Abstract
Protein lysine acetylation, one of the most abundant post-translational modifications in eukaryotes, occurs in prokaryotes as well. Despite the evidence of lysine acetylation in bacterial RNA polymerases (RNAPs), its function remains unknown. We found that the housekeeping sigma factor (HrdB) was acetylated throughout the growth of an actinobacterium, Streptomyces venezuelae, and the acetylated HrdB was enriched in the RNAP holoenzyme complex. The lysine (K259) located between 1.2 and 2 regions of the sigma factor, was determined to be the acetylated residue of HrdB in vivo by LC–MS/MS analyses. Specifically, the label-free quantitative analysis revealed that the K259 residues of all the HrdB subunits were acetylated in the RNAP holoenzyme. Using mutations that mimic or block acetylation (K259Q and K259R), we found that K259 acetylation enhances the interaction of HrdB with the RNAP core enzyme as well as the binding activity of the RNAP holoenzyme to target promoters in vivo. Taken together, these findings provide a novel insight into an additional layer of modulation of bacterial RNAP activity.
INTRODUCTION
Protein lysine acetylation, one of the major post-translational modifications (PTMs), occurs in all three domains of life (1,2). Recent proteomic analyses in bacteria showed that substantial parts of proteins are acetylated in organisms (2–4). These acetylated proteins are distributed broadly in biological processes and functions (5,6). Since lysine acetylation results in the increase in the net negative charge of proteins, the characteristics of acetylated proteins can be modulated in terms of metabolic activity (7–12), RNA metabolism (13,14), DNA replication (15,16), cell shape (17), chemotaxis (18–20) and gene expression (21–26). Regulation of transcription by acetylation of RNA polymerase (RNAP)-associated proteins has been shown for cAMP-receptor protein (CRP) in Escherichia coli and CshA in Bacillus subtilis (27–29).
Lysine acetylation of transcription factors in the DNA-binding domains generally hinders ionic interaction with DNA and inhibits their DNA-binding activity (21–26). RscB, a response regulator in E. coli, is the first such example: the acetylation of K154 and K180 residues of RcsB impairs its DNA binding activity toward the target genes, at which K154 acetylation leads to increased motility and sensitivity to acidic stress (21,22,30). In the case of HilD in Salmonella, lysine acetylation modulates protein stability in addition to its DNA binding activity (25). Acetylation of the CRP in E. coli results in either enhanced or reduced transactivation, depending on the target promoters (27). Additionally, lysine acetylation of a protein can affect the activity of other transcription factors. For example, an isoform of glutamine synthetase GlnA acts as a chaperone for a transcription factor GlnR in the actinobacterium Saccharopolyspora erythraea, whose lysine acetylation enhanced the DNA binding ability of GlnR (8).
Recent proteome research revealed that the bacterial RNAP subunits are acetylated in a broad range of bacteria: Proteobacteria (30–38), Firmicutes (17,39,40), Actinobacteria (41–44) and Cyanobacteria (45). Despite numerous observations of lysine acetylation in RNAP subunits, its physiological impact has not been well elucidated, except for RpoA acetylation in E. coli (31,32). Escherichia coli cells grown in the presence of glucose showed extensive protein acetylation, and cpxP gene expression required the acetylation of K298 in the C-terminal domain of RpoA (31). Acetylation of K291 in the same domain inhibited cpxP expression under conditions of excess glucose and acetyl-phosphate accumulation (31,32).
Streptomyces venezuelae ATCC15439, a soil actinobacterium producing pikromycin, undergoes sporulation in both solid and liquid media (46). It contains a genome of 9.05 Mb harboring 8,080 protein-coding genes (47). It is predicted to encode 43 sigma factors: a housekeeping sigma factor HrdB (homolog of RpoD), two Group 2 sigma factors (HrdA and HrdD), six Group 3 sigma factors and 34 Group 4 (ECF) sigma factors, based on domain analysis (48,49). In 1990, hrdB was first discovered to be a gene for essential housekeeping sigma factor in Streptomyces coelicolor (50). To date, HrdB target genes have been identified based on in vitro transcription, S1-nuclease mapping, and ChIP-seq analysis (50–53). The transcription of HrdB target genes is modulated by RbpA and CarD, which are RNA polymerase-associated proteins to stabilize the transcription initiation complex in Actinobacteria (54,55). The hrdB gene expression is under the control of two ECF sigma factors, ShbA and SigR (56,57). However, PTMs of HrdB have not been reported thus far.
In this study, we present evidence of HrdB acetylation in S. venezuelae and its role in transcription.
MATERIALS AND METHODS
Strains and growth conditions
Streptomyces venezuelae strain ATCC15439 (Sven15439) and its derivatives were grown and maintained according to standard procedures (46,58). The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Supplementary Tables S1 and S2. Spores of S. venezuelae were inoculated in MYM liquid media containing 0.4% (w/v) maltose, 0.4% (w/v) yeast extract and 1% (w/v) malt extract (59) and cultured with shaking (at 180 rpm) at 30°C. All the experimental replicates were done by using the independently prepared samples.
Construction of the strain (JE04) with His-tagged RpoC
The scheme for constructing the strain that encodes RpoC with a C-terminal His-tag is summarized in Supplementary Figure S1. For this purpose, the C-terminal region of the rpoC gene (from –333 to –1 codons from the stop codon; AQF52_4787) ligated with 6× His-tag was generated from the genomic DNA of Sven15439 by PCR using the primer pair rpo/pKC-F and rpo/his-R. The rpoC downstream fragment (from the stop codon to downstream 1008 nt) was also generated by PCR using the primer pair rpo/his-F and rpo/pKC-R. The two fragments were cloned into pKC1139 plasmid (60) digested with HindIII/EcoRV, via Gibson assembly. The resulting recombinant plasmid was introduced into Sven15439. The His-tagged-RpoC strain (JE04) generated by double crossover was selected by apramycin sensitivity and confirmed by screening by PCR using primer pair rpoC-F and His-R, followed by nucleotide sequencing.
Immunoprecipitation and western blot
For immunoprecipitation, cells were disrupted by ultrasonication in lysis buffer containing 20 mM Tris (pH 7.9), 10% (v/v) glycerol, 5 mM EDTA, 10 mM MgCl2, 150 mM NaCl, 0.1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail (S8830, Sigma), 20 mM nicotinamide and 3 μM trichostatin A. Cell lysates (2 mg proteins) were incubated with protein A/G agarose beads (sc-2003, Santa Cruz) for 1 h at 4°C. After removing the beads, lysates were incubated overnight at 4°C with 3 μl of anti-HrdB polyclonal antibody generated using purified His-tagged HrdB proteins (AbClon Inc., Seoul, Korea) and then incubated with protein A/G agarose beads for 4 h at 4°C. The precipitated beads were washed twice in lysis buffer and boiled for 10 min at 100°C to separate proteins from the beads. For western blot analysis, cell lysates, immunoprecipitated samples, or purified proteins were separated by 8% SDS-PAGE. Following electro-transfer of proteins from the gel onto nitrocellulose membrane for 1.5 h at 175 mA, the blots were blocked with 5% (w/v) BSA in TBST (10 mM Tris [pH 7.4], 0.9% [w/v] NaCl, 0.1% [v/v] Tween-20). Primary antibodies against HrdB (1:5000), RpoB (sc-56766, Santa Cruz, 1:5000), acetyl-lysine (ab190479, Abcam, 1:2000) and FLAG (M185, MBL, 1:5000) were used. For secondary antibodies, HRP-conjugated goat anti-rabbit (1:5000) or anti-mouse (1:5000) antibodies were used. The immunoblot was exposed to EZ-western Lumi Femto (DG-WF100, DoGen) and analyzed by WSE-6200H LuminoGraph II (ATTO).
