To the Editor:
Pneumonia is a major contributor to infectious disease mortality (1). Blood cultures in patients with pneumonia return positive in only 4–18% of cases, presumably because of distance from the site of active infection or antibiotic sterilization before sample collection (2). The degree to which microbial DNA circulates in the blood of patients with pneumonia is unknown, and this information may improve our understanding of disease pathophysiology and clinical diagnosis.
Detection of circulating microbial DNA in plasma via metagenomic next-generation sequencing (mNGS) is a recently described strategy for culture-independent assessments of infectious diseases (3–5). We hypothesized that plasma mNGS might be able to detect respiratory pathogen DNA in the bloodstream of patients with pneumonia, including those with negative blood cultures. To address this issue, we assessed 25 critically ill adults (18 with pneumonia [four with concurrent bloodstream infections] and seven with noninfectious acute respiratory illnesses). We then asked whether mNGS of plasma cell-free DNA could detect culture-confirmed respiratory pathogens, and we compared the results against those obtained by mNGS of respiratory fluid, which has been established as an accurate diagnostic method for pneumonia (6).
Subjects were enrolled within the first 72 hours of ICU admission for acute respiratory failure as part of an ongoing prospective cohort study (approved by the Institutional Review Board of the University of California, San Francisco, #10-02701) (Figure 1). Adjudication by two physicians, based on retrospective medical record review (blinded to mNGS results) and the CDC surveillance case definitions of pneumonia (7), was used to identify 18 subjects with culture-confirmed bacterial pneumonia, including two who later developed clinically diagnosed ventilator-associated pneumonia (VAP), and 7 subjects with respiratory illnesses due to noninfectious etiologies (Tables 1 and 2). The first 18 pneumonia-positive and first 7 pneumonia-negative subjects enrolled in the study with available matched plasma (from an indwelling line, central or arterial) and respiratory samples available for study underwent mNGS. Before enrollment, 83% of the subjects (13/18 pneumonia-positive, 7/7 pneumonia-negative) had received empiric broad-spectrum antibiotics.
Figure 1.
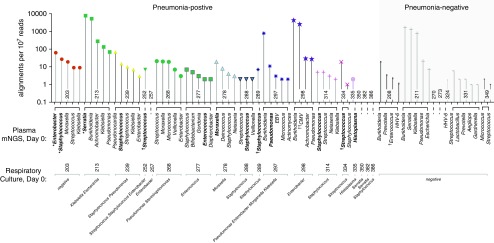
Microbes detected by metagenomic next-generation sequencing (mNGS) of plasma cell-free DNA performed on the day of study enrollment (Day 0). The y-axis corresponds to taxon abundance measured by sequencing alignments per 107 reads, stratified by subjects listed in order of the study ID number. Respiratory culture–identified microbes are indicated below and also highlighted in bold font. *Detected by respiratory culture performed on study Day 4 for clinically suspected ventilator-associated pneumonia. Each color/symbol combination represents a unique patient plasma sample. †Detected by blood culture. ††Detected by clinical PCR of blood. CMV = cytomegalovirus; EBV = Epstein-Barr virus; HHV-6 = human herpes virus 6.
Table 1.
Clinical and Demographic Characteristics of the Study Cohort
| Total | Pneumonia-Positive | Pneumonia-Negative | |
|---|---|---|---|
| Total enrolled, n | 25 | 18 | 7 |
| Age, yr, mean | 57 | 56 | 57 |
| Sex, F | 8 (32%) | 6 (33%) | 2 (29%) |
| African American | 3 (12%) | 3 (17%) | 1 (14%) |
| Asian | 2 (8%) | 2 (11%) | 1 (14%) |
| Caucasian | 12 (48%) | 12 (67%) | 4 (57%) |
| Other race | 4 (8%) | 1 (6%) | 1 (14%) |
| Hispanic ethnicity | 1 (4%) | 1 (6%) | 0 (0%) |
| CAP | 9 (36%) | 9 (50%) | — |
| HAP/VAP | 9 (36%) | 9 (50%) | — |
| Subsequent VAP | 2 (8%) | 2 (11%) | — |
| Immunosuppression* | 11 (44%) | 6 (33%) | 5 (83%) |
| Prior antibiotic use | 13 (83%) | 13 (72%) | 7 (100%) |
| Bacteremia/viremia | 4 (16%) | 4 (22%) | 0 (0%) |
| Mortality, 30 d | 5 (20%) | 5 (28%) | 0 (0%) |
Definition of abbreviations: CAP = community-acquired pneumonia; HAP = hospital-acquired pneumonia; VAP = ventilator-associated pneumonia.
Data are shown as n (%) unless otherwise indicated.
Includes history of solid organ transplant, hematopoietic stem cell transplant, or treatment with a biologic immune-modulating agent, corticosteroid, or cancer chemotherapy.
Table 2.
