Abstract
Objective
The clinical outcomes of gastric diseases such as chronic gastritis, peptic ulcer, and gastric cancer have been attributed to the interplay of virulence factors of Helicobacter pylori (H. pylori), host genetic susceptibility, and host immune responses. This study investigated the presence of cagA, vacA, iceA2, babA2, and oipA genes and their association with clinical outcomes.
Methods
Chronic gastritis, atrophic gastritis, and intestinal metaplasia specimens were obtained from patients who underwent endoscopy and surgical resection between January 2017 and December 2018; specimens from gastric cancer patients treated between January 2014 and December 2018 were also added. H. pylori), host genetic susceptibility, and host immune responses. This study investigated the presence of cagA, vacA, iceA2, babA2, and oipA genes and their association with clinical outcomes. H. pylori), host genetic susceptibility, and host immune responses. This study investigated the presence of
Results
H. pylori), host genetic susceptibility, and host immune responses. This study investigated the presence of vacA, babA2, and oipA genes and their association with clinical outcomes. vacA, babA2, and oipA genes and their association with clinical outcomes. P=0.033, OR = 2.64; 95% CI = 1.44–4.82, P=0.033, OR = 2.64; 95% CI = 1.44–4.82, P=0.033, OR = 2.64; 95% CI = 1.44–4.82, H. pylori vacA+/babA2, and oipA genes and their association with clinical outcomes. P=0.033, OR = 2.64; 95% CI = 1.44–4.82,
Conclusion
In this present study, we reported on the virulence genes of H. pylori infection to reveal their association with increased risk of chronic gastritis, precancerous gastric lesions, and gastric cancer. Precancerous gastric lesions with H. pylori vacA+/babA2+/oipA+ genotype increased the risk of gastric cancer.H. pylori), host genetic susceptibility, and host immune responses. This study investigated the presence of H. pylori vacA+/babA2, and oipA genes and their association with clinical outcomes.
1. Introduction
Helicobacter pylori (H. pylori) is a spiral-shaped Gram-negative bacterium that selectively colonizes the gastric mucosa of the humans in more than half of the world's population [1]. H. pylori infection has been proven to be highly associated with the development of a variety of gastric diseases such as chronic gastritis, peptic ulcer disease (PUD), mucosal associated lymphoid tissue (MALT), and gastric cancer (GC) [2–4]. These different clinical outcomes have been attributed to the interplay of several factors, including virulence factors of H. pylori, host genetic susceptibility, and host immune responses to H. pylori infection [5–7]. Several virulence factors have been proposed for H. pylori infections; they include adapting to different tissues like urease and flagella, using adhesins such as blood-antigen binding protein A (BabA) and outer inflammatory protein A (OipA), and toxins that damage host tissues such as cytotoxin-associated gene A (CagA) and vacuolating cytotoxin (VacA). Another putative virulence factor is that induced by contact with epithelium A (IceA) [8, 9].
The BabA is a protein for Lewis b binding activity on human gastric epithelial cells. Three bab alleles; babA1, babA2, and babB genes have been identified, but only the babA2 gene product is functional [10, 11]. BabA2 is associated with an increased risk of PUD and GC [12–14]. The OipA is one of the porin proteins and proinflammatory proteins associated with severe neutrophil infiltration in IL-8 induction and gastric colonization [15]. It contributes to the pathogenesis and is associated with the elevated risks of PUD and GC [16–18]. The cagA gene is located at the end of the cag pathogenicity island (cagPAI), encodes a type IV secretion system (T4SS) that is functional for translocating bacterial effectors into host cytoplasm, and triggers the manipulation of cell signaling pathways and also the induction of the proinflammatory cytokines, specifically interleukin IL-8 [19, 20]. It is associated with severe clinical diseases in PUD and GC [21–24]. The VacA is a pore-forming toxin, which causes progressive vacuolation and injury to the gastric epithelium [25]. It is associated with an increased risk of PUD and GC [26, 27]. Specific allelic types in the vacA gene are signal (s1, s2) and the middle regions (m1, m2) due to sequence heterogeneity [28]. The variation in cytotoxic activities in relation to H. pylori-related diseases and gastric mucosal changes is considered the cause of different strains [29]. The iceA gene is induced by contact with epithelium and has been considered as a marker for PUD [30]. It has two main allelic variants, iceA1 and iceA2 [31]. It has been found that iceA1 is the predominant subtype in East Asia, while iceA2 is the predominant subtype in the USA and Columbia [32]. Although several studies have reported different results, the iceA2 gene was detected to be the predominant genotype [33, 34].
