Abstract
The respective effects of tissue alarmins interleukin (IL)-15 and interferon beta (IFNβ), and IL-21 produced by T cells on the reprogramming of cytotoxic T lymphocytes (CTLs) that cause tissue destruction in celiac disease is poorly understood. Transcriptomic and epigenetic profiling of primary intestinal CTLs showed massive and distinct temporal transcriptional changes in response to tissue alarmins, while the impact of IL-21 was limited. Only anti-viral pathways were induced in response to all the three stimuli, albeit with differences in dynamics and strength. Moreover, changes in gene expression were primarily independent of changes in H3K27ac, suggesting that other regulatory mechanisms drive the robust transcriptional response. Finally, we found that IL-15/IFNβ/IL-21 transcriptional signatures could be linked to transcriptional alterations in risk loci for complex immune diseases. Together these results provide new insights into molecular mechanisms that fuel the activation of CTLs under conditions that emulate the inflammatory environment in patients with autoimmune diseases.
Keywords: Autoimmune disease, Cytotoxic T lymphocytes, Intraepithelial lymphocytes, IFNβ, IL-15, IL-21, Tissue-resident lymphocytes
Highlights
-
•
We report the epigenetic and transcriptomic response of IE-CTLs to tissue-derived alarmins and adaptive cytokine.
-
•
IL-15 and IFNβ induce massive and distinct transcriptional changes in IE-CTLs, while the impact of IL-21 was limited.
-
•
Genes associated with immune-mediated intestinal diseases are enriched in genes responding to IL-15, IFNβ and IL-21 in IE-CTLs.
1. Introduction
Tissue-resident cytotoxic T lymphocytes (CTLs) have been shown to play a critical role in mediating tissue destruction in organ-specific autoimmune disorders [1,2]. Understanding the molecular mechanisms that underlie CTL activation within the tissue environment will help identify new therapeutic strategies. CTL differentiation, activation and function are greatly influenced by the tissue microenvironment, where they sense and respond to microbial products, cytokines and alarmins [3,4]. Specifically, chronic production of innate immune or epithelial cell-derived cytokines (e.g. IFNβ and IL-15, also referred to as tissue alarmins) and T-cell-derived cytokines (e.g. IL-21) have been shown to modulate the expansion and/or effector functions of CTLs and to contribute to the development of tissue-specific immune-mediated disorders including celiac disease (CeD), rheumatoid arthritis (RA), inflammatory bowel disease (IBD) and type I diabetes (T1D) [[5], [6], [7], [8]]. However, because it is difficult to collect materials from human tissues from the site of disease, our understanding of the contribution of tissue-derived alarmins versus the role of cytokines produced by antigen-specific T cells in the development of autoimmune disorders and activation of tissue-resident CTLs remains poorly understood [9,10].
Intraepithelial CTL (IE-CTLs) are tissue-resident cytotoxic T lymphocytes that are located in between intestinal epithelial cells. IE-CTLs protect against infection directly, by killing infected cells, and indirectly, by promoting the immune response via production of cytokines such as IFNγ [10,11]. They are also the key effector cell type in mediating the tissue destruction central to the pathogenesis of some autoimmune diseases, including CeD and T1D8, [12]. Type-1 IFNs (IFN-1), IL-15 and IL-21 are all upregulated in active CeD, a complex T-cell-mediated enteropathy with an autoimmune component that is induced by dietary gluten and characterized by the presence of villous atrophy in the intestinal mucosa [8]. Interestingly, IL-15 and IL-21 are absent in patients who have lost tolerance to dietary gluten but who have not yet developed villous atrophy (a condition called “potential CeD”), indicating that these alarmins may play a crucial part in tissue destruction [[13], [14], [15]]. IFNβ [16] and IL-15 [[17], [18], [19]] are upregulated in intestinal epithelial cells and antigen presenting cells, and IL-21 is produced by gluten-specific CD4+ T cells [20], but the effect of IFNβ, IL-15 and IL-21 in the activation of IE-CTLs remains to be determined.
Here we report the first comparative characterization of the dynamics of transcriptomic and epigenetic changes in primary-tissue-resident IE-CTL lines upon exposure to tissue-derived alarmins and adaptive cytokines involved in disease pathophysiology. We stimulated CD8+ TCRαβ IE-CTLs isolated from intestinal biopsies with IFN-1 (IFNβ), IL-15 and IL-21 at multiple time points. This allowed us to characterize the overall changes in gene expression and the pathways affected under these different inflammatory conditions. We analyzed genome-wide H3K27ac signatures to identify epigenetic changes critical for the modulation of transcriptional responses to these cytokines.
2. Material and methods
Human subjects: Control individuals who were undergoing endoscopies and biopsies for functional intestinal disorders of non-celiac origin and CeD patients who were diagnosed based on the presence of elevated anti-transglutaminase antibodies in serum, expression of HLA DQ2 or DQ8, presence of increased IE-CTLs, partial or total duodenal villous atrophy, crypt hyperplasia on duodenal biopsy and clinical response to a gluten free diet. All subjects gave written informed consent and all protocols were approved by the University of Chicago Institutional Review Board.
IE-CTL cultures: TCRαβ+ CD8+ short-term IE-CTL cell lines derived from 4 to 6 duodenal biopsies from CeD patients (n = 4) or healthy control individuals (n = 4) were generated as previously described [18,21]. Briefly, cells from the intraepithelial lymphocyte compartment were isolated via 1 h of mechanical disruption in media containing 1% dialyzed fetal bovine serum (Biowest), 2 mM EDTA (Corning) and 1.5 mM MgCl2 (Thermo Fisher Scientific). Up to 10,000 cells expressing both TCRαβ (IP26, BioLegend) and CD8α (RPAT-8, BioLegend) were collected via fluorescence-activated cell sorting (FACS) on a FACSAria II cell sorter (BD Biosciences) and expanded in vitro with a mixture of irradiated heterologous peripheral blood mononuclear cells (PBMCs) from 2 donors and EBV (Epstein Barr virus) transformed B cells in RPMI 1640 (GIBCO) medium supplemented with 1 μg/ml PHA-L (Calbiochem) and 10% human serum albumin (Atlanta Biologicals) and maintained with 100 units/ml IL-2 (NIH) over the course of expansion. After 14–21 days of expansion, aliquots of cells were frozen for future use to ensure all experiments could be done on the full set of cell lines to avoid experimental batch effects. Representative plots of the gating strategy for the ex-vivo isolation of TCRαβ+ CD8+ IE-CTLs and the cell purity checking after in vitro expansion are depicted in Supplementary Fig. 1.
IE-CTL stimulation: IE-CTLs were thawed and expanded for 12 days as described above. To ascertain cell viability (>80%), cells were analyzed using LIVE/DEAD fixable Aqua Cell Stain Kit (Life Technologies), and cell activation (<10%) was determined by forward and sideward scatter analysis using the LSR Fortessa™ (BD Biosciences) flow cytometer. After 12 days of culture, cells were washed, starved for IL-2 for 48 h, and subsequently stimulated for 30 min, 3, 4 or 24 h with IL-15 (20 ng/ml, Biolegend, cat 570304), IL-21 (3 ng/ml, Biolegend, cat 571204) or IFNβ (300 ng/ml, Pbl Assay science, cat 11410-2). Unstimulated IE-CTLs were included as control. Prior to the assay, the induction of granzyme B (GZMB), IFNG and BCL2 was measured by qPCR, as a proxy for IE-CTL activation [22,23] to determine the optimal cytokine concentration to induce stable expression of the proxies. After stimulation for the indicated periods, cells were collected by centrifugation (10 min at 400 g) and pellets were lysed in buffer RTL of the RNeasy plus mini kit (Qiagen) for subsequent RNA isolation. A fraction of control cells and cells stimulated for 3 h with IL-15, IL-21 and IFNβ were collected for subsequent chromatin immunoprecipitation. Importantly, all IE-CTL lines from controls and CeD patients were generated and cultured using the same protocols. Of note, IE-CTLs are effector memory T cells, and the short-term lines preserve their tissue-resident status as assessed by their CD103+CD69+ and CD62− expression pattern (Meresse, B. et al. [24] and Supplementary figure 2A). Furthermore, the expression levels of IFNAR, IL2RB, and IL21R, through which IFNβ, IL15, and IL-21 signal, respectively, were comparable in ex vivo IE-CTLs and primary IE-CTLs cell lines (Supplementary Figure 2B).
RNA sequencing (RNA-seq): RNA from unstimulated or stimulated IE-CTLs was isolated with the RNeasy plus mini kit. RNA concentration and quality were measured using the Nanodrop 1000 Spectrophotometer (Thermo Scientific) and the high-sensitivity RNA analysis kit (Experion, software version 3.0, Bio-Rad). RNA-seq libraries were prepared from 1 μg of total RNA using the Quant seq 3′ kit (Lexogen). The libraries were sequenced on the Nextseq500 (Illumina) yielding at least fifteen million sequence reads per sample. The fastQ files were then trimmed for low quality reads, adaptors and poly-A tails. Trimmed fastQ files were aligned to build a human_g1k_v37 ensemble Release 75 reference genome using hisat [25] with default settings and sorted using SAMtools [26]. After alignment, gene level quantification was performed by HTSeq-count [27] using default mode = union. A modified Ensembl version 75 gtf file mapping only to the last 5’ 500 bps per gene was used as a gene annotation database to prevent counting of reads mapping to intra-genic A-repeats. Differential expression analysis between time points was performed using the DESeq2 [28] package in R. We defined differentially expressed genes as genes with an absolute log2 fold change >1 and a False Discovery Rate (FDR) ≤ 0.01 between untreated controls and cytokine-treated samples. Principal component analysis (PCA) and heatmaps of hierarchical clustering were done using R base functions (v3.4) to identify the most relevant transcriptional and epigenomic profiles. Gene set enrichment analysis (GSEA) [29] and Reactome pathways [30] were used to detect which biological processes and pathways were enriched in the groups of differentially expressed genes (DEGs). Co-expression analysis and enrichment analysis of co-expression networks was performed using Gene Network v2.0 (www.genenetwork.nl) [31].