Real-time RT-PCR (RT-qPCR)
Total RNAs were isolated from Sven15439 cells according to the standard procedure with slight modifications (58). Cells were disrupted in Kirby mixture using an ultrasonicator at 20% of the amplitude (5 cycles of 1-s burst and 1-s pause). Following phenol/chloroform extraction and isopropanol precipitation, RNA pellets were dissolved in RNase-free water. Contaminated DNAs were removed with a DNA-free kit (AM1906, Ambion) with RNase inhibitor (N8080119, Applied Biosystems). cDNAs were synthesized from the RNA sample (1 μg) using SuperScript™ III Reverse Transcriptase (18080093, Invitrogen) and random hexamers, as recommended by the manufacturer. RT-qPCR was carried out for the hrdB gene (AQF52_5901) and the 16S rRNA genes using gene-specific primers (Supplementary Table S2) and TOPreal qPCR 2× PreMIX (RT500, Enzynomics) on a quantitative real-time PCR machine (Stratagene MX300P, Agilent Technologies). For amplification, each sample was pre-incubated at 95°C for 15 min, followed by 40 cycles of 95°C for 10 s, 60°C for 15 s and 72°C for 30 s. Analysis of RT-qPCR was performed as described previously (61).
Purification of RNAP holoenzyme
RNAP was purified from the JE04 cells with a slight modification of the procedures for His-tagged RNAP in S. coelicolor (52,62). The JE04 cells harvested from 500 ml MYM medium of exponential phase culture or from 250 ml culture for transition or stationary phase were resuspended in lysis buffer (3 ml/g of cell wet weight) with lysozyme (1 mg/ml final) and incubated for 30 min on ice. The cell suspension was disrupted by sonication at 25% of the amplitude (5-s burst and 25-s pause) for 30 min until the viscosity disappeared. Following polyethyleneimine (0.3%) precipitation, the pellet was washed in 3 ml of TGED buffer (10 mM Tris–HCl [pH 7.9], 0.1 mM EDTA, 0.1 mM dithiothreitol, 10% glycerol) containing 0.5 M NaCl. The pellet was then resuspended in 2 ml of TGED buffer containing 1 M NaCl. Proteins were precipitated with saturated ammonium sulfate at 4°C for 30 min. The pellet was dissolved in buffer I (10 mM Tris–HCl [pH 7.9], 0.1 M NaCl, 1 mM β-mercaptoethanol, 5% [v/v] glycerol) with 2.5 mM imidazole and loaded onto His-Bind agarose resin (EBE-1031, ELPis). The resin was washed with buffer I with 20 mM imidazole. RNAP was eluted with buffer I containing 300 mM imidazole, and concentrated and buffer exchanged into storage buffer (20 mM Tris–HCl [pH 7.5], 0.1 mM EDTA, 1 mM DTT, 100 mM NaCl) using a centrifugal filter (UFC901024, Merck Millipore). For LC–MS/MS analysis, RNAP was buffer-exchanged with urea buffer (8 M urea, 50 mM NH4HCO3).
LC–MS/MS analysis and label-free quantification (LFQ) of lysine acetylation
HrdB peptides were generated either by tryptic digestion of purified RNAP samples or by in-gel digestion following SDS-PAGE. For in-gel digestion, gel slices corresponding to HrdB were destained in 50% acetonitrile solution of 25 mM ammonium bicarbonate (ABC) buffer for 10 min and then followed by in-gel alkylation of cysteine residues with dithiothreitol and iodoacetamide. The resulting samples were washed three times with 25 mM ABC, then were digested with sequencing-grade trypsin at a ratio of 1:50 (w/w) overnight at 37°C. The purified RNAP samples were subjected to reduction and alkylation as in the in-gel digestion, followed by dilution with 50 mM ABC buffer containing 1 M urea for protein digestion by trypsin under the same conditions as in the in-gel digestion.
The digested peptides were subjected to C18-SPE cleanup and sample concentration in speed-vac followed by LC–MS/MS analysis. The final peptide samples were resuspended in 25 mM ABC for LC–MS/MS analysis. LC–MS/MS experiments with HCD fragmentation mode were performed on Q-Exactive mass spectrometry (Thermo Fisher Scientific) coupled with nanoACQUITY UPLC (Waters), equipped with an in-house packed capillary trap column (150 μm internal diameter, 3 cm) and analytical column (75 μm internal diameter, 100 cm) with 3 μm Jupiter C18 particles (Phenomenex) at a flow rate of 300 nl/min. A linear gradient (100 min) was applied for each biological replicate.
The acquired datasets were subjected to MaxQuant (v. 1.5.3.30) with Andromeda search engine at 10 ppm precursor ion mass tolerance against the S. venezuelae proteome database at <1% of protein false discovery rate. The following search parameters were applied: tryptic digestion, fixed carbaminomethylation on cysteine, dynamic oxidation of methionine, and dynamic acetylation of lysine. LFQ mode was turned on and applied for the quantitative analysis of acetylation level. The extracted ion chromatogram (XIC) was plotted by Qual Browser in Xcalibur software (Thermo Scientific).
Construction of the hrdB mutants
The scheme for construction of the FLAG-tagged HrdB mutants (JE02, JE02-Q, and JE02-R) is summarized in Supplementary Figure S2. The full-length hrdB gene (from –533 to +1545 nt from the start codon) without the stop codon was generated from Sven15439 genomic DNA using the primer pair pSET/HrdB-F and pSET/HrdB-R. This amplicon was cloned into pSET152F (49) digested with XbaI/StuI via Gibson assembly, generating pSETF-hrdB (Supplementary Figure S2A). For mutation of the lysine (either K259Q or K259R), we assembled the mutagenized fragments via Gibson assembly (Supplementary Figure S2B). Primers were designed to substitute K259 with the changed codons underlined: for K259Q, 259Q-F and 259Q-R; for K259R, 259R-F and 259R-R. The mutagenized fragments were amplified using the appropriate primer sets (Supplementary Figure S2B) and then cloned into pSET152F digested with XbaI/StuI via Gibson assembly, generating pSETF-HrdB (K259Q) and pSETF-HrdB (K259R). Then, each plasmid was introduced into Sven15439, generating JE02, JE02-Q or -R.
Chromatin immunoprecipitation (ChIP)
Cells grown in 50 ml of MYM medium were fixed with 1% formaldehyde for 30 min. Glycine was added (125 mM) and incubated for 5 min at room temperature. Following centrifugation at 5000 × g at 4°C, cells were washed twice in PBS buffer (pH 7.4), resuspended in 0.5 ml of lysis buffer with lysozyme (14 mg/ml), and incubated at 37°C for 25 min. An additional 0.5 ml of lysis buffer was added, and cells were disrupted by sonication at 20% of the amplitude (6 cycles of 20-s burst and 60-s pause). Following centrifugation to remove the cell debris, aliquots of lysates were incubated with 20 μl of protein A/G agarose beads (sc-2003, Santa Cruz) for 2 h at 4°C. An aliquot with 50 μg DNA was used for the input DNA control. After removing the beads by centrifugation at 1000 × g at 4°C for 1 min, the lysates were incubated with 2 μl of anti-FLAG antibody (M185, MBL) and 20 μl of protein A/G agarose beads for overnight at 4°C, with gentle mixing by rotation. Following centrifugation at 1000 × g at 4°C for 1 min, beads were washed twice with ChIP wash buffer I (20 mM Tris [pH 7.5], 0.15 M NaCl) and then twice with ChIP wash buffer II (20 mM Tris [pH 7.5], 0.15 M NaCl, 0.05% Tween-20). Beads and the input DNA control were incubated in the ChIP elution buffer (10 mM Tris [pH 7.4], 1 mM EDTA, 1% SDS) at 65°C for 30 min. After removing beads, eluates were incubated with RNase A (1 μg / 200 μl) at 37°C for 1 h, and then with proteinase K (0.1 mg/200 μl) at 55°C for 2 h. DNA was purified by phenol–chloroform and chloroform extraction and quantified by qPCR using the primer sets described in Supplementary Table S2.