In intestinal types of GC, H. pylori infection triggers a multistep progression from chronic gastritis, atrophic gastritis (AG), intestinal metaplasia (IM), and finally to GC [35]. Several studies suggest that AG and IM are the major precursor gastric lesions of intestinal-type GC and elevate the risk of GC [36–38]. Moreover, AG and IM increased the risk of intestinal-type GC exponentially when compared with other risk factors [39]. From this background, H. pylori infection is thought to be involved in chronic gastritis, precancerous gastric lesions, and GC; however, the relationship between virulence status and its association with these clinical outcomes has not been well reported and are not fully understood in Asian countries. Thus, the aim of this study was to investigate the H. pylori virulence genes including cagA, vacA, iceA2, babA2, and oipA of patients with chronic gastritis, precancerous gastric lesions, and GC, and to determine whether the virulence genes are associated with the risk of chronic gastritis, precancerous gastric lesions, and GC.
2. Material and Methods
2.1. Patients and Specimens
Patients were subjected to esophagogastroduodenoscopy (EGD) (Olympus Corp., Tokyo, Japan), which was carried out using an upper GI video endoscope (Olympus EVIS EXERA III, CV-190). Gastric tissue biopsies of chronic gastritis, AG, and IM were obtained between January 2017 and December 2018, and GC biopsies were obtained between January 2014 and December 2018. Surgical resection was performed at the Suranaree University of Technology Hospital, Buriram Hospital Medical center, or Surin Hospital Medical center in the Northeastern region of Thailand. Written informed consent was obtained from all patients, and the study protocol was approved by the Ethics Committee for Research Involving Human Subjects, Suranaree University of Technology (EC-59-45 and EC 16-2560). The whole stomach was examined and biopsies were conducted using the site-specific biopsy technique [40]. All biopsies were directly tested for H. pylori infection by using the rapid urease test (RUT) kit (Pentland Medical, Edinburgh, UK). The methods were carried out in accordance with good clinical practice and the guidelines of the Declaration of Helsinki [41]. Histological determinations were subsequently examined by a pathologist. The patient retrospective cohort included 70 cases, which were used to analyze the association between overall survival (OS) and genotype combinations.
2.2. DNA Extraction
DNA extraction from fresh tissues of chronic gastritis, AG, and IM was performed using the QIAamp DNA mini kit (Qiagen, Düsseldorf, Germany), and the tissues of GC were formalin-fixed and paraffin-embedded (FFPE) using xylene and hydrate in 100% ethanol and subsequently by using QIAamp DNA FFPE tissue kit (Qiagen, Düsseldorf, Germany) according to the manufacturer's instructions. Genomic DNA was purified from the tissue lysate using the QIAamp spin column and eluted. The DNA concentration and purity were determined using a DS-11+ spectrophotometer (Denovix, Wilmington, Delaware, USA) and stored at −20°C.
2.3. Real-Time Polymerase Chain Reaction (Real-Time PCR)
H. pylori infection 16s rRNA and ureA genes were identified. H. pylori-positive samples were used to determine the virulence genes (cagA, vacA, iceA2, babA2, and oipA) using real-time PCR. The primers for 16s rRNA, ureA, cagA, vacA, iceA2, babA2, and oipA (Integrated DNA Technologies, Coralville, IA, USA) are shown in Table 1. Briefly, DNA samples were used as templates in the amplification reactions. The real-time PCR was performed according to the manufacturer's protocol in a final volume of 20 μL containing DNA template, 2X SYBR Green PCR Master Mix (Roche Applied Science, Mannheim, Germany), and 50 pmol of each primer using a Light Cycler® 480 Instrument (Roche diagnostics, Neuilly sur Seine, France). The PCR conditions used in this study were as follows: preincubation at 95°C for 5 min, 45 cycles of amplification (10 s of denaturation at 95°C, 10 s of annealing at Tm of specific primer, and 10 s of extension at 72°C). Each sample was performed in duplicates reactions for standard. All data were analyzed using the Light Cycler 480 software, version 1.5 (Roche diagnostics, Neuilly sur Seine, France).
Table 1.
Primers used for the amplification of H. pylori genes.
| Primer | Forward | Reverse | Reference |
|---|---|---|---|
| 16s rRNA | GGAGTACGGTCGCAAGATTAAA | CTAGCGGATTCTCTCAATGTCAA | [42] |
| UreA | CGTGGCAAGCATGATCCAT | GGGTATGCACGGTTACGAGTTT | [43] |
| CagA | GAGTCATAATGGCATAGAACCTGAA | TTGTGCAAGAAATTCCATGAAA | [44] |
| VacA | CTCCAGAAGGCACACCAATAA | TGGCTTCCACTTCCCCATTAA | [45] |
| IceA2 | GTTGTCGTTGTTTTAATGAA | GTCTTAAACCCCACGATTAAA | [46] |
| BabA2 | CCAAACGAAACAAAAAGCGT | GCTTGTGTAAAAGCCGTCGT | [47] |
| OipA | GTTTTTGATGCATGGGATTT | GTGCATCTCTTATGGCTTT | [48] |
2.4. Statistical Analysis
The differences between the virulence genes of H. pylori infection for the patient's demographic data were determined using ANOVA. The associations between virulence genes and clinical outcomes and risks of GC were evaluated using the univariate regression model analysis. Odds ratios (OR) and 95% confidence intervals (CI) were calculated using the multivariate regression model analysis. Survival analysis was performed using the Kaplan–Meier method and the overall survival differences were analyzed using the log-rank test. A P value of less than 0.05 was considered statistically significant. All statistical analyses were carried out using SPSS for Windows (version 20.0; IBM Corp., Armonk, NY, USA).
