To maintain cellular homeostasis, the endoplasmic reticulum (ER) necessitates a continuous removal of ER fragments via a selective, receptor‐mediated, form of autophagy known as ER‐phagy. In this issue of The EMBO Journal, Jiang et al (2020) shed light on how the best characterized autophagy receptor FAM134B mediates ER membrane fragmentation, the earliest event during ER‐phagy. They propose a dynamic model for FAM134B protein oligomerization and ER membrane scission, which are driven by CAMK2B‐mediated phosphorylation of the receptor and are altered in sensory neuropathy.
Subject Categories: Autophagy & Cell Death; Post-translational Modifications, Proteolysis & Proteomics
A new study reveals that oligomerization of the autophagy receptor FAM134B drives endoplasmic reticulum fragmentation and ER‐phagy, two processes deregulated in hereditary neuropathies.
The endoplasmic reticulum (ER) is the largest cellular organelle, forming a continuous membrane network of cisternae and tubules spreading from the nucleus to the plasma membrane (Schwarz & Blower, 2016). The ER plays fundamental cellular functions, including calcium storage, protein and lipid biosynthesis (Schwarz & Blower, 2016). In order to maintain cellular homeostasis, the ER adapts its shape and size in response to cellular stress conditions (Walter & Ron, 2011). In mammals, “ER‐phagy” is as a selective form of ER degradation through the engulfment of ER fragments into autophagosomes for lysosomal delivery (Khaminets et al, 2015; De Leonibus et al, 2019). This process needs ER‐phagy receptors, which are ER‐resident proteins that harbour a LC3 interaction motif (LIR) enabling the binding of the cargo to LC3/GABARAP family proteins on autophagosomal membranes (Stolz et al, 2014). FAM134B was the first and so far the best characterized ER‐phagy receptor (Khaminets et al, 2015). FAM134B is an intramembrane ER‐resident protein that contains a LIR domain and a reticulon homology domain (RHD), which consists of two membrane‐embedded hydrophobic regions bridged by a flexible cytoplasmic linker (Khaminets et al, 2015; Bhaskara et al, 2019). In addition to ER turnover, FAM134B‐mediated ER‐phagy exerts ER quality control functions by facilitating the degradation of inappropriately folded ER clients (Cui et al, 2019; Forrester et al, 2019).
What is currently missing in the field of ER‐phagy, and specifically in selective autophagy, is the understanding of the intracellular signals that govern these types of processes. Jiang et al describe a series of molecular events that trigger FAM134B‐mediated ER‐phagy. First, they demonstrated that the RHD of FAM134B drives receptor oligomerization and that this event promotes ER fragmentation prior to LIR‐mediated ER‐phagy. Similar findings have been recently reported by another group using molecular modelling and dynamics simulations (Bhaskara et al, 2019). Second, the authors identified three potential phosphorylation sites [serine149, serine151 and serine153 (S149, S151 and S153)] within the flexible cytoplasmic linker that bridges the two intermembrane hairpins of the RHD. Using site‐directed mutagenesis, they demonstrated that the serine residues S149, S151 and S153 regulate FAM134B oligomerization and, in turn, ER fragmentation during ER‐phagy. Third, Jiang and co‐workers identified the Calcium/Calmodulin Dependent Protein Kinase II Beta (CAMK2B) as a putative kinase responsible for the phosphorylation of FAM134B at S151. Consistently, CAMK2B activators (ionomycin and EB1089) or inhibitors (KN‐93 or CAM2KB‐specific knockdown), respectively, stimulated or repressed ER fragmentation and ER‐phagy in a FAM134B‐dependent manner. Taken together, these data demonstrate the existence of specific signalling events that induce ER‐phagy by favouring the oligomerization of FAM134B.
Loss‐of‐function mutations in FAM134B were identified to be responsible for the pathogenesis of a hereditary sensory and autonomic neuropathy (HSAN‐II) (Kurth et al, 2009). Jiang et al studied a disease‐associated FAM134B missense variant (FAM134BG216R) in the RHD with unknown pathogenic significance (Davidson et al, 2012). Surprisingly, FAM134BG216R exhibited gain‐of‐function activity by inducing FAM134B oligomerization, ER scission and ER‐phagy more efficiently than the wild‐type form. Notably, they demonstrated that FAM134BG216R expression affected the survival of dorsal root ganglion (DRG) sensory neurons. Together, these data suggest that FAM134BG216R acts as a gain‐of‐function mutation that might trigger sensory neuronal cell death by inducing excessive ER fragmentation and ER‐phagy (Fig 1). Interestingly, chemical inhibition of CAMK2B activity partly rescued the cytotoxicity in DRG neurons infected with FAM134BG216R variant, suggesting that targeting FAM134B oligomerization might be beneficial for a subset of patients affected by HSAN‐II.