RESULTS
Expression of the hrdB gene in S. venezuelae ATCC15439
The ortholog of E. coli rpoD (hrdB) and its two paralogs (hrdA and hrdD) were identified from the complete genome sequence of S. venezuelae ATCC15439 (Sven15439) (47). Alignment of the amino acid sequences of the housekeeping sigma factors (HrdBs) from Streptomyces species revealed variations in the predicted N-terminus of the HrdB proteins (Supplementary Figure S3). The predicted start codon of HrdB in Sven15439 and S. venezuelae ATCC10712 (Sven 10712) lies two (S2) and 52 (S3) amino acids upstream from the conserved start codon (S1), respectively, in the same reading frame (Supplementary Figure S4A). Thus, to define the translational start site of the hrdB gene in Sven15439, we constructed a series of translational fusions using a β-glucuronidase (gus)-based reporter plasmid (pGUS-TL). The pG-HrdB-T series include fusions with mutations at S1 and S3 sites (Supplementary Figure S4B). As shown in Supplementary Figure S4C, the conserved S1 is the initiation codon used for translating Sven15439 HrdB. This corresponds to the initiation codon of the S. coelicolor HrdB, whose N-terminus was determined by peptide sequencing (63). We therefore changed the numbering of the amino acid sequences of the Sven15439 HrdB, consisting of 515 amino acids from the GTG initiation codon (Met1) that had been previously annotated as Val3 (GenBank ALO11494.1).
We then determined the expression level of the hrdB gene during the growth of Sven15439 in liquid MYM medium. Both RNA and protein levels were determined by RT-qPCR and western blot, respectively. As shown in Figure 1A, the transcript level gradually decreased throughout growth. The amount of the hrdB transcripts at the stationary phase was ∼30% of that at the early exponential phase. The HrdB protein level in the cell lysates decreased in accordance with the RNA levels (Figure 1B).
Figure 1.
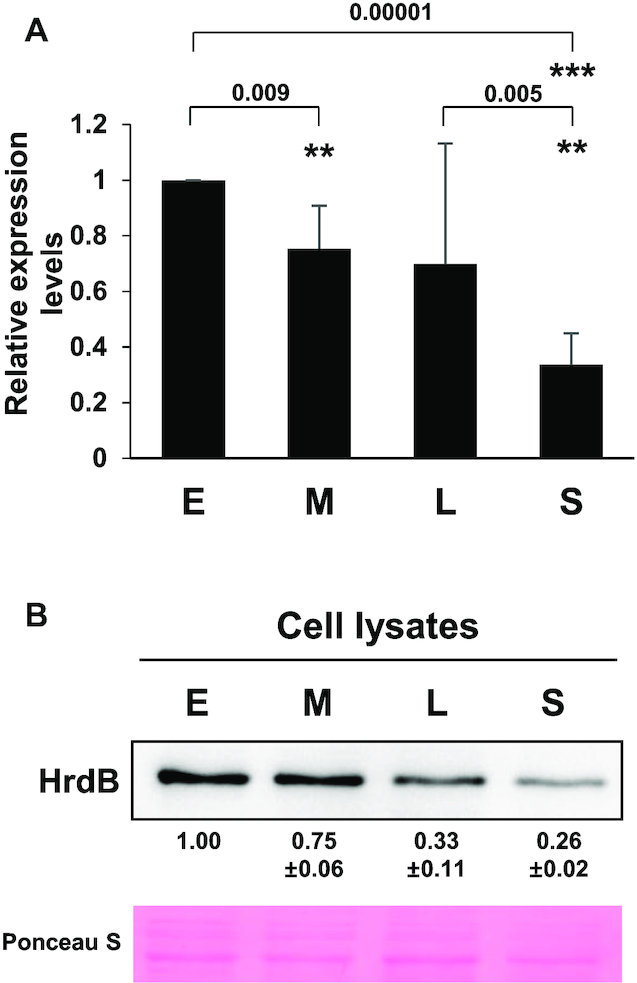
Expression of HrdB during growth. The RNA (A) and protein (B) levels of the hrdB gene expression during the growth of Sven15439. Cells were grown in MYM medium to early- (E), mid- (M) and late- (L) exponential and stationary (S) phases. The hrdB RNA levels were determined by RT-qPCR and presented as the relative values to the level at E phase. The HrdB protein levels from Sven15439 cell lysates (5 μg) were measured by densitometric analyses of the western blots using anti-HrdB antibody. The data represent the average values from independent experiments (n = 4 for RT-qPCR and n = 3 for western analysis), and the standard errors are represented by error bars (A) or values (B). Statistical significance between the groups in A is indicated, based on P values of less than 0.01 (**) or 0.001 (***) by using Student's t-test (n = 4).
HrdB is lysine-acetylated throughout growth
Prompted by several proteomic studies that reported the acetylation of bacterial RNAP subunits (17,30–44), we investigated whether HrdB is acetylated in Streptomyces. We first immunoprecipitated HrdB proteins from cell lysates prepared from different growth phases, and then detected acetylated proteins using monoclonal antibodies against acetyl-lysine (AcK). As shown in Figure 2, the amount of immunoprecipitated HrdB proteins gradually decreased throughout the growth phases, consistent with the level in whole cell lysates (Figure 1B). It is notable that a similarly decreasing trend in AcK was observed in the immunoprecipitated HrdB samples. This result indicates that HrdB proteins are lysine-acetylated throughout the growth phases and, more importantly, that the proportion of HrdB acetylation does not significantly change during growth.
Figure 2.
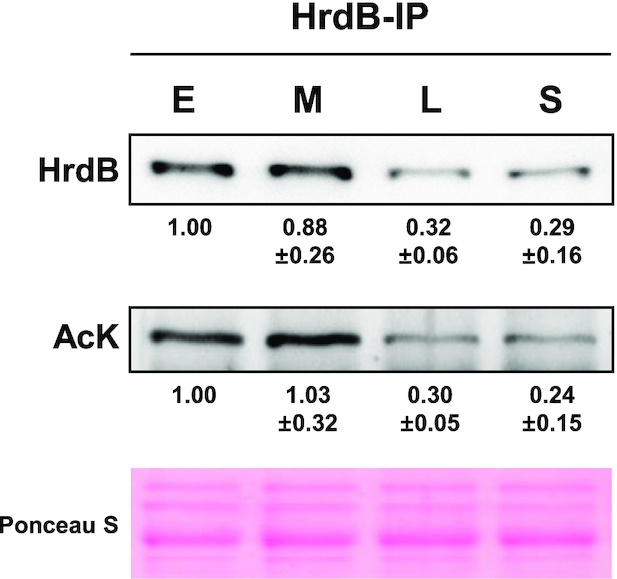
Lysine-acetylation of HrdB during growth. The HrdB proteins were immunoprecipitated with anti-HrdB antibody from Sven15439 cell extracts (2 mg) prepared at early- (E), mid- (M) and late- (L) exponential and stationary (S) phases. Precipitated proteins (HrdB-IP) (3 μl for HrdB and 15 μl for AcK) were analyzed by western blot using antibodies against HrdB or acetylated-lysine (AcK). For each immunoblot, the levels were measured by densitometric analyses of the western blots and presented as the relative values to the level at E phase. The data represent average values with standard errors from four independent experiments.
RNAP-bound HrdB is acetylated
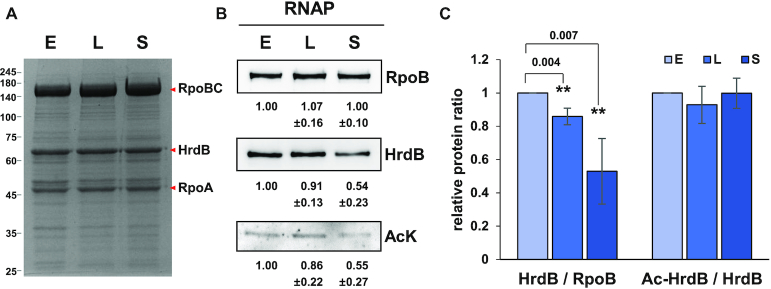
We wondered whether lysine acetylation in HrdB could affect its interaction with the RNAP core enzyme. To investigate this possibility, we prepared RNAP from S. venezuelae JE04 cells that express His-tagged RpoC. The RNAP samples prepared from cells at three growth phases (E, L, S) all contained stoichiometric amounts of β (RpoB), β′ (RpoC), and α (RpoA) subunits (Figure 3A) as judged by densitometric tracing, indicating the good quality of RNAP preparations from JE04 cells. Unlike the core RNAP subunits, however, determination of HrdB stoichiometry was not possible due to co-migration of multiple abundant proteins such as GroEL with HrdB (Supplementary Table S3).