Chromatin immunoprecipitation and library preparation: Cell pellets from unstimulated or 3 h stimulated IE-CTLs were cross-linked with 1% formaldehyde at room temperature for 5 min. Nuclei were isolated with the truChIP chromatin shearing kit (Covaris). Chromatin was sheared for 6 min by sonication in the S220 sonicator (Covaris; 140 W, 5% duty factor, 200 CPB). Chromatin immunoprecipitation and ChIP-seq library preparation were performed according to protocols described by the Blueprint consortium (http://www.blueprint-epigenome.eu). In brief, 22 μl of chromatin was incubated overnight at 4 °C with 1 μg of H3K27ac antibody (Diagenode, cat C15410196), followed by 1 h incubation with protein A- and G-coated beads (Invitrogen, Dynabeads. Catalog 1003D and 1004D). The chromatin was reverse crosslinked for 4 h at 65 °C in the presence of proteinase K (0.1 μg/μl, Roche, cat 3115887001), then purified using the QIAquick MinElute PCR Purification Kit (Qiagen). The libraries were prepared using the Kapa Hyper Prep Kit (Kapa biosystem). After end repair, tailing and Nexflex adapter ligation (Nextera), the libraries were amplified for 10 cycles, then purified with the QIAquick MinElute PCR Purification Kit. DNA fragments 300 bp in size were purified with LabChip Caliper XT 700 electrophoresis system (PerkinElmer). Library concentration and fragment size distribution were analyzed using the High sensitivity NGS fragment analysis kit using the Fragment Analyzer (Advanced Analytical). ChIP-seq libraries were sequenced (50 bp single end reads) using the Nextseq500 sequencer (Illumina; >15 million reads per sample). As a control for background noise, input DNA (sheared chromatin that underwent all the crosslinking and sonication steps, but not immunoprecipitation) was included. Sequence alignment was performed as described above for the RNA-seq reads. The H3K27ac peaks of each sample were determined using MACS 2.0 [32] applying standard parameters, while controlling for experimental and technical bias. Differential peak analysis between unstimulated samples and cytokine-stimulated samples was performed using DiffBind in DESeq2 mode to detect differential peaks with an FDR ≤0.01. H3K27ac profiles of representative genes were visualized using the integrative genomics viewer after normalization of bed files by total read count per sample [33].
2.1. Differential expression analysis on CeD and IBD biopsies
The CeD dataset consisted of 5 control (healthy) colonoscopy biopsies, and 6 samples derived from CeD patients. Library preparation was performed using the Illumina TruSeq stranded total RNA library kit, including a riboZero rRNA depletion step, and subsequently sequenced using HiSeq 2500. The resulting fastQ files were filtered for low quality reads and adaptors. Alignment was done using hisat [25] against the human_g1k_v37 ensemble Release 75 reference genome with default settings and sorted using SAMtools [26]. Gene level quantification was done with HTSeq-count [27] using default mode = union. After filtering out all genes with 0 reads in all samples, differential expression analysis between controls and CeD patients was performed using DESeq228. DEGs were defined by using a threshold of adjusted P value ≤ 0.01.
Summary statistics for the differential expression analysis on the (unpublished data) IBD data were provided by the authors. In short, the cohort contained only IBD patients with inflamed (n = 112) and non-inflamed (n = 176) tissue. The differential expression analysis was performed using a generalized mixed linear model [34], using as covariates the first 18 principal components (minus PC2 which was significantly associated with inflammation) while considering for random effects using multiple measurements per biopsies.
The summarized count table of gene expression levels for the GSE111889 dataset was downloaded directly from GEO. Genes and samples with a total sum of 0 reads across all genes or samples were removed from the analysis. After normalizing for library size using the log2 transformed trimmed mean for M counts (TMM), a PCA performed at a sample level to find any potential outliers or mislabeled samples. A total of 4 samples labeled as ileum that were clustering within colon/rectum samples or vice versa were removed from further analysis. Samples where then separated into ileum and colon/rectum samples and a differential expression analysis across IBD and non-IBD samples was carried out independently between different tissues using the R package DESeq228.
2.2. Enrichment of CeD and IBD DEGs in DEG clusters of IE-CTLs upon cytokine stimulation
We used available RNA-seq data from intestinal biopsies obtained from a CeD cohort consisting of 5 healthy controls and 6 patients. Two IBD cohorts where also included in the analysis, one including only IBD patients with inflamed (n = 112) and non-inflamed (n = 176) tissue (unpublished data), and a second one that contains tissue biopsies from colon and ileum from IBD and non IBD patients (GSE111889)35. To identify DEGs between patients and their respective controls in each cohort we applied a filter of adjusted p value ≤ 0.01. We then assessed if any of the transcriptional clusters upon IL-15, IFNβ and IL-21 stimulations where enriched for DEG between disease and controls (CeD and IBD) and between inflamed and non-inflamed tissue (IBD) using a fisher exact test. For this analysis, we used as background set the 25,862 genes that remained after filtering the genes in our current IE-CTL data set (genes non-expressed, with variance = 0 and less than 10 counts across all samples).
2.3. Association between risk loci for autoimmune diseases and DEG clusters in IE-CTLs
ImmunoChip summary statistics were downloaded from https://www.immunobase.org/and https://www.ibdgenetics.org/for several autoimmune disease traits: CeD, T1D, multiple sclerosis (MS), psoriasis (PSO), primary biliary cirrhosis (PBC), ankylosing spondylitis (AS), RA, IBD, ulcerative colitis (UC) and Crohn's Disease (CD) [[36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]] For each trait, we pruned all genome-wide significant (5 × 10−8) single nucleotide polymorphisms (SNPs) by chromosomal proximity (2 Mb) while selecting only the one with the lowest p-value per locus, which we defined as top-SNP. Next, we filtered out all SNPs from non-autosomal chromosomes because not all of the genome-wide association studies (GWAS) had assessed the X chromosome. Then we defined all neighboring genes located within a window of 125 kilo bases (kb) on both sides of each top-SNP as GWAS-genes. Overlapping these GWAS-genes with the differentially expressed gene clusters from IE-CTL stimulation (as described in Fig. 2), allowed us to calculate the number of top-SNPs for which GWAS-genes were present in each gene cluster. This frequency was converted to percentages by dividing by the total number of GWAS top-SNPs per autoimmune disease trait.
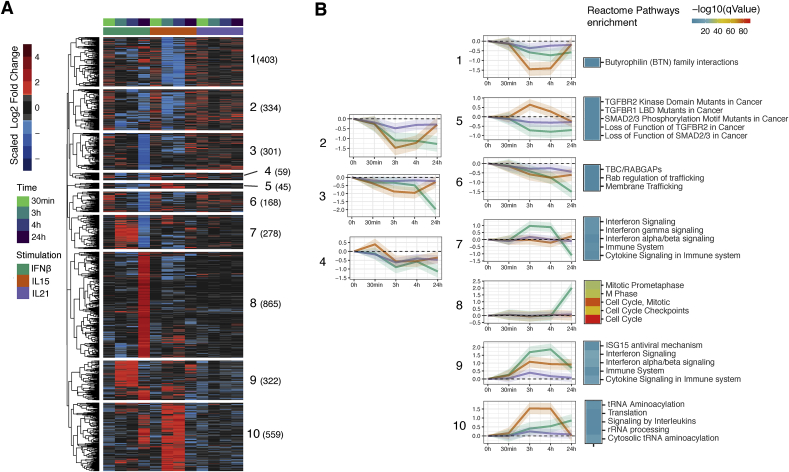
Fig. 2.
Dynamic profile of DEGs in IE-CTLs in response to stimulation with tissue alarmins and adaptive cytokine. (A) Unsupervised hierarchical clustering of all DEGs (|log2 fold change|>1, FDR ≤ 0.01) across stimulations (n = 8 samples per timepoint and condition). Ten major clusters were defined as biologically relevant. Colors indicate scaled log2 fold change in expression with respect to unstimulated samples, with red and blue indicative of a two-fold or more increase or decrease in expression, respectively. (B) Line plots illustrating the mean log2 fold changes in expression of genes in a given cluster. Shades represent the standard error of the log2 fold changes. Top 5 most significant pathways from GSA identified by Reactome (q-value ≤ 0.01) in DEGs (only clusters where enrichment was found are depicted). Colors (key) indicate significance. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To ascertain the statistical significance of the percentage of GWAS top-SNPs represented in a given gene cluster, we computed an empirical null distribution of the expected percentage GWAS top-SNPs using randomized sets of genes. This empirical null distribution was calculated using 10,000 random gene sets, matching the exact same number of genomic regions as the query gene set (since genes are not randomly distributed throughout the genome). Finally, we calculated the percentage of GWAS top-SNPs represented in each of these random gene sets. Using this empirical null distribution, we were able to ascertain an empirical p-value by taking the number of times the null distribution had a higher percentage of overlaps with GWAS top-SNPs per autoimmune disease trait than the observed percentage with the queried gene clusters, divided by the total number of random gene sets (n = 10,000). This empirical p-value was then corrected for multiple testing using the FDR. By applying this method, we assessed the statistical significance of the overlapped percentage without bias for the number for genome wide loci in each trait and the number of genes per each transcriptional cluster.
