Abstract
The human Microprocessor complex cleaves primary microRNA (miRNA) transcripts (pri-miRNAs) to initiate miRNA synthesis. Microprocessor consists of DROSHA (an RNase III enzyme), and DGCR8. DROSHA contains two RNase III domains, RIIIDa and RIIIDb, which simultaneously cleave the 3p- and 5p-strands of pri-miRNAs, respectively. In this study, we show that the internal loop located in the lower stem of numerous pri-miRNAs selectively inhibits the cleavage of Microprocessor on their 3p-strand, thereby, facilitating the single cleavage on their 5p-strand. This single cleavage does not lead to the production of miRNA but instead, it downregulates miRNA expression. We also demonstrate that by manipulating the size of the internal loop in the lower stem of pri-miRNAs, we can alter the ratio of single-cut to double-cut products resulted from the catalysis of Microprocessor, thus changing miRNA production in the in vitro pri-miRNA processing assays and in human cells. Therefore, the oscillating level of the single cleavage suggests another way of regulation of miRNA expression and offers an alternative approach to miRNA knockdown.
INTRODUCTION
Double-stranded RNAs (dsRNAs) are formed by the base pairing of complement sequences. They are essential in biological systems as they play important roles in various cellular processes, such as gene silencing (1–5), repair of DNA breaks (6,7), mRNA stability (8–10), and other RNA metabolic processes (11,12). Human cells produce single-stranded RNA (ssRNA) molecules, and these can generate dsRNAs via two distinct pathways. First, ssRNAs can form intramolecular base-pairs to create a stem–loop structure like that of primary miRNA transcripts (pri-miRNAs) (1,2) or Alu RNA elements (13). Alternatively, two ssRNA strands that share complementary sequences can form an intermolecular dsRNA (3–5,8,9,14–19).
DsRNA cleavage is catalyzed by members of the RNase III ribonuclease family, which were first discovered in Escherichia coli ∼50 years ago (20,21). All RNase IIIs share a highly conserved RNase III domain (RIIID) and execute the concerted cleavage of both RNA strands at specific target sites (22–24). RNase IIIs are present in bacteria and eukaryotes, but not in archaea (22,23), and whereas those from lower organisms (such as bacteria and yeast) usually contain one RIIID, those from higher eukaryotic organisms (such as Giardia intestinalis and humans) often possess two RIIIDs (22). The single RIIID-containing RNase III enzymes function as homodimers in which two monomers share an extensive subunit interface. RNase IIIs containing two RIIIDs, such as DROSHA and DICER in humans, exhibit an intramolecular dimerization between the two domains. In general, each RIIID dimer forms a single catalytic center at which point each RIIID cleaves one of the dsRNA strands, thus producing double cuts on dsRNAs. RNase IIIs recognize different features of dsRNAs to identify and interact with the specific cleavage sites (22–34).
The dsRNA cleavage activity of the human RNase III enzymes, DROSHA and DICER, plays essential roles in multiple cellular RNA pathways (1,2,5). For example, during the biogenesis of miRNA, they sequentially process pri-miRNAs to generate miRNAs that primarily function in gene silencing. DROSHA and its cofactor, DGCR8, which exists as a dimer, form the trimeric Microprocessor complex (28,32,33,35–39). In the nucleus, Microprocessor makes double cuts on pri-miRNAs to produce miRNA precursors, called pre-miRNAs, which are then exported to the cytoplasm. Subsequently, in the cytoplasm, DICER also creates double cuts on pre-miRNAs to generate miRNAs. Apart from its primary cellular substrates (i.e. pri-miRNAs), Microprocessor can also generate double cuts on stem–loop-containing mRNAs (40–47).
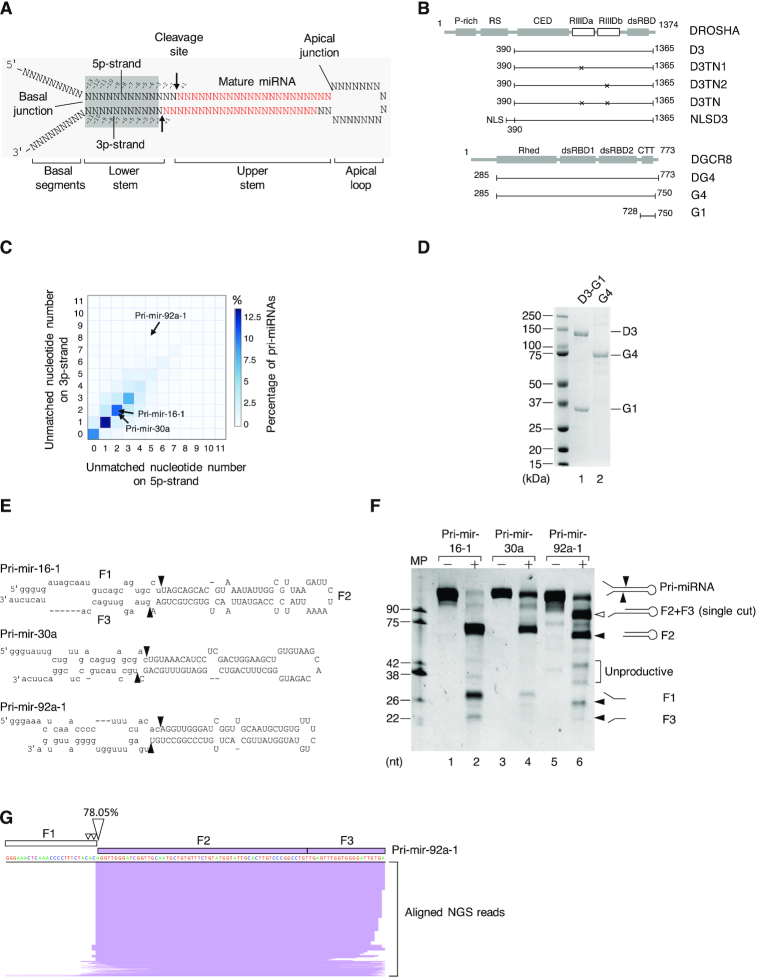
Human pri-miRNAs contain a dsRNA region of ∼35 base pairs (bp), called the stem (48). One end of the stem is flanked by two ssRNA regions (basal 5p- and 3p-RNA segments), whereas the other end connects to the ssRNA apical loop. The boundaries between the dsRNA stem and the ssRNA regions are referred to as the basal and apical junctions (Figure 1A). The stem has two strands, namely, the 5p- and 3p-strands, which are linked to the basal 5p- and 3p-RNA segments, respectively (Figure 1A). In addition, Microprocessor has two RIIIDs, called ‘a’ and ‘b’, which are located in the C-terminal region of DROSHA (Figure 1B), and these cleave the 3p-strand and 5p-strand of pri-miRNAs, respectively. Mutations within the consensus sequence of either of the RIIIDs selectively block one of these cleavages, whereas those in both RIIIDs completely abolish the Microprocessor activity (28,32,33). The Microprocessor complex recognizes various features of pri-miRNAs, and it interacts with and places the RIIIDa and RIIIDb cutting sites approximately 11 and 13 nucleotides (nt) from the basal junction, respectively (2,5,32,33,39,48–51). As a result, Microprocessor makes double cuts on the dsRNA stem of pri-miRNAs, generating pre-miRNAs with 2-nt overhangs at the 3′-end. The correct positioning of Microprocessor on pri-miRNAs is also mediated by a cofactor, called SRSF3, which interacts with the CNNC motif in the 3p-RNA segment of pri-miRNAs, and recruits DROSHA to the basal junction (49,52,53). The double-cut activity executed by the simultaneous actions of both RIIIDa and RIIIDb of Microprocessor is crucial for miRNA biogenesis. Thus, this activity is tightly controlled by multiple regulatory mechanisms (2,5,51,54,55). However, mechanisms that differentially regulate RIIIDa and RIIIDb are still unknown.
Figure 1.
The Microprocessor complex executes a single cleavage on the 5p-strand of pri-miRNAs. (A) Schematic illustration of the pri-miRNA structure. The mature miRNA region is shown in red. The arrows indicate the cleavage sites of Microprocessor. (B) The protein domain structure of DROSHA and DGCR8. P-rich: Proline-rich domain; RS: Arginine/serine-rich domain; CED: central domain; RIIIDa and RIIIDb: RNase III (a and b) domains; dsRBD: double-stranded RNA-binding domain; Rhed: RNA-binding heme domain; CTT: C-terminal tail region; and NLS: Nuclear localization sequence. (C) The percentage of human pri-miRNAs containing different numbers of unmatched nt in their lower stems. The unmatched nt for the 5p- and 3p-strands of the pri-miRNAs were quantified as described in the Materials and Methods. (D) The purified D3-G1 complex and the G4 dimer analyzed by SDS-PAGE. (E) Schematic illustration of pri-mir-16-1, pri-mir-30a, and pri-mir-92a-1. (F) The pri-miRNA processing assays. Five pmol of each pri-miRNA were incubated with 8 pmol each of D3-G1 and the G4 dimer for 2 h at 37°C. (G) The long product (F2+F3) in (F) was cloned and sequenced using next-generation sequencing (NGS), after which NGS reads were aligned with the pri-mir-92a-1 sequence and visualized by IGV. MP: Microprocessor.