Figure 3.
Lysine-acetylation of HrdB in RNAP holoenzyme. RNAP holoenzyme was purified from JE04 cells at early- (E) and late- (L) exponential and stationary (S) phases as described in Materials and Methods. Proteins in the RNAP samples (0.5, 1 and 5 μg for RpoB, HrdB and AcK, respectively) were electrophoresed on SDS-PAGE gel and stained with Coomassie brilliant blue (A) and analyzed by western blots using antibodies against RpoB, HrdB or AcK (B). Representative images are shown with the average values and the standard errors measured from three independent experiments. (C) The quantification of the data in (B). The ratios of HrdB/RpoB and acetylated HrdB (Ac-HrdB)/total HrdB relative to values at E phase are presented with error bars representing the standard errors. Statistical significance between the groups is indicated, based on P values <0.01 (**) by using Student's t-test.
We measured the level of RpoB, HrdB and AcK in each RNAP sample by western analysis and compared the HrdB/RpoB and AcK/HrdB ratios during growth relative to the ratios of the exponential phase sample that were set to 1.0. The relative ratio of HrdB/RpoB decreased to ∼54% in the stationary phase sample (Figure 3B and C). Considering that the level of HrdB in whole cell lysate decreased to ∼26% in the stationary phase (Figure 1), enrichment of HrdB was significantly observed in the RNAP sample relative to the cell lysate sample. The level of AcK decreased similarly to the level of HrdB in the RNAP samples during growth, showing the ratio of AcK/HrdB to be nearly unchanging (Figure 3C).
We then compared the levels of AcK relative to HrdB in the cell lysate and in RNAP obtained from exponential phase cells by western analysis. The AcK/HrdB ratio in the RNAP sample was higher by about 2-fold compared with the ratio in cell lysates as shown in Figure 4. This indicates that the HrdB in RNAP is acetylated to a higher extent than in whole cell lysate and thus suggests that the lysine acetylation of HrdB may have a positive effect on the interaction between HrdB and the RNAP core enzyme.
Figure 4.
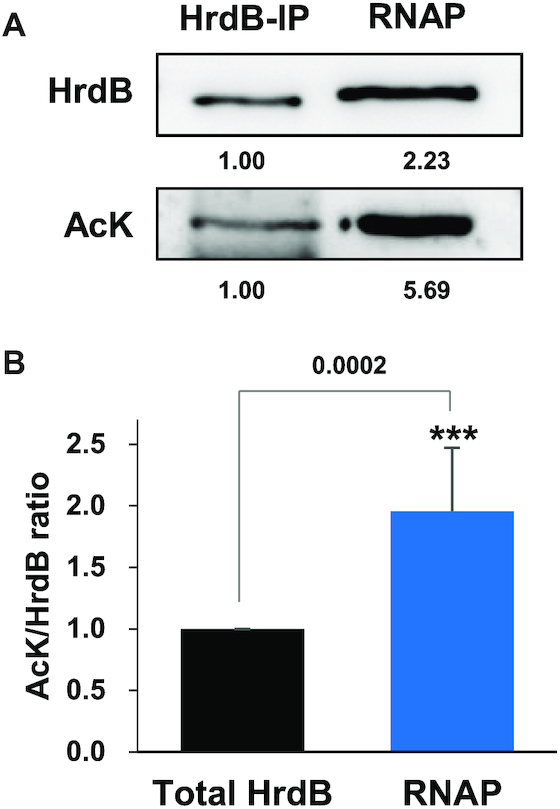
Lysine-acetylation of the total and the RNAP-associated HrdB proteins at the exponential growth phase. (A) Lysine-acetylation of the total and the RNAP-associated HrdB proteins. Cell lysates (2 mg) prepared from JE04 cells at early exponential phase were subjected either to immunoprecipitation with anti-HrdB antibody to collect total HrdB or to purification of RNAP as described in Materials and Methods. Immunoprecipitate (HrdB-IP) (3 μl for HrdB and 15 μl for AcK) and purified RNAP samples (1 μg for HrdB and 5 μg for AcK) were analyzed by western blot using either anti-HrdB or anti-AcK antibodies. Representative images from seven independent experiments are shown. (B) Relative ratio of HrdB acetylation in total vs. RNAP-bound HrdB. The lysine-acetylation ratio of HrdB in the RNAP was compared with that of the total immunoprecipitated HrdB, which was set to 1.0. Quantification from seven independent experiments is presented with an average value and standard error. Statistical significance with a P value of <0.001 (***) is indicated.
K259 residue in RNAP-associated HrdB is fully acetylated
As an independent approach to the detailed analysis of HrdB acetylation, we carried out LC–MS/MS analyses of total HrdB immunoprecipitated from cell lysates and RNAP-associated HrdB samples. Total HrdB samples were prepared from the gel after immunoprecipitation of Sven15439 cell lysates harvested at different growth phases (early-, late-exponential phase and stationary phase), whereas RNAPs were purified from JE04 cells grown to corresponding phases. Each sample was analyzed by LC–MS/MS following trypsin digestion. The representative MS spectrum data are shown in Supplementary Figure S5. Acetylation of the four lysine residues (K228, K259, K279 and K481) among 45 lysine residues of HrdB was detected in total HrdB samples, whereas only K259 was acetylated in RNAP-associated HrdB samples (Figure 5A). Label-free quantitative (LFQ) analysis of the identified acetylated lysine site was performed to measure the acetylation ratio of each lysine and its change during growth. Interestingly, the LFQ intensity ratios of acetylated peptides including K259 in total HrdB samples were over 0.8 at all growth phases, whereas those for the other lysines were <0.1 (Supplementary Figure S6), suggesting that K259 is the preponderant acetylated residue in total HrdB. It is notable that K259 in the RNAP-associated HrdB was 100% acetylated at all growth phases (Figure 5B). These results imply that K259 of HrdB is the predominant acetylated residue and only K259-acetylated HrdB is associated with RNAP. This result coincides with the western blot results in which lysine acetylation was enriched in the RNAP fraction, compared with the level in the cell lysate (Figure 4).
Figure 5.
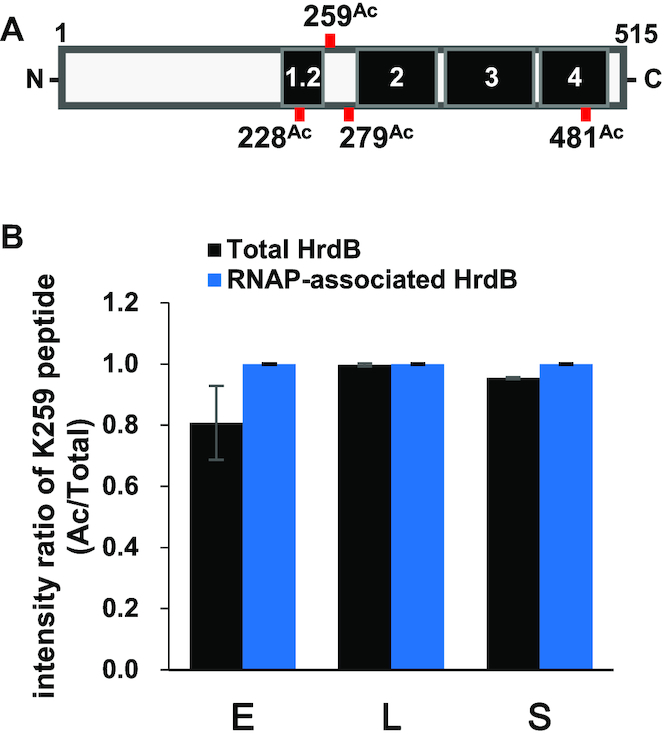
Determination of the AcK residues in HrdB. (A) Schematic representation of the acetylated lysine residues of the total and the RNAP-associated HrdB proteins (see Supplementary Figures S5 and S6), with 4 lysine residues deemed acetylated (228Ac, 259Ac, 279Ac and 481Ac). The functional conserved regions (1.2, 2, 3 and 4) in HrdB are shown in black. (B) K259 acetylation ratio (acetylated vs total) of HrdB in the immunoprecipitates (total HrdB, black) and the purified RNAP samples (RNAP-associated HrdB, blue) at early- (E) and late- (L) exponential and stationary (S) phases as determined from LC–MS/MS after trypsin digestion. The data represent the average values with standard errors from four (for total HrdB) and three (for RNAP-associated HrdB) independent experiments.