3. Results
3.1. Tissue alarmins and adaptive cytokines induce distinct and shared responses in IE-CTLs
To study the temporal transcriptional changes in CD8+ IE-CTLs, we profiled IE-CTLs before and after stimulation with IFNβ, IL-15 or IL-21 at four time points (30 min, 3, 4 and 24 h). To ensure that optimal concentrations of cytokines were used, we performed a titration experiment to select the minimum concentration of cytokines that induced maximal expression of a set of known marker genes (Supplementary Fig. 3). Using these optimal concentrations, we observed specific and distinct temporal transcriptional responses upon stimulation for all three stimuli (IFNβ, IL-15 and IL-21). Of note, we observed in a transcriptome-wide PCA that celiac and control derived samples clustered together by projecting principal component (PC) 1, PC2 and PC3 (Supplementary Fig. 4A and B). Moreover, we evaluated the correlation of log2 fold changes in gene expression of differentially expressed genes upon cytokine stimulations between CeD and healthy controls; we observed a high spearman correlation coefficient across all the sets of DEGs for CeD or control derived samples (time points and stimulations, mean ρ = 0.81, Supplementary Fig. 4C). Thus, since the differences in their transcriptome were minimal, we combined all samples to perform differential gene expression analysis to increase power for subsequent analyses (Fig. 1A and Supplementary Table.1).
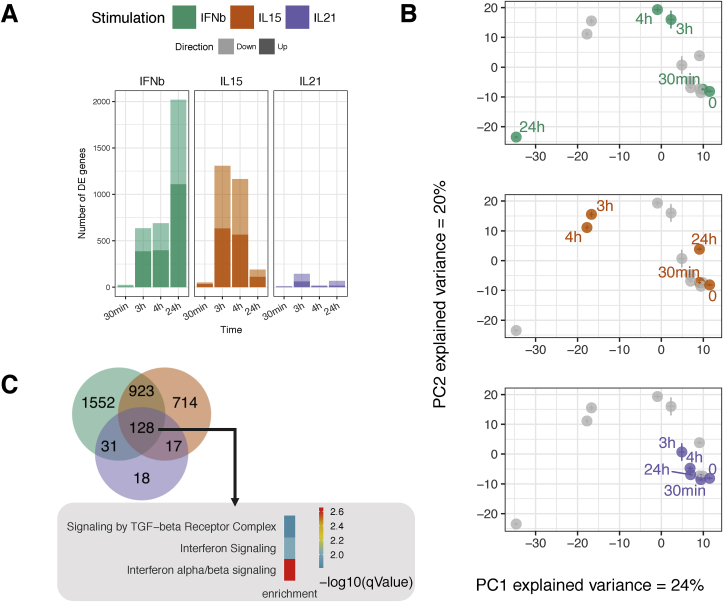
Fig. 1.
Distinct and common transcriptional response of IE-CTLs in response to tissue alarmins and adaptive cytokine. After 12 days in culture, IE-CTLs were starved for 48 h, then stimulated for 30 min, 3 h, 4 h or 24 h with IFNβ, IL-15 or IL-21. Unstimulated cells (0 h) were used as control. n = 8 samples per time point and condition. The transcriptome was analyzed by RNA-seq. (A) Number of DEGs after cytokine treatment in comparison with the unstimulated samples at each time point (|log2 fold change|>1, FDR ≤ 0.01). Light colors indicate up-regulated genes. Dark colors indicate down-regulated genes. (B) Centroids of all the samples using Principal Components 1 and 2 from all DEGs reveal different time course patterns of gene expression upon IFNβ (green), IL-15 (orange) or IL-21 (purple) stimulation on IE-CTLs. (C) Venn diagram depicting the number of unique and overlapping genes across the stimulations and time points. The most significantly enriched pathways (determined by Reactome) of shared genes between the three stimulations are depicted. Color (key) indicates level of significance. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
After 30 min of stimulation with IFNβ and IL-15, the expression of a relatively low number of genes was affected (27 and 50 DEGs, respectively; |log2 fold change| >1, FDR ≤ 0.01). In contrast, after 3 and 4 h of stimulation, both IFNβ and IL-15 promoted a strong transcriptional response (688 and 1308 DEGs, respectively). However, the number of DEGs for IL-21 was very limited (only 144 genes after 3 h). Furthermore, while the IE-CTLs returned to a basal state after 24 h of IL-15 stimulation, IFNβ signaling was exacerbating after 24 h, with 2018 DEGs (Fig. 1A). To visualize the overall dynamics of gene expression, we performed PCA using all the DEGs across all stimulations and time points (3383 genes) (Fig. 1B). Consistent with the bar plots, the PCA revealed a modest change in gene expression upon IL-21 stimulation at all time points and strong but distinct responses upon IL-15 and IFNβ stimulation as indicated by the distance between the unstimulated and stimulated samples in PC1 and PC2. Each stimulation thus resulted in a unique gene expression profile, with the most dramatic effects induced by the tissue alarmins IL-15 and IFNβ.
Next, we evaluated the number of shared and unique genes modulated by each cytokine at each time point (Fig. 1C and Supplementary Table 2). We identified 128 common DEGs in response to the three different stimulations. Among these, we found genes implicated in the type 1 IFN response, including MX1, IFIT3 and STAT1; cytotoxicity genes like GZMB and NKTR; and genes contributing to proinflammatory pathways like TNF, TNFSF13B, LTB4R and the AP-1 transcription factor family members JUNB and BATF3 (Supplementary Table 2). The largest overlap, with 923 shared DEGs, was observed between IL-15 and IFNβ, and these included additional antiviral response genes and cytokine and chemokine genes (e.g. MX2, OAS1, IFNG, IL18 and CCR5). Stimulus-specific DEGs were associated with various functions. For instance, some genes involved in T cell activation and coding for transcription factors were differentially induced by IL-15 (CD69, EGR1, LTA and LTB) and IFNβ (CD44, STAT2 and IL15). Furthermore, IL-21 specifically induced genes involved in general metabolic processes (SNRPA1 and CSDE1) and T cell activation (CD300A) (See supplementary Table 2 for a complete overview).
In summary, we demonstrate that cytokines up-regulated in the tissues targeted in organ-specific autoimmune disorders alter the expression of thousands of genes in IE-CTLs, even though these cells are terminally differentiated. Remarkably, tissue alarmins and IL-21 activate both shared and distinct transcriptional responses in IE-CTLs, with tissue alarmins having fundamentally a strong yet specific impact on the transcriptional program of IE-CTLs.
3.2. Alarmins and adaptive cytokines induce dynamic DEGs profiles in IE-CTLs
To characterize dynamic gene expression changes in response to cytokines over time, for each stimulation we conducted unsupervised clustering analysis using the log2 fold changes observed between unstimulated samples and all time points. Furthermore, to understand the main biological processes induced upon stimulation with tissue alarmins and IL-21 in IE-CTLs, we performed gene set enrichment analysis (GSEA) [29] using Reactome Pathway analysis [30]. Ten gene clusters were identified encompassing genes that followed distinct patterns (Fig. 2A and B and Supplementary Table 3). In general, we observed that time-dependent response patterns correspond to distinct biological pathways (Fig. 2B and Supplementary Table 4). Cluster 4 was marked by immediate early response gene (e.g. JUNB, IER2, FOS and EGR1) [47] activation at 30 min that was IL-15-specific, suggesting that these transcription factors may regulate the IE-CTL response to IL-15 at 3 and 4 h. Clusters 1 (Butyrophilin family interactions), 5 (TGF-β signaling) and 10 (protein synthesis and unfolded protein response) were primarily determined by genes responding to IL-15 as early as 3 h. In contrast, cluster 7 (antiviral pathways) and cluster 8 (cell cycle and proliferation) were genes primarily regulated by IFNβ at 3 and 24 h, respectively. Cluster 5 was the only cluster where we observed changes in gene expression in opposite directions, with genes being activated upon IL-15 stimulation and inhibited by IL-21 and IFNβ (Fig. 2B and Supplementary Table 4). Finally, cluster 9 (interferon and cytokine signaling) was characterized by DEGs responding to all three cytokines. No clusters specific to IL-21 stimulation were found (Fig. 2A and B), likely due to the minimal response of IE-CTLs to IL-21 stimulation (Fig. 1A). Of note, no pathway enrichment was found for clusters 2 and 3, which were consistently downregulated. However, some of the genes in these clusters are cytokine receptors and signal transducers (e.g. IL6ST, IL21R, UBASH3A and DGKZ), suggesting the existence of a negative feedback loop upon stimulation (Fig. 2B).
Together, this analysis identified 10 distinct clusters of gene expression patterns that are associated with specific biological pathways.