3. Results
3.1. Detection of H. pylori cagA, vacA, iceA2, babA2, and oipA Virulence Genes in Chronic Gastritis, Precancerous Gastric Lesions, and Gastric Cancer
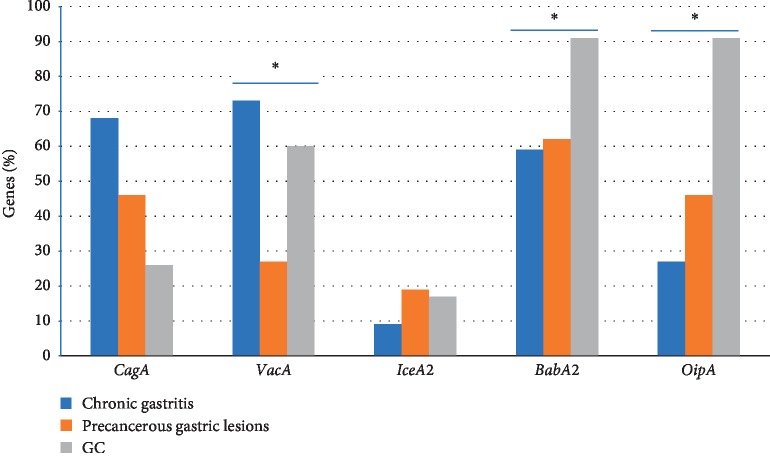
A total of 200 H. pylori-positive samples was examined for 16s rRNA and ureA gene detection; out of which 166 patients (83%) were positive for both 16s rRNA and ureA genes. These patients were divided into three groups: chronic gastritis (n = 44), precancerous gastric lesions including AG and IM (n = 52), and GC (n = 70). All patients were not significantly different in age and gender. The patient's demographic data are summarized in Table 2. The vacA (73%) gene was mainly present in chronic gastritis followed by cagA (68%), babA2 (59%), oipA (27%), and iceA2 (9%). The babA2 (62%) gene was mainly present in precancerous gastric lesions followed by cagA and oipA (46%), vacA (27%), and iceA2 (19%). Interestingly, babA2 and oipA (91%) were almost present in GC followed by vacA (60%), cagA (26%), and iceA2 (16%). The iceA2 gene was the lowest frequency of detection in all clinical outcomes. The frequency of cagA, vacA, iceA2, babA2, and oipA genes is shown in Figure 1. The presence of vacA, babA2, and oipA genes was significantly different between clinical outcomes (P=0.036, 0.042 and 0.039, respectively).
Table 2.
Demographic characteristics.
| Chronic | Precancerous lesions | Gastric cancer | P value | |
|---|---|---|---|---|
| (n = 44) | (n = 52) | (n = 70) | ||
| Age: mean | 43 ± 1.6 | 46 ± 2.4 | 52 ± 1.2 | 0.192 |
| Gender: male/female (%) | 63.6/32.4 | 46.2/43.8 | 45.2/44.8 | 0.082 |
| Pathological characteristic of gastric cancer patients (n = 70) | ||||
| Location of tumor n (%) | ||||
| Upper | 12 (17.15) | |||
| Middle | 36 (51.42) | |||
| Lower | 22 (31.43) | |||
| Tumor size n (%) | ||||
| <70 mm | 18 (25.72) | |||
| ≥70 mm | 52 (74.28) | |||
| Histologic type n (%) | ||||
| Differentiated | 54 (77.14) | |||
| Undifferentiated | 16 (22.86) | |||
| Lymphatic invasion n (%) | ||||
| Absent | 46 (65.71) | |||
| Present | 24 (34.29) | |||
| Vascular invasion n (%) | ||||
| Absent | 62 (88.57) | |||
| Present | 8 (11.43) | |||
| Pathological T stage n (%) | ||||
| T1-T2 | 22 (31.43) | |||
| T3-T4 | 48 (68.57) | |||
| Pathological TNM stage n (%) | ||||
| I | 8 (11.43) | |||
| II | 14 (20.0) | |||
| III | 36 (51.43) | |||
| IV | 12 (17.14) | |||
| Residual tumor n (%) | ||||
| No residual tumor | 52 (74.28) | |||
| Microscopic | 6 (8.57) | |||
| Gross (unresectable) | 12 (17.14) | |||
| CEA n (%) | ||||
| <5.0 (ng/ml) | 42 (60.0) | |||
| ≥5.0 (ng/ml) | 28 (34.29) | |||
Comparisons between the groups were done by using ANOVA. P < 0.05 considered as statistically significant.
Figure 1.
The frequency of cagA, vacA, iceA2, babA2, and oipA genes in each clinical outcome.
