (ii) to measure the length of cells on phase-contrast images using the Medial-Axis mode of detection option; (iii) to detect and count fluorescent foci in cells on fluorescence images using the Foci-mode of detection option (foci were counted if their signal intensity was minimum two fold above the signal of the cell cytoplasm).
qPCR analysis to evaluate Cori/ter ratios
Genomic DNA was extracted from C. crescentus cells using the PureGene genomic DNA extraction kit (QIAGEN, GER) and stored in dH2O at 4°C. Quantification and purity of DNA samples were analyzed using a NanoDrop UV–Vis Spectrophotometer (ThermoFisher, USA): 260/280 nm ratios were ≥1.8 and 260/230 nm ratios were ≥2.0. A qPCR calibration curve was performed using different genomic DNA dilutions (10, 1, 0.1, 0.01, 0.001 and 0.0001 ng). qPCR reactions were then performed in a final volume of 10 μl containing: 1 μl of genomic DNA at 0.1 ng/μl, 5 μl of 2× GoTaq q-PCR master mix (Promega #A6001, USA) and 0.6 μM of AF77/78 or AF79/80 primer pairs (HPLC purified from Microsynth, CH) targeting a region close to the Cori or to the terminus (ter) of the chromosome of C. crescentus, respectively. The lengths of the Cori and ter amplification products were 166 bp (C. crescentus genome NC_011916.1, nt:1410:1576), and 174 bp (C. crescentus genome NC_011916.1, nt:1971139:1971313), respectively. qPCR reactions were run using a Rotor Gene-Q instrument (QIAGEN, GER), using the following cycling conditions: 2 min at 95°C (Hot start DNA polymerase activation), 15 s at 95°C (denaturation), 1 min at 60°C (annealing and extension); Denaturation, annealing and extension steps were repeated 40 times. To quantify the relative abundance of Cori and ter chromosomal regions in each sample, a calibrator normalized relative analysis was performed.
Co-immunoprecipitation assays
C. crescentus strains used for co-immunoprecipitation experiments were first cultivated over-night with shaking in 20 ml of PYE at 30°C. Once cultures reached stationary phase the next day, they were diluted into 200 ml of PYE and cultivated with shaking for 90 min at 30°C, until cells reached exponential phase again. Xylose and/or vanillate were then added into the medium and cells were incubated with shaking for four more hours at 30°C. Exponentially growing cells were then harvested by centrifugation at 6000 × g for 20 min at 4°C, washed with 25 ml of PBS (Phosphate Buffered Saline pH 7.4) and centrifuged again at 6000 × g for 10 min at 4°C. Cells were then resuspended in 10 ml of PBS supplemented with 2 mM DTSP (3,3′-dithiodipropionic acid di(N-hydroxysuccinimide ester) dissolved in DMSO, which is the crosslinking agent D3669 from Sigma, USA) and incubated for 30 min at 30°C with shaking. To stop the crosslinking reaction, 1.5 ml of 1 M Tris–HCl (150 mM final) was added and incubation at 30°C with shaking was continued for 30 more min. Cells were then centrifuged at 6000 × g for 10 min at 4°C, washed with 30 ml PBS and centrifuged at 6000 × g for 10 min at 4°C. The resulting pellet was carefully resuspended in 20 ml CoIP buffer (20 mM HEPES, 150 mM NaCl, 20% glycerol, 80 mM EDTA pH 8 and complete EDTA-free protease inhibitor cocktail according to instructions (Roche, CH)), centrifuged at 6000 × g for 20 min at 4°C (note that the pellet was sometimes frozen over-night at that step) and resuspended in 2 ml of NP40 lysis buffer (1 × CellLytic B (Sigma C8740, USA), 50 U DNAse I (Fermentas, USA), 75 000 U ReadyLyse (Epicentre, USA), 10 mM MgCl2, 2% NP40 (Merck Millipore, USA), 1× protease inhibitor cocktail) and incubated at 25°C for 30 min with end-over-end rotation. Cells were then subjected to sonication (2-minute pulse at medium power using a Telsonic Ultrasonic) and centrifuged at 13 362 × g for 15 min at 4°C. The resulting cell extract was used to determine the total protein concentration using a Bradford assay (Biorad, USA). A small aliquot (150 μl) of the cell extract was collected at that step (‘IN’ samples). For the co-immunoprecipitation, 10 mg of total proteins from the cell extract were mixed with 20 μl of Magnetic anti-FLAG beads (M8823 from Sigma, USA) and incubated over-night with end-over-end rotation at 4°C. Beads were then washed six times with 1 ml of CoIP buffer + 1% Triton X100. Lastly, beads were resuspended in 50 μl of 1× Laemmli Sample Buffer (Biorad) and incubated at 70°C for 10 min (‘IP’ samples). Samples (IN/IP) were heated at 95°C for 10 min before loading 25 μl on SDS-PAGE gels for immunoblotting.
Genetic screen to isolate suppressors of the toxicity associated with hdaB expression in C. crescentus
pHPV414 is a himar1 transposon (Tn::km) delivery plasmid (29) and it was introduced by conjugation into NA1000 cells carrying the pBX-HdaBAe plasmid and cultivated into PYE + glucose medium. More than 8000 random mutants were then picked and transferred in parallel onto PYEA plates containing either glucose or xylose. Only two transposon mutants were found to grow on both media; these were named JC 2077 and JC2078. For each of these two mutants, the pBX-HdaBAe plasmid was extracted and introduced back into NA1000 (WT) cells, to confirm that presence of these plasmids still led to toxicity in WT cells, thereby indicating that chromosomal mutations in JC2077 and JC2078 suppress the toxicity associated with hdaBAe expression from pBX-HdaBAe. Transposon mutations from strains JC2077 and JC2078 were transduced into NA1000 cells using ΦCr30 and kanamycin selection, giving JC2080 and JC2079, respectively. Strains JC2080 and JC2079 were then used to map the transposon insertion sites in the chromosome of these mutants of interest. Their chromosomal DNA was extracted and digested with SacII. DNA fragments were then self-ligated before transformation of E. coli S17–1 λpir with the ligation mix. The transposon insertion sites were then identified by sequencing using primer himup2 (29) and comparison with the NA1000 genome available on NCBI (NC_011916): strain JC2080 carried a dnaK::Tn insertion at position 9955 of this genome, while strain JC2079 carried a recQ::Tn insertion at position 3735008 of this genome. To further verify that dnaK::Tn or recQ::Tn insertions were sufficient to suppress the toxicity associated with the expression of different HdaB proteins, pBX-HdaB plasmids were introduced back into JC2079 and JC2080 prior to phenotype analysis.