Recognition of the basal junction and interaction with the lower stem of pri-miRNAs is known to be crucial for the Microprocessor activity (32,33,48–50). The lower stem of pri-miRNAs is usually an almost perfect dsRNA region, accepting a few mismatches or small bulges (48–50,56). In this study, we investigated the enzymatic activity of Microprocessor on numerous pri-miRNAs that contain multiple mismatches in the lower stem. Interestingly, we demonstrated that the asymmetric internal loops (AIL) in the lower stem of pri-miRNAs prohibited the cleavage of Microprocessor on their 3p-strand, thus permitting Microprocessor to produce a single cleavage on their 5p-strand. In addition, we showed that the size of AIL determined the single cleavage level and, in turn, it affected the pre-miRNA production in the in vitro pri-miRNA processing and miRNA expression in human cells. The 5p-strand single cleavage controlled by the AIL, therefore, suggests an alternative way by which miRNA expression is regulated in human cells.
MATERIALS AND METHODS
Counting the number of unmatched nucleotides in the lower stem of pri-miRNAs
1855 pri-miRNA sequences were extracted from the human genome assembly provided by UCSC (hg38, https://genome.ucsc.edu/) using the genome annotation from miRBase v.21 (57). Each pri-miRNA sequence obtained comprises its pre-miRNA as well as 20-nt extensions on either end of pre-miRNA.
We used the RNAfold program (default settings) from ViennaRNA Package version 2.4.9 (58) to predict RNA secondary structure. In addition, the minimum free energy structure for each pri-miRNA was selected for further calculation. We excluded 295 pri-miRNAs, which contain multiple loops, and counted the number of unmatched nt in the lower stem of the remaining 1,560 pri-miRNAs. The number of unmatched nt was shown in Supplementary Table S1. In addition, the diagrams showing the structures of pri-mir-16-1, pri-mir-30a, and pri-mir-92a-1 (drawn using VARNA) (59) were presented in Supplementary Figure S1A. The positional entropy was estimated for both the 5p- and 3p-strands of the lower stem of each of pri-mir-16-1, pri-mir-30a, and pri-mir-92a-1 using the RNAfold program (Supplementary Figure S1B).
Protein purification of D3-G1, D3TN1-G1, D3TN2-G1, and D3TN-G1
The G1 fragment (amino acids 728–750) of DGCR8 was fused to the C-terminal CFP and 10xHis-tag in the pXC-G1 vector (32). In addition, the D3 fragment (amino acids 390–1,365) of DROSHA was fused to protein G in the pXab-D3 vector (32). The pXC-G1 and pXab-D3 vectors were both gifts from Dr. Narry Kim (Seoul National University). pXab-D3TN1 and pXab-D3TN2 were generated by mutating the glutamic acid residues at positions 1045 and 1222 to glutamine and lysine residues, respectively. The pXG-D3TN vector (containing mutations at positions 1045 and 1222) was also kindly provided by Dr. Narry Kim.
To purify the D3-G1 complex, pXC-G1 and pXab-D3 were co-transfected into 50 dishes (100 mm in diameter) of HEK293E cells. The transfected cells were harvested after 3 days and lysed with 40 ml T500 lysis buffer (20 mM Tris-HCl (pH 7.5), 500 mM NaCl, 4 mM β-mercaptoethanol), supplemented with 2 μg/ml RNase A and protease inhibitor cocktail. The harvested cells were lysed by sonication, and the cell lysate was subsequently collected by centrifugation. The clear lysate obtained was bound with 3 ml Ni-NTA resin. The resin was washed with 50 mL T500 supplemented with 20 mM imidazole, and the proteins were eluted with T500 containing 300 mM imidazole. The resin-eluted proteins were pooled and diluted five times with T0 buffer (20 mM Tris–HCl (pH 7.5), 4 mM β-mercaptoethanol), to reduce the NaCl concentration to 100 mM. The diluted proteins were loaded onto 2 ml Bio-Rad UNOsphere Q beads. The Q beads were washed with T150 buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 4 mM β-mercaptoethanol) and eluted with T500 containing 2 mM DTT. The eluted proteins were subjected to a Superdex 200 Increase 10/300 GL gel filtration column. The peak fractions were collected from the column and glycerol was added to a final concentration of 10%, after which the samples were stored at −80°C. The D3TN1-G1, D3TN2-G1 and D3TN-G1 complexes were purified in a similar way as that described for D3-G1.
Protein purification of the D3-DG4 and NLSD3-DGCR8 complexes
The DG4 fragment of DGCR8 (amino acids 285–773) was fused with the C-terminal GFP tag in the pXG-DG4 vector. pXab-D3 and pXG-DG4 were co-transfected in 150 dishes (100 mm in diameter) of HEK293E cells. The transfected cells were harvested after 4 days, after which the D3-DG4 complex was purified using Ni-NTA resin and Bio-Rad UNOsphere Q, as described for D3-G1.
The NLS sequence encoding 7 amino acids (i.e. PKKKRKV) was inserted upstream of the D3-encoding sequence in the pXab-NLSD3 vector. The DNA coding sequence of full-length DGCR8 was fused to the C-terminal GFP in the pXG-DGCR8 vector. The pXab-NLSD3 and pXG-DGCR8 vectors were then co-transfected in 50 dishes (100 mm in diameter) of HEK293E cells. The transfected cells were harvested after 4 days. The cell pellet was lysed with T10 buffer (20 mM Tris–HCl (pH 7.5), 10 mM NaCl, 4 mM β-mercaptoethanol), supplemented with 0.2% IGEPAL® CA-630 (Sigma). After centrifugation and removal of the cytoplasmic portion of the extract, the nuclear pellet was resuspended in T500 lysis buffer, sonicated, and centrifuged. The clear nuclear extract was then purified using Ni-NTA resin and Bio-Rad UNOsphere Q, as described for D3-G1. The full-length DROSHA-DGCR8 complex was expressed in HEK293E cells, which were transfected with the pCK-DROSHA and pXG-DGCR8 plasmids. The DROSHA in pCK plasmid was tagged with the Flag tag at its C-terminus. The full-length complex was purified using Ni-NTA resin and the anti-Flag-agarose beads.
The dimer form of the G4 fragment of DGCR8 (amino acids 285–750) was purified as described in the previous studies (32,39).
Preparation of the substrates
The pri-miRNAs were synthesized by in vitro transcription (IVT) in 20 μl reaction mixture containing 200 ng double-stranded DNA (dsDNA) templates using the MEGAscript T7 Kit (Invitrogen, AMB13345). The DNA templates were prepared by PCR using the primers shown in Supplementary Table S2. The PCR templates are either the genomic DNAs or the pri-miRNA-containing vectors. The IVT mixture was incubated at 37°C overnight. The reaction was then stopped by the addition of 20 μl 2× TBE-Urea sample buffer (20 mM Tris–HCl (pH 7.5), 20 mM EDTA, 8 M Urea), and heating to 75°C for 5 min. The RNAs were then separated by 10% Urea-PAGE. The RNAs were gel-eluted overnight, precipitated by the addition of isopropanol, washed with 80% ethanol, dried, and finally dissolved in distilled water. The RNA concentration was measured using a Nanodrop spectrophotometer, and the RNA quality was assessed by 10% urea–PAGE before usage. The purified RNAs were stored at −80°C.
Pri-miRNA processing assay
The pri-miRNA processing assay was carried out at 37°C in 10 μl of a buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 10% glycerol, 0.2 μg/μl BSA, 1 mM DTT and 2 mM MgCl2. Five pmol of each of the RNA substrates were used, and the enzyme concentrations and incubation times used were indicated in the figures. The reaction was stopped by the addition of 10 μl 2× TBE–urea sample buffer and it was immediately chilled on ice. The stopped reaction mixture was treated with 20 μg proteinase K at 37°C for 15 min, and then at 50°C for 15 min, and finally at 95°C for 10 min, after which it was quickly chilled on ice before being loaded onto a pre-run 12% urea–PAGE in 1× TBE buffer. The gel was run at 300 V for 40 min and then it was stained with SYBR™ Green II RNA Gel Stain (Invitrogen, S7564) for 5 min and images were captured using the Bio-Rad Gel Doc XR+ system. The RNA band intensities were estimated using Image Lab 3.0.
RNA sequencing for single-cut products of pri-mir-92a-1
The long RNA fragments that resulted from the cleavage of pri-mir-92a-1 by Microprocessor were extracted from the gel and purified with isopropanol. The purified RNAs were ligated with the 3′-adapter (4N-RA3, /5rApp/NN NNT GGA ATT CTC GGG TGC CAA GG/3ddC/) using T4 RNA Ligase 2, truncated KQ (NEB, M0373L). The ligated products were then reverse-transcribed (RT) with the cirRTP primer (/5Phos/NNN NNN GA TCG TCG GAC TGT AGA ACT CTG AAC /iSp18/CC TTG GCA CCC GAG AAT TCC A) using Superscript IV reverse transcriptase (Invitrogen, 18090050). The RT mixture was treated with 0.1 mM NaOH at 95°C for 5 min to remove RNAs. The resulting cDNAs were circularized with the CircLigase™ ssDNA Ligase (Epicentre, CL4115K), after which the circular DNAs were visualized via 18% Urea-PAGE with EtBr and gel-eluted. The purified circular DNAs were amplified by PCR using a pair of primers (RP1, AAT GAT ACG GCG ACC ACC GAG ATC TAC ACG TTC AGA GTT CTA CAG TCC GA and RPI4, CAA GCA GAA GAC GGC ATA CGA GAT TGG TCA GTG ACT GGA GTT CCT TGG CAC CCG AGA ATT CCA), using Phusion High-Fidelity DNA polymerase (Thermo Scientific). The DNAs were sequenced with the Illumina platform (PE150 v2 Kit).