3.2. Association between the Presence of vacA, babA2, and oipA Genes and Clinical Outcomes
The association between the presence of vacA, babA2, and oipA genes and clinical outcomes was assessed. Among chronic gastritis, vacA was present in H. pylori infection and was associated with significantly increased risk of chronic gastritis (OR = 2.14, 95% CI = 1.62–4.46, P=0.036). The vacA, babA2, and oipA genes were present in patients, but they were not associated with increased risk of precancerous gastric lesions. Additionally, vacA, babA2, and oipA genes were associated with increased risk of GC (OR = 1.23; 95% CI = 1.13–3.32; P=0.033, OR = 2.64; 95% CI = 1.44–4.82, P=0.024 and OR = 2.79; 95% CI = 1.58–5.41; P=0.031, respectively) (Table 3).
Table 3.
Virulence gene in association with clinical outcomes.
| Gastric mucosa pathology/Virulence gene | Chronic | Precancerous | OR; 95% CI | P value | Precancerous | GC | OR (95% CI) | P value |
|---|---|---|---|---|---|---|---|---|
| (n = 44) | (n = 52) | (n = 52) | (n = 70) | |||||
| VacA | 32 (73%) | 14 (27%) | 2.14 (1.62–4.46) | 0.036 | 14 (27%) | 42 (60%) | 1.23 (1.13–3.32) | 0.033 |
| BabA2 | 26 (59%) | 32 (62%) | 0.77 (0.56–0.94) | 0.833 | 32 (62%) | 64 (91%) | 2.64 (1.44–4.82) | 0.024 |
| OipA | 12 (27%) | 24 (46%) | 0.69 (0.49–0.82) | 0.546 | 24 (46%) | 64 (91%) | 2.79 (1.58–5.41) | 0.031 |
Multivariate regression model analysis used to analyze the data. OR: odds ratio; CI: confidence interval. Significance is set at P < 0.05.
3.3. H. pylori vacA+/babA2+/oipA+ Genotype Conferred Increased Risk of GC
We examined the virulence combinations based on the analysis of vacA, babA2, and oipA genotypes. Precancerous gastric lesions infected with vacA+/babA2+, vacA+/oipA+, babA2+/oipA+, and vacA+/bibA2+/oipA+ genotypes were 3.85%, 3.85%, 26.92%, and 11.54%, whereas GC were 2.86%, 2.86%, 34.29%, and 51.43%, respectively (Table 4). Interestingly, precancerous gastric lesions infected with H. pylori genotype combination of vacA+/bibA2+/oipA+ were highly significantly associated with increased risk of GC (OR = 3.85, 95% CI = 1.67–5.77, P=0.021), but not vacA+/babA2+, vacA+/oipA+, and bibA2+/oipA+ genotypes. Chronic gastritis was not associated with the development of precancerous gastric lesions or GC when infected with any H. pylori genotype combination (data not shown).
Table 4.
Virulence genotype combination in association with clinical outcomes.
| Virulence gene | Precancerous gastric lesion (%) | GC (%) | OR (CI 95%) | P value | ||
|---|---|---|---|---|---|---|
| VacA | BabA2 | OipA | ||||
| + | + | − | 2 (3.85) | 2 (2.86) | 0.72 (0.42–0.97) | 0.634 |
| + | − | + | 2 (3.85) | 2 (2.86) | 0.72 (0.42–0.97) | 0.634 |
| − | + | + | 14 (26.92) | 24 (34.29) | 0.7 (0.37–0.96) | 0.091 |
| + | + | + | 6 (11.54) | 36 (51.43) | 4.28 (1.82–7.41) | 0.021 |
Multivariate regression model analysis used to analyze the data. OR: odds ratio; CI: confidence interval. Significance is set at P < 0.05.
3.4. Overall Survival of GC Patients with H. pylori Genotype Combination Infections
We examined the overall survival of GC patients that were infected with H. pylori two-genotype strains (vacA+/babA2+, vacA+/oipA+, and babA2+/oipA+) and H. pylori three-genotype strain (vacA+/babA2+/oipA+). The mean survival time for patients with H. pylori two-genotype infection was 69.52 ± 3.72 months and with H. pylori three-genotype infection was 56.16 ± 3.23 months. However, the overall survival of patients infected with vacA+/babA2+/oipA+ genotype strain was decreased, but there was no statistically significant difference between the two groups (P=0.148; Figure 2).
Figure 2.