RESULTS
HdaB is a conserved DnaA-related protein
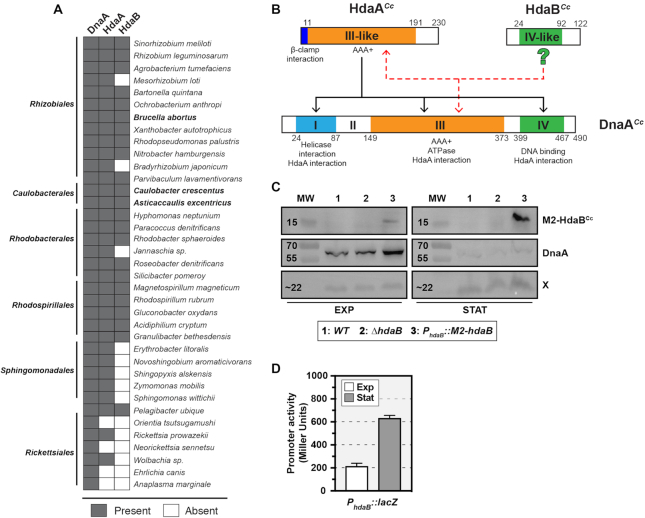
We noticed that the majority of Alphaproteobacteria that possess a HdaA protein also encode a second protein that shows similarities with DnaA (Figure 1A) and thus logically named such proteins HdaB (homologous to DnaA B). HdaB proteins are short proteins resembling the domain IV of DnaA proteins. One example is the uncharacterized CCNA_01111 protein of the C. crescentus NA1000 strain (HdaBCc), which displays 32% of identity with DnaACc domain IV (Figure 1B and Supplementary Figure S1A). Considering that the DnaA domain IV is involved in DNA binding and in interactions with Hda/HdaA during the RIDA process (2,10), we suspected that HdaB proteins might be involved in the control of chromosome replication in certain Alphaproteobacteria. Indeed, we now provide evidences that HdaB proteins from minimum three different Alphaproteobacteria can interfere with the RIDA process and affect replication frequency in the C. crescentus model bacterium.
Figure 1.
HdaB is a conserved DnaA-related protein that accumulates during stationary phase. (A) Distribution of DnaA, HdaA and HdaB proteins in Alphaproteobacteria. The presence or the absence of a homolog of each protein was estimated using bi-directional best-blast-hit (BBH) using the C. crescentus genome as reference. HdaB from species in bold have been used in this study. (B) Similarities between DnaACc domains and HdaACc or HdaBCc. The predicted (from (2)) or demonstrated (from (12,13,18,30)) function of each protein motif or domain is indicated under its position. Black arrows indicate interactions demonstrated between HdaEc and DnaAEc in E. coli (2). Red dashed arrows indicate putative interactions between HdaBCc and HdaACc or DnaACc. Numbers indicate amino-acid positions on each protein. (C) Immunoblots showing the intracellular levels of M2-HdaBCc and DnaACc in C. crescentus cells cultivated in exponential (EXP) or stationary (STAT) phase. Strains NA1000 (WT), JC1456 (ΔhdaB) and JC2106 (PhdaB::M2-hdaB) were cultivated in PYE medium and samples were collected at an OD660nm of ∼0.3 (EXP) and after an over-night culture (STAT). Proteins were analyzed by SDS-PAGE and detected by immunoblotting using anti-M2 (for M2-HdaB) or anti-DnaA (for DnaA and the unknown X protein that serves as a loading control here) antibodies. MW stands for the approximate molecular weight in kD. (D) Activity of the hdaBCc promoter in cells cultivated in exponential or stationary phase. The placZ290-PhdaB plasmid was introduced into the NA1000 (WT) strain. Cells were then cultivated in PYE medium and β-galactosidase assays were performed when cultures reached an OD660 nm of ∼0.3 (EXP) and after an over-night incubation (STAT). Error bars correspond to standard deviations from minimum three experiments.
HdaB accumulates in stationary phase C. crescentus cells
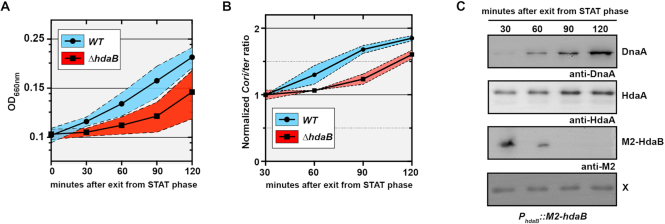
To get a first indication on the potential role of HdaB proteins during the life cycles of Alphaproteobacteria, we compared the expression of HdaBCc in C. crescentus cells cultivated in different growth conditions. We used cells expressing HdaBCc with a M2 (FLAG) epitope at its N-terminus (M2-HdaBCc) from the native hdaB promoter to facilitate detection by immunoblotting. Surprisingly, we found that M2-HdaBCc was hardly detectable in exponentially-growing cells, while it accumulated at much higher levels in stationary-phase cells (Figure 1C). This observation suggested that the synthesis of HdaBCc may be stimulated upon entry into stationary phase. To test this possibility more directly, we constructed a plasmid carrying a reporter where lacZ transcription is under the control of the hdaBCc promoter region and introduced this plasmid into C. crescentus cells. Cells were then cultivated from exponential to stationary phase and β-galactosidase assays were performed to compare the activity of the hdaBCc promoter. Consistent with our previous findings (Figure 1C), we found that the hdaBCc promoter was ∼3-fold more active in stationary phase cells than in exponentially growing cells (Figure 1D). Altogether, these results suggested that HdaBCc could play a role during stationary phase or during exit from stationary phase, rather than during exponential growth when HdaB levels remain extremely low. Consistent with this proposal, we found that an in-frame deletion of the hdaBCcgene did not affect the generation time of C. crescentus cells during exponential growth: 96.8 ± 6.6 min for ΔhdaB cells, compared to 95.2 ± 10.7 min for wild-type cells in complex PYE medium (Supplementary Figure S1B). Instead, deletion of the hdaB gene significantly expanded the duration of the lag phase during exit from stationary phase (Figure 2A). Interestingly, this growth delay appeared to coincide with a delay in replication restart since the origin/terminus (Cori/ter) ratio estimated by quantitative real-time PCR (qPCR) increased earlier in wild-type than in ΔhdaB populations of cells (Figure 2B).
Figure 2.
HdaB promotes replication initiation when cells exit stationary phase. (A) Growth of a ΔhdaB mutant upon exit from stationary phase. C. crescentus strains NA1000 (WT) and JC1456 (ΔhdaB) were cultivated into PYE medium until stationary phase (over-night) and cultures were diluted back into fresh PYE medium to reach an OD660nm of 0.1 at time 0. Blue or red shaded areas represent standard deviations from 4 independent cultures of each strain. (B) Cori/ter ratio in ΔhdaB cells during exit from stationary phase. Caulobacter crescentus strains NA1000 (WT) and JC1456 (ΔhdaB) were cultivated into PYE medium until stationary phase (over-night) and cultures were diluted back into fresh PYE medium to reach an OD660nm of 0.1 at time 0. gDNA was extracted at the indicated time points and Cori/ter ratio were estimated by qPCR. Results were then normalized so that the Cori/ter ratio of WT cells at time 30 min equals 1. Blue or red shaded areas represent standard deviations from two independent cultures of each strain. (C) Immunoblots showing intracellular levels of DnaA, HdaA and M2-HdaB as JC2106 (PhdaB::M2-hdaB) cells exit stationary phase. Cells were cultivated into PYE medium until stationary phase (overnight) and cultures were diluted back into fresh PYE medium to reach an OD660nm of 0.1 at time 0. Samples were collected every 30 min for a total of 120 min and proteins were analyzed by SDS-PAGE. DnaA, HdaA and M2-HdaB were detected by immunoblotting using anti-DnaA (for DnaA and the unknown X protein that serves as a loading control here), anti-HdaA or anti-M2 antibodies, respectively.