The 3′- and 5′-adapters in the raw reads were removed using cutadapt (cutadapt -a TGG AAT TCT CGG GTG CCA AGG -A GAT CGT CGG ACT GTA GAA CTC TGA AC) (60). We then used fastq-join (default parameter) to join the paired-end reads before removing low quality reads and deduplicating the reads with the same barcodes using fastq_quality_filter (-q 20 -p 90) and fastx_collapser (default parameter) from FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html, version 0.0.13), respectively. After trimming the barcode sequences from both ends (6-nt random sequence at the 5′-end, and 4-nt random sequence at the 3′-end), the trimmed reads were mapped to the pri-mir-92a-1 sequence using BWA (61). Only perfect alignments were visualized by IGV (62).
Some DNAs which were sent for next-generation sequencing were also cloned into the pCR-Blunt II-TOPO vector using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen, 450245). The cloned vectors were subsequently sequenced by Sanger sequencing using the M13 forward (-20) primer (GTA AAA CGA CGG CCA G).
RNA sequencing for single-cut products of other pri-miRNAs
The long RNA fragments that resulted from the Microprocessor cleavage assays were excised from the gel and purified. The purified RNAs were first ligated to the 4N-RA3 using T4 RNA ligase 2, truncated KQ (NEB, M0373L). The RA3-ligated RNAs were separated from the unligated RNAs and 4N-RA3 by 10% urea–PAGE and then gel-purified. The RA3-ligated RNAs were further ligated to the 5′-adapter (RA5-4N, GUU CAG AGU UCU ACA GUC CGA CGA UCN NNN) using T4 RNA ligase I (NEB, M0204L). These two-adapter ligated RNAs were then reverse transcribed using the R-RA3 primer (CCT TGG CAC CCG AGA ATT CCA) and Superscript IV reverse transcriptase (Invitrogen, 18090050), and the synthesized cDNAs were amplified using RP1 and RPI14 (CAA GCA GAA GAC GGC ATA CGA GAT GGA ACT GTG ACT GGA GTT CCT TGG CAC CCG AGA ATT CCA) primers. The final PCR products were subjected to Illumina Nextseq 500 sequencer using High Output 150 cycles kit. The next-generation sequencing (NGS) reads were then processed as described above for the single-cut products of pri-mir-92a-1, except that only read 1 was used. The single-cut products of these pri-miRNAs were selected and visualized by IGV (62). The NGS data were deposited in NCBI Gene Expression Omnibus with accession number GSE138950.
DROSHA fCLIP data processing
The raw sequence libraries of the DROSHA fCLIP-seq were downloaded from GSE93651 (43). The 5′- and 3′-adapters of the raw reads were removed using cutadapt (–O 5 -g TAC AGT CCG ACG ATC -A TGG AAT TCT CGG GTG CCA AGG) (60). We used fastq-join (–p 5) to merge the paired ends and fastx_collapser to remove duplicates (using http://hannonlab.cshl.edu/fastx_toolkit/index.html, version 0.0.13). The 4-nt random barcodes at both ends of each read were also removed. The reference we used for mapping was 1,855 human pri-miRNA sequences in which each pri-miRNA contains its pre-miRNA and 100-nt extensions at both ends of pre-miRNA. The reads were mapped using BWA (61), and then the unique mapped alignments containing fewer than five mismatches were selected. The fCLIP-seq libraries collected from the same cell line were then merged. We examined groups of pri-miRNAs that had cleavage sites consistently identified in the DROSHA fCLIP study and presented in miRbase (43). We regarded a read that shared the 5′-end nucleotide identity with that of pre-miRNAs and extended at least 5 nt from the 3′-end of pre-miRNA as being a potential single-cut product of DROSHA. We only found these potential single-cut products in pri-mir-92a-1 and pri-mir-15a in HEK293T cells with 38 and 10 read counts, respectively.
Constructing the knock-in cells
We created pri-mir-92a-1 AIL3/9 knock-in cells from parental HCT116 cells. We transfected sgRNA (CAA UCC CCA CCA AAC UCA ACG UUU UAG AGC UAG AAA UAG CAA GUU AAA AUA AGG CUA GUC CGU UAU CAA CUU GAA AAA GUG GCA CCG AGU CGG UGC UUU UUU, where the underlined sequence is complementary to the genomic DNA target region) and donor ssDNA (TCT ACA CAG GTT GGG ATC GGT TGC AAT GCT GTG TTT CTG TAT GGT ATT GCA CTT GTC CCG GCC TGT TGA GTT TT TTG GTG GGG ATT GTG ACC AGA AGA TTT AAA ATT AAA TAT TAC TGA AGA TTT CGA CTT CCA CTG TTA AAG TAC, where the underlined sequence shows the inserted nt) using electroporation. A mixture containing 10 μg Cas9 (Thermo scientific, A36498), 5 μg sgRNA, and 50 pmol donor ssDNA was electroporated into 200,000 HCT116 cells. After three days, the electroporated cells were sorted into single cells and cultured for 2 more weeks. The genomic DNAs of the single cells were then isolated, and their edited region was amplified using PCR with F-4-5 (CCA ATC AAA CTG TCC TGT TAC) and R-4-11 (CAC TAA CTC CAA AGA AAG). The DNA sequences of the PCR products were confirmed using Sanger sequencing and next-generation sequencing.
Vector construction, transfection, and real-time PCR
The DNA regions encoding the pri-miRNA sequences were cloned into the pcDNA3 vector. The cloning details were presented in Supplementary Table S3. The sequences of the cloned pcDNA3 vectors were confirmed by Sanger sequencing. Two μg of wild-type (WT) or mutant pri-miRNA vector were co-transfected with 0.5 μg pcDNA3-pri-mir-16-1 in one well of HCT116 cells in a six-well plate using Lipofectamine 3000 transfection reagent (Thermo Scientific, L3000015). The total RNAs were extracted from the transfected cells one day after transfection using the miRNeasy Mini Kit (Qiagen).
Fifty nanograms of total RNA were used in the reverse transcription (RT) step using stem–loop RT primers that were designed for each miRNA, according to the reported method (63). The qPCR for miRNAs was performed using the iTaq Universal SYBR Green Supermix (Bio-rad, 1725121). One μg of total RNA was added to the RT mixture containing 1 μM of each target-specific reverse primer to synthesize cDNA. qPCR was then performed using the iTaq Universal SYBR Green Supermix. The primers used for the RT and qPCR were presented in Supplementary Table S4.
RESULTS
Microprocessor produces the 5p-strand single cleavage products from pri-miRNAs due to the inefficient cleavage on the 3p-strand
Since Microprocessor recognizes and measures ∼11 bp from the basal junction of pri-miRNAs to detect the site of cleavage, it is the lower stem region of the pri-miRNA structure, which is essential for its activity (32,33,48–50). We therefore, hypothesized that Microprocessor might exhibit a different enzymatic activity on those pri-miRNAs that contain many mismatches in the lower stem. Thus, we investigated the possible influence of mismatches in the lower stem of pri-miRNAs on the activity of Microprocessor. We first created a library of human pri-miRNA sequences by extending 20 nt from the 5′- and 3′-ends of 1,855 pre-miRNA sequences. We then predicted the secondary structure of the pri-miRNA sequences using RNAfold (58). Finally, using the folded pri-miRNA structures, we counted the number of unmatched nt on the 5p- (i.e. from positions –3 to –13) or 3p- (i.e. from positions –1 to –11) strand of the lower stem for each pri-miRNA (Figure 1A). As shown in Figure 1C, the majority of pri-miRNAs contain up to three unmatched nt on either strand. This calculation is consistent with previous estimations (48,50,56). However, we observed that many pri-miRNAs contain a large number of (i.e. ranging from 7 to 14) unmatched nt in both strands of the lower stem (Figure 1C). We investigated pri-mir-92a-1 in further detail, and showed that it has 5 and 8 unmatched nt in the 5p- and 3p-strands, respectively. We also estimated the entropy at each nucleotide position in the lower stem using RNAfold (58), and found that in the majority of these positions pri-mir-92a-1 has a significantly higher entropy than that calculated for either pri-mir-16-1 or pri-mir-30a, which both have 4 unmatched nt in their lower stems (Supplementary Figure S1A and B). Such a high number of unmatched nt indicates instability of the secondary structure of pri-miRNAs, and this in turn, might affect the processing of pri-miRNA by Microprocessor.
We examined the cleavage of pri-mir-92a-1 by Microprocessor in further detail. The D3-G1 complex, which consisted of the DROSHA fragment (D3, amino acids 390–1,365) and the DGCR8 fragment (G1, amino acids 728–750), was expressed in HEK293E cells and then purified as described above (Figure 1B and D). The DGCR8 dimer (G4, amino acids 285–750) was purified as described in the previous studies (Figure 1B and D) (32,39). The Microprocessor complex (D3-G4) was reconstituted by mixing the D3-G1 complex with the G4 dimer. Noted that the G4 dimer can efficiently displace the G1 fragment from D3 as previously reported (32,33). Pri-miRNA processing assays were carried out as described above. Consistent with previous studies, we demonstrated that Microprocessor cleaved pri-mir-16-1 and pri-mir-30a to generate three fragments, F1, F2 (pre-miRNA), and F3. This suggests that the reconstituted Microprocessor has a typical RNase III enzyme activity in that it cleaves both of the pri-miRNA strands (Figure 1E and F). We also found that Microprocessor makes double cuts on pri-mir-92a-1, again producing three fragments, F1, F2 (pre-mir-92a-1), and F3 (Figure 1E and F). Microprocessor also exhibited weakly unproductive cleavages on these substrates due to its misorientation on pri-miRNAs (51).
