Abstract
Adiponectin is a highly abundant protein hormone secreted by adipose tissue. It elicits diverse biological responses, including anti-diabetic, anti-inflammatory, anti-tumor, and anti-atherosclerotic effects. Adiponectin consists of a globular domain and a collagen-like domain, and it occurs in three major oligomeric forms that self-assemble: trimers, hexamers, and high-molecular-weight oligomers. Adiponectin has been reported to bind to two seven-transmembrane domain receptors, AdipoR1 and AdipoR2, as well as to the protein T-cadherin, which is highly expressed in the cardiovascular system and binds only