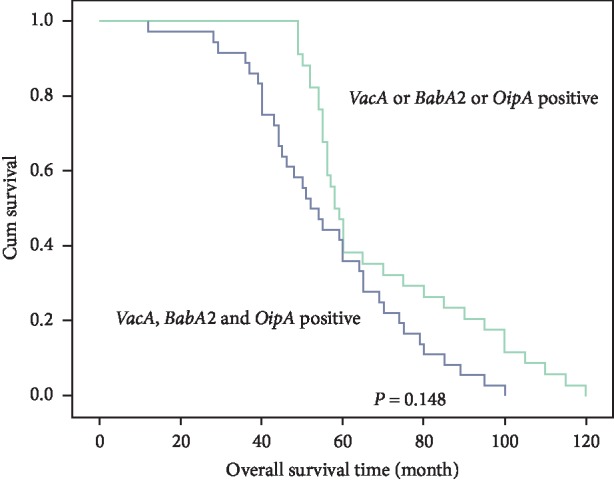
Overall survival time of gastric cancer patients infected with H. pylori two-genotypes combination (vacA+/babA2+, vacA+/oipA+ and babA2+/oipA+) and H. pylori three-genotypes combination (vacA+/babA2+/oipA+).
4. Discussion
Our study was a cross-sectional study, which investigated the presence of cagA, vacA, iceA2, babA2, and oipA genes in H. pylori infected patients with chronic gastritis, precancerous gastric lesions, and GC. The rate of H. pylori infection in this study was 83%, which was comparable with a retrospective study in the Northeastern region of Thailand [49] and a prospective study in Japan [36]. This study was the first report on the associations between virulence genes and the risk of chronic gastritis, precancerous gastric lesions, and GC in Thailand. Our results revealed that the vacA gene was associated with chronic gastritis whereas vacA, babA2, and oipA genes were associated with increased risk of GC. These indicated that vacA and babA2 genes influenced chronic gastritis and precancerous gastric lesions, respectively. Meanwhile, the babA2 and oipA genes had virulence potential on GC development. The babA2 and oipA genes were present mostly in H. pylori-positive GC; it seems vacA, babA2, and oipA genes exhibited different levels of virulence. It is probable that vacA alone was not directly associated with gastric carcinogenesis. Although, vacA effects on disruption of gastric epithelial barrier function and modulation of the inflammatory response, vacA also suppresses the activation of ERK1/2 mitogen-activated protein (MAP) kinase suggesting that H. pylori can avoid the induction of excess cellular damage and maintain long-term colonization [5]. Therefore, vacA individual may develop chronic gastritis.
H. pylori infection induced cell-mediated immunity. Th1 cells play a central role in H. pylori immune response. Predominant Th17 expression was positively correlated with the degree of immunopathologic reactions resulting in peptic ulcers [6]. Furthermore, increasing of T-bet + cells and the mucosal INF-γ expression related to the degree of H. pylori density in infected patients can lead to ulcer or GC [7]. In addition, the roles of babA2 and oipA proteins might have the potential to exert pressure on H. pylori by enhancing the production of free radicals that cause mutations in target cells and the neoplastic clones are established [39]. Tumor necrosis factor alpha (TNF-α) plays major roles in the growth, invasion, and metastasis of neoplasm called a perigenetic pathway [50]. The TNF-α inducing protein (Tipα) from H. pylori binds to and enters the nucleus through a specific binding molecule, which acts as a carcinogen [51] and contributes to GC. Although, cagA is recognized as an oncoprotein and confers oncogenesis, the presence of cagA was not associated with any clinical outcome, suggesting that the cagA gene present in East Asian strains might not be influential of its risk enough.
Interestingly, genotype combination was associated with GC. The precancerous gastric lesions with H. pylori vacA+/babA2+/oipA+ genotype infection had high association with 4.3-fold increased risk of GC development. The babA2 gene has been strongly associated with vacA and increased risk of GC development [24]; however, in this study, the babA2+/vacA+ genotype did not have increased risk of GC. Taken together, oipA was associated with higher neutrophil activity and IL-8 secretion and showed toxic effects by an apoptosis-triggered cascade via signaling that affected the Bax/Bcl-2 protein ratio and cleaved-caspase 3 level, leading to a mitochondrial apoptotic cascade [52–54]. These findings suggest that the vacA+/babA2+/oipA+genotype may contribute to the genotoxicity caused by DNA damage and aberrant methylation of genes in H. pylori-related gastric carcinogenesis. A cohort study with long-term follow-up demonstrated that infection with cagA genotype was associated with increased risk of precancerous gastric lesions progression [55, 56]. Therefore, vacA, babA2, and oipA have been implicated in the development of GC. Regarding precancerous gastric lesions after H. pylori eradication, H. pylori-induced chronic inflammation can provide the seed of cascade leading to GC, which can continuously progress even in the absence of H. pylori [57]. The patients with H. pylori and IM have more than 6.4-fold increased risk of GC than that of the patients with H. pylori but without IM [36]. However, 2.9% GC was developed in individuals during the mean follow-up of 7.8 years [36]. Therefore, precancerous gastric lesions patients infected with the H. pylori vacA+/babA2+/oipA+ genotype were prone to developing GC compared with patients infected with other combination genotypes. However, in the present study, the overall survival time of GC patients with the H. pylori vacA+/babA2+/oipA+ genotype infection was not reduced.