Interestingly, when it cleaved pri-mir-92a-1, Microprocessor created a fragment that was longer than the F2 fragment or pre-mir-92a-1 (Figure 1F). We identified this long fragment by RNA cloning followed by next-generation sequencing (NGS). Most of the reads obtained from the NGS mapped to the region covering F2 and F3, and approximately 78% of them shared the 5′-end nucleotide identity with pre-mir-92a-1 (Figure 1G). We also cloned the long fragment in the pCR Blunt II-TOPO vector and then sequenced it by Sanger sequencing. We obtained 8 out of 10 fragments, which had sequences covering F2 and F3 (Supplementary Figure S1C). This indicates that the longer fragment resulted from a single cleavage of Microprocessor on the 5p-strand of pri-mir-92a-1. Because Microprocessor cleaved pri-mir-92a-1 in two ways, (i.e. a single-cut on the 5p-strand and double cuts on both strands), this suggests that it cleaved the 3p-strand less efficiently than the 5p-strand of this pri-miRNA.
We also tested the activity of Microprocessor on three other pri-miRNAs; pri-mir-217, pri-mir-654, and pri-mir-200b, which all contain multiple unmatched nt in the lower stem. Supplementary Figure S1D and E show that for each of these pri-miRNAs, Microprocessor also produced fragments that were longer than their corresponding pre-miRNAs. This supports the idea that the 5p-strand single cleavage of Microprocessor might occur for various pri-miRNAs that share the common feature of having many unmatched nt in their lower stem.
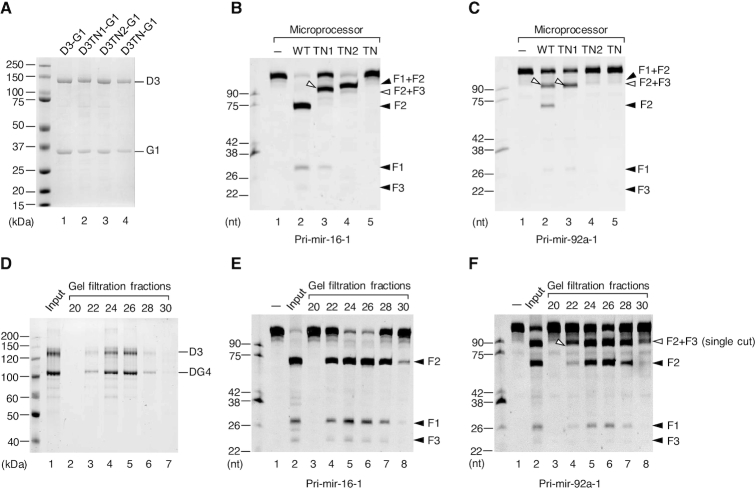
Microprocessor, not contaminant proteins, catalyzes the single cleavage on the 5p-strand of pri-miRNAs
To demonstrate if the 5p-strand single cleavage products observed were resulted from the catalysis of Microprocessor rather than contaminant proteins, we purified the mutant D3-G1 complexes, D3TN1-G1, D3TN2-G1, ands D3TN-G1, which contain mutations at RIIIDa alone, RIIIDb alone, and both RIIIDa and RIIIDb, respectively (Figure 1B). D3TN1 only cleaves the 5p-strand of the pri-miRNA stem using RIIIb, and D3TN2 solely cuts the 3p-strand using RIIIDa, whereas D3TN cannot cleave pri-miRNAs at all. The three mutant D3-G1 complexes were purified as described for the WT D3-G1 complex (Figure 2A). The mutant D3-G1 complexes showed the expected cleavage sites for pri-mir-16-1 (Supplementary Figure S2A), consistent with the previous studies (32,33). To make the mutant Microprocessor complex, we mixed the mutant D3-G1 complex with purified G4 dimer proteins. The three mutant Microprocessor complexes also displayed the expected cleavage pattern with pri-mir-16-1 as shown in Figure 2B. In the case of pri-mir-92a-1, Microprocessor-TN1 produced a fragment (F2+F3) that was equivalent to the size of the longer fragment produced by Microprocessor-WT (Figure 2C). This further confirms that this longer fragment was yielded from the 5p-strand single cleavage by RIIIDb of Microprocessor-WT. In addition, the 3p-strand cleavage activity of the Microprocessor-TN2 complex on pri-mir-92a-1 was only detected when a higher amount of protein was used (Supplementary Figure S2B), indicating the inefficient cleavage on the 3p-strand by RIIIDa. Like pri-mir-92a-1, the Microprocessor-TN1 complex also cleaved other pri-miRNAs, (i.e. pri-mir-217, pri-mir-654, and pri-mir-200b) to generate F2+F3 fragments that have the same length as the long fragments released from the Microprocessor-WT complex (Supplementary Figure S2C). This further supports that the cleavage of Microprocessor on the 3p-strand of these pri-miRNAs was also inefficient.
Figure 2.
The 5p-strand single cleavage products of pri-miRNAs are intrinsically resulted from Microprocessor catalysis. (A) The purified wild-type (WT) and mutant D3-G1 complexes analyzed by SDS-PAGE. (B and C) Pri-miRNA processing assays. Five pmol of pri-mir-16-1 (B) or pri-mir-92a-1 (C) were incubated with 8 pmol each of D3-G1 and the G4 dimer for 2 h at 37°C. (D) The purified D3-DG4 complex was fractionated by a gel filtration column (Superdex 200 Increase 10/300 GL) in the NGC system (Bio-Rad) and was analyzed by SDS-PAGE. (E and F) Pri-miRNA processing assays. Five pmol of pri-mir-16-1 (E) or pri-mir-92a-1 (F) were incubated with 3 μl of each D3-DG4 fraction from (D) for 2 h at 37°C.
Next, we purified the Microprocessor complex (D3-DG4) by co-expressing DROSHA (D3) and DGCR8 (DG4, amino acids 285–773) in human HEK293E cells. The complex was purified using Ni-NTA and UNOsphere Q, as described above, after which the proteins were fractionated on a gel filtration column (Superdex 200 Increase 10/300 GL) using a Bio-Rad NGC system. The protein fractions obtained from the gel filtration assay were analyzed by SDS-PAGE (Figure 2D) and assayed for pri-mir-16-1 (Figure 2E) and pri-mir-92a-1 (Figure 2F). As shown in Figure 2D–F, the amount of pre-miRNA released from each pri-miRNA and the long products of pri-mir-92a-1 correlated with that of the Microprocessor proteins in the gel filtration fractions. This indicates that the single-cut products were resulted from Microprocessor, not contaminant proteins.
Finally, we compared the cleavage pattern of pri-mir-92a-1 with different reconstituted Microprocessor complexes; D3-DG4, D3-G4, and NLSD3-DGCR8. The NLSD3-DGCR8 complex was expressed in the nucleus of HEK293E cells. The NLSD3 was achieved by adding the NLS sequence to the N-terminal of the D3 fragment so that DROSHA (NLSD3) could be expressed in the nucleus (Figure 1B). The NLSD3-DGCR8 complex was then purified as described above. As the NLSD3 and DGCR8 are similar in size, they migrated the same distance by SDS-PAGE (Supplementary Figure S2D). Supplementary Figure S2E showed that all three types of reconstituted Microprocessor complexes released the single-cut fragment, demonstrating that these Microprocessor complexes also exhibited a weak 3p-strand cleavage on pri-mir-92a-1.
In order to determine if the single-cut product is an on-pathway intermediate or an aberrant product of the double cuts by Microprocessor, we attempted to detect the products released due to the action of this complex. We carried out pri-mir-92a-1 processing assays with Microprocessor, and separated the free RNAs and the Microprocessor-RNA complex via native PAGE (Supplementary Figure S3A and B). The free RNAs of the native PAGE were cut, gel-purified, and analyzed via denaturing Urea-PAGE (Supplementary Figure S3C). In the free RNAs, we found both pre-mir-92a-1 and single-cut products. This indicates that the single-cut products are likely to be Microprocessor by-products. Alternatively, we immobilized the Microprocessor complex on IgG beads and carried out pri-mir-92a-1 cleavage assay. We detected both single-cut products and pre-miR-92a-1 in the supernatant fraction. The ratios of single-cut/double-cut (sc/dc) products were similar when comparing the supernatant fractions of the on-bead assay and the in-solution cleavage assay (Supplementary Figure S3D and E). This suggests that the single-cut products were released from Microprocessor as pre-miRNAs. We also analyzed the DROSHA-associated pri-miRNAs from the DROSHA formaldehyde crosslinking immunoprecipitation data (43), and found the existence of single-cut products of pri-mir-92a-1 (Supplementary Figure S3F).