Ectopic expression of HdaB proteins inhibits cell division and leads to cell death
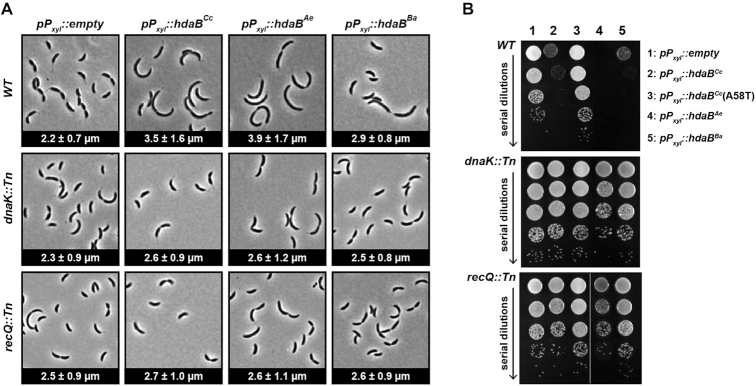
Considering that hdaBCc is hardly expressed in fast-growing cells, we anticipated that looking at the impact of hdaB expression in fast-growing cells may turn out more informative than characterizing hdaB deletion strains. Thus, we cloned hdaB genes from three different Alphaproteobacteria under the control of the xylose-inducible xylX promoter (Pxyl) of C. crescentus on a medium copy number plasmid and introduced these plasmids into wild-type C. crescentus cells. Strikingly, in all three cases, actively growing cells expressing hdaB homologs progressively became elongated with frequent failure during the division process compared to control cells (Figure 3A and Supplementary Figure S2A and B), showing that expression of hdaBCc, hdaBAe (from Asticaccaulis excentricus) or hdaBBa (from Brucella abortus) interferes directly or indirectly with cell division. Moreover, cell elongation upon hdaB expression rapidly lead to cell death. In liquid medium, up to ∼ 50% of the cells died after just 4 h of induction (Supplementary Figure S2C). On solid medium containing the xylose inducer, very few cells expressing hdaBCc or hdaBBa could form visible colonies, while nearly no cell expressing hdaBAe, except potential suppressors, could form visible colonies (Figure 3B), demonstrating the strong impact that an ectopic expression of hdaB homologs can have on C. crescentus proliferation. Importantly, none of these phenotypes were observed when a mutant HdaBCc(A58T) protein was expressed (from a plasmid randomly isolated during the cloning procedure to construct pBX-HdaBCc) in identical conditions (Figure 3B and Supplementary Figure S2B–D), showing that they were not simply connected to a proteotoxic stress generated by protein over-production.
Figure 3.
hdaB expression interferes with cell division and leads to cell death in C. crescentus. (A) Cellular morphology of C. crescentus cells expressing hdaBCc from C. crescentus, hdaBAe from A. excentricus or hdaBBa from B. abortus. Medium copy number vectors (pBXMCS-4 derivatives) expressing or not hdaB genes from a Pxyl promoter were introduced into NA1000 (WT), JC2080 (dnaK::Tn) or JC2079 (recQ::Tn) cells. The resulting strains were cultivated into PYEG medium until mid-exponential phase and xylose was added for 4 h before microscopy images were acquired. The numbers indicated at the bottom of each image correspond to the median medial axis length of cells in μm with standard deviations from three independent experiments. (B) Growth of colonies of C. crescentus cells expressing hdaBCc, hdaBCc(A58T), hdaBAe or hdaBBa. The strains described in panel (A) were cultivated in PYE medium until mid-exponential phase and cultures were serially diluted (10-fold each time) before plating onto PYEA medium containing xylose to induce expression of hdaB genes. Note that data obtained using dnaK::Tn and recQ::Tn cells is only described in the last section of the Results.
Expression of HdaB proteins leads to over-initiation of chromosome replication in growing cells
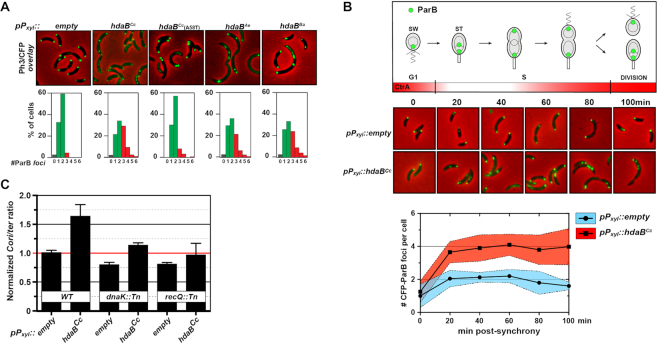
Since HdaB shows similarities with the domain IV of DnaA (Figure 1B and Supplementary Figure S1A) and since its ectopic expression leads to cell division defects and cell death (Figure 3 and Supplementary Figure S2), like cells displaying DNA replication defects (12,18,24,30), we hypothesized that HdaB may play a direct role in replication control. To test this possibility, we first introduced the previously described plasmids expressing hdaB homologs from the Pxyl promoter into a C. crescentus strain where the parB gene was exchanged with a cfp-parB construct, expressing a functional CFP-tagged ParB protein (31). ParB proteins bind to parS sequences, which are located next to the Cori. Thus, CFP-ParB can be used as a proxy to visualize Cori in single cells by fluorescence microscopy: in C. crescentus, it forms a single fluorescent focus at the flagellated cell pole before the onset of DNA replication and then forms two fluorescent foci located at each cell pole right after the onset of DNA replication (31) (Figure 4, cell cycle schematic). When cells were cultivated with the xylose inducer for 4 h, we observed that cells expressing each of the three hdaB homologs often accumulated more than two foci per cell (up to six foci per cell), which was only rarely observed with cells carrying the empty control vector or expressing the inactive HdaBCc(A58T) protein (Figure 4A). This first observation showed that the initiation of DNA replication can still take place in actively growing C. crescentus cells expressing hdaB homologs, but that the control of its frequency seems disturbed, leading to over-initiation. To bring more light on this observation, we also isolated G1 swarmer cells from the cfp-parB strains carrying the empty vector or the hdaBCc-expressing plasmid and followed their differentiation into S phase stalked cells and their division during time-course fluorescence microscopy experiments. Strikingly, as early as 20 min after the addition of the xylose inducer, when cells differentiated into stalked cells, we already observed many cells with more than two CFP-ParB foci when hdaBCc was expressed (Figure 4B). While control cells typically displayed only two CFP-ParB foci during the S phase of the cell cycle, cells expressing hdaBCc displayed an average of approximately four distinct CFP-ParB foci, showing that such cells can restart DNA replication multiple times during the same cell cycle, at least during the time period when CtrA is inactive. To further confirm that hdaBCc expression leads to over-initiation, we also measured the Cori/ter ratio by qPCR using un-synchronized populations of cells and found that it increased by a factor of ∼1.6 in wild-type cells expressing hdaBCc for 4 h compared to control cells (Figure 4C).