3.3. Alarmins and adaptive cytokine induce stimulus specific epigenetic profiles
Having found that tissue alarmins induce massive transcriptional changes in IE-CTLs, we sought to determine whether transcriptomic responses were associated with widespread changes in epigenetic profiles by performing H3K27ac ChIP-seq in unstimulated IE-CTLs and after 3 h of stimulation. H3K27ac is a mark associated with active promoters and enhancers, and is therefore indicative of the epigenetic activation state and regulation of gene expression [48]. We identified 20,840H3K27ac peaks in unstimulated samples, while IFNβ, IL-15 or IL-21 treatment changed 2,147, 1,114, and 165H3K27ac peaks significantly, respectively (FDR < 0.01, Fig. 3A and Supplementary Table 5), with 491, 208, and 60 of these being promoters and 908, 1657, and 106 of these defined as enhancers. Thus, gene expression changes were accompanied by significant changes in H3K27ac levels, but only in a subset of DEGs. Even though not all DEGs showed significant changes in H3K27ac levels, the PCA for peaks with differential levels of H3K27ac showed a clear distinction between stimulations, showing that these epigenetic profiles are indicative of the unique response of IE-CTLs to stimulations (Fig. 3B).
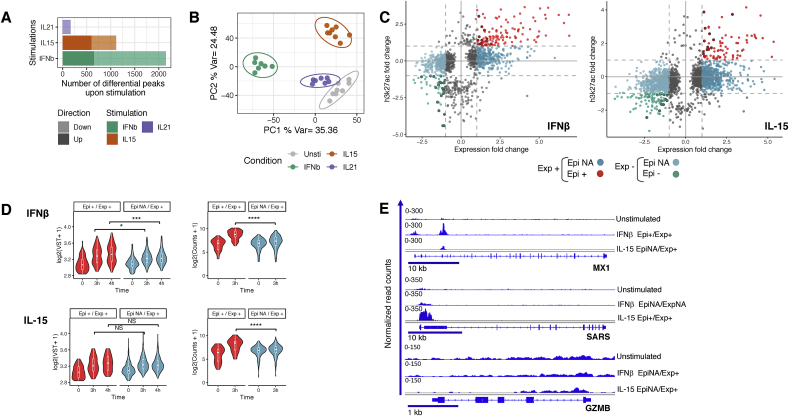
Fig. 3.
Specific epigenomic profiles induced by alarmins and adaptive cytokine. All the differential H3K27ac peaks after 3 h of stimulation with IFNβ, IL-15 or IL-21 (FDR ≤ 0.01) are depicted in (A) barplots and (B) used for PCA to depict epigenetic responses of IE-CTLs upon stimulation. (C) Scatter plot displaying the association between changes in H3K27ac (Y-axis) and expression (X-axis) upon stimulation. Gene-peak pairs were grouped based on the direction of the fold change in expression (Exp+ and Exp-) and the direction (or lack of changes) in H3K27ac after stimulation (Epi+, Epi - and Epi NA). Red dots (Epi + Exp+) indicate a fold change >1 in both expression and H3K27ac occupancy. Green dots (Epi- Exp-) indicate a fold change < 1 in both expression and H3K27ac occupancy. Dark blue (Exp + EpiNA) and light blue (Exp- EpiNA) are gene-peak pairs with fold change >1 or <1 in expression, respectively, and no change in H3K27ac. (D) Violin plots showing the distribution of gene expression levels (left, log2(VST+1)) and H3K27ac occupation (right, log2(Counts+1)) in up-regulated genes responding to IFNβ (upper panel) and IL-15 (lower panel) stimulation. Upregulated genes with H3K27ac |fold change| >1 (Epi+/Exp+) are shown in red and those without H3K27ac changes (EpiNA/Exp+) are shown in blue. Significant differences were assessed by Wilcoxon test (*p < 0.05; ***p < 0.01; ****p < 0.001, NS non-significant). (E) Representative H3K27ac tracks illustrating the concordance between epigenomics and gene expression after IFNβ or IL-15 treatment. n = 8 samples per time point and condition. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As the changes were most robust after stimulation with tissue alarmins, we focused our analysis in epigenetic changes upon IFNβ and IL-15 stimulation by linking H3K27ac promoter peaks with changes in gene expression (Fig. 3C). Subsequently, we found that DEGs could be subdivided, based on significant changes in H3K27ac levels, into increased (Epi+), decreased (Epi-) and absence (EpiNA) of epigenetic changes (Fig. 3C–E and Supplementary Table 6). Strikingly, the strongest and most robust upregulation in gene expression upon IFNβ stimulation was associated with increased levels of H3K27ac (Fig. 3D, top panels, and Fig. 3E). Conversely, for genes downregulated upon IFNβ and IL-15 stimulation, a decrease in gene expression was accompanied by a reduction in H3K27ac levels (Supplementary Fig. 5A and B). However, an increase in gene expression upon IL-15 stimulation was de-coupled from changes in levels of H3K27ac (p-values between gene expression levels of Epi+ and EpiNA genes were non-significant; Fig. 3D, bottom panels). This difference in behavior in H3K27ac levels of upregulated genes upon IFNβ and IL-15 stimulation could not be explained by epigenetic priming before stimulation (as assessed by H3K27ac levels at baseline; Supplementary Fig. 5C). Alternatively, gene expression could be regulated by non-coding RNAs (ncRNAs) [49,50]. Indeed, at 3 and 4 h of IL-15 stimulation, more ncRNAs were downregulated than upregulated, which suggests that ncRNAs stabilize gene expression of IL-15-inducible genes under unstimulated conditions. This was not observed after 3 and 4 h of IFNβ stimulation (Supplementary Fig. 6).
Taken together, our data suggests that gene expression effects are accompanied by significant changes in H3K27ac levels in promoters and enhancers for only a subset of DEGs and particularly upon IFNβ stimulation. This suggests that additional regulatory mechanisms need to be invoked.
3.4. IL-15 and IFNβ induce specific transcriptional signatures though epigenetic changes in promoter regions
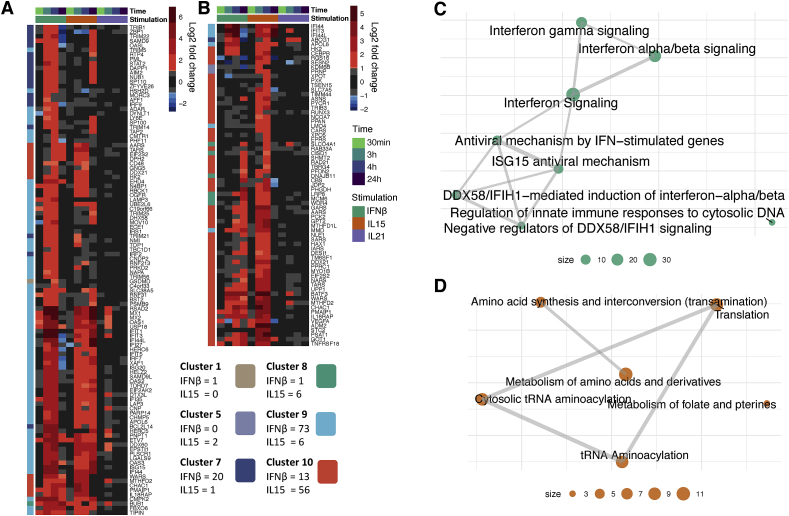
We next wanted to see if the genes and pathways under strong epigenetic control were the same as those expressed upon stimulation with IFNβ and IL-15 (Epi + genes in red, Fig. 3C). Genes under epigenetic control upon stimulation with IFNβ or IL-15 were mostly unique per stimulus (with only 10 genes shared between IFNβ and IL-15), confirming that epigenetic changes are invoked differently between the two stimuli (Fig. 4A and B). Strikingly, genes under epigenetic control were strongly enriched in certain DEG clusters, with epigenetically controlled genes upon IFNβ and IL-15 stimulation belonging predominantly to clusters 9 and 10, respectively (Fig. 4A and B). This was also reflected in the enriched pathways for genes under epigenetic control. IFNβ Epi + genes were strongly enriched for antiviral, innate and interferon response pathways (Fig. 4C), whereas IL-15 Epi + genes primarily play a role in pathways associated with protein synthesis (Fig. 4D).
Fig. 4.
Alarmin-induced genes and biological pathways potentially regulated by epigenetic modifications. Heatmap showing the differential expression at all time points of cytokine stimulation of (A) IFNβ and (B) IL-15 upregulated genes (log2 fold change >1, FDR ≤ 0.01) with concordant epigenomic changes (Epi+/Exp+). The number of Epi+/Exp + genes per cluster are indicated. (n = 8 samples per time point and condition). Network-based representation of enriched biological pathways identified by GSEA on Epi+/Exp + genes responding to (C) IFNβ or (D) IL-15 stimulation.
Thus, distinct transcriptional and regulatory programs are initiated upon stimulation of IE-CTLs with tissue alarmins.