K259 acetylation facilitates the association between HrdB and the RNAP core enzyme
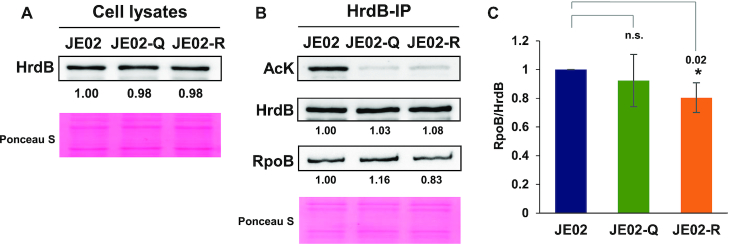
To elucidate the function of K259 acetylation of HrdB, we constructed hrdB variants with the C-terminal FLAG-tag and a K259 substitution. Since hrdB is an essential gene, we introduced the variant hrdB genes to the att site in the chromosome, resulting in merozygotic strains (JE02 series) with two copies of the hrdB (i.e. the wild type and the variant) genes. The K259 residue was substituted either with glutamine (K259Q) that mimics the constitutively acetylated form or arginine (K259R) that mimics the non-acetylated form, as widely exploited in lysine acetylation studies (64,65). Similar levels of HrdB-FLAG protein expression in the JE02, JE02-Q and JE02-R cells were verified by western blot analysis (Figure 6A). To examine the effect of K259 acetylation on the association of HrdB with the RNAP core enzyme, we first measured the amount of RpoB co-precipitated with the HrdB variants by using anti-FLAG antibody for immunoprecipitation of the cell lysates. The precipitated proteins were analysed by western blot with antibodies against AcK, FLAG and RpoB. As shown in Figure 6B, the AcK signals in JE02-Q and JE02-R samples were drastically reduced, substantiating that K259 is the major acetylation site in HrdB as indicated from MS results. The relative ratios of RpoB/HrdB in the immunoprecipitated samples from the wild type (JE02) and HrdB-K259 variant cells were estimated from three independent experiments. The results in Figure 6C show that the amount of co-precipitated RpoB was reduced by ∼20% in JE02-R compared with the wild type. This supports the possibility that K259 acetylation of HrdB facilitates the association between HrdB and the RNAP core enzyme.
Figure 6.
Effect of K259 mutations on HrdB binding to RNAP core enzyme. (A) Expression levels of FLAG-tagged HrdB proteins with a K259 mutation to either glutamine (JE02-Q) or arginine (JE02-R) in comparison with that of the wild type (JE02). Cell extracts (5 μg) at the early exponential phase were analyzed by western blot using anti-FLAG antibody. (B) Effect of mutations on the association of HrdB with the RNAP core enzyme. The same cell samples of A were lysed and immunoprecipitated with anti-FLAG to collect HrdB and its associated proteins. Precipitates (HrdB-IP) (4, 2 and 0.5 μg for AcK, HrdB and RpoB, respectively) were analysed by western blots using anti-AcK, anti-FLAG, or anti-RpoB antibodies. Representative images from three independent experiments are shown. (C) The quantification of the data in (B). The RpoB/HrdB ratios relative to that in JE02 are shown with error bars representing the standard errors from three independent experiments. Statistical significance between the groups is indicated, based on a P value of <0.05 (*) by using Student's t-test. The difference between JE02 and JE02-Q is not statistically significant (n.s.).
We next investigated the effect of K259 acetylation on the binding of HrdB-containing RNAP holoenzyme to the target promoters in vivo by chromatin immunoprecipitation (ChIP) analysis. The rpoB gene and the atpL gene encoding ATP synthase protein I were selected as the representative target genes of the HrdB regulon (53,54,66). JE02 series cells that had been grown to the exponential phase were treated with formaldehyde to cross-link the RNAP and the DNA in physical proximity followed by ChIP with anti-FLAG antibody as described in Materials and Methods. The amount of rpoB and atpL promoter DNAs in the precipitated chromosomal DNA was analyzed by qPCR. Figure 7 shows that the degree of promoter association of the HrdB-containing RNAP was significantly reduced by ∼40% in the K259R mutation for both promoters. Under assumption that the K259R mutant HrdB was defective only in acetylation, but functional as a non-acetylated variant, these and the above results together suggest that K259 acetylation of HrdB enhances the activity of the housekeeping RNAP through the facilitation of HrdB–RNAP core interaction and binding to target promoters.
Figure 7.
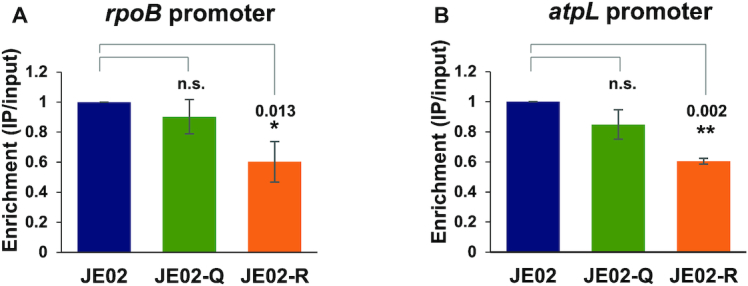
Effect of K259 mutations of HrdB on housekeeping promoter binding in vivo. Cells (JE02, JE02-Q, and JE02-R) were grown to the early exponential phase and subjected to ChIP with anti-FLAG antibody against FLAG-tagged HrdB. Gene-specific primers for the rpoB (A) and the atpL (B) promoters were used to amplify the qPCR amplicons to measure the binding of HrdB-containing RNAP to its cognate promoters. The amplicon enrichment ratios relative to that in JE02 are shown with error bars representing the standard errors from three independent experiments. Statistical significance between the groups is indicated, based on a P value of less than 0.05 (*) or 0.01 (**) by using Student's t-test. The difference between JE02 and JE02-Q is not statistically significant (n.s.).
DISCUSSION
In the present study, we demonstrated that K259 of the housekeeping sigma factor HrdB is constitutively acetylated during the growth of S. venezuelae and that K259 acetylation facilitates the association between HrdB and the RNAP core enzyme and subsequent promoter binding by the housekeeping RNAP holoenzyme. More importantly, based on LC-MS/MS analysis, almost all of the K259 residues of the total HrdB proteins were acetylated throughout growth, despite the slightly lower level (i.e. ∼80%) of K259 acetylation observed at the early exponential growth phase in the cell lysate. Having been examined on two target genes (rpoB and atpL) of HrdB, it is evident that K259-acetylation enhances promoter binding of the housekeeping RNAP holoenzyme. These findings might provide an insight into a new regulatory module to enhance and/or fine-tune the activity of the bacterial sigma factors, via PTM.