Figure 4.
hdaB expression leads to fast over-replication in C. crescentus. (A) Microscopy detection of ParB-CFP foci in C. crescentus cells expressing hdaB genes. Vectors expressing or not hdaBCc, hdaBCc(A58T), hdaBAe or hdaBBafrom the Pxyl promoter were introduced into the MT190 strain (PparB::cfp-parB). The resulting strains were cultivated into PYEG medium to mid-exponential phase and xylose was then added for four more hours prior to fluorescence microscopy. Representative images corresponding to phase contrast and fluorescence (colored in green on images) overlays are shown. The number of CFP-ParB foci per cell was estimated using ∼200 cells on multiple images acquired using each strain. Results are shown as a percentage of cells displaying each number of CFP-ParB foci in each population and are averages of minimum three independent experiments. (B) Time-course microscopy images to detect CFP-ParB foci in isolated swarmer cells expressing hdaBCc. MT190 strains carrying the empty vector or the vector expressing hdaBCc from Pxyl were cultivated until mid-exponential phase in PYEG medium before synchronization. Isolated swarmer cells were released into fresh PYE medium containing xylose and Ph3/CFP images (overlays are shown) were acquired at the indicated time points. The number of CFP-ParB foci per cell was estimated using ∼200 cells acquired using each strain at each time point (lower panel). The blue or red-shaded areas correspond to standard deviations from two independent cultures of each strain post-synchrony. The schematic above the images shows where ParB is expected to localize in wild-type cells as a function of the cell cycle (31). It also indicates when the CtrA repressor of chromosomal replication is accumulating in cells (50). (C) Comparison of the Cori/ter ratio in populations of NA1000 (WT), JC2080 (dnaK::Tn) or JC2079 (recQ::Tn) cells expressing or not hdaBCc. Cori/ter ratios were measured by qPCR using genomic DNA extracted from cells cultivated to mid-exponential phase in PYEG medium and in the presence of the xylose inducer for 4 h. Results were normalized so that the Cori/ter ratio of WT cells carrying the empty vector equals 1 (highlighted with a red line) to facilitate comparisons. Note that data obtained using dnaK::Tn and recQ::Tn cells is only described in the last section of the Results.
Altogether, these results demonstrated that HdaB homologs from three different Alphaproteobacteria can disturb the frequency of the initiation of chromosome replication in C. crescentus, which is normally controlled by the RIDA process.
HdaB interacts with HdaA to inhibit the RIDA process
In C. crescentus, the RIDA process controls DnaA activity and DnaA levels, by stimulating the ATPase activity of DnaA and its subsequent degradation by ATP-dependent proteases (4,12,14,18,30). It is mediated by the conserved HdaA protein (12,13). In E. coli, the RIDA process needs both an interaction between the DnaAEc domain IV and Hda, and an interaction between the AAA+ domain of the two proteins (2,10). Supporting this model, it was shown that the Leu-422 residue of the DnaAEc domain IV is required for RIDA-dependent ATP hydrolysis (10). Considering that HdaB proteins show similarities with the DnaA domain IV (Figure 1B and Supplementary Figure S1A), we hypothesized that HdaB could interact with HdaA to block the RIDA process in Alphaproteobacteria, leading to higher intracellular levels of active DnaA and subsequent over-initiation events leading to cell death. Consistent with this proposal, we observed that the Leu-422 residue of the DnaAEc domain IV, which is critical for the DnaAEc/Hda interaction (10), is present in HdaBCc (Supplementary Figure S1A), while the Arg-399 residue, which is necessary for DnaAEc binding to DnaA boxes (32), is not.
As a first step to test this hypothesis, we probed whether HdaA and HdaB can interact with one another in C. crescentus cells and whether this interaction is required for the toxic function of HdaB. To do so, we first engineered two constructs expressing either M2-HdaBCc or M2-HdaBCc(A58T) from the Pxyl promoter into a medium copy number plasmid and introduced these constructs into wild-type C. crescentus cells. As expected, we found that the ectopic expression of M2-HdaBCc in the presence of xylose for 4 h had as much impact on cell viability and cell morphology (Supplementary Figure S2C and D) as that of HdaBCc (Figure 3A and Supplementary Figure S2C), showing that the addition of a M2 tag at the N-terminus of HdaBCc does not interfere with the toxic effect of HdaB. We then used the two strains to perform anti-M2 immunoprecipitation experiments followed by anti-HdaA immunoblots to get a first insight on the potential interaction between native HdaA and M2-tagged HdaB proteins. We found that the functional M2-HdaBCc protein co-immunoprecipitated with HdaA, while the non-functional M2-HdaBCc(A58T) protein did not (Supplementary Figure S3). Importantly, HdaA was not immunoprecipitated when we used extracts of cells that expressed an un-related M2-CdnL protein (to even greater levels then M2-HdaBCc) or simply in the absence of M2-tagged proteins (Supplementary Figure S3), indicating that M2-HdaBCc forms a specific complex with HdaA when it is expressed in C. crescentus cells. Since control cells lacking hdaA could not be used in such experiments as hdaA is an essential gene (12), we also engineered two more constructs expressing either the functional M2-HdaBCc or the non-functional M2-HdaBCc(A58T) proteins from the vanillate-inducible Pvan promoter into a medium copy number plasmid. In the presence of these plasmids, cells exposed to the vanillate inducer for 4 h accumulated M2-tagged HdaBCc proteins at levels similar to cells expressing these proteins from the Pxyl promoter instead (Figure 5, IN), leading to similar phenotypes (data not shown). These two constructs or the empty control vector were then introduced into a C. crescentus strain expressing a functional GFP-tagged HdaA protein from the native chromosomal Pxyl promoter (12,13). These strains were cultivated in the presence of xylose and vanillate inducers for 4 h and cell extracts were used to perform anti-M2-immunoprecipitation experiments followed by anti-GFP and anti-HdaA immunoblots. We found that the functional M2-HdaBCc protein co-immunoprecipitated with the HdaA protein and with the functional GFP-HdaA protein, while the non-functional M2-HdaBCc(A58T) protein did not (Figure 5, IP). Importantly, neither HdaA, nor GFP-HdaA, were immunoprecipitated in the absence of M2-tagged proteins in cell extracts (cells with the empty control vector). Worth noticing, we also found that none of the M2-tagged HdaBCc proteins co-immunoprecipitated with DnaA (Figure 5, IP), suggesting that HdaBCc does not target DnaA directly. Altogether, these results provide a strong indication that HdaB must interact with HdaA to exert its toxic function.
Figure 5.