3.5. IE-CTLs transcriptional clusters are enriched in DEGs in immune mediated gastro intestinal diseases
To identify if transcriptomic response of IE-CTLs upon stimulation with cytokines associated with autoimmune disease captures in vivo responses in disease, we performed an enrichment analysis of the DEGs from clusters in DEGs of biopsy material from CeD patients and controls (6 patient versus 5 control biopsies; n = 1443 DEGs), and of inflamed and non-inflamed tissue in IBD patients (176 inflamed versus 112 non-inflamed; n = 20,953 DEGs). Significant enrichment of clusters 7, 8 and 9 were observed in both CeD and IBD disease-specific samples, whereas cluster 1, 2, 3 and 10 were primarily enriched in IBD (Fig. 5A). These enrichments were stronger when looking only at concordant effects in biopsies and the clusters (Supplementary Fig. 7A), indicating that the clusters are representative of biological effects in patient derived tissue. While some enrichments of clusters were stronger in IBD derived biopsies, interaction plots of changes in gene expression in CeD and IBD indicate that the effects are significantly correlated for all clusters, except for 3, 4 and 5 (red lines, Supplementary Fig. 7B). Moreover, the greater enrichment in IBD samples of clusters 1,2, 3 and 10 are likely due to sample size differences between the CeD and IBD (Supplementary Fig. 7A). Finally, to further support the relevance of IE-CTLs upon stimulation, the enrichment of IE-CTLs in transcriptional signatures of biopsies obtained for IBD (inflamed vs non-inflamed) was replicated using the publicly available RNA-Seq dataset from the Human Microbiome in IBD project [35]. This dataset contains transcriptional profiles from colon/rectum and ileum, from IBD and non IBD individuals and we identified a total of 3045 DEGs in colon biopsies, and 540 in ileum. Using these DEG sets, we observed that the IE-CTLs transcriptional clusters 7,8,9 and 10 were enriched in colon biopsies, whereas an enrichment for clusters 9 and 10 was observed in ileum biopsies (Supplementary Figure 8B), further implying the role of IFN and IL-15 signaling (cluster 9 and 10, respectively) in IBD etiology. Thus, the pathways that are enriched in the dynamic response of IE-CTLs upon cytokine stimulation reflect intestinal disease specific pathways.
Fig. 5.
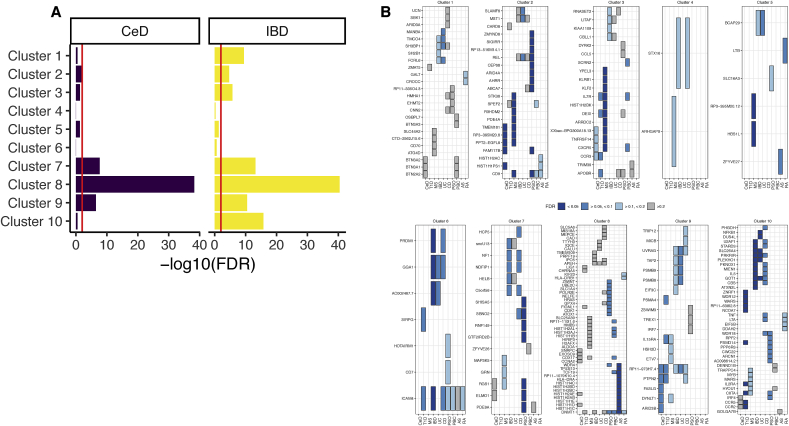
DEGs in immune mediated gastro intestinal diseases and risk loci for autoimmune diseases are enriched in IE-CTLs transcriptional clusters. (A) The enrichment for DEGs in CeD (left) or IBD (right) on each of the DEG clusters from IE-CTLs stimulated with cytokines was assessed by fisher exact test. The significance of the enrichment (-log10(FDR)) is shown in the X-axis (B) The overlap between autoimmune disease risk loci and the transcriptional response of IE-CTLs exposed to cytokines was calculated. Colors indicate the significance of the overlap between DEGs of clusters (Fig. 2) and genes in autoimmune risk loci. Diseases are shown on the X-axis and genes contributing to the overlap on the Y-axis. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Autoimmune disease loci are enriched with DEGs of IE-CTLs stimulated with the alarmins IL-15 and IFNβ and adaptive cytokine IL-21
To see if the gene expression enrichment of the clusters can also inform us about the causes for immune mediated disease, we also performed enrichment analysis of the clusters in GWAS loci (Fig. 5B). To calculate the enrichment of DEGs in autoimmune disease loci, we tested whether our 10 clusters of DEGs (Fig. 2) contained genes that also reside in loci associated with ten common autoimmune diseases (CeD, T1D, MS, PSO, PBC, AS, RA, IBD, UC and CD; all loci identified by ImmunoChip analysis) [[36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]]. We found significant overlap with one or more autoimmune diseases for all our DEG clusters except cluster 4 (Supplementary Fig. 9A and B). The large overlap between DEG clusters and autoimmune disease loci is not surprising as genetic loci associated with autoimmune diseases are often shared [51] and enriched for immune-related genes. Moreover, CTLs have been implicated in multiple autoimmune diseases, including CeD, MS and T1D [[52], [53], [54]]. We noted that the genes participating in IFN-1 signaling (clusters 7 and 9) were enriched in loci associated with several autoimmune diseases, including CeD and IBD, which is consistent with the pivotal role this pathway plays in the development and progression of immune mediated disorders [55]. Conversely, no strong enrichment in autoimmune disease loci was observed for cluster 8. This is in contrast to the enrichment analysis of clusters in DEGs from IBD and CeD patient-derived tissues or control tissues, which shows the strongest enrichment for cluster 8, and significant, but less pronounced enrichment for clusters 7 and 9 (Fig. 5A). Thus, IFN signaling pathways is likely causative to disease onset, while proliferation and cell cycle responses enriched in cluster 8 may be a consequence of the inflamed state observed in patient derived tissue.
Because the enrichment of DEGs in autoimmune risk loci can be driven by the same or by different genes per disease, we determined which individual genes overlap with autoimmune risk loci. Indeed, we found that cluster 2 DEGs were enriched in CeD, MS and PSO risk loci, but that these enrichments were caused by different genes (Fig. 5B). Conversely, some immune regulators and cytokine/chemokine genes (e.g. REL, TNFRSF14, PTPN2 and CXCR5) were shared amongst multiple autoimmune disease loci, suggesting their involvement in common inflammatory processes that may trigger or maintain these diseases. Interestingly, we also found several non-immune related genes (e.g. TMEM181, ARHGAP9 and DNMT1) and ncRNAs (e.g. HOTAIRM1, RP11-335O4.3 and RP13-516M14.1) that were shared by many loci, implicating a role for these genes in autoimmune diseases. Other examples of DEGs in IE-CTLs that are associated with multiple autoimmune diseases are ICAM4 and ncRNA RP11-973H7.4. Interestingly, genetic association of ICAM-4 levels in systemic lupus erythematosus [56] has been described previously, and RP11-973H7.4 has been implicated in the activation of T cells via modulation of the expression of MAP2K4 and the transcription factor ZEB1 [57]. Co-expression analysis also indicated a role for this ncRNA in T cell activation and cellular defense responses (Supplementary Fig. 10A and B).
In summary, our data has identified several novel potential gene targets that may play a unique and specific role in IE-CTLs and autoimmune disease etiology.
4. Discussion
This study aimed to compare dynamic transcriptional responses to two key tissue alarmins, IL-15 and IFNβ, and IL-21, a cytokine produced by CD4+ T cells, in human-tissue-resident IE-CTLs. This is particularly relevant in the context of organ-specific autoimmune disorders like CeD, where upregulation of the three cytokines is associated with tissue-resident CTL-mediated tissue destruction [1,8]. Two previous reports have studied the transcriptomic profile in freshly isolated IE-CTLs under resting conditions [9,24]. Furthermore, the transcriptional response to IL-15 and IL-21 had been analyzed in peripheral T cells in humans or mice, but not in tissue-resident CTLs. Finally, while a role for IFNβ in the activation of CTLs has been reported previously [58], to our knowledge no genome-wide transcriptome analysis of IE-CTLs upon type-1 IFN stimulation had been performed.
Overall, our analysis indicates that although tissue alarmins and adaptive cytokines both modulate the transcriptomic profile in IE-CTLs, the gene expression changes induced by the tissue alarmins IFNβ and IL-15 significantly exceed the changes caused by IL-21. Furthermore, IFNβ, IL-15 and IL-21 have the capacity to mediate distinct epigenetic and transcriptional changes in IE-CTLs, suggesting that they may play non-redundant, complementary roles in CTL-mediated tissue destruction. These findings need to be considered when designing therapies, as targeting one cytokine alone may only show partial efficacy in the prevention of tissue destruction in diseases such as Ce [59,60] that can involve upregulation of all three cytokines [16,61]. Finally, all three cytokines regulate a common core of genes involved in viral and interferon pathways that are enriched in autoimmune disease risk loci.
In this study, cell lines from biopsies were derived and expanded under controlled and validated experimental procedures. To ensure that in vitro expansion of IE-CTLs did not result in extensive changes of their biological properties, we performed several analyses to ensure that the cells were similar to ex vivo IE-CTLs (supplementary figures 1, 2 and 4). Moreover, Ciszewski, C et al. has shown that the overall transcriptional response of the in vitro expanded cell lines remains similar compared to their ex vivo counterpart [61]. Thus, IE-CTL cell lines may be slightly altered compared to ex vivo IE-CTLs, yet they provide a reliable approximation.