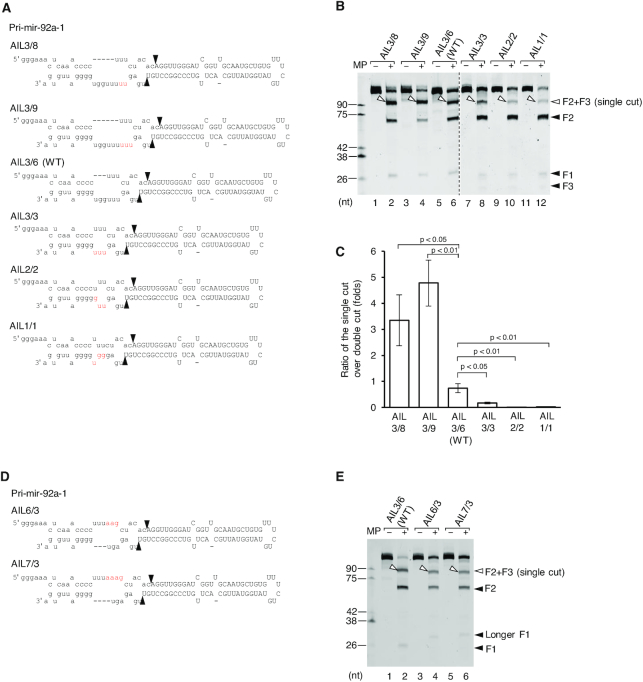
The large-sized asymmetric internal loop (AIL) in the lower stem of pri-miRNAs inhibits the 3p-strand cleavage of Microprocessor
Since Microprocessor generates the 5p-strand single cleavage products from those pri-miRNAs that possess the asymmetric internal loop (AIL) in their lower stem, we hypothesized that the AILs with different sizes might halt the 3p-strand cleavage at the various levels. Therefore, we made different mutations in the lower stem of pri-mir-92a-1 and tested the mutated pri-mir-92a-1 variants with Microprocessor in a series of processing assays (Figure 3A). We named a pri-miRNA variant in this study ‘AILx/y’ whereby x and y are the numbers of nt in the internal loop on the 5p- and 3p-strands, respectively. We observed that the different-sized AILs in the lower stem showed distinct ratios of single-cut to double-cut (sc/dc) products (Figure 3B and C). The introduction of a few additional nt on the 3p-strand to enlarge the internal loop at the mismatched site, significantly increased the 5p-strand single-cut products. As shown in Figure 3B and C, the sc/dc ratios of AIL3/9, AIL3/8, and AIL3/6 (WT) were 4.8, 3.3 and 0.7, respectively. In contrast, removal of the internal loop significantly decreased the sc/dc ratio such that the sc/dc ratio of AIL3/3 was ∼0.2, compared with that of AIL3/6, which was 0.7 (Figure 3B and C). In addition, the pri-mir-92a-1 variants containing only 1 (AIL1/1) or 2 (AIL2/2) mismatches almost entirely lost their single-cut activity (i.e. with sc/dc ratios ∼0.0, Figure 3B and C). This suggests that a high number of mismatches and an asymmetric internal loop (AIL) in the lower stem effectively inhibit the 3p-strand cleavage of Microprocessor, thereby, releasing the 5p-strand cleavage products. The similar effects of the AIL size on the level of the 5p-strand single cleavage were also observed with Microprocessor (NLSD3-DGCR8) at different concentrations of MgCl2 (Supplementary Figure S3G) or with the full-length Microprocessor complex (Supplementary Figure S3H and I). In addition to the short pri-mir-92a-1, we also examined the longer pri-mir-92a-1 variants that are 176 nt in length (i.e. F1: 47 nt, F2: 59 nt, F3: 70 nt). We found that the Microprocessor-WT complex also exhibited different levels of the 5p-strand cleaved-products for these long pri-mir-92a-1 variants, again depending on the AIL size (Supplementary Figure S3J).
Figure 3.
The size of asymmetric internal loop in the lower stem affects the level of the 5p-strand single cleavage of Microprocessor. (A and D) Diagram of pri-mir-92a-1 and its variants. The mutated nucleotides are in red. (B and E) Pri-miRNA processing assays. Five pmol of each pri-miRNA were incubated with 8 pmol each of D3-G1 and the G4 dimer for 2 h at 37°C. (C) Quantification of the pri-miRNA processing data from (B). The pri-miRNA processing assays in (B) were repeated three times. The single-cut (sc) to double-cut (dc) activity ratio (sc/dc ratio) was estimated as the ratio of the single-cut to double-cut product (F2) band density in the gel. The p-values of the one-tailed t-test for the sc/dc ratios estimated from three replicates were shown. MP: Microprocessor.
Next, we switched the asymmetry from the 3p- to the 5p-strand of pri-mir-92a-1 (Figure 3D), and found that Microprocessor still preferentially cleaved the 5p-strand of these pri-mir-92a-1 variants. Noted that, the single-cut products of pri-mir-92a-1 AIL6/3 and AIL7/3 were shorter than that of pri-mir-92a-1 WT, since 3 nt were deleted from the 3p-strand of the lower stem (i.e. compare lanes 4 or 6 with lane 2, Figure 3E). In contrast, the 3 nt and 4 nt-addition in the 5p-strand of AIL6/3 and AIL7/3, respectively, resulted in longer F1 (i.e., Figure 3E, compare lane 4 or 6 with lane 2). This suggests that the AIL hinders the 3p-strand cleavage of Microprocessor regardless of its asymmetric directions toward the 5p- or 3p-strand.
The 3p-strand cleavage of Microprocessor is restrained on numerous pri-miRNAs containing a large AIL in their lower stem
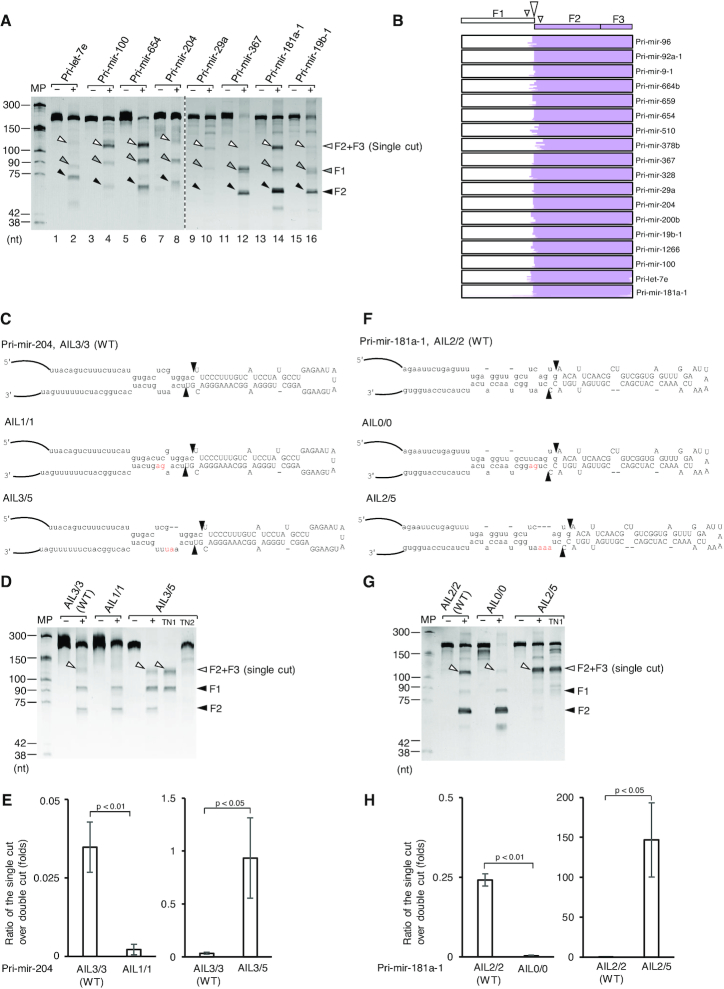
In order to determine if an AIL in the lower stem of pri-miRNAs might suppress the 3p-strand cleavage on various pri-miRNA backbones, we compared the activity of Microprocessor with 17 more pri-miRNAs containing an AIL (AIL-pri-miRNAs) with that of 4 more pri-miRNAs, which do not contain an AIL (nonAIL-pri-miRNAs) in the lower stem. We found that Microprocessor only showed double-cut products (pre-miRNAs) from nonAIL-pri-miRNAs (Supplementary Figure S4A). In contrast, Microprocessor produced pre-miRNAs and longer fragments from AIL-pri-miRNAs (Figure 4A and Supplementary Figure S4B). The size of these long fragments is similar to that of the F2+F3 fragments resulted from the 5p-strand single cleavage of Microprocessor-TN1 (Supplementary Figure S4C). We cloned and sequenced the long RNA fragments and found that the majority of the NGS reads were mapped to the F2+F3 fragments (Figure 4B and Supplementary Figure S4D), suggesting that they were the 5p-strand cleaved-products of Microprocessor. Noted that the 3p-strand single cleavage by Microprocessor-TN2 seemed to be weaker than the 5p-strand single cleavage by Microprocessor-TN1 on these tested AIL-pri-miRNAs (Supplementary Figure S4C). These data indicate that AIL is an essential element, which inhibits the 3p-strand cleavage of Microprocessor, and thus, augments the 5p-strand cleaved-products from numerous pri-miRNAs. Next, we decreased or increased the size of the AILs of pri-mir-204 and pri-mir-181a-1, pri-mir-217, and pri-mir-133-1, and tested these pri-miRNA variants with Microprocessor. We demonstrated that the larger and smaller AILs augmented and reduced the sc/dc ratios, respectively (Figure 4C–H and Supplementary Figure S4E, F).
Figure 4.
Microprocessor displays the 5p-strand single cleavage on many AIL-harboring pri-miRNAs. (A) Pri-miRNA processing assays. Five pmol of each pri-miRNA were incubated with 6 pmol of Microprocessor (NLSD3-DGCR8) for 2 h at 37°C. (B) The long product was cut from the gel (in A and Supplementary Figure S4B) and cloned, after which next-generation sequencing (NGS) reads were aligned with pri-miRNA sequences. The single-cut products were selected and visualized by IGV. (C and F) Diagram of pri-mir-204, pri-mir-181a-1, and their variants. The mutated nucleotides are in red. (D and G) Pri-miRNA processing assays. Five pmol of each pri-miRNA were incubated with 6 pmol Microprocessor (NLSD3-DGCR8) for 2 h at 37°C. (E and H) Quantification of the pri-miRNA processing data from (D) and (G), respectively. The pri-miRNA processing assays in (D and G) were repeated three times. The sc/dc ratio was estimated as the ratio of the single-cut to double-cut product (F2) band density in the gel. The p-values of the one-tailed t-test for the sc/dc ratios estimated from three replicates were shown. MP: Microprocessor.