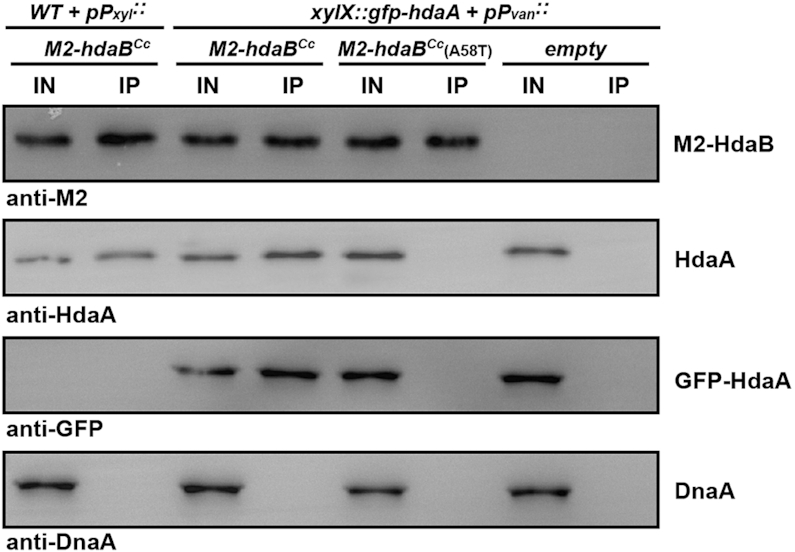
HdaB interacts with HdaA but not DnaA in C. crescentus. Probing the in vivo interaction(s) between M2-HdaBCc/M2-HdaBCc(A58T) and HdaACc/GFP-HdaACc/DnaACc by co-immunoprecipitation experiments. NA1000 (WT) cells carrying the pBX-M2-HdaBCc vector were cultivated to mid-exponential phase in PYEG medium and xylose was then added into the medium for 4 h prior to preparing cell extracts. JC208 (PxylX::gfp-hdaA at the xylX locus) cells carrying pBVMCS-4, pBV-M2-HdaBCc or pBV-M2-HdaBCc(A58T) were cultivated to mid-exponential phase in PYE medium. Xylose and vanillate were then added into the medium for 4 h prior to preparing cell extracts. M2-tagged HdaBCc proteins were immunoprecipitated from cell extracts using anti-M2 antibodies (IP) and loaded into the same SDS-PAGE gel as the cell extracts (IN) prior to immunoblotting using anti-M2, anti-HdaA, anti-GFP or anti-DnaA antibodies. A representative image of each immunoblot is shown.
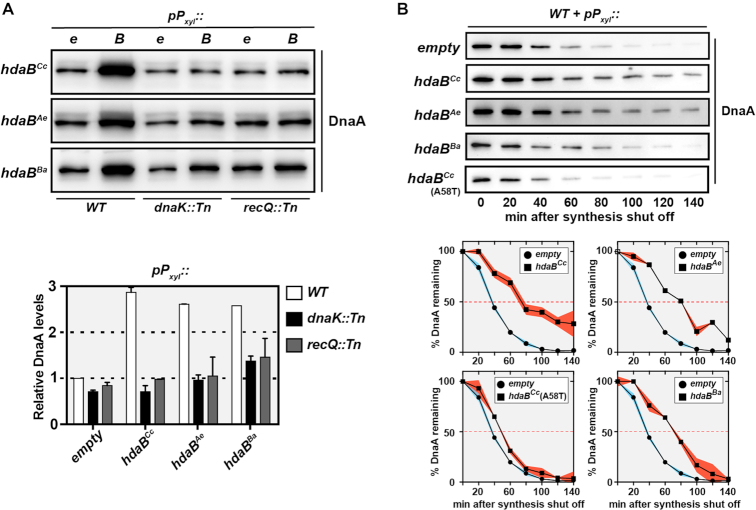
As a second step to test this hypothesis, we probed whether the ectopic expression of HdaB proteins leads to higher DnaA accumulation in C. crescentus, as expected if they target HdaA and inhibit the RIDA process. We performed immunoblot experiments using extracts of wild-type C. crescentus cells expressing any of the three selected hdaB homologs of Alphaproteobacteria from a Pxyl promoter on a plasmid and found that DnaA accumulated at higher intracellular levels in all three cases, compared to control cells carrying the empty vector (Figure 6A). Four hours after the addition of xylose into the medium, cells expressing hdaB homologs accumulated at least 2.5-fold more DnaA than control cells (Figure 6A). Since the RIDA process inhibits DnaA accumulation by promoting its degradation, we also compared the stability of DnaA in hdaB-expressing cells and in control cells by immunoblotting using samples collected at different times after an addition of chloramphenicol to stop DnaA synthesis (Figure 6B). We found that the half-life of DnaA roughly doubled (from ∼40 min to ∼80 min) when wild-type HdaB proteins were expressed from Pxyl for 4 h, compared to control cells. Importantly, a similar expression of the inactive HdaB(A58T)Cc protein had essentially no impact on DnaA stability (Figure 6B).
Figure 6.
HdaB proteins stabilize DnaA in C. crescentus. (A) Relative levels of DnaA in hdaB-expressing cells. NA1000 (WT), JC2080 (dnaK::Tn) or JC2079 (recQ::Tn) cells carrying the empty (e) control vector (pBXMCS-4) or derivatives (B) expressing hdaBCc, hdaBAe or hdaBBa from the Pxyl promoter were cultivated to mid-exponential phase in PYEG medium and xylose was added for the last 4 h. Cultures were normalized to the same OD660nm and cell extracts were then prepared for immunoblotting using anti-DnaA antibodies (and anti-FtsZ antibodies as a loading control shown in Supplementary Figure S4). Images of representative blots are shown in the upper panel. Relative quantifications of DnaA levels from three independent experiments are shown in the lower panel. Error bars correspond to standard deviations. Note that data obtained using dnaK::Tn and recQ::Tn cells is only described in the last section of the Results. (B) DnaA stability upon expression of hdaB homologs. NA1000 (WT) cells carrying the empty control vector (pBXMCS-4) or derivatives expressing hdaBCc, hdaBAe, hdaBBa or hdaBCc(A58T) from the Pxyl promoter were cultivated to mid-exponential phase in PYEG medium and xylose was added for 4 h before chloramphenicol (2 μg/ml) was added to shut-off protein synthesis. Cultures were normalized to the same OD660 nm and cell extracts were then prepared at the indicated time points for immunoblotting using anti-DnaA antibodies. Images of representative blots are shown in the upper panel. Relative quantifications of DnaA levels at each time point are shown in the lower panel: DnaA levels were set to an arbitrary value of 100 at the time when chloramphenicol was added for each culture to better compare the speed at which DnaA is degraded in cells carrying different vectors. Red- or blue-shaded areas correspond to standard deviations between three independent experiments.
Collectively, this data supports a model where HdaB interacts with HdaA to block the RIDA process and stimulate the initiation of DNA replication by DnaA.
HdaB and HdaA are simultaneously present when DnaA levels are low in cells exiting stationary phase
Since ΔhdaB cells appear to restart the replication of their chromosome with a slight delay compared to wild-type cells during the period when they exit stationary phase (Figure 2B), we looked more carefully at DnaA, HdaA and M2-HdaB (expressed from its native promoter) intracellular levels during this period (for up to 120 min, corresponding to more than one generation time). Immunoblot experiments (Figure 2C) showed that: (i) HdaA is detectable during the whole exit period; (ii) DnaA levels are very low right after cells exit stationary phase, but gradually increase to higher levels toward the end of this exit period; (iii) M2-HdaB is detectable for 60 min and is then rapidly cleared from the cells after replication re-initiates. These observations indicate that HdaB could inhibit HdaA to stimulate DnaA activity at the time when DnaA levels are still extremely low, potentially helping cells to re-start chromosome replication when they exit stationary phase. This interaction may also transiently increase DnaA stability to promote the re-accumulation of DnaA during this transition period, since DnaA levels appear slightly higher in wild-type compared to ΔhdaB cells between 60 and 90 min after the exit from stationary phase (Supplementary Figure S1C). Overall, these findings provide interesting information on when HdaB might be useful to inhibit the RIDA process during the life of C. crescentus cells.