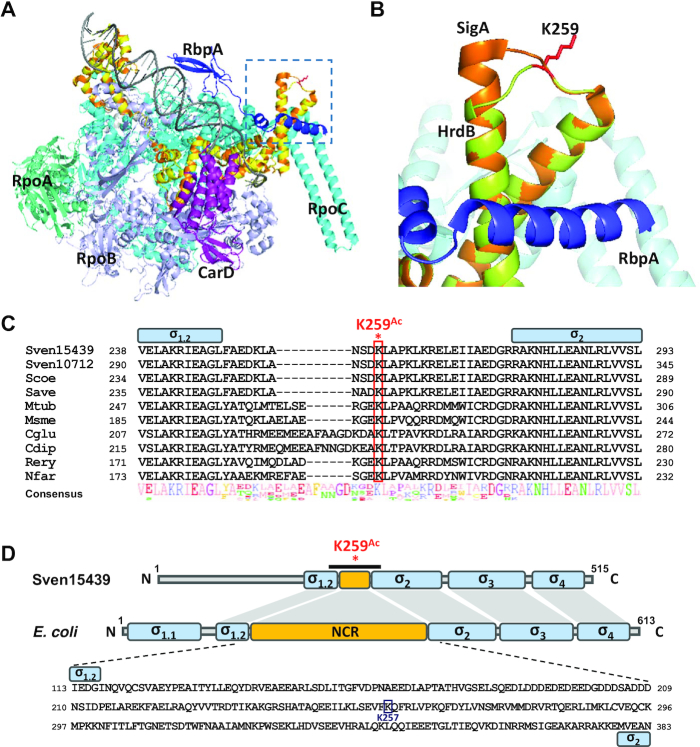
In Actinobacteria, transcription initiation complexes appear less stable than those in E. coli, so that they require auxiliary factors such as RbpA and CarD, presumably for the structural stability necessary for optimal activity (54,55). We have mapped the position of K259 in HrdB by structural modeling based on the housekeeping RNAP complex from M. smegmatis (PDB ID 5TW1) and Thermus aquaticus (PDB ID 4XLR) (55,67), which resides in a loop between two helices involved in RbpA binding (Figure 8A and B). In this structural model (Figure 8B), the K259 residue of HrdB is positioned on the surface facing outward in the RNAP complex, with its distance to RbpA too great to allow direct interaction with RbpA. RbpA and CarD have been proposed to facilitate the formation of open promoter complex from the closed one formed between promoter DNA and RNA polymerase holoenzyme during transcription initiation (67). Since K259 acetylation facilitates interaction between HrdB and RNAP that occurs prior to the closed complex formation, its role differs from those of RbpA and CarD. However, it is open to question whether the action of RbpA and/or CarD depends on K259 acetylation or not. It will be interesting to unravel this relationship in the future.
Figure 8.
Location and conservation of the K259 residue. (A) Partial structure of the RNAP complex modeled by SWISS-MODEL. The model is based on the structure of the transcription initiation complexes in Mycobacterium smegmatis and Thermus aquaticus from RSC Protein Data Bank (5TW1 and 4XLR) as the template. The Sven15439 HrdB is shown in yellow, which almost overlaps with the SigA (orange) of M. smegmatis. RpoA, RpoB, RpoC, CarD, and RpbA of M. smegmatis are shown in green–cyan, light-purple, cyan, purple and blue, respectively, with DNA in gray. The dotted box denotes the structure containing the K259, as magnified in (B). (B) Location of K259 in the modeled RNAP complex. The K259 residue of HrdB is shown as a red-stick. (C) Sequence alignment of the NCR regions in Actinobacteria. Multiple sequence alignment of the amino acids of the HrdB NCR is shown: Sven15439, S. venezuelae ATCC15439; Sven10712, S. venezuelae ATCC10712; Scoe, S. coelicolor M145; Save, S. avermitilis ATCC31267; Mtub, Mycobacterium tuberculosis H37Rv; Msme, M. smegmatis MC2 155; Cglu, Corynebacterium glutamicum R; Cdip, C. diphtheriae ATCC700971; Rery, Rhodococcus erythropolis PR4; Nfar, Nocardia farcinica IFM10152. The conserved Lys is indicated in red box and NCR in yellow box. (D) Domain similarity of Sven15439 HrdB and E. coli RpoD. Each domain and NCR are shown in blue and yellow boxes, respectively. K259Ac indicates the acetylated residue in the NCR of HrdB. The black bar indicates the region whose sequences are shown in (C). The amino acid sequences of NCR in E. coli are represented with the acetylated residue in the NCR of RpoD indicated.
Based on amino acid sequence alignment (Figure 8C), the Lys residue corresponding to K259 in the non-conserved region (NCR) between regions 1.2 and 2 of HrdB is well conserved in Streptomyces as well as other Actinobacteria (Corynebacterium, Mycobacterium, Nocardia and Rhodococcus). Based on this conservation, it is tempting to propose that the Lys residue in the housekeeping SigA/HrdB might also be acetylated in other Actinobacteria as well. Whether K259-type acetylation occurs in the short NCR of housekeeping sigma factors and thereby contributes to the enhancement of RNAP activity in other Actinobacteria warrants further investigation. Careful investigation of the acetylome data produced from E. coli studies enabled us to find that RpoD K257 is acetylated (30,33,34,68–70) and resides in the long NCR of RpoD (Figure 8D). Interestingly, both K257 and K259 residues appear in a similar position on the surface facing outside in the RNAP holoenzyme (Supplementary Figure S7). However, the abundance and possible role of the acetylation of K257 in RpoD of E. coli has not been revealed.
A recent cryo-EM study on the RNAP open complex revealed that the RpoD NCR could interact with DNA, suggesting that this interaction presumably affects RNAP activity during open complex formation (71). Although the exact role of the NCR in the function of housekeeping sigma factors remains to be discovered, our study proposes that lysine acetylation in NCR might be an important determinant in the function of housekeeping RNAP, especially in Actinobacteria that require auxiliary factors for RNAP to initiate transcription. Further investigation is needed to reveal the mechanism of how K259 acetylation of HrdB facilitates RNAP core-sigma association, promoter selection, and transcription initiation.
Protein lysine acetylation is catalyzed enzymatically by lysine acetyltransferases and/or non-enzymatically by acetyl donors such acetyl phosphate (AcP) (72). Based on the sequence homology, 54 putative acetyltransferases have been found on the Sven15439 genome, which include PatA (AQF52_5916), PatB (AQF52_1268), AcuA (AQF52_3408), RimI (AQF52_4872) and Eis (AQF52_2862 and AQF52_4352), with two potential deacetylases such as CobB (AQF52_6342) and AcuC (AQF52_3609). Furthermore, the MYM medium contains enough sugar to support non-enzymatic acetylation through AcP, since Sven15439 possesses the genes encoding phosphotransacetylase (Pta, encoding as AQF52_5523) and acetate kinase (AckA, AQF52_5522) for AcP metabolism. Thus, it needs to be further verified which acetylation mechanism(s) might preponderantly work for the lysine acetylation of HrdB during the growth.
A variety of sigma factors need to be orchestrated to ensure the complex life cycle of S. venezuelae that sporulates and produces antibiotics when cells enter the stationary phase. Competition with a differing array of alternative sigma factors to bind core RNAP will balance the level of the housekeeping RNAP holoenzyme to the optimal level to support sporulation and antibiotic production when nutrients become depleted. Thus, the interplay of PTM, especially lysine acetylation, with the sigma-core interaction among multiple sigma factors, will be a challenging puzzle to solve to understand gene regulation in antibiotic production, stress response, and environmental adaption.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD015174.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Andriy Luzhetskyy (Helmholtz Institute for Pharmaceutical Research) for providing the pGUS plasmid. We also thank Jeesoo Kim (Center for RNA Research, IBS) for LC–MS analysis.
Notes
Present address: Ji-Eun Kim, Department of Microbiology and Immunology, Yonsei University College of Medicine, Seoul 03722, Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intelligent Synthetic Biology Center of Global Frontier Project Grant [2011-0031960]; National Research Foundation of Korea (NRF) Grant [NRF-2017R1A2B3005239]. Funding for open access charge: National Research Foundation of Korea (NRF) Grant.
Conflict of interest statement. The authors declare no conflict of interest. The funding sponsors had no role in any of the following: the design of the study, the collection, analyses or interpretation of data, the writing of the manuscript and the decision to publish the results.