The introduction of an AIL to non-AIL-harboring pri-miRNAs enhances the level of 5p-strand cleaved-products from Microprocessor
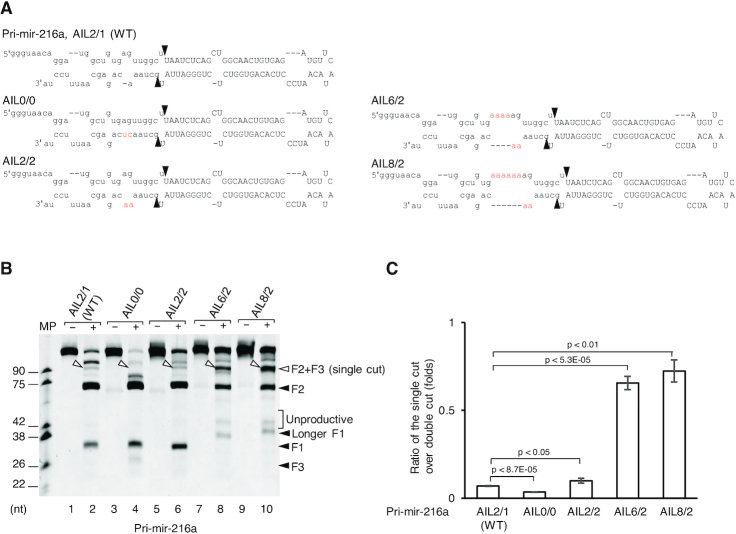
We investigated the effect of mismatches and AIL on the lower stem of the pri-miRNAs, which are resistant to the single cleavage of the Microprocessor complex. We used pri-mir-216a, which has less than three unmatched nt on each strand of the lower stem (Figure 5A), and showed that Microprocessor generated a negligible amount of the 5p-strand cleaved-product (Figure 5B, Supplementary Figure S5A, lane 2). The removal (AIL0/0) or the introduction of a mismatch (AIL2/2) slightly altered the 5p-strand single cleavage level. For example, when compared with the WT (AIL2/1), pri-mir-216a-AIL0/0 resulted in less while pri-mir-216a-AIL2/2 led to marginally more single cleavage product (Figure 5B, compare lane 4 or 6 with lane 2, and 5C). In contrast, the addition of a large AIL profoundly enhanced the single cleavage products of Microprocessor, but largely diminished its double-cut products from pri-mir-216a-AIL6/2 and AIL8/2 (Figure 5B, compare lane 8 or 10 with lane 2, 5C, and Supplementary Figure S5A). Consistent with the pri-mir-92a-1 results shown in Figure 3, the inclusion of mismatches and AIL in pri-mir-216a considerably increased the sc/dc ratios (Figure 5C). In addition to the short pri-mir-216a variants, we also tested the longer pri-mir-216a variants (F1: 80 nt, F2: 61 nt, F3: 100 nt), and found the single cleavage products of both to be similar (Supplementary Figure S5B). We also observed similar effects of the AIL addition in the lower stem of pri-mir-9-1 (Supplementary Figure S5C and D) and pri-mir-30a (Supplementary Figure S5E and F). These observations demonstrate that the addition of an AIL to the lower stem of various non-AIL pri-miRNAs can block the cleavage of Microprocessor on the 3p-strand, thereby facilitating the single cleavage on the 5p-strand of these AIL-introduced pri-miRNAs.
Figure 5.
The addition of a large AIL facilitates the 5p-strand single cleavage of Microprocessor on non-AIL-harboring pri-miRNAs. (A) Diagram of pri-mir-216a and its variants. The mutated nucleotides are in red. (B) Pri-miRNA processing assays. Five pmol of each pri-miRNA were incubated with 8 pmol each of D3-G1 and the G4 dimer for 2 h at 37°C. (C) Quantification of the pri-miRNA processing data from (B). The pri-miRNA processing assays in (B) were repeated three times. The sc/dc ratio was estimated as the ratio of the single-cut to double-cut product (F2) band density in the gel. The P-values of the one-tailed t-test for the sc/dc ratios estimated from three replicates were shown. MP: Microprocessor.
Knocking down miRNA expression using AILs
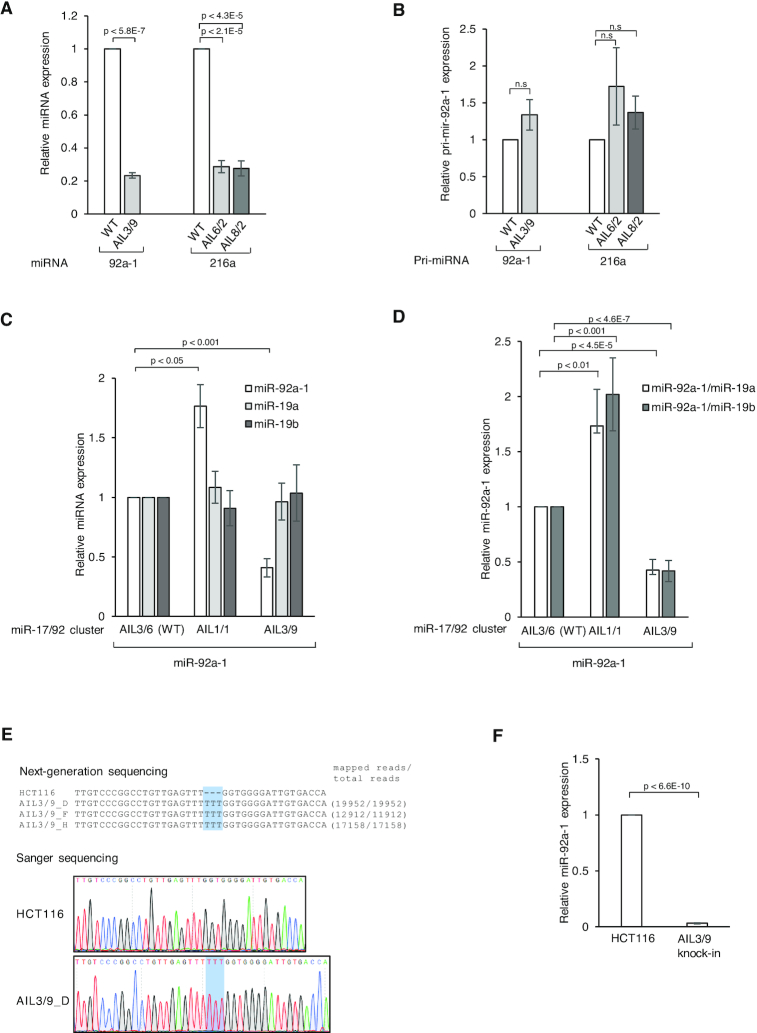
We noticed that the addition of an AIL to the lower stem of pri-miRNAs did not affect the miRNA region but it did reduce pre-miRNA production in vitro. Therefore, we proposed to make use of this phenomenon in miRNA knockdown assays. To test this idea, we generated pCDNA3 vectors, which expressed pri-mir-92a-1, pri-mir-216a, and their AIL-containing variants, thus: pri-mir-92a-1-AIL3/9, pri-mir-216a-AIL6/2 and pri-mir-216a-AIL8/2. We co-transfected two vectors expressing one of the pri-miRNA variants along with pri-mir-16-1, and then measured the expression of miRNA in the transfected cells by qPCR. The expression of miRNA from the different pri-miRNA variants was normalized to the level of miR-16-1. As shown in Figure 6A, the AILs significantly reduced the level of miR-92a-1 and miR-216a expression. This is consistent with our in vitro processing data, which shows that the AIL-containing pri-miRNAs generated less pre-miRNA product than WT (i.e., for pri-mir-92a-1, compare lane 4 with lane 6 of Figure 3B, and for pri-mir-216a, compare lane 8 or 10 with lane 2 of Figure 5B). In addition, pri-mir-92a-1-AIL3/9 and pri-mir-216a-AIL6/2 or pri-mir-216a-AIL8/2 insignificantly showed higher expression levels than those of pri-mir-92a-1 WT and pri-mir-216a WT in human cells, respectively (Figure 6B and Supplementary Figure S6A), suggesting that the reduction in miRNA expression was likely due to the changes in the sc/dc ratio, not the overall reduction of pri-miRNA processing. Overall, these data suggest that the addition of an AIL offers an alternative way to knockdown miRNA expression in human cells.
Figure 6.
miRNA knockdown experiments based on the addition of a large AIL to pri-miRNAs. (A) The DNA sequence encoding pri-mir-92a-1 WT, pri-mir-92a-1-AIL3/9, pri-mir-216a WT, pri-mir-216a-AIL6/2, or pri-mir-216a-AIL8/2 was cloned in the pcDNA3 vector. Each cloned vector was co-transfected with pcDNA3 expressing pri-mir-16-1 into HCT116 cells. The RNA transcription in human cells was carried out with the CMV promoter of the pcDNA3 vector. The expression of miRNAs was quantified by qPCR and normalized against that of miR-16-1. (B) The expression of pri-miRNAs in (A) was quantified by qPCR and normalized against that of pri-mir-16-1. (C) The DNA sequence encoding the miR-17/92 cluster that contains WT, AIL1/1, or AIL3/9 of pri-mir-92a-1 was cloned in the pcDNA3 vector. Each cloned vector was co-transfected with pcDNA3 expressing pri-mir-16-1 into HCT116 cells. The expression of miRNAs was quantified by qPCR and normalized against that of miR-16-1. (D) The expression of miR-92a-1 relative to that of miR-19a or miR-19b-1 was quantified by qPCR and normalized against that of miR-19a or miR-19b-1. (E) Genomic DNA sequences of the pri-mir-92a-1 AIL3/9 knock-in cells. The genomic DNA region covering the knock-in site in pri-mir-92a-1 was amplified by PCR, and after which the PCR products were sequenced by both Sanger sequencing and NGS. Three knock-in cell lines were generated. (F) The expression of miR-92a-1 in HCT116 (parental cells) and AIL3/9 cells was quantified by qPCR and normalized against that of U6. Three biological replicates for each experiment and three technical replicates for each biological replicate were performed. The P-values of the one-tailed t-test were shown.