Mutations in dnaK and recQ can bypass the toxicity of HdaB
As described earlier, most cells expressing HdaB proteins rapidly loose viability when actively growing in liquid or on solid media (Figure 3 and Supplementary Figure S2). If this toxicity is due to the over-initiation of DNA replication (Figure 4), as anticipated if the target of HdaB is HdaA (Figure 5), one would expect that suppressors of this toxicity should carry mutations that reduce the levels of active DnaA and/or that reduce initiation frequency and/or that restore viability despite excessive chromosome replication (33). To test if this was indeed the case, we isolated random transposon mutants that could bypass the toxicity associated with the expression of HdaBAe from Pxyl on a medium copy number vector in C. crescentus (Figure 3B). Out of a total of ∼8000 mutants that were screened (Supplementary Figure S5), two extragenic suppressors acquired the capacity to grow and form colonies despite the presence of the xylose inducer (Figure 3 and Supplementary Figure S2).
The first suppressor mutant carried a transposon insertion in the dnaK gene (Supplementary Figure S6). Strikingly, dnaK mutations had already been randomly isolated previously and shown to bypass the lethality associated with the over-production of DnaA in C. crescentus (15). DnaK is an essential chaperone activated by its non-essential co-chaperone DnaJ (34,35). DnaKJ inhibits the transcription of lon, itself encoding the main protease that degrades DnaA during exponential phase (Supplementary Figure S7A) (15). In the mutant we isolated, the transposon was inserted in the disruptable 3′ region of the dnaK open reading frame (ORF) (Supplementary Figure S6) located upstream of the co-transcribed dnaJ ORF (34,36). Thus, the transposon most likely decreases DnaK activity and shuts off dnaJ transcription through a polar effect. Indeed, the use of a transcriptional reporter to compare the activity of the lon promoter showed that it was ∼2-fold more active in the dnaK::Tn than in the wild-type strain (Supplementary Figure S7B). Consistent with this finding, immunoblotting experiments then revealed that the dnaK::Tn mutation prevented the excessive accumulation of DnaA in C. crescentus cells expressing HdaBCc, HdaBAe or HdaBBa (Figure 6A), most likely by stimulating DnaA degradation by Lon. Finally, qPCR experiments to measure the Cori/ter ratio (Figure 4C) and microscopy experiments (Figure 3 and Supplementary Figure S2) revealed that the isolated dnaK::Tn mutation largely attenuated the over-initiation and the cell elongation phenotypes associated with the ectopic expression of HdaB proteins in C. crescentus. Altogether, these results are fully consistent with the proposal that actively growing C. crescentus cells die as a consequence of elevated DnaA levels when HdaB proteins are expressed.
The second suppressor mutant carried a transposon insertion in the recQ gene (Supplementary Figure S6) encoding a DNA helicase involved in DNA repair (37). Interestingly, RIDA deficiencies in E. coli were shown to provoke cell death through the accumulation of DNA strand breaks connected with the accumulation of reactive oxygen species (ROS) (11,33,38) and RecQ has been shown to be necessary for such ROS-dependent death as also seen during the so-called ‘thymineless death’ (TLD) process (39,40). Then, we tested whether C. crescentus cells over-expressing HdaBCc accumulated more double-strand breaks (DSB) than control cells using a RecA-CFP reporter expressed from the endogenous Pvan promoter and fluorescence microscopy. As expected if HdaBCc inhibits the RIDA process, we found that cells over-expressing HdaBCc displayed RecA-CFP foci far more frequently than control cells (∼36% of the cells instead of ∼2% in Supplementary Figure S8), indicating that potentially lethal DSBs accumulate. More surprisingly, we found that the recQ::Tn mutation when HdaBCc is expressed in exponential phase lead to an inhibition of DnaA accumulation (Figure 6A) and to a reduction of the over-initiation (Figure 4C) and lethal (Figure 3 and Supplementary Figure S2C) phenotypes, in a manner very similar to the dnaK::Tn mutation. This observation suggested that RecQ and DnaKJ may promote DnaA accumulation through the same Lon-dependent pathway. In agreement with this proposal, lon transcription was as much induced in recQ::Tn (Supplementary Figure S7B) or ΔrecQ (data not shown) mutants than in the dnaK::Tn mutant. Noteworthy, we also found that RecQ promotes the accumulation of another well-known substrate of the Lon protease: the CcrM DNA methyltransferase (Supplementary Figure S7C). Thus, this lucky finding also uncovered the existence of a novel RecQ-dependent pathway controlling Lon-dependent degradation in C. crescentus.
DISCUSSION
Lag phase is the most poorly understood phase of the bacterial growth cycle and how bacteria can leave this phase to start proliferating remains obscure. In this study, we identified a novel and conserved small protein that shows similarities with DnaA domain IV (Figure 1B). We found that HdaBCc interacts with HdaACc (Figure 5 and Supplementary Figure S3), most likely titrating HdaACc away from DnaACc and leading to a transient inhibition of the RIDA process when HdaBCc is present (Figure 7). Thus, the ectopic expression of hdaB genes in exponentially growing cells promotes DnaA activity (Figure 4) and represses DnaA degradation by Lon (Figure 6), leading to over-initiation events and cell death in C. crescentus (Figure 3 and Supplementary Figure S2). Supporting this model, we found that HdaB toxicity can be bypassed by suppressor mutations in dnaK or recQ that stimulate Lon-dependent degradation (Supplementary Figure S7), restoring normal DnaA levels (Figure 6) and eliminating uncontrolled initiation events (Figure 4).
Figure 7.
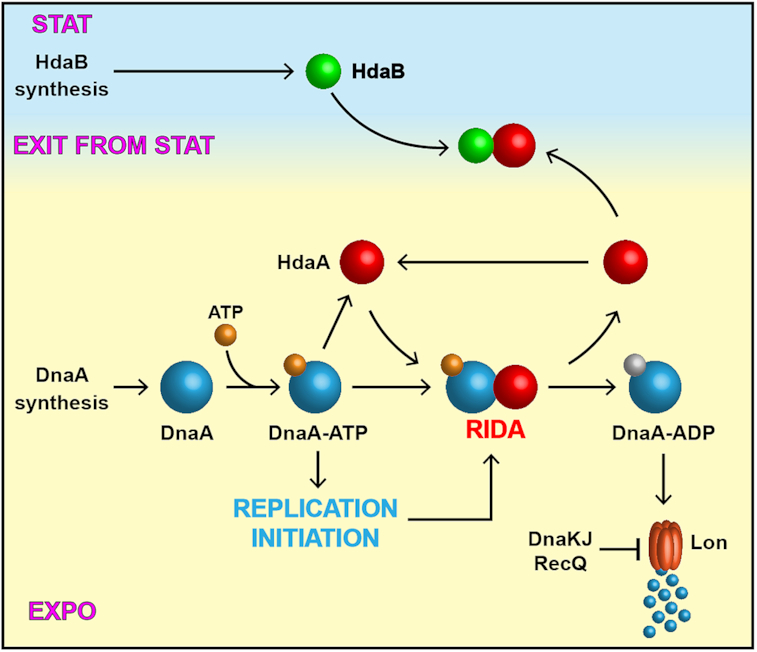
Model for the biological function of HdaB in C. crescentus. DnaA synthesis mostly occurs during exponential phase and DnaA associates with ATP soon after its synthesis, giving an active DnaA-ATP complex that can initiate chromosome replication. Once replication has started, HdaA binds to DnaA and stimulates its ATPase activity. The resulting DnaA-ADP complex is actively degraded by the Lon protease as part of the Regulated Inactivation and degradation of DnaA (RIDA) process. Upon entry into stationary phase, DnaA synthesis is strongly repressed, leading to an inhibition of replication initiation and a cell cycle arrest in G1 or G2 phase. Meanwhile, HdaB accumulates through a stimulation of its synthesis. During exit form stationary phase, DnaA synthesis restarts while the RIDA process is inhibited by a sequestration of HdaA by HdaB, leading to optimum levels of DnaA-ATP for rapid replication restart.