The limitations of the present study were that subanalysis for the precancerous gastric lesions was not performed and the number of patients involved was small and there was no regular follow-up on them. Furthermore, several virulence factors of H. pylori were not investigated. The expression of vacA, babA2, and oipA protein should be evaluated in a future study of GC carcinogenesis to determine the underlying mechanisms associated with GC development in precancerous gastric lesions patients.
5. Conclusion
This study provided important information regarding the presence of virulence genes in different clinical outcomes of H. pylori infection. Precancerous gastric lesions of patients infected with H. pylori vacA+/babA2+/oipA+ genotype infection have an increased risk of GC. The H. pylori vacA+/babA2+/oipA+ genotype might prove helpful in predicting individuals in the high-risk group of GC in the Thai population.
Acknowledgments
This research was supported by a grant from Suranaree University of Technology (SUT) and by Office of the Higher Education Commission under NRU project of Thailand.
Data Availability
No data were used to support this study.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Covacci A., Telford J. L., Del Giudice G., Parsonnet J., Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284(5418):1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 2.Polk D. B., Peek R. M., Jr. Helicobacter pylori: gastric cancer and beyond. Nature Reviews Cancer. 2010;10(6):403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peek R. M., Jr., Blaser M. J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature Reviews Cancer. 2002;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 4.Kao C. Y., Sheu B. S., Wu J. J. Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomedical Journal. 2016;39(1):14–23. doi: 10.1016/j.bj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wroblewski L. E., Peek R. M., Jr., Wilson K. T. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clinical Microbiology Reviews. 2010;23(4):713–739. doi: 10.1128/cmr.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagheri N., Razavi A., Pourgheysari B., et al. Up-regulated Th17 cell function is associated with increased peptic ulcer disease in Helicobacter pylori-infection. Infection, Genetics and Evolution. 2018;60:117–125. doi: 10.1016/j.meegid.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Bagheri N., Shirzad H., Mirzaei Y., et al. T-bet+ cells polarization in patients infected with Helicobacter pylori increase the risk of peptic ulcer development. Archives of Medical Research. 2019;50(3):113–121. doi: 10.1016/j.arcmed.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Yamaoka Y., Graham D. Y. Helicobacter pylorivirulence and cancer pathogenesis. Future Oncology. 2014;10(8):1487–1500. doi: 10.2217/fon.14.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saeidi Y., Pournajaf A., Gholami M., et al. Determination of Helicobacter pylori virulence-associated genes in duodenal ulcer and gastric biopsies. Medical Journal of the Islamic Republic of Iran. 2017;31:555–559. doi: 10.14196/mjiri.31.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pride D. T., Meinersmann R. J., Blaser M. J. Allelic variation within Helicobacter pylori babA and babB. Infection and Immunity. 2001;69(2):1160–1171. doi: 10.1128/iai.69.2.1160-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Posselt G., Backert S., Wessler S. The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces a multi-step process in pathogenesis. Cell Communication and Signaling. 2013;11(1) doi: 10.1186/1478-811x-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miftahussurur M., Yamaoka Y. Helicobacter pylori virulence genes and host genetic polymorphisms as risk factors for peptic ulcer disease. Expert Review of Gastroenterology & Hepatology. 2015;9(12):1535–1547. doi: 10.1586/17474124.2015.1095089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyanova L., Yordanov D., Gergova G., Markovska R., Mitov I. Association of iceA and babA genotypes in Helicobacter pylori strains with patient and strain characteristics. Antonie Van Leeuwenhoek. 2010;98(3):343–350. doi: 10.1007/s10482-010-9448-y. [DOI] [PubMed] [Google Scholar]
- 14.Lehours P., Menard A., Dupouy S., et al. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphoma. Infection and Immunity. 2004;72(2):880–888. doi: 10.1128/iai.72.2.880-888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaoka Y., Kikuchi S., El–Zimaity H. M. T., Gutierrez O., Osato M. S., Graham D. Y. Importance of Helicobacter pylori OipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123(2):414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 16.Souod N., Sarshar M., Dabiri H., et al. The study of the OipA and DupA genes in Helicobacter pylori strains and their relationship with different gastroduodenal diseases. Gastroenterology and Hepatology from Bed to Bench. 2015;8(Suppl 1):S47–S53. [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., He C., Chen M., Wang Z., Xing C., Yuan Y. Association of presence/absence and on/off patterns of Helicobacter pylori OipA gene with peptic ulcer disease and gastric cancer risks: a meta-analysis. BMC Infectious Diseases. 