Next, we investigated the expression of miR-92a-1 in the context of the miR-17/92 cluster containing six different miRNAs, miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1 (64). We expected to adjust the expression of miR-92a-1 alone by mutating its AIL, and not influence the expression of the other miRNAs in the same cluster. Here, we showed that by removing the AIL (AIL1/1) or by adding three nt (AIL3/9) to enlarge the AIL of pri-mir-92a-1, we could solely change the expression of miR-92a-1. As shown in Figure 6C, the expression of miR-92a-1 increased in the AIL1/1 cluster but decreased in the AIL3/9 cluster, when compared with that from the WT cluster. In contrast, the expression of miR-17, miR-18a, miR-19a, miR-19b-1, and miR-20a was not significantly different among the WT, AIL1/1, and AIL3/9 clusters (Figure 6C and Supplementary Figure S6B). As expected, we observed an altered ratio of miR-92a-1 to any miRNA member in the same cluster, such as miR-19a or miR-19b-1 in the mutated clusters (Figure 6D). In addition, the expression level of pri-miRNAs was not significantly different, suggesting that the discrimination in miRNA expression might be mainly caused by the changes in the sc/dc ratio rather than the overall alteration of pri-miRNA processing (Supplementary Figure S6C and D). Altogether, our data suggest that altering the AIL region might be a novel approach for regulating the expression of a specific miRNA without affecting the expression of others in the same cluster.
To exclude the possibility that the ectopic expression of AIL-containing pri-miR-92a-1 might exert a dominant-negative effect on endogenous miRNA expression, we carried out qPCR for miR-16-1, miR-30a, and let-7d. We found that their expression was not affected by the expression of AIL or non-AIL pri-mir-92a-1 (Supplementary Figure S7A). Furthermore, we compared the processing efficiency of long pri-miR-92a-1 WT in the three different combinations with each of short pri-miR-92a-1 WT (AIL3/6), AIL1/1, or AIL3/9. Because long and short pri-mir-92a-1 resulted in the same length of pre-mir-92a-1, their processing of long and short pri-mir-92a-1 was estimated using the single-cut products with different lengths (Supplementary Figure S7B and C). We showed that Microprocessor cleaved long pri-miR-92a-1 WT with a similar efficiency regardless of the presence of any short pri-mir-92a-1 (Supplementary Figure S7B and C). This indicates that AIL-containing pri-mir-92a-1 did not have a dominant-negative effect on the processing of pri-mir-92a-1 WT.
Finally, we edited the genomic DNAs of HCT116 cells using the CRISPR-Cas9 technique and generated knock-in cells, which produced a larger AIL-containing pri-mir-92a-1 (AIL3/9). The authenticity of the knock-in cells was confirmed by both NGS and Sanger sequencing (Figure 6E). Our qPCR data showed that the large AIL reduced the miRNA expression of miR-92a-1 specifically, whereas it did not affect the expression of miRNA in or out of the 17/92 cluster (Figure 6F and Supplementary Figure S7D).
DISCUSSION
Since its discovery in 2004, the Microprocessor complex, which contains a typical RNase III enzyme, DROSHA, has long been thought to create the standard double cuts on pri-miRNAs and mRNAs (2,5,40–43). Multiple mechanisms involving RNA elements and protein factors have been proposed to regulate the double-cut activity of Microprocessor to control miRNA expression (2,5,51,55,65,66). Here, we demonstrate that the asymmetric internal loop (AIL) in the lower stem of pri-miRNAs differentially affects the cleavage of pri-miRNA on each strand. The AIL somehow biasedly inhibits the cleavage of Microprocessor on the 3p-strand of many pri-miRNAs, thereby, allowing it to make a single cleavage on their 5p-strand. This single cleavage on the 5p-strand competes with the double-cut on both stands for the same pri-miRNA substrate, resulting in a reduction of pre-miRNA production and thereby downregulation of miRNA expression. This fluctuation in the level of the 5p-strand cleavage indicates another layer of regulation for miRNA expression. For example, RNA modifications or single nucleotide polymorphisms that reshape the pri-miRNA structure, or any auxiliary factors that influence the interaction between Microprocessor and pri-miRNAs, might change the single-cut-to-double-cut activity ratio, and thus alter the expression of miRNAs.
Previous studies indicate that a bacterial RNase III displays a single cleavage (22,23,67,68). However, this single cleavage seems to be different from that of Microprocessor revealed in this study in two ways. First, the single cleavage of the bacterial RNase III occurs on the 3p-strand of RNA duplexes and second, it is stimulated by the AIL located at the cleavage sites. In contrast, we showed that Microprocessor exhibits its single cleavage using RIIIDb on the 5p-strand of the pri-miRNAs tested. Moreover, the 5p-strand cleavage is enhanced by mismatches and the AIL located a few base pairs distant from the cleavage sites. This suggests that the mismatches and AIL in the lower stem of pri-miRNAs somehow restrict cleavage on the 3p-strand by RIIIDa of Microprocessor. Perhaps, these RNA elements prohibit the interaction between RIIIDa and the RNA duplex, facilitating the release of Microprocessor after cleavage of the 5p-strand, and preventing the enzyme from cleaving the 3p-strand. Since RIIIDa is closer to the AIL than RIIIDb, it is likely to be more affected by this structure. It will be intriguing to explore how the mismatches and AIL in the stem affect the interaction between Microprocessor and its various RNA substrates at the atomic level, and if other RNase III enzymes might also share a single-cut activity similar to Microprocessor.
Microprocessor might use the single cleavage as a specific ssRNA endonuclease, which cleaves substrates containing a stem–loop structure. This suggests that Microprocessor might have a larger substrate repertoire, beyond the known pri-miRNAs and mRNAs (2,5,40–43). In the future, it would be interesting to investigate if novel stem–loop RNA structures in various different types of RNA, such as rRNAs, mRNAs and long non-coding RNAs, might also be substrates for the single-cut activity of Microprocessor.
The abnormal expression of miRNAs is associated with numerous human diseases (69,70). Therefore, being able to specifically and accurately manipulate the biogenesis of miRNAs is essential for the development of therapies to correct these expression defects and thus treat such diseases. Complementary oligos are widely used to antagonize the function of miRNAs (71). Alternatively, gene-editing technology is also exploited to mutate the regions of genomic DNA that encode miRNA sequences, and thereby block miRNA synthesis (72–75). However, the former method has proven to be challenging because the level of miRNA knockdown needs to be accurately controlled, whereas the latter might lead to the production of altered miRNA sequences. In addition, both of these methodologies might also lead to the complete knockdown of miRNAs that are required for the normal function of the cell. Targeting the AIL suggests a novel method to knockdown the expression of miRNAs in a specific manner, while still retaining a certain level of miRNA expression, and thus preserving the normal cellular functions of miRNAs. In addition, our study also indicates the usefulness of the Microprocessor single-cut activity to manipulate the expression of a specific miRNA in a cluster without affecting the neighbouring miRNAs in the same cluster; for example, miR-92a-1 in the miR-17/92 cluster. It is the balance of miRNA expression in this cluster that is thought to be crucial for normal cellular functions. For example, a decrease in the miR-92a-1/miR-19 ratio likely favors the suppression of c-Myc-mediated apoptosis and hence advances oncogenesis (76). Here, we showed that we could increase the miR-92a-1/miR-19 ratio, and this might antagonize the promotion of oncogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Narry V. Kim and Dr. Jaesung Woo for sharing with us the essential materials including the vectors, antibodies, and cell lines. We thank Dr. Karl Herrup, Dr. Sarah E. Webb, and Ms. Phuong Thao Phan for critical reading. We appreciate our lab members for discussion and technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Hong Kong University of Science and Technology (HKUST) [R9380, in part]; Croucher Foundation [CIA17SC03, in part]; T.L.N. and T.D.N. are recipients of postgraduate scholarships at the HKUST. Funding for open access charge: [CIA17SC03].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kim V.N., Han J., Siomi M.C.. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009; 10:126–139. [DOI] [PubMed] [Google Scholar]
- 2. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 3. Meister G., Tuschl T.. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004; 431:343–349. [DOI] [PubMed] [Google Scholar]
- 4. Friedman R.C., Farh K.K.H., Burge C.B., Bartel D.P.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel D.P. Metazoan MicroRNAs. Cell. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Storici F., Tichon A.E.. RNA takes over control of DNA break repair. Nat. Cell Biol. 2017; 19:1382–1384. [DOI] [PubMed] [Google Scholar]
- 7. Michelini F., Pitchiaya S., Vitelli V., Sharma S., Gioia U., Pessina F., Cabrini M., Wang Y., Capozzo I., Iannelli F. et al.. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017; 19:1400–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gong C., Tang Y., Maquat L.E.. mRNA–mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2013; 20:1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park E., Maquat L.E.. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA. 2013; 4:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gong C., Maquat L.E.. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011; 470:284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henras A.K., Plisson-chastang C.. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA. 2015; 6:225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hawley B.R., Lu W., Wilczynska A., Bushell M.. The emerging role of RNAs in DNA damage repair. Cell Death Differ. 2017; 24:580–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen L.L., Yang L.. ALUternative regulation for gene expression. Trends Cell Biol. 2017; 27:480–490. [DOI] [PubMed] [Google Scholar]