Comparison of Cell-Free Plasma mNGS versus Respiratory mNGS for Detection of Clinically Confirmed Pneumonia Pathogens, with Microbes Ranked by Abundance (Reads per Million)
| Pneumonia-Positive, Blood Culture/PCR-Positive Cases | ||||
|---|---|---|---|---|
| ID | Respiratory Culture, Day 0 | Blood Culture or PCR, Day 0 | Plasma mNGS Rank, Day 0 | Respiratory mNGS Rank, Day 0 |
| 252 | Streptococcus | Streptococcus | 1 | 2 |
| Enterobacter | ND | 3 | ||
| Staphylococcus | ND | >10 | ||
| 289 | Staphylococcus | Staphylococcus | 1 | 2 |
| 298 | Enterobacter* | Cytomegalovirus† | ND*, 2† | 1*, >10† |
| 334 | Streptococcus | Streptococcus | 1 | 1 |
|
Pneumonia-Positive Cases | ||||
| ID | Respiratory Culture, Day 0 | Blood Culture or PCR, Day 0 | Plasma mNGS Rank, Day 0 | Respiratory mNGS Rank, Day 0 |
| 213‡ | Klebsiella | — | 4 | 4 |
| Escherichia | — | ND | 7 | |
| 239 | Staphylococcus | — | 2 | 4 |
| Pseudomonas | — | ND | 1 | |
| 257 | Enterobacter | — | ND | 1 |
| 268 | Pseudomonas | — | ND | 1 |
| Stenotrophomonas | — | ND | 2 | |
| 278 | Moraxella | — | 1 | 1 |
| 277 | Enterococcus | — | 4 | 1 |
| 288 | Staphylococcus | — | 2 | 2 |
| 297 | Pseudomonas | — | 2 | 2 |
| Enterobacter | — | ND | 3 | |
| Morganella | — | ND | 4 | |
| Klebsiella | — | ND | 7 | |
| 314 | Staphylococcus | — | 1 | 2 |
| Streptococcus | — | 2 | 4 | |
| 335 | Histoplasma§ | — | 1 | 1 |
| 350 | Serratia | — | ND | 1 |
| 382 | Serratia | — | ND | 1 |
| 386 | Staphylococcus | — | ND | 2 |
|
VAP Cases | ||||
| ID | VAP Respiratory Culture, Day 4 | Plasma mNGS Rank, Day 0 | Respiratory mNGS Rank, Day 0 | Respiratory mNGS Rank, Day 2 |
| 203 | Enterobacter | 1 | ND | 1 |
| Staphylococcus | 2 | ND | 2 | |
| 213‡ | Serratia | 1 | 7 | n/a |
Definition of abbreviations: mNGS = metagenomic next-generation sequencing; n/a = respiratory mNGS data not available; ND = not detected; VAP = ventilator-associated pneumonia. Pathogen detection was performed using respiratory mNGS as previously described (4).
Culture-identified pneumonia pathogen.
Detected by clinical PCR of blood.
Patient developed Serratia VAP 4 days after enrollment.
Detected by serum antigen testing.
Plasma specimens were collected in citrate tubes on the day of study enrollment (Day 0) and underwent DNA extraction (ZR-Duet kit; Zymo Research) followed by sequencing library preparation (NEBNext; New England BioLabs) according to previously described methods (6). After paired-end Illumina sequencing, we used ID-Seq v3.2 to detect microbes from raw sequence data (8). Matched respiratory samples underwent paired DNA and RNA sequencing, and microbes that were detected concordantly in both nucleic acid types were aggregated to the genus level and ranked by abundance (reads per million reads mapped [rpM]) as previously described (6). The sequencing data are available via BioProject accession number PRJNA525157 (http://www.ncbi.nlm.nih.gov/bioproject/525157).
For plasma samples, we performed pathogen assessments and background contaminant correction using a previously described Bayesian scoring metric (9): score = [(|genus nt Z| × species nt Z × species nt rpM) + (|genus nr Z| × species nr Z × species nr rPM)], where nr = nonredundant and nt = nucleotide. We optimized the metric for plasma by incorporating the following rules: 1) percent identity of nt alignment ≥95%; 2) nt reads >10; 3) log(1/e) ≥ 75; 4) sequencing read length >135 nt; 5) score + 0.05 > 0; and 6) exclusion of background environmental microbiota representing common mNGS library preparation contaminants (such as skin flora), as described in Reference 10. Z-scores were calculated using a cohort-optimized background model, and microbes were aggregated to the genus level and ranked by rpM, with the top five most abundant microbes in each patient carried forward for analysis (10).
We first examined positive control subjects with culture-confirmed bacteremia (n = 3) or cytomegalovirus viremia (n = 1) and found that plasma mNGS identified the etiologic bloodstream pathogen in every one of these cases (Table 1). We next assessed the ability of plasma mNGS to detect pathogens identified by lower-respiratory culture (mini-BAL or tracheal aspirate) or serum antigen testing from the patients with pneumonia. Plasma mNGS identified one or more clinically confirmed pneumonia pathogens in 67% (12/18) of cases, and in 44% (8/18) of cases the pathogen was the most abundant microbe detected. These results compared with 100% and 61% of cases, respectively, for mNGS of respiratory samples (6). Notably, in addition to bacterial pathogens, plasma mNGS also identified the invasive fungal pathogen Histoplasma capsulatum in a patient with disseminated disease (Tables 1 and 2).