A role for HdaB during exit from stationary phase?
The intracellular accumulation of HdaBCc is mostly occurring during stationary phase (Figure 1C), apparently due to a strong activation of hdaBCc transcription during this period (Figure 1D). When cells are diluted back into fresh medium, HdaBCc remains detectable (Figure 2C) for >1 h, when many cells initiate the replication of their chromosome (Figure 2B) despite very low levels of the DnaA initiator (Figure 2C). This observation suggests that the biological function of HdaB consists in blocking the activity of HdaA, and thereby boost DnaA activity to promote the initiation of DNA replication when cells exit stationary phase (Figure 7), even if DnaA levels are still very low during this period (Figure 2C). Supporting this model, we found that chromosome replication restart is delayed in ΔhdaBCc compared to wild-type C. crescentus cells during the exit from stationary phase (Figure 2B). Interestingly, this replication delay is coincident with a growth delay (Figure 2A), indicating that HdaB may also promote the activity of DnaA when it acts as a master transcriptional regulator activating the transcription of many genes in C. crescentus (41).
Finally, if HdaB promotes DnaA activity through an inhibition of HdaA activity when cells are getting ready to enter S phase but are still in G1 phase (Figure 7), it implies that HdaA may also be active in non-replicating cells. Indeed, although the interaction of HdaA with the β-clamp was shown to play an important role in stimulating HdaA activity during the S phase of the cell cycle (13), there are also evidences that HdaA promotes the degradation of DnaA in stationary phase C. crescentus cells that are not actively replicating their DNA (14). Accordingly, we propose that HdaB may be able to boost DnaA activity even in cells that have not yet restarted the replication of their chromosome.
Functional conservation for HdaB proteins
HdaB homologs are found in a majority of Alphaproteobacteria (Figure 1A), suggesting that they play an important role in this class of bacteria, which includes a large number of pathogens (42). It is possible that HdaB contributes to infectious processes, notably when intracellular pathogens must restart their cell cycle following internalization (43). Here, we provide evidence that HdaB homologs from three distantly-related Alphaproteobacteria (Figure 1A), including the B. abortus pathogen, can promote DnaA accumulation (Figure 6) and replication initiation in C. crescentus (Figure 4), indicating a strong functional conservation. Importantly, every bacterium that has a HdaB protein also has a HdaA protein (Figure 1A), consistent with the proposal that HdaB targets HdaA to inhibit the RIDA process.
Ccrescentus death during over-initiation
Similar to what was previously observed when the RIDA process was inhibited by depleting hdaACc (12) or by over-expressing a hyper-active DnaACc(R357A) protein (18,30), the ectopic expression of hdaB genes in C. crescentus lead to over-initiation events coincident with cell division defects and cell death (Figure 3, 4 and Supplementary Figure S2). The accumulation of multiple origins perturbed the subcellular localization of ParB (Figure 4A and B), itself disturbing the bi-polar recruitment of MipZ (data not shown) during S phase, which plays a critical role in Z-ring positioning and cell division (31,44). Cell constriction might be further inhibited by an SOS response activated in response to RecA recruitment to ssDNA. Indeed, we found that many cells displayed RecA-CFP foci (Supplementary Figure S8) before they died (Figure 3), indicating the presence of ssDNA zones/lesions on the chromosome prior to cell death (45). Such regions can become substrates for the production of double-stranded DNA breaks (DSB) by reactive oxygen species (ROS) leading to cell death in bacteria (11,33). Interestingly, such rapid death in response to replication defects is reminiscent of the so-called thymineless death (TLD) process that underlies the action of several bactericidal drugs such as trimethoprim or sulfamethoxazole (40,46,47).
A novel regulatory function for the RecQ helicase
It was recently shown that RecQ is required for TLD in E. coli (39,40). However, in our study, we found that recQ mutations did not rescue over-initiating C. crescentus cells by increasing their tolerance for over-initiation, but instead, by reducing replication initiation (Figure 4C). Further characterization of the process showed that RecQ promotes DnaA accumulation not only in hdaB over-expressing cells (Figure 6), but also in wild-type cells cultivated in exponential phase (Supplementary Figure S7C), through a repression of lon transcription (Supplementary Figure S7B). This newly discovered regulatory process has not only an impact on DnaA levels, but also on the levels of other Lon substrates such as the cell cycle dependent DNA methyltransferase CcrM (Supplementary Figure S7C) which plays a key role in the network controlling the cell cycles of Alphaproteobacteria (48,49). Understanding how RecQ inhibits Lon-dependent proteolysis and may affect genome maintenance or cell cycle progression in Alphaproteobacteria provides a very interesting outlook for this study.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Laurent Casini for his help during the genetic screen, Regis Hallez for advices to perform immunoprecipitation assays and the Genomic Technology Facility of the University of Lausanne for advices for qPCR analysis. We thank Diego Gonzalez for the gift of pBX-M2-CdnL. We thank members of the Collier/Viollier/Genevaux/Falquet laboratories for helpful discussions during the project. Finally, we thank Stefano Sanselicio, Clément Gallay, Tiancong Chai, Laurent Casini and Noémie Matthey for their feedback on the manuscript.
Notes
Present address: Antonio Frandi and Justine Collier, Department of Fundamental Microbiology, Faculty of Biology and Medicine, University of Lausanne, Lausanne CH-1015, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation (SNSF) [CRSII3_160703 to J.C.]. Funding for open access charge: SNSF; University of Lausanne.
Conflict of interest statement. None declared.