The observation that the tissue alarmins IFNβ and IL-15 have a more profound effect on IE-CTL than IL-21 suggests that tissue stress signals have a more direct impact on the regulation of IE-CTLs than cytokines produced by antigen-specific T cells. IL-15 regulates immediate-early response genes and temporarily induces metabolic, unfolded protein response (UPR) and amino-acid synthesis pathways. The UPR can trigger inflammatory signals and has been reported to occur in T cells activated by infection or excessive inflammation [62,63]. IFNβ induces a distinct transcriptional signature with an early type-1 interferon response, followed by a more sustained upregulation of cell cycle genes at 24 h. The strong cell cycle and proliferative response to IFNβ after 24 h is in line with the massive expansion of CTLs observed in tissue destruction in autoimmune diseases [8,52,53]. How the IFNβ-mediated type-1 interferon response impacts the function of IE-CTLs in tissue destruction remains to be determined. In contrast, IL-21 induced only 18 unique genes, which suggests that it mainly acts in cooperation with the tissue alarmins. The fact that IL-21 induces a limited reprogramming in IE-CTLs (relative to IL-15 and IFNβ) and its absence in potential CeD, where intestinal morphology is conserved [15], suggest that this adaptive cytokine may not be involved at disease onset but in the development of tissue destruction via cooperation with tissue alarmins.
Epigenetic changes in H3K27ac levels at enhancers and promoters were very modest after 3 h of stimulation with IFNβ, IL-15 and IL-21. Indeed, IL-15 induces the strongest transcriptional response after 3 and 4 h of stimulation, yet the majority of these DEGs did not display striking changes in the levels of H3K27ac. This contrasts sharply to naïve CD8+ T cells, where profound changes in epigenetic marks are readily observed upon stimulation [64], implying that mechanisms other than epigenetic changes are necessary to induce dynamic transcriptional programs in terminally differentiated effector IE-CTLs. Possible alternative regulatory mechanisms include regulation by ncRNAs. Indeed, a recent study identified ncRNAs uniquely expressed in different subsets of human CD8+ T cells upon activation [49]. Similarly, we found several differentially expressed ncRNA genes, including some located in genomic loci associated with autoimmune disease risk. Of note, we identified significant downregulated ncRNAs upon 3 and 4 h of IL-15 stimulation, which suggests that ncRNAs may have a specific role in IL-15-induced gene regulation that is independent of H3K27ac epigenetic modifications.
We further observed that all three cytokines promoted a common core of IFN immune response genes. Strikingly, these genes are also present in genomic risk loci associated with CeD, T1D, MS, IBD and UC. This suggests a critical role for IFN signaling in these autoimmune disorders and implies that all three cytokines could cooperate to fully activate these pathways and endow IE-CTLs with the ability to promote tissue destruction. Moreover, ncRNAs were found to contribute to the enrichment of DEGs in autoimmune risk loci. Although the role of most ncRNAs is unknown, we inferred potential functions of these ncRNAs by performing co-expression network analysis with GeneNetwork [31]. For instance, the ncRNA RP11-973H7.4 was found to be involved in immune activation pathways that include TNF and MAPK signaling [65]. These findings further underscore the relevance of these transcripts in the regulation of gene expression and IE-CTL activation.
Overall, our approach elucidates the transcriptional responses of IE-CTLs to individual alarmins and cytokines, that are pivotal to the inflammation and tissue-destruction associated with autoimmune disease. We show that the alarmins IFNβ and IL-15 released from injured, infected or stressed epithelial cells may, in turn, trigger massive and numerous non-redundant changes in the transcriptional program of IE-CTLs. This suggests that they may play a critical role regulating IE-CTLs, and more generally tissue-resident CTLs, in organ-specific autoimmune disorders. Conversely, IL-21, a cytokine produced by CD4+ T helper cells, may work collaboratively with alarmins on IE-CTLs. These findings underline the need for in vivo studies determining whether these cytokines play a redundant role and which cytokines are required for tissue destruction. Our study also emphasizes the need to investigate the mechanisms that underlie transcriptional changes in terminally differentiated tissue-resident CTLs, as our results suggest that these differ from those that govern the regulation of gene expression from naïve and memory CD8+ T cells. Our approach teases apart specific responses and mechanisms regulated by these crucial alarmins and cytokines and provides unique insights into the interplay between IE-CTLs and cytokines present in the diseased tissue in organ-specific autoimmune disorders. Finally, our findings suggest that there may be a value to stratifying patients based on their intestinal inflammatory profile in clinical trials targeting cytokines to prevent tissue destruction, and that it may be necessary to target several cytokines to conserve or restore tissue integrity.
Authors contributions
M.M.Z., R.A.G, I.J, Y.L, S.W, TM, B.J and C.W. envisioned this project. M.M.Z., R.A.G, I.J, Y.L, S.W, TM, B.J and C.W. coordinated the research with input from all authors. C.C. provided cell line expansions and assistance in the experiments. M.M.Z. and TM performed cell line expansion and stimulations. MMZ performed sample downstream processing for RNA-seq and ChIP-seq libraries. R.A.G. and Y.L. performed the bioinformatics analysis and interpreted the data. M.M.Z., R.A.G, C.W, B.J. and I.J. wrote the manuscript with input from all co-authors. All authors have read and agreed on the manuscript.
Declaration of competing interest
B. J. is an advisor to Immusan T, BIONIZ therapeutics and Actobio therapeutics.
Acknowledgments
We thank Kate McIntyre for editorial assistance and Morris Swertz for data storage. M.Z. is supported by an FP7/2007–2013/European Research Council (ERC) Advanced Grant (agreement 2012–322698). R.G.A. is supported by is supported by CONACYT-I2T2 scholarship (382117). T.M. is supported by the US National Institutes of Health (T32 AI07090). S.W. is supported by the Netherlands Organization for Scientific Research (NWO) Gravity Grant ‘NOCI’ 670229 (file: 024.003.001). Y.L. is supported by a Radboud University Medical Centre Hypatia Tenure Track Grant [2018]. C.W. is supported a FP7/2007–2013/ERC Advanced Grant (agreement 2012–322698), the Stiftelsen K.G. Jebsen Celiac Disease Research Centre (Oslo, Norway), an NWO Spinoza Prize (NWO SPI 92–266), the NWO Gravity Grant ‘NOCI’ 670229 (file: 024.003.001) and a UEG research prize (801449). B.J. Is supported by a Cancer Center support grant (P30CA014599), the Digestive Diseases Research Core Center at the University of Chicago (P30 DK42086) and the US National Institutes of Health (RO1 DK67180 and R01 DK098435). I.J. is supported by a Rosalind Franklin Fellowship from the University of Groningen and an NWO VIDI grant (016.171.047).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jaut.2020.102422.
Contributor Information
Bana Jabri, Email: bjabri@bsd.uchicago.edu.
Iris H Jonkers, Email: i.h.jonkers@umcg.nl.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Gating strategy and cell purity. (A) Intestinal derived cells were collected via FACS sorting to separate TCRαβ+ CD8α+ IE-CTLs from the remaining population. Representative flow plots showing TCRαβ+ CD8α+ expression on gated CD3+ cells. (B) Cells were subsequently expanded in vitro, and after 12 days in culture the cell purity was assessed by flow cytometry. Representative plots show the percentage of TCRαβ+ CD8α+ cells within the living lymphocyte gate.
Short-term IE-CTLs express tissue resident markers and receptors for IFNβ, IL-15 and IL-21. Intestinal derived TCRαβ+ CD8α+ IE-CTLs were collected via FACS sorting (ex vivo) and expanded (in vitro) for 12 days. Bar plots of RNA-seq data showing FPKM (Fragments Per Kilobase Million) expression of (A) typical tissue-resident markers (CD103, CD69, CCR7, CD62L) and (B) receptors for IFNβ (IFNAR1), IL-15 (IL2RB) and IL-21 (IL21). Data of one representative sample is shown.
Cytokine titration. Following 12 days in culture, the IE-CTLs were starved for 48 h, then stimulated for 3 h with different concentrations of IFNβ, IL-15, or IL-21 (ng/ml). The gene expression of marker genes was evaluated by qPCR (IFNG, GRB, BCL2). The expression of each gene was normalized against GAPDH. The ΔΔ CT values are shown as percentages in the Y-axis, using the unstimulated cells as reference (100% expression). Error bars indicate the standard deviation of technical triplicates. Results are representative of at least two independent experiments. The concentrations of cytokines used for the RNA-seq and ChIP-seq experiments are indicated with a dotted line.
Transcriptional profile in CeD patients and healthy controls. PCA showing the overall genome-wide transcriptome of all IE-CTL samples derived from CeD patients and healthy controls across the different time points and cytokine treatments. (n = 8 samples per condition and time point) (A, B). (C) Correlation of changes in expression of DEGs (log2 fold change) upon stimulations at multiple time points between healthy controls (Y-axis) and CeD patient (X-axis) derived IE-CTLs. Spearman correlation coefficient is show in each scatter plot on the bottom right corner.
H3K27ac in down-regulated genes upon cytokine stimulation. (A) Distribution of gene expression levels (left, log2 (VST+1)) and H3K27ac levels (right, log2(Counts+1)) in down-regulated genes responding to IFNβ (upper panel) and IL-15 (lower panel). Downregulated genes with fold change <1 (Epi-/Exp-) are shown in green and those with no change (EpiNA/Exp-) in H3K27ac are shown in blue. Significant differences were assessed by Wilcoxon test (*p < 0.05; ***p < 0.01; ****p < 0.001). (B) Representative H3K27ac tracks illustrating the concordance between epigenomics and gene expression after IFNβ or IL-15 treatment. (C) Distribution of H3K27ac levels in promoter peak of DEGs upon 3 and 4 h stimulation with IFNβ (green) or IL-15 (orange) at baseline conditions. P-value calculated by Wilcoxon test. (n = 8 samples per time point and condition).