Two patients in the study received a clinical diagnosis of VAP after enrollment. In these cases, plasma cell-free mNGS detected the culture-confirmed bacterial pathogens at the time of study enrollment, 4 days before the VAP diagnosis (Table 1). The first of these cases was a patient hospitalized for intracranial hemorrhage who developed VAP secondary to Enterobacter cloacae and Staphylococcus aureus, both of which were detectable by plasma mNGS performed at the time of enrollment. The second patient developed VAP secondary to Serratia marcescens while undergoing ceftriaxone treatment for a primary pneumonia due to Escherichia coli and Klebsiella pneumoniae. mNGS of plasma cell-free DNA obtained 4 days before the VAP diagnosis identified Serratia as the most abundant microbe. In addition, plasma mNGS detected the SRT-1 β-lactamase gene, which is known to confer inducible resistance to ceftriaxone (8), providing a potential molecular explanation for emergence of the VAP pathogen.
As negative controls, we sequenced seven critically ill pneumonia-negative subjects with respiratory failure. In two subjects (29%), no microbes were identified (Table 2). Microbes identified in the remaining subjects included human herpes virus 6, which may be incidentally present in healthy individuals and reactivate in the setting of sepsis (9), as well as several potential pathogens and commensals. These results suggest that circulating microbial DNA is detectable in the bloodstream of critically ill patients, even in the setting of negative blood cultures. Microbes detected in such cases could represent true but missed pneumonia pathogens or circulating microbial DNA in the bloodstream derived from gut translocation, the respiratory tract, skin, or other sources.
In summary, we report that plasma mNGS can identify circulating DNA from respiratory pathogens in critically ill patients with culture-confirmed bacterial pneumonia. Our results suggest that release of pathogen DNA into the bloodstream may be more common than is appreciated in patients with pneumonia and negative blood cultures, and that plasma mNGS, although less informative than respiratory mNGS, may have value for detecting pneumonia pathogens when respiratory specimens are unavailable. Future studies in a larger cohort will be needed to assess the generalizability of these findings, evaluate the diagnostic performance of plasma mNGS, and determine its utility for pathogen detection in culture-negative pneumonia cases.
Supplementary Material
Footnotes
Supported by NIH grants HL140026 (C.S.C.) and K23HL138461-01A1 (C.L.).
Originally Published in Press as DOI: 10.1164/rccm.201904-0905LE on October 24, 2019
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.World Health Organization. The 10 leading causes of death in the world [accessed 2019 Jul 1] Available from: http://www.who.int/mediacentre/factsheets/fs310/en/
- 2.Metersky ML, Ma A, Bratzler DW, Houck PM. Predicting bacteremia in patients with community-acquired pneumonia. Am J Respir Crit Care Med. 2004;169:342–347. doi: 10.1164/rccm.200309-1248OC. [DOI] [PubMed] [Google Scholar]
- 3.De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci USA. 2015;112:13336–13341. doi: 10.1073/pnas.1517494112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langelier C, Kalantar KL, Moazed F, Wilson MR, Crawford ED, Deiss T, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci USA. 2018;115:E12353–E12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramesh A, Nakielny S, Hsu J, Kyohere M, Byaruhanga O, de Bourcy C, et al. Metagenomic next-generation sequencing of samples from pediatric febrile illness in Tororo, Uganda. PLoS One. 2019;14:e0218318. doi: 10.1371/journal.pone.0218318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramesh A, Nakielny S, Hsu J, Kyohere M, Byaruhanga O, de Bourcy C, et al. Etiology of fever in Ugandan children: identification of microbial pathogens using metagenomic next-generation sequencing and IDseq, a platform for unbiased metagenomic analysis [preprint] bioRxiv. 2018. [accessed 2019 Jul 1]. Available from: https://www.biorxiv.org/content/10.1101/385005v2.
- 7.Langelier C, Zinter MS, Kalantar K, Yanik GA, Christenson S, O’Donovan B, et al. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am J Respir Crit Care Med. 2018;197:524–528. doi: 10.1164/rccm.201706-1097LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iguchi A, Nagaya Y, Pradel E, Ooka T, Ogura Y, Katsura K, et al. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol. 2014;6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong DSY, Bonten MJM, Spitoni C, Verduyn Lunel FM, Frencken JF, Horn J, et al. Molecular Diagnosis and Risk Stratification of Sepsis Consortium. Epidemiology of multiple herpes viremia in previously immunocompetent patients with septic shock. Clin Infect Dis. 2017;64:1204–1210. doi: 10.1093/cid/cix120. [DOI] [PubMed] [Google Scholar]
- 10.Langelier C, Fung M, Caldera S, Deiss T, Lyden A, Moazed F, et al. Supplemental online methods and protocols for: detection of pneumonia pathogens from plasma cell-free DNA [accessed 2019 Nov 1] doi: 10.1164/rccm.201904-0905LE. Available from: https://www.protocols.io/view/supplemental-online-methods-and-protocols-for-dete-6vyhe7w. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.