2013;13:p. 555. doi: 10.1186/1471-2334-13-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Y. L., Huang H. L., Huang B. S., et al. Combination of OipA, BabA, and SabA as candidate biomarkers for predicting Helicobacter pylori-related gastric cancer. Scientific Reports. 2016;6:p. 36442. doi: 10.1038/srep36442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alm R. A., Ling L. S., Moir D. T., et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397(6715):176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 20.Odenbreit S., Puls J., Sedlmaier B., Gerland E., Fischer W., Haas R. Translocation of Helicobacter pylori cagA into gastric epithelial cells by type IV secretion. Science. 2000;287(5457):1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 21.Argent R. H., Kidd M., Owen R. J., Thomas R. J., Limb M. C., Atherton J. C. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology. 2004;127(2):514–523. doi: 10.1053/j.gastro.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Azuma T. Helicobacter pylori CagA protein variation associated with gastric cancer in Asia. Journal of Gastroenterology. 2004;39(2):97–103. doi: 10.1007/s00535-003-1279-4. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host & Microbe. 2014;15(3):306–316. doi: 10.1016/j.chom.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Dadashzadeh K., Peppelenbosch M. P., Adamu A. I. Helicobacter pylori pathogenicity factors related to gastric cancer. Canadian Journal of Gastroenterology and Hepatology. 2017;2017:6. doi: 10.1155/2017/7942489.7942489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cover T. L., Blanke S. R. Helicobacter pylori vacA, a paradigm for toxin multifunctionality. Nature Reviews Microbiology. 2005;3(4):320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 26.Palframan S. L., Kwok T., Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Frontiers in Cellular and Infection Microbiology. 2012;2:p. 92. doi: 10.3389/fcimb.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakhti S. Z., Latifi-Navid S., Mohammadi S., et al. Relevance of Helicobacter pylori vacA 3′-end region polymorphism to gastric cancer. Helicobacter. 2016;21(4):305–316. doi: 10.1111/hel.12284. [DOI] [PubMed] [Google Scholar]
- 28.Kabamba E. T., Tuan V. P., Yamaoka Y. Genetic populations and virulence factors of Helicobacter pylori. Infection, Genetics and Evolution. 2018;60:109–116. doi: 10.1016/j.meegid.2018.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogiwara H., Sugimoto M., Ohno T., et al. Role of deletion located between the intermediate and middle regions of the Helicobacter pylori vacA gene in cases of gastroduodenal diseases. Journal of Clinical Microbiology. 2009;47(11):3493–3500. doi: 10.1128/jcm.00887-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peek R. M., Jr., Thompson S. A., Donahue J. P., et al. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, IceA, that is associated with clinical outcome. Proceedings of the Association of American Physicians. 1998;110(6):531–544. [PubMed] [Google Scholar]
- 31.van Doorn L. J., Figueiredo C., Sanna R., et al. Clinical relevance of the CagA, VacA, and IceA status of Helicobacter pylori. Gastroenterology. 1998;115(1):58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 32.Yamaoka Y., Kodama T., Gutierrez O., Kim J. G., Kashima K., Graham D. Y. Relationship between Helicobacter pylori IceA, CagA, and VacA status and clinical outcome: studies in four different countries. Journal of Clinical Microbiology. 1999;37(7):2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aghdam S. M., Sardari Z., Safaralizadeh R., et al. Investigation of association between Oipa and IceA1/IceA2 genotypes of Helicobacter pylori and gastric cancer in Iran. Asian Pacific Journal of Cancer Prevention. 2014;15(19):8295–8299. doi: 10.7314/apjcp.2014.15.19.8295. [DOI] [PubMed] [Google Scholar]
- 34.Biernat M. M., Gościniak G., Iwańczak B. Prevalence of Helicobacter pylori Caga, Vaca, Icea, Baba2 genotypes in Polish children and adolescents with gastroduodenal disease. Postępy Higieny I Medycyny Doświadczalnej. 2014;68:1015–1021. doi: 10.5604/17322693.1118211. [DOI] [PubMed] [Google Scholar]
- 35.Siurala M., Varis K., Wiljasalo M. Studies of patients with atrophic gastritis: a 10–15-year follow-up. Scandinavian Journal of Gastroenterology. 1966;1(1):40–48. doi: 10.1080/00365521.1966.11800612. [DOI] [PubMed] [Google Scholar]
- 36.Uemura N., Okamoto S., Yamamoto S., et al. Helicobacter pylori infection and the development of gastric cancer. New England Journal of Medicine. 2001;345(11):784–789. doi: 10.1056/nejmoa001999. [DOI] [PubMed] [Google Scholar]
- 37.Ohata H., Kitauchi S., Yoshimura N., et al. Progression of chronic atrophic gastritis associated with Helicobacter pylori infection increases risk of gastric cancer. International Journal of Cancer. 2004;109(1):138–143. doi: 10.1002/ijc.11680. [DOI] [PubMed] [Google Scholar]
- 38.Kim N., Park R. Y., Cho S.-I., et al. Helicobacter pylori infection and development of gastric cancer in Korea. Journal of Clinical Gastroenterology. 2008;42(5):448–454. doi: 10.1097/mcg.0b013e318046eac3. [DOI] [PubMed] [Google Scholar]