Non-coding and protein coding genes responding to cytokine stimulation. Bar-plots showing the percentage of protein coding and non-coding genes among all DEGs per cytokine stimulation. Red and blue indicate up-regulated or down-regulated genes, respectively.
DEGs from immune mediated gastro intestinal diseases and risk loci for autoimmune diseases are enriched in IE-CTLs transcriptional clusters. (A) The enrichment for DEGs in CeD (left) or IBD (right (first IBD cohort)) on each of the DEG clusters from IE-CTLs stimulated with cytokines was assessed using the fisher exact test. The enrichment was evaluated in all the DEGs (upper panel) or in the set of the down- and upregulated genes (middle and lower panel, respectively). The significance (-log 10 FDR) is shown in the X-axis. (B) Interaction plots classify the DEGs in CeD and IBD biopsies that are present in each of the transcriptional IE-CTLs clusters. Genes that are uniquely DE in either CeD or IBD biopsies are in light and dark purple, respectively, and shared DE genes are shown in green.
Risk loci for IBD are enriched in clusters of DEGs responding to cytokine stimulation in IE-CTLs. (A) PCA showing the TMM at sample level in the second IBD cohort [34]. We removed a total of 6 samples from the analysis, 2 of them due to having zero levels of expression among all genes, and 4 more due to potential mislabeled samples. We then performed differential expression analysis and found a total of 3045 DEGs in colon biopsies, and 540 in ileum. (B) The enrichment for DEGs in colon (left) or ileum (right) on each of the DEG clusters responding to cytokine stimulation in IE-CTLs was evaluated by fisher exact test. The enrichments were assessed including all DEGs (upper panel) and by considering the down- and up-regulated genes (middle or lower panel, respectively). The significance of the enrichment (-log10(FDR)) is depicted in the X-axis, the red line marks 0.01 FDR.
Risk loci for autoimmune diseases are enriched in clusters of DEGs responding to cytokine stimulation in IE-CTLs. (A) The overlap between genetic risk loci associated with ten common autoimmune diseases and the transcriptional response of IE-CTLs exposed to cytokines is indicated as a percentage for each of the ten DEG clusters and disease traits. Colors indicate significance. X-axis shows the different autoimmune diseases with their corresponding number of risk loci and total number of genes in a 250 kb window across all risk loci. Y-axis shows the number of genes present in the DEG cluster. (B) Empirical null distribution of a random set of genes representative for each of the DEG clusters. Blue lines indicate the observed enrichment for autoimmune risk loci within DEGs. The diseases and DEG clusters (c1-c10) are depicted on the X- and Y-axis, respectively.
RP11-973H7.4 associated biological processes and co-expressed genes. (A) Significant biological process (GO terms) associated with RP11-973H7.4 and co-expressed genes. (B) Co-expression network of genes contributing to the cellular defense response GO term from figure A, extracted from Gene Network.
References
- 1.Jabri, B. & Abadie, V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat. Rev. Immunol. doi:10.1038/nri3919. [DOI] [PMC free article] [PubMed]
- 2.Jabri B., Meresse B. NKG2 receptor-mediated regulation of effector CTL functions in the human tissue microenvironment. Curr. Top. Microbiol. Immunol. 2006;298:139–156. doi: 10.1007/3-540-27743-9_7. [DOI] [PubMed] [Google Scholar]
- 3.Sheridan B.S., Lefrançois L. Intraepithelial lymphocytes: to serve and protect. Curr. Gastroenterol. Rep. 2010;12:513–521. doi: 10.1007/s11894-010-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan X., Rudensky A.Y. Hallmarks of tissue-resident lymphocytes. Cell. 2016;164:1198–1211. doi: 10.1016/j.cell.2016.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moudgil K.D., Choubey D. Cytokines in autoimmunity: role in induction, regulation, and treatment. J. Interferon Cytokine Res. 2011;31:695–703. doi: 10.1089/jir.2011.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechomska M., Skalska U. Targeting interferons as a strategy for systemic sclerosis treatment. Immunol. Lett. 2018;195:45–54. doi: 10.1016/j.imlet.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Yuan F.-L. Targeting interleukin-21 in rheumatoid arthritis. Mol. Biol. Rep. 2011;38:1717–1721. doi: 10.1007/s11033-010-0285-x. [DOI] [PubMed] [Google Scholar]
- 8.Jabri B., Sollid L.M. Tissue-mediated control of immunopathology in coeliac disease. Nat. Rev. Immunol. 2009 doi: 10.1038/nri2670. [DOI] [PubMed] [Google Scholar]
- 9.Raine T., Liu J.Z.J.Z., Anderson C.A.C.A., Parkes M., Kaser A. Generation of primary human intestinal T cell transcriptomes reveals differential expression at genetic risk loci for immune-mediated disease. Gut. 2015;64:250–259. doi: 10.1136/gutjnl-2013-306657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoytema van Konijnenburg D.P., Mucida D. Intraepithelial lymphocytes. Curr. Biol. 2017;27 doi: 10.1016/j.cub.2017.05.073. [DOI] [PubMed] [Google Scholar]
- 11.Mayassi T., Jabri B. Human intraepithelial lymphocytes. Mucosal Immunol. 2018 doi: 10.1038/s41385-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai S., Shameli A., Santamaria P. CD8+ T cells in type 1 diabetes. Adv. Immunol. 2008;100:79–124. doi: 10.1016/S0065-2776(08)00804-3. [DOI] [PubMed] [Google Scholar]
- 13.Setty M. Distinct and synergistic contributions of epithelial stress and adaptive immunity to functions of intraepithelial killer cells and active celiac disease. Gastroenterology. 2015;149:681–691. doi: 10.1053/j.gastro.2015.05.013. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrelli M. In the intestinal mucosa of children with potential celiac disease IL-21 and IL-17A are less expressed than in the active disease. Am. J. Gastroenterol. 2016;111:134–144. doi: 10.1038/ajg.2015.390. [DOI] [PubMed] [Google Scholar]
- 15.Sperandeo M.P. Potential celiac patients: a model of celiac disease pathogenesis. PloS One. 2011;6 doi: 10.1371/journal.pone.0021281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Sabatino A. Evidence for the role of interferon-alfa production by dendritic cells in the Th1 response in celiac disease. Gastroenterology. 2007;133:1175–1187. doi: 10.1053/j.gastro.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Maiuri L. Interleukin 15 mediates epithelial changes in celiac disease. Gastroenterology. 2000 doi: 10.1053/gast.2000.18149. [DOI] [PubMed] [Google Scholar]
- 18.Jabri B. Selective expansion of intraepithelial lymphocytes expressing the HLA-E- specific natural killer receptor CD94 in celiac disease. Gastroenterology. 2000;118:867–879. doi: 10.1016/S0016-5085(00)70173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mention J.J. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology. 2003 doi: 10.1016/s0016-5085(03)01047-3. [DOI] [PubMed] [Google Scholar]
- 20.Bodd M. HLA-DQ2-restricted gluten-reactive T cells produce IL-21 but not IL-17 or IL-22. Mucosal Immunol. 2010;3 doi: 10.1038/mi.2010.36. [DOI] [PubMed] [Google Scholar]
- 21.Roberts A.I. Cutting edge: NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J. Immunol. 2001 doi: 10.4049/jimmunol.167.10.5527. [DOI] [PubMed] [Google Scholar]
- 22.Berard M., Brandt K., Paus S.B., Tough D.F. IL-15 promotes the survival of naive and memory phenotype CD8+ T cells. J. Immunol. 2003;170:5018–5026. doi: 10.4049/jimmunol.170.10.5018. [DOI] [PubMed] [Google Scholar]
- 23.Pouw N. Combination of IL-21 and IL-15 enhances tumour-specific cytotoxicity and cytokine production of TCR-transduced primary T cells. Cancer Immunol. Immunother. 2010;59:921–931. doi: 10.1007/s00262-010-0818-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meresse B. Reprogramming of CTLs into natural killer-like cells in celiac disease. J. Exp. Med. 2006;203:1343–1355. doi: 10.1084/jem.20060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S., Pyl P.T., Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. Unit. States Am. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haw R., Hermjakob H., D'Eustachio P., Stein L. Reactome pathway analysis to enrich biological discovery in proteomics data sets. Proteomics. 2011;11:3598–3613. doi: 10.1002/pmic.201100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deelen P. bioRxiv; 2018. Improving the Diagnostic Yield of Exome-Sequencing, by Predicting Gene-Phenotype Associations Using Large-Scale Gene Expression Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson J.T. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X., Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd-Price J. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trynka G. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onengut-Gumuscu S. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franke A. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beecham A.H. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsoi L.C. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J.Z. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 2012;44:1137–1141. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortes A. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eyre S. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J.Z. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jostins L. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goyette P. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet. 2015;47:172–179. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bahrami S., Drabløs F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016;62:37–49. doi: 10.1016/j.jbior.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 48.Core L.J. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014 doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudson W.H. Expression of novel long noncoding RNAs defines virus-specific effector and memory CD8+ T cells. Nat. Commun. 2019;10 doi: 10.1038/s41467-018-07956-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Almo M.M., Sousa I.G., Maranhão A.Q., Brigido M.M. The role of long noncoding RNAs in human T CD3+ cells. J. Immunol. Sci. 2018;2:32–36. [Google Scholar]