REFERENCES
- 1. Drazic A., Myklebust L.M., Ree R., Arnesen T.. The world of protein acetylation. Biochim. Biophys. Acta. 2016; 1864:1372–1401. [DOI] [PubMed] [Google Scholar]
- 2. Christensen D.G., Baumgartner J.T., Xie X., Jew K.M., Basisty N., Schilling B., Kuhn M.L.. Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio. 2019; 10:e02708-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hentchel K.L., Escalante-Semerena J.C.. Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 2015; 79:321–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ren J., Sang Y., Lu J., Yao Y.F.. Protein acetylation and its role in bacterial virulence. Trends Microbiol. 2017; 25:768–779. [DOI] [PubMed] [Google Scholar]
- 5. Carabetta V.J., Cristea I.M.. Regulation, function, and detection of protein acetylation in bacteria. J. Bacteriol. 2017; 199:e00107-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. VanDrisse C.M., Escalante-Semerena J.C.. Protein acetylation in bacteria. Annu. Rev. Microbiol. 2019; 73:111–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gardner J.G., Grundy F.J., Henkin T.M., Escalante-Semerena J.C.. Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD+ involvement in Bacillus subtilis. J. Bacteriol. 2006; 188:5460–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. You D., Yin B.C., Li Z.H., Zhou Y., Yu W.B., Zuo P., Ye B.C.. Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:6653–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vergnolle O., Xu H., Tufariello J.M., Favrot L., Malek A.A., Jacobs W.R. Jr., Blanchard J.S.. Post-translational acetylation of mbta modulates mycobacterial siderophore biosynthesis. J. Biol. Chem. 2016; 291:22315–22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ishigaki Y., Akanuma G., Yoshida M., Horinouchi S., Kosono S., Ohnishi Y.. Protein acetylation involved in streptomycin biosynthesis in Streptomyces griseus. J. Proteomics. 2017; 155:63–72. [DOI] [PubMed] [Google Scholar]
- 11. Starai V.J., Escalante-Semerena J.C.. Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 2004; 61:2020–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakayasu E.S., Burnet M.C., Walukiewicz H.E., Wilkins C.S., Shukla A.K., Brooks S., Plutz M.J., Lee B.D., Schilling B., Wolfe A.J. et al.. Ancient regulatory role of lysine acetylation in central metabolism. mBio. 2017; 8:e01894-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones S.E., Leong V., Ortega J., Elliot M.A.. Development, antibiotic production, and ribosome assembly in Streptomyces venezuelae are impacted by RNase J and RNase III deletion. J. Bacteriol. 2014; 196:4253–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song L., Wang G., Malhotra A., Deutscher M.P., Liang W.. Reversible acetylation on Lys501 regulates the activity of RNase II. Nucleic Acids Res. 2016; 44:1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Q., Zhou A., Li S., Ni J., Tao J., Lu J., Wan B., Li S., Zhang J., Zhao S. et al.. Reversible lysine acetylation is involved in DNA replication initiation by regulating activities of initiator DnaA in Escherichia coli. Sci. Rep. 2016; 6:30837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Q., Zhou Y.N., Jin D.J., Tse-Dinh Y.C.. Deacetylation of topoisomerase I is an important physiological function of E. coli CobB. Nucleic Acids Res. 2017; 45:5349–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carabetta V.J., Greco T.M., Tanner A.W., Cristea I.M., Dubnau D.. Temporal regulation of the Bacillus subtilis Acetylome and evidence for a role of MreB Acetylation in cell wall growth. mSystems. 2016; 1:e00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barak R., Prasad K., Shainskaya A., Wolfe A.J., Eisenbach M.. Acetylation of the chemotaxis response regulator CheY by acetyl-CoA synthetase purified from Escherichia coli. J. Mol. Biol. 2004; 342:383–401. [DOI] [PubMed] [Google Scholar]
- 19. Baron S., Eisenbach M.. CheY acetylation is required for ordinary adaptation time in Escherichia coli chemotaxis. FEBS Lett. 2017; 591:1958–1965. [DOI] [PubMed] [Google Scholar]
- 20. Li R., Gu J., Chen Y.Y., Xiao C.L., Wang L.W., Zhang Z.P., Bi L.J., Wei H.P., Wang X.D., Deng J.Y. et al.. CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol. Microbiol. 2010; 76:1162–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thao S., Chen C.S., Zhu H., Escalante-Semerena J.C.. Nepsilon-lysine acetylation of a bacterial transcription factor inhibits Its DNA-binding activity. PLoS One. 2010; 5:e15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu L.I., Chi B.K., Kuhn M.L., Filippova E.V., Walker-Peddakotla A.J., Basell K., Becher D., Anderson W.F., Antelmann H., Wolfe A.J.. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J. Bacteriol. 2013; 195:4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang H., Sha W., Liu Z., Tang T., Liu H., Qin L., Cui Z., Chen J., Liu F., Zheng R et al.. Lysine acetylation of DosR regulates the hypoxia response of Mycobacterium tuberculosis. Emerg. Microbes Infect. 2018; 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wei W., Liu T., Li X., Wang R., Zhao W., Zhao G., Zhao S., Zhou Z.. Lysine acetylation regulates the function of the global anaerobic transcription factor FnrL in Rhodobacter sphaeroides. Mol. Microbiol. 2017; 104:278–293. [DOI] [PubMed] [Google Scholar]
- 25. Sang Y., Ren J., Qin R., Liu S., Cui Z., Cheng S., Liu X., Lu J., Tao J., Yao Y.F.. Acetylation regulating protein stability and DNA-binding ability of HilD, thus modulating Salmonella typhimurium virulence. J. Infect. Dis. 2017; 216:1018–1026. [DOI] [PubMed] [Google Scholar]
- 26. Ren J., Sang Y., Tan Y., Tao J., Ni J., Liu S., Fan X., Zhao W., Lu J., Wu W. et al.. Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog. 2016; 12:e1005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis R., Ecija-Conesa A., Gallego-Jara J., de Diego T., Filippova E.V., Kuffel G., Anderson W.F., Gibson B.W., Schilling B., Canovas M. et al.. An acetylatable lysine controls CRP function in E. coli. Mol. Microbiol. 2018; 107:116–131. [DOI] [PubMed] [Google Scholar]
- 28. Guebel D.V., Torres N.V.. Influence of glucose availability and CRP acetylation on the genome-wide transcriptional response of Escherichia coli: assessment by an optimized factorial microarray analysis. Front. Microbiol. 2018; 9:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogura M., Asai K.. Glucose induces ECF sigma factor genes, sigX and sigM, Independent of cognate anti-sigma factors through acetylation of CshA in Bacillus subtilis. Front. Microbiol. 2016; 7:1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Castano-Cerezo S., Bernal V., Post H., Fuhrer T., Cappadona S., Sanchez-Diaz N.C., Sauer U., Heck A.J., Altelaar A.F., Canovas M.. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol. Syst. Biol. 2014; 10:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lima B.P., Antelmann H., Gronau K., Chi B.K., Becher D., Brinsmade S.R., Wolfe A.J.. Involvement of protein acetylation in glucose-induced transcription of a stress-responsive promoter. Mol. Microbiol. 2011; 81:1190–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lima B.P., Thanh Huyen T.T., Basell K., Becher D., Antelmann H., Wolfe A.J.. Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J. Biol. Chem. 2012; 287:32147–32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weinert B.T., Iesmantavicius V., Wagner S.A., Scholz C., Gummesson B., Beli P., Nystrom T., Choudhary C.. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell. 2013; 51:265–272. [DOI] [PubMed] [Google Scholar]
- 34. Schilling B., Christensen D., Davis R., Sahu A.K., Hu L.I., Walker-Peddakotla A., Sorensen D.J., Zemaitaitis B., Gibson B.W., Wolfe A.J.. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 2015; 98:847–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y et al.. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010; 327:1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pan J., Ye Z., Cheng Z., Peng X., Wen L., Zhao F.. Systematic analysis of the lysine acetylome in Vibrio parahemolyticus. J. Proteome Res. 2014; 13:3294–3302. [DOI] [PubMed] [Google Scholar]