Figure 1. Proposed model of FAM134B oligomerization and regulation.
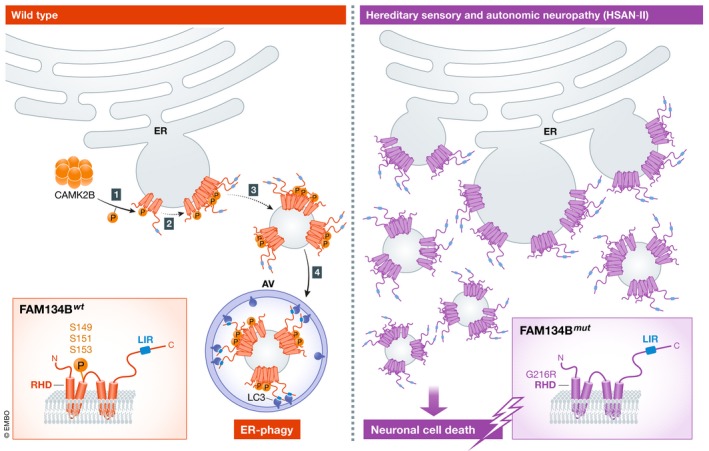
CAMK2B‐mediated phosphorylation of FAM134B (1) enhances its self‐association and oligomerization (2), thus promoting ER membrane fragmentation (3) and the ER‐phagy process (4). FAM134B oligomerization can be pathologically enhanced in type II HSAN syndrome due to G216R mutation in the RHD of FAM134B. As a consequence, an excessive ER fragmentation and ER‐phagy may promote neuronal cell death through unknown mechanisms.
In summary, this work offers novel insights into the understanding of FAM134B function. It further opens new avenues on intracellular signals that govern selective forms of autophagy and on the dysfunctional cellular mechanisms beneath the pathogenesis of sensory neuropathy.
The EMBO Journal (2020) 39: e104546
See also: https://doi.org/10.15252/embj.2019102608 (March 2020)
References
- Bhaskara RM, Grumati P, Garcia‐Pardo J, Kalayil S, Covarrubias‐Pinto A, Chen W, Kudryashev M, Dikic I, Hummer G (2019) Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER‐phagy. Nat Commun 10: 2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Parashar S, Zahoor M, Needham PG, Mari M, Zhu M, Chen S, Ho HC, Reggiori F, Farhan H et al (2019) A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science (New York, NY) 365: 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G, Murphy S, Polke J, Laura M, Salih M, Muntoni F, Blake J, Brandner S, Davies N, Horvath R et al (2012) Frequency of mutations in the genes associated with hereditary sensory and autonomic neuropathy in a UK cohort. J Neurol 259: 1673–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leonibus C, Cinque L, Settembre C (2019) Emerging lysosomal pathways for quality control at the endoplasmic reticulum. FEBS Lett 593: 2319–2329 [DOI] [PubMed] [Google Scholar]
- Forrester A, De Leonibus C, Grumati P, Fasana E, Piemontese M, Staiano L, Fregno I, Raimondi A, Marazza A, Bruno G et al (2019) A selective ER‐phagy exerts procollagen quality control via a Calnexin‐FAM134B complex. EMBO J 38: e99847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang X, Ding X, Du M, Li B, Weng X, Zhang J, Li L, Tian R, Zhu Q et al (2020) FAM134B oligomerization drives endoplasmic reticulum membrane scission for ER‐phagy. EMBO J 39: e102608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N et al (2015) Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522: 354–358 [DOI] [PubMed] [Google Scholar]
- Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, Rotthier A, Baets J, Senderek J, Topaloglu H, Farrell SA et al (2009) Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet 41: 1179–1181 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Blower MD (2016) The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci 73: 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I (2014) Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16: 495–501 [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science (New York, NY) 334: 1081–1086 [DOI] [PubMed] [Google Scholar]