- 14. Shendure J., Church G.M.. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol. 2002; 3:RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009; 457:413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Danielsen J.M.R., Yang Y.G., Qi Y.. A role for small RNAs in DNA double-strand break repair. Cell. 2012; 149:101–112. [DOI] [PubMed] [Google Scholar]
- 17. D’Adda di Fagagna F. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014; 24:171–178. [DOI] [PubMed] [Google Scholar]
- 18. Portal M.M., Pavet V., Erb C., Gronemeyer H.. Human cells contain natural double-stranded RNAs with potential regulatory functions. Nat. Struct. Mol. Biol. 2015; 22:89–97. [DOI] [PubMed] [Google Scholar]
- 19. Nguyen T.C., Zaleta-Rivera K., Huang X., Dai X., Zhong S.. RNA, action through interactions. Trends Genet. 2018; 34:867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robertson H.D., Webster R.E., Zinder N.D.. A nuclease specific for double-stranded RNA. Virology. 1967; 32:718–719. [DOI] [PubMed] [Google Scholar]
- 21. Robertson H.D., Webster R.E., Zinder N.D.. Purification and properties of ribonuclease III from Escherichia coli. J. Biol. Chem. 1968; 243:82–91. [PubMed] [Google Scholar]
- 22. Court D.L., Gan J., Liang Y.-H., Shaw G.X., Tropea J.E., Costantino N., Waugh D.S., Ji X.. RNase III: genetics and function; structure and mechanism. Annu. Rev. Genet. 2013; 47:405–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nicholson A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA. 2014; 5:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X.. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006; 124:355–366. [DOI] [PubMed] [Google Scholar]
- 25. Lamontagne B., Elela S.A.. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 2004; 279:2231–2241. [DOI] [PubMed] [Google Scholar]
- 26. Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X.. Intermediate states of ribonuclease III in complex with double-stranded RNA. Structure. 2005; 13:1435–1442. [DOI] [PubMed] [Google Scholar]
- 27. Sinha N.K., Iwasa J., Shen P.S., Bass B.L.. Dicer uses distinct modules for recognizing dsRNA termini. Science. 2018; 359:329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N.. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004; 18:3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W.. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004; 118:57–68. [DOI] [PubMed] [Google Scholar]
- 30. MacRae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A.. Structural basis for double-stranded RNA processing by Dicer. Science. 2006; 311:195–198. [DOI] [PubMed] [Google Scholar]
- 31. Park J., Heo I., Tian Y., Simanshu D.K., Chang H., Jee D., Patel D.J., Kim V.N.. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011; 475:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen T.A., Jo M.H., Choi Y.G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.S.. Functional anatomy of the human microprocessor. Cell. 2015; 161:1374–1387. [DOI] [PubMed] [Google Scholar]
- 33. Kwon S.C., Nguyen T.A., Choi Y.G., Jo M.H., Hohng S., Kim V.N., Woo J.S.. Structure of human DROSHA. Cell. 2016; 164:81–90. [DOI] [PubMed] [Google Scholar]
- 34. Liu Z., Wang J., Cheng H., Ke X., Sun L., Zhang Q.C., Wang H.W.. Cryo-EM structure of human dicer and its complexes with a Pre-miRNA substrate. Cell. 2018; 173:1191–1203. [DOI] [PubMed] [Google Scholar]
- 35. Denli A.M., Tops B.B.J., Plasterk R.H.A., Ketting R.F., Hannon G.J.. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004; 432:231–235. [DOI] [PubMed] [Google Scholar]
- 36. Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R.. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004; 432:235–240. [DOI] [PubMed] [Google Scholar]
- 37. Landthaler M., Yalcin A., Tuschl T.. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004; 14:2162–2167. [DOI] [PubMed] [Google Scholar]
- 38. Herbert K.M., Sarkar S.K., Mills M., Delgado De la Herran H.C., Neuman K.C., Steitz J.A.. A heterotrimer model of the complete Microprocessor complex revealed by single-molecule subunit counting. RNA. 2016; 22:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen T.A., Park J., Dang T.L., Choi Y.-G., Kim V.N.. Microprocessor depends on hemin to recognize the apical loop of primary microRNA. Nucleic Acids Res. 2018; 46:5726–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han J., Pedersen J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H., Yang W.Y., Haussler D., Blelloch R., Kim V.N.. Posttranscriptional crossregulation between drosha and DGCR8. Cell. 2009; 136:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Triboulet R., Chang H.M., Lapierre R.J., Gregory R.I.. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009; 15:1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seong Y., Lim D.H., Kim A., Seo J.H., Lee Y.S.i., Song H., Kwon Y.S.. Global identification of target recognition and cleavage by the Microprocessor in human ES cells. Nucleic Acids Res. 2014; 42:12806–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim B., Jeong K., Kim V.N.. Genome-wide mapping of DROSHA cleavage sites on primary MicroRNAs and noncanonical substrates. Mol. Cell. 2017; 66:258–269. [DOI] [PubMed] [Google Scholar]
- 44. Kadener S., Rodriguez J., Abruzzi K.C., Khodor Y.L., Sugino K., Marr M.T 2nd, Nelson S., Rosbash M.. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009; 15:537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Macias S., Plass M., Stajuda A., Michlewski G., Eyras E., Caceres J.F.. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat. Struct. Mol. Biol. 2012; 19:760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knuckles P., Vogt M.A., Lugert S., Milo M., Chong M.M.W., Hautbergue G.M., Wilson S.A., Littman D.R., Taylor V.. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 2012; 15:962–969. [DOI] [PubMed] [Google Scholar]
- 47. Marinaro F., Marzi M.J., Hoffmann N., Amin H., Pelizzoli R., Niola F., Nicassio F., De Pietri Tonelli D.. MicroRNA-independent functions of DGCR8 are essential for neocortical development and TBR1 expression. EMBO Rep. 2017; 18:603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N.. Molecular basis for the recognition of primary microRNAs by the drosha-DGCR8 complex. Cell. 2006; 125:887–901. [DOI] [PubMed] [Google Scholar]
- 49. Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P.. Beyond secondary structure: primary-sequence determinants license Pri-miRNA hairpins for processing. Cell. 2013; 152:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fang W., Bartel D.P.. The menu of features that define primary microRNAs and enable de novo design of MicroRNA genes. Mol. Cell. 2015; 60:131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nguyen H.M., Nguyen T.D., Nguyen T.L., Nguyen T.A.. Orientation of human Microprocessor on primary microRNAs. Biochemistry. 2018; 58:189–198. [DOI] [PubMed] [Google Scholar]
- 52. Fernandez N., Cordiner R.A., Young R.S., Hug N., Macias S., Caceres J.F.. Genetic variation and RNA structure regulate microRNA biogenesis. Nat. Commun. 2017; 8:15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim K., Duc Nguyen T., Li S., Anh Nguyen T.. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA. 2018; 24:892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Michlewski G., Cáceres J.F.. Post-transcriptional control of miRNA biogenesis. RNA. 2019; 25:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Treiber T., Treiber N., Meister G.. Publisher correction: regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019; 20:321. [DOI] [PubMed] [Google Scholar]
- 56. Roden C., Gaillard J., Kanoria S., Rennie W., Barish S., Cheng J., Pan W., Liu J., Cotsapas C., DIng Y., Lu J.. Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res. 2017; 27:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Griffiths-Jones S. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006; 34:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Darty K., Denise A., Ponty Y.. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10. [Google Scholar]
- 61. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R. et al.. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005; 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mogilyansky E., Rigoutsos I.. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013; 20:1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Michlewski G., Cáceres J.F.. Post-transcriptional control of miRNA biogenesis. RNA. 2019; 1–16.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Treiber T., Treiber N., Meister G.. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2018; 20:5–20. [DOI] [PubMed] [Google Scholar]
- 67. Saito H., Richardson C.C.. Processing of mRNA by ribonuclease III regulates expression of gene 1.2 of bacteriophage T7. Cell. 1981; 27:533–542. [DOI] [PubMed] [Google Scholar]
- 68. Nicholson A.W., Niebling K.R., McOsker P.L., Robertson H.D.. Accurate in vitro cleavage by RNase III of phosphorothioate-substituted RNA processing signals in bacteriophage T7 early mRNA. Nucleic Acids Res. 1988; 16:1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ardekani A.M., Naeini M.M.. The role of MicroRNAs in human diseases. Avicenna J. Med. Biotechnol. 2010; 2:161–179. [PMC free article] [PubMed] [Google Scholar]
- 70. Li Y., Kowdley K.V.. MicroRNAs in common human diseases. Genomics Proteomics Bioinforma. 2012; 10:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rupaimoole R., Slack F.J.. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017; 16:203–222. [DOI] [PubMed] [Google Scholar]
- 72. Kim Y.K., Wee G., Park J., Kim J., Baek D., Kim J.S., Kim V.N.. TALEN-based knockout library for human microRNAs. Nat. Struct. Mol. Biol. 2013; 20:1458–1464. [DOI] [PubMed] [Google Scholar]
- 73. Jing W., Zhang X., Sun W., Hou X., Yao Z., Zhu Y.. CRISPR/CAS9-Mediated genome editing of miRNA-155 inhibits proinflammatory cytokine production by RAW264.7 cells. Biomed Res. Int. 2015; 2015:326042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chang H., Yi B., Ma R., Zhang X., Zhao H., Xi Y.. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 2016; 6:22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huo W., Zhao G., Yin J., Ouyang X., Wang Y., Yang C., Wang B., Dong P., Wang Z., Watari H. et al.. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J. Cancer. 2017; 8:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Olive V., Sabio E., Bennett M.J., De Jong C.S., Biton A., McGann J.C., Greaney S.K., Sodir N.M., Zhou A.Y., Balakrishnan A. et al.. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. Elife. 2013; 2:e00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.