REFERENCES
- 1. Katayama T., Ozaki S., Keyamura K., Fujimitsu K.. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010; 8:163–170. [DOI] [PubMed] [Google Scholar]
- 2. Katayama T., Kasho K., Kawakami H.. The DnaA cycle in escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 2017; 8:2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jameson K.H., Wilkinson A.J.. Control of initiation of DNA replication in bacillus subtilis and escherichia coli. Genes (Basel). 2017; 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frandi A., Collier J.. Multilayered control of chromosome replication in Caulobacter crescentus. Biochem. Soc. Trans. 2019; 47:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Felletti M., Omnus D.J., Jonas K.. Regulation of the replication initiator DnaA in Caulobacter crescentus. Biochim. Biophys. Acta Gene Regul. Mech. 2018; 1862:697–705. [DOI] [PubMed] [Google Scholar]
- 6. Kato J., Katayama T.. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001; 20:4253–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawakami H., Su’etsugu M., Katayama T.. An isolated Hda-clamp complex is functional in the regulatory inactivation of DnaA and DNA replication. J. Struct. Biol. 2006; 156:220–229. [DOI] [PubMed] [Google Scholar]
- 8. Su’etsugu M., Shimuta T.R., Ishida T., Kawakami H., Katayama T.. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J. Biol. Chem. 2005; 280:6528–6536. [DOI] [PubMed] [Google Scholar]
- 9. Kim J.S., Nanfara M.T., Chodavarapu S., Jin K.S., Babu V.M.P., Ghazy M.A., Chung S., Kaguni J.M., Sutton M.D., Cho Y.. Dynamic assembly of Hda and the sliding clamp in the regulation of replication licensing. Nucleic Acids Res. 2017; 45:3888–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keyamura K., Katayama T.. DnaA protein DNA-binding domain binds to Hda protein to promote inter-AAA+ domain interaction involved in regulatory inactivation of DnaA. J. Biol. Chem. 2011; 286:29336–29346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charbon G., Campion C., Chan S.H., Bjorn L., Weimann A., da Silva L.C., Jensen P.R., Lobner-Olesen A.. Re-wiring of energy metabolism promotes viability during hyperreplication stress in E. coli. PLos Genet. 2017; 13:e1006590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collier J., Shapiro L.. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J. Bacteriol. 2009; 191:5706–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fernandez-Fernandez C., Grosse K., Sourjik V., Collier J.. The beta-sliding clamp directs the localization of HdaA to the replisome in Caulobacter crescentus. Microbiology. 2013; 159:2237–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wargachuk R., Marczynski G.T.. The Caulobacter crescentus homolog of DnaA (HdaA) also regulates the proteolysis of the replication initiator protein DnaA. J. Bacteriol. 2015; 197:3521–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jonas K., Liu J., Chien P., Laub M.T.. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell. 2013; 154:623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu J., Zeinert R., Francis L., Chien P.. Lon recognition of the replication initiator DnaA requires a bipartite degron. Mol. Microbiol. 2019; 111:176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quon K.C., Yang B., Domian I.J., Shapiro L., Marczynski G.T.. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jonas K., Chen Y.E., Laub M.T.. Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr. Biol. 2011; 21:1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leslie D.J., Heinen C., Schramm F.D., Thuring M., Aakre C.D., Murray S.M., Laub M.T., Jonas K.. Nutritional control of DNA replication initiation through the proteolysis and regulated translation of DnaA. PLos Genet. 2015; 11:e1005342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gorbatyuk B., Marczynski G.T.. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 2005; 55:1233–1245. [DOI] [PubMed] [Google Scholar]
- 21. Wortinger M.A., Quardokus E.M., Brun Y.V.. Morphological adaptation and inhibition of cell division during stationary phase in Caulobacter crescentus. Mol. Microbiol. 1998; 29:963–973. [DOI] [PubMed] [Google Scholar]
- 22. Heinrich K., Leslie D.J., Morlock M., Bertilsson S., Jonas K.. Molecular basis and ecological relevance of caulobacter cell filamentation in freshwater habitats. mBio. 2019; 10:e01557-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Evinger M., Agabian N.. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 1977; 132:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gorbatyuk B., Marczynski G.T.. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 2001; 40:485–497. [DOI] [PubMed] [Google Scholar]
- 25. Zweiger G., Marczynski G., Shapiro L.. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 1994; 235:472–485. [DOI] [PubMed] [Google Scholar]
- 26. Miller J.H. Experiments in Molecular Genetics. 1972; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 27. Mouammine A., Eich K., Frandi A., Collier J.. Control of proline utilization by the Lrp-like regulator PutR in Caulobacter crescentus. Sci. Rep. 2018; 8:14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ducret A., Quardokus E.M., Brun Y.V.. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 2016; 1:16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Viollier P.H., Thanbichler M., McGrath P.T., West L., Meewan M., McAdams H.H., Shapiro L.. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9257–9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fernandez-Fernandez C., Gonzalez D., Collier J.. Regulation of the activity of the Dual-Function DnaA protein in caulobacter crescentus. PLoS One. 2011; 6:e26028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thanbichler M., Shapiro L.. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006; 126:147–162. [DOI] [PubMed] [Google Scholar]
- 32. Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., Yokoyama S.. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003; 31:2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Charbon G., Riber L., Lobner-Olesen A.. Countermeasures to survive excessive chromosome replication in Escherichia coli. Curr. Genet. 2018; 64:71–79. [DOI] [PubMed] [Google Scholar]
- 34. Christen B., Abeliuk E., Collier J.M., Kalogeraki V.S., Passarelli B., Coller J.A., Fero M.J., McAdams H.H., Shapiro L.. The essential genome of a bacterium. Mol. Syst. Biol. 2011; 7:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schramm F.D., Heinrich K., Thuring M., Bernhardt J., Jonas K.. An essential regulatory function of the DnaK chaperone dictates the decision between proliferation and maintenance in Caulobacter crescentus. PLos Genet. 2017; 13:e1007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Avedissian M., Lessing D., Gober J.W., Shapiro L., Gomes S.L.. Regulation of the Caulobacter crescentus dnaKJ operon. J. Bacteriol. 1995; 177:3479–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bernstein K.A., Gangloff S., Rothstein R.. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010; 44:393–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simmons L.A., Breier A.M., Cozzarelli N.R., Kaguni J.M.. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol. Microbiol. 2004; 51:349–358. [DOI] [PubMed] [Google Scholar]
- 39. Hastings P.J., Rosenberg S.M.. A radical way to die. Nat. Microbiol. 2017; 2:1582–1583. [DOI] [PubMed] [Google Scholar]
- 40. Hong Y., Li L., Luan G., Drlica K., Zhao X.. Contribution of reactive oxygen species to thymineless death in Escherichia coli. Nat. Microbiol. 2017; 2:1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hottes A.K., Shapiro L., McAdams H.H.. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 2005; 58:1340–1353. [DOI] [PubMed] [Google Scholar]
- 42. Collier J. Cell cycle control in Alphaproteobacteria. Curr. Opin. Microbiol. 2016; 30:107–113. [DOI] [PubMed] [Google Scholar]
- 43. Deghelt M., Mullier C., Sternon J.F., Francis N., Laloux G., Dotreppe D., Van der Henst C., Jacobs-Wagner C., Letesson J.J., De Bolle X.. G1-arrested newborn cells are the predominant infectious form of the pathogen Brucella abortus. Nat. Commun. 2014; 5:4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Collier J. Cell division control in Caulobacter crescentus. Biochim. Biophys. Acta. 2018; 1862:685–690. [DOI] [PubMed] [Google Scholar]
- 45. Badrinarayanan A., Le T.B., Laub M.T.. Rapid pairing and resegregation of distant homologous loci enables double-strand break repair in bacteria. J. Cell Biol. 2015; 210:385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matic I. The major contribution of the DNA damage-triggered reactive oxygen species production to cell death: implications for antimicrobial and cancer therapy. Curr. Genet. 2018; 64:567–569. [DOI] [PubMed] [Google Scholar]
- 47. Giroux X., Su W.L., Bredeche M.F., Matic I.. Maladaptive DNA repair is the ultimate contributor to the death of trimethoprim-treated cells under aerobic and anaerobic conditions. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:11512–11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mouammine A., Collier J.. The impact of DNA methylation in Alphaproteobacteria. Mol. Microbiol. 2018; 110:1–10. [DOI] [PubMed] [Google Scholar]
- 49. Gonzalez D., Kozdon J.B., McAdams H.H., Shapiro L., Collier J.. The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res. 2014; 42:3720–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Domian I.J., Quon K.C., Shapiro L.. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997; 90:415–424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.