- 37. Ouidir T., Cosette P., Jouenne T., Hardouin J.. Proteomic profiling of lysine acetylation in Pseudomonas aeruginosa reveals the diversity of acetylated proteins. Proteomics. 2015; 15:2152–2157. [DOI] [PubMed] [Google Scholar]
- 38. Crosby H.A., Pelletier D.A., Hurst G.B., Escalante-Semerena J.C.. System-wide studies of N-lysine acetylation in Rhodopseudomonas palustris reveal substrate specificity of protein acetyltransferases. J. Biol. Chem. 2012; 287:15590–15601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kosono S., Tamura M., Suzuki S., Kawamura Y., Yoshida A., Nishiyama M., Yoshida M.. Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One. 2015; 10:e0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu L., Wang G., Song L., Lv B., Liang W.. Acetylome analysis reveals the involvement of lysine acetylation in biosynthesis of antibiotics in Bacillus amyloliquefaciens. Sci. Rep. 2016; 6:20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xie L., Wang X., Zeng J., Zhou M., Duan X., Li Q., Zhang Z., Luo H., Pang L., Li W. et al.. Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int. J. Biochem. Cell Biol. 2015; 59:193–202. [DOI] [PubMed] [Google Scholar]
- 42. Guo J., Wang C., Han Y., Liu Z., Wu T., Liu Y., Liu Y., Tan Y., Cai X., Cao Y. et al.. Identification of lysine acetylation in Mycobacterium abscessus using LC-MS/MS after immunoprecipitation. J. Proteome Res. 2016; 15:2567–2578. [DOI] [PubMed] [Google Scholar]
- 43. Liao G., Xie L., Li X., Cheng Z., Xie J.. Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J. Proteomics. 2014; 106:260–269. [DOI] [PubMed] [Google Scholar]
- 44. Huang D., Li Z.H., You D., Zhou Y., Ye B.C.. Lysine acetylproteome analysis suggests its roles in primary and secondary metabolism in Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 2015; 99:1399–1413. [DOI] [PubMed] [Google Scholar]
- 45. Mo R., Yang M., Chen Z., Cheng Z., Yi X., Li C., He C., Xiong Q., Chen H., Wang Q. et al.. Acetylome analysis reveals the involvement of lysine acetylation in photosynthesis and carbon metabolism in the model cyanobacterium Synechocystis sp. PCC 6803. J. Proteome Res. 2015; 14:1275–1286. [DOI] [PubMed] [Google Scholar]
- 46. Kim J.-E., Choi J.S., Roe J.H.. Growth and differentiation properties of pikromycin-producing Streptomyces venezuelae ATCC15439. J. Microbiol. 2019; 57:388–395. [DOI] [PubMed] [Google Scholar]
- 47. Song J.Y., Yoo Y.J., Lim S.K., Cha S.H., Kim J.E., Roe J.H., Kim J.F., Yoon Y.J.. Complete genome sequence of Streptomyces venezuelae ATCC 15439, a promising cell factory for production of secondary metabolites. J. Biotechnol. 2016; 219:57–58. [DOI] [PubMed] [Google Scholar]
- 48. Helmann J.D. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002; 46:47–110. [DOI] [PubMed] [Google Scholar]
- 49. Kim J.-E. Expression and Regulation of RNA Polymerase Sigma Factors in Steptomyces venezuelae. 2019; Doctor of Philosophy, Seoul National University. [Google Scholar]
- 50. Buttner M.J., Chater K.F., Bibb M.J.. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2). J. Bacteriol. 1990; 172:3367–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hahn M.Y., Bae J.B., Park J.H., Roe J.H.. Isolation and characterization of Streptomyces coelicolor RNA polymerase, its sigma, and antisigma factors. Methods Enzymol. 2003; 370:73–82. [DOI] [PubMed] [Google Scholar]
- 52. Kang J.G., Hahn M.Y., Ishihama A., Roe J.H.. Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2). Nucleic Acids Res. 1997; 25:2566–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smidova K., Zikova A., Pospisil J., Schwarz M., Bobek J., Vohradsky J.. DNA mapping and kinetic modeling of the HrdB regulon in Streptomyces coelicolor. Nucleic Acids Res. 2019; 47:621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tabib-Salazar A., Liu B., Doughty P., Lewis R.A., Ghosh S., Parsy M.L., Simpson P.J., O’Dwyer K., Matthews S.J., Paget M.S.. The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase. Nucleic Acids Res. 2013; 41:5679–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bae B., Chen J., Davis E., Leon K., Darst S.A., Campbell E.A.. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. eLife. 2015; 4:e08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Otani H., Higo A., Nanamiya H., Horinouchi S., Ohnishi Y.. An alternative sigma factor governs the principal sigma factor in Streptomyces griseus. Mol. Microbiol. 2013; 87:1223–1236. [DOI] [PubMed] [Google Scholar]
- 57. Kim M.S., Dufour Y.S., Yoo J.S., Cho Y.B., Park J.H., Nam G.B., Kim H.M., Lee K.L., Donohue T.J., Roe J.H.. Conservation of thiol-oxidative stress responses regulated by SigR orthologues in actinomycetes. Mol. Microbiol. 2012; 85:326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A.. Practical Streptomyces Genetics. 2000; Norwich: John Innes Foundation. [Google Scholar]
- 59. Bibb M.J., Domonkos A., Chandra G., Buttner M.J.. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by sigmaBldN and a cognate anti-sigma factor, RsbN. Mol. Microbiol. 2012; 84:1033–1049. [DOI] [PubMed] [Google Scholar]
- 60. Bierman M., Logan R., O’Brien K., Seno E.T., Rao R.N., Schoner B.E.. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992; 116:43–49. [DOI] [PubMed] [Google Scholar]
- 61. Kim H.M., Ahn B.E., Lee J.H., Roe J.H.. Regulation of a nickel-cobalt efflux system and nickel homeostasis in a soil actinobacterium Streptomyces coelicolor. Metallomics. 2015; 7:702–709. [DOI] [PubMed] [Google Scholar]
- 62. Babcock M.J., Buttner M.J., Keler C.H., Clarke B.R., Morris R.A., Lewis C.G., Brawner M.E.. Characterization of the rpoC gene of Streptomyces coelicolor A3(2) and its use to develop a simple and rapid method for the purification of RNA polymerase. Gene. 1997; 196:31–42. [DOI] [PubMed] [Google Scholar]
- 63. Brown K.L., Wood S., Buttner M.J.. Isolation and characterization of the major vegetative RNA polymerase of Streptomyces coelicolor A3(2); renaturation of a sigma subunit using GroEL. Mol. Microbiol. 1992; 6:1133–1139. [DOI] [PubMed] [Google Scholar]
- 64. Wang X., Hayes J.J.. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol. Cell. Biol. 2008; 28:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. He M., Zhang L., Wang X., Huo L., Sun L., Feng C., Jing X., Du D., Liang H., Liu M. et al.. Systematic analysis of the functions of lysine acetylation in the regulation of tat activity. PLoS One. 2013; 8:e67186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Landick R., Krek A., Glickman M.S., Socci N.D., Stallings C.L.. Genome-Wide mapping of the distribution of CarD, RNAP sigmaA, and RNAP beta on the Mycobacterium smegmatis chromosome using chromatin immunoprecipitation sequencing. Genomics Data. 2014; 2:110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hubin E.A., Fay A., Xu C., Bean J.M., Saecker R.M., Glickman M.S.. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. eLife. 2017; 6:e22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu B.J., Kim J.A., Moon J.H., Ryu S.E., Pan J.G.. The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 2008; 18:1529–1536. [PubMed] [Google Scholar]
- 69. Zhang K., Zheng S., Yang J.S., Chen Y., Cheng Z.. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res. 2013; 12:844–851. [DOI] [PubMed] [Google Scholar]
- 70. Kuhn M.L., Zemaitaitis B., Hu L.I., Sahu A., Sorensen D., Minasov G., Lima B.P., Scholle M., Mrksich M., Anderson W.F. et al.. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One. 2014; 9:e94816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Narayanan A., Vago F.S., Li K., Qayyum M.Z., Yernool D., Jiang W., Murakami K.S.. Cryo-EM structure of Escherichia coli sigma70 RNA polymerase and promoter DNA compleux revealed a role of sigma non-conserved region during the open complex formation. J. Biol. Chem. 2018; 293:7367–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Christensen D.G., Xie X., Basisty N., Byrnes J., McSweeney S., Schilling B., Wolfe A.J.. Post-translational protein acetylation: an elegant mechanism for bacteria to dynamically regulate metabolic functions. Front. Microbiol. 2019; 10:1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD015174.