- 39.Park Y. H., Kim N. Review of atrophic gastritis and intestinal metaplasia as a premalignant lesion of gastric cancer. Journal of Cancer Prevention. 2015;20(1):25–40. doi: 10.15430/jcp.2015.20.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tongtawee T., Dechsukhum C., Leeanansaksiri W., et al. Improved detection of Helicobacter pylori infection and premalignant gastric mucosa using “site specific biopsy”: a randomized control clinical trial. Asian Pacific Journal of Cancer Prevention. 2015;16(12):8487–8490. doi: 10.7314/apjcp.2015.16.12.4885. [DOI] [PubMed] [Google Scholar]
- 41.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 42.Boonjakuakul J. K., Canfield D. R., Solnick J. V. Comparison of Helicobacter pylori virulence gene expression in vitro and in the rhesus macaque. Infection and Immunity. 2005;73(8):4895–4904. doi: 10.1128/iai.73.8.4895-4904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schabereiter-Gurtner C., Hirschl A. M., Dragosics B., et al. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. Journal of Clinical Microbiology. 2004;42(10):4512–4518. doi: 10.1128/jcm.42.10.4512-4518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loh J. T., Shaffer C. L., Piazuelo M. B., et al. Analysis of cagA in Helicobacter pylori strains from Colombian populations with contrasting gastric cancer risk reveals a biomarker for disease severity. Cancer Epidemiology Biomarkers & Prevention. 2011;20(10):2237–2249. doi: 10.1158/1055-9965.epi-11-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boonjakuakul J. K., Syvanen M., Suryaprasad A., Bowlus C. L., Solnick J. V. Transcription profile of Helicobacter pyloriin the human stomach reflects its physiology in vivo. The Journal of Infectious Diseases. 2004;190(5):946–956. doi: 10.1086/423142. [DOI] [PubMed] [Google Scholar]
- 46.Essawi T., Hammoudeh W., Sabri I., Sweidan W., Farraj M. A. Determination of Helicobacter pylori virulence genes in gastric biopsies by PCR. ISRN Gastroenterol. 2013;2013:4. doi: 10.1155/2013/606258.606258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bibi F., Alvi S. A., Sawan S. A., et al. Detection and genotyping of Helicobacter pylori among gastric ulcer and cancer patients from Saudi Arabia. Pakistan Journal of Medical Sciences. 2017;33(2):320–324. doi: 10.12669/pjms.332.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sohrabi M., Khashei R., Alizadeh M., et al. Low rate of babA2 genotype among Iranian Helicobacter pylori clinical isolates. Journal of Clinical and Diagnostic Research. 2017;11(7):DC32–DC36. doi: 10.7860/jcdr/2017/24810.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tongtawee T., Wattanawongdon W., Simawaranon T. Effects of periodontal therapy on eradication and recurrence of Helicobacter pylori infection after successful treatment. Journal of International Medical Research. 2019;47(2):875–883. doi: 10.1177/0300060518816158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuji S., Kawai N., Tsujii M., Kawano S., Hori M. Review article: inflammation-related promotion of gastrointestinal carcinogenesis-a perigenetic pathway. Alimentary Pharmacology and Therapeutics. 2003;18(s1):82–89. doi: 10.1046/j.1365-2036.18.s1.22.x. [DOI] [PubMed] [Google Scholar]
- 51.Suganuma M., Yamaguchi K., Ono Y., et al. TNF-α-inducing protein, a carcinogenic factor secreted from H. pylori, enters gastric cancer cells. International Journal of Cancer. 2008;123(1):117–122. doi: 10.1002/ijc.23484. [DOI] [PubMed] [Google Scholar]
- 52.Salimzadeh L., Bagheri N., Zamanzad B., et al. Frequency of virulence factors in Helicobacter pylori-infected patients with gastritis. Microbial Pathogenesis. 2015;80:67–72. doi: 10.1016/j.micpath.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 53.Franco A. T., Johnston E., Krishna U., et al. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Research. 2008;68(2):379–387. doi: 10.1158/0008-5472.can-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teymournejad O., Mobarez A. M., Hassan Z. M., Talebi Bezmin Abadi A. Binding of the Helicobacter pylori OipA causes apoptosis of host cells via modulation of bax/bcl-2 levels. Scientific Reports. 2017;7(1):p. 8036. doi: 10.1038/s41598-017-08176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demirturk L., Ozel A. M., Yazgan Y., et al. CagA status in dyspeptic patients with and without peptic ulcer disease in Turkey: association with histopathologic findings. Helicobacter. 2001;6(2):163–168. doi: 10.1046/j.1523-5378.2001.00024.x. [DOI] [PubMed] [Google Scholar]
- 56.Saruc M., Demir M. A., Kucukmetin N., Kandiloglu A. R., Akarca U. S., Yuceyar H. Histological and clinical predictive value of determination of tissue cagA status by PCR in Helicobacter pylori infected patients; results of the large population based study in western Turkey. Hepatogastroenterology. 2002;49(45):878–881. [PubMed] [Google Scholar]
- 57.Kato S., Matsukura N., Tsukada K., et al. Helicobacter pylori infection-negative gastric cancer in Japanese hospital patients: incidence and pathological characteristics. Cancer Science. 2007;98(6):790–794. doi: 10.1111/j.1349-7006.2007.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used to support this study.