- 51.Ricaño-Ponce I., Wijmenga C. Mapping of immune-mediated disease genes. Annu. Rev. Genom. Hum. Genet. 2013;14:325–353. doi: 10.1146/annurev-genom-091212-153450. [DOI] [PubMed] [Google Scholar]
- 52.Salou M., Nicol B., Garcia A., Laplaud D.-A. Involvement of CD8(+) T cells in multiple sclerosis. Front. Immunol. 2015;6:604. doi: 10.3389/fimmu.2015.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pugliese A. Autoreactive T cells in type 1 diabetes. J. Clin. Invest. 2017;127:2881–2891. doi: 10.1172/JCI94549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gianfrani C. Celiac disease association with CD8+ T cell responses: identification of a novel gliadin-derived HLA-A2-restricted epitope. J. Immunol. 2003;170:2719–2726. doi: 10.4049/jimmunol.170.5.2719. [DOI] [PubMed] [Google Scholar]
- 55.Hall, J. C. & Rosen, A. Type I interferons: crucial participants in disease amplification in autoimmunity. doi:10.1038/nrrheum.2009.237. [DOI] [PMC free article] [PubMed]
- 56.Kim K. Variation in the ICAM1-ICAM4-ICAM5 locus is associated with systemic lupus erythematosus susceptibility in multiple ancestries. Ann. Rheum. Dis. 2012;71:1809–1814. doi: 10.1136/annrheumdis-2011-201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houtman M. T cells are influenced by a long non-coding RNA in the autoimmune associated PTPN2 locus. J. Autoimmun. 2018;90:28–38. doi: 10.1016/j.jaut.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 58.Mescher M.F. Signals required for programming effector and memory development by CD8+ T cells. Immunol. Rev. 2006;211:81–92. doi: 10.1111/j.0105-2896.2006.00382.x. [DOI] [PubMed] [Google Scholar]
- 59.Cellier C. Safety and efficacy of AMG 714 in patients with type 2 refractory coeliac disease: a phase 2a, randomised, double-blind, placebo-controlled, parallel-group study. Lancet Gastroenterol. Hepatol. 2019 doi: 10.1016/S2468-1253(19)30265-1. [DOI] [PubMed] [Google Scholar]
- 60.Lähdeaho M.L. Safety and efficacy of AMG 714 in adults with coeliac disease exposed to gluten challenge: a phase 2a, randomised, double-blind, placebo-controlled study. Lancet Gastroenterol. Hepatol. 2019 doi: 10.1016/S2468-1253(19)30264-X. [DOI] [PubMed] [Google Scholar]
- 61.Ciszewski C. Identification of a γc receptor Antagonist that prevents reprogramming of human tissue-resident cytotoxic T cells by IL15 and IL21. Gastroenterology. 2019 doi: 10.1053/j.gastro.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamimura D., Bevan M.J. Endoplasmic reticulum stress regulator XBP-1 contributes to effector CD8+ T cell differentiation during acute infection. J. Immunol. 2008;181:5433–5441. doi: 10.4049/jimmunol.181.8.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grootjans J., Kaser A., Kaufman R.J., Blumberg R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016;16:469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cuddapah S., Barski A., Zhao K. Epigenomics of T cell activation, differentiation, and memory. Curr. Opin. Immunol. 2010;22:341–347. doi: 10.1016/j.coi.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sabio G., Davis R.J. TNF and MAP kinase signalling pathways. Semin. Immunol. 2014;26:237–245. doi: 10.1016/j.smim.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gating strategy and cell purity. (A) Intestinal derived cells were collected via FACS sorting to separate TCRαβ+ CD8α+ IE-CTLs from the remaining population. Representative flow plots showing TCRαβ+ CD8α+ expression on gated CD3+ cells. (B) Cells were subsequently expanded in vitro, and after 12 days in culture the cell purity was assessed by flow cytometry. Representative plots show the percentage of TCRαβ+ CD8α+ cells within the living lymphocyte gate.
Short-term IE-CTLs express tissue resident markers and receptors for IFNβ, IL-15 and IL-21. Intestinal derived TCRαβ+ CD8α+ IE-CTLs were collected via FACS sorting (ex vivo) and expanded (in vitro) for 12 days. Bar plots of RNA-seq data showing FPKM (Fragments Per Kilobase Million) expression of (A) typical tissue-resident markers (CD103, CD69, CCR7, CD62L) and (B) receptors for IFNβ (IFNAR1), IL-15 (IL2RB) and IL-21 (IL21). Data of one representative sample is shown.
Cytokine titration. Following 12 days in culture, the IE-CTLs were starved for 48 h, then stimulated for 3 h with different concentrations of IFNβ, IL-15, or IL-21 (ng/ml). The gene expression of marker genes was evaluated by qPCR (IFNG, GRB, BCL2). The expression of each gene was normalized against GAPDH. The ΔΔ CT values are shown as percentages in the Y-axis, using the unstimulated cells as reference (100% expression). Error bars indicate the standard deviation of technical triplicates. Results are representative of at least two independent experiments. The concentrations of cytokines used for the RNA-seq and ChIP-seq experiments are indicated with a dotted line.
Transcriptional profile in CeD patients and healthy controls. PCA showing the overall genome-wide transcriptome of all IE-CTL samples derived from CeD patients and healthy controls across the different time points and cytokine treatments. (n = 8 samples per condition and time point) (A, B). (C) Correlation of changes in expression of DEGs (log2 fold change) upon stimulations at multiple time points between healthy controls (Y-axis) and CeD patient (X-axis) derived IE-CTLs. Spearman correlation coefficient is show in each scatter plot on the bottom right corner.
H3K27ac in down-regulated genes upon cytokine stimulation. (A) Distribution of gene expression levels (left, log2 (VST+1)) and H3K27ac levels (right, log2(Counts+1)) in down-regulated genes responding to IFNβ (upper panel) and IL-15 (lower panel). Downregulated genes with fold change <1 (Epi-/Exp-) are shown in green and those with no change (EpiNA/Exp-) in H3K27ac are shown in blue. Significant differences were assessed by Wilcoxon test (*p < 0.05; ***p < 0.01; ****p < 0.001). (B) Representative H3K27ac tracks illustrating the concordance between epigenomics and gene expression after IFNβ or IL-15 treatment. (C) Distribution of H3K27ac levels in promoter peak of DEGs upon 3 and 4 h stimulation with IFNβ (green) or IL-15 (orange) at baseline conditions. P-value calculated by Wilcoxon test. (n = 8 samples per time point and condition).
Non-coding and protein coding genes responding to cytokine stimulation. Bar-plots showing the percentage of protein coding and non-coding genes among all DEGs per cytokine stimulation. Red and blue indicate up-regulated or down-regulated genes, respectively.
DEGs from immune mediated gastro intestinal diseases and risk loci for autoimmune diseases are enriched in IE-CTLs transcriptional clusters. (A) The enrichment for DEGs in CeD (left) or IBD (right (first IBD cohort)) on each of the DEG clusters from IE-CTLs stimulated with cytokines was assessed using the fisher exact test. The enrichment was evaluated in all the DEGs (upper panel) or in the set of the down- and upregulated genes (middle and lower panel, respectively). The significance (-log 10 FDR) is shown in the X-axis. (B) Interaction plots classify the DEGs in CeD and IBD biopsies that are present in each of the transcriptional IE-CTLs clusters. Genes that are uniquely DE in either CeD or IBD biopsies are in light and dark purple, respectively, and shared DE genes are shown in green.
Risk loci for IBD are enriched in clusters of DEGs responding to cytokine stimulation in IE-CTLs. (A) PCA showing the TMM at sample level in the second IBD cohort [34]. We removed a total of 6 samples from the analysis, 2 of them due to having zero levels of expression among all genes, and 4 more due to potential mislabeled samples. We then performed differential expression analysis and found a total of 3045 DEGs in colon biopsies, and 540 in ileum. (B) The enrichment for DEGs in colon (left) or ileum (right) on each of the DEG clusters responding to cytokine stimulation in IE-CTLs was evaluated by fisher exact test. The enrichments were assessed including all DEGs (upper panel) and by considering the down- and up-regulated genes (middle or lower panel, respectively). The significance of the enrichment (-log10(FDR)) is depicted in the X-axis, the red line marks 0.01 FDR.
Risk loci for autoimmune diseases are enriched in clusters of DEGs responding to cytokine stimulation in IE-CTLs. (A) The overlap between genetic risk loci associated with ten common autoimmune diseases and the transcriptional response of IE-CTLs exposed to cytokines is indicated as a percentage for each of the ten DEG clusters and disease traits. Colors indicate significance. X-axis shows the different autoimmune diseases with their corresponding number of risk loci and total number of genes in a 250 kb window across all risk loci. Y-axis shows the number of genes present in the DEG cluster. (B) Empirical null distribution of a random set of genes representative for each of the DEG clusters. Blue lines indicate the observed enrichment for autoimmune risk loci within DEGs. The diseases and DEG clusters (c1-c10) are depicted on the X- and Y-axis, respectively.
RP11-973H7.4 associated biological processes and co-expressed genes. (A) Significant biological process (GO terms) associated with RP11-973H7.4 and co-expressed genes. (B) Co-expression network of genes contributing to the cellular defense response GO term from figure A, extracted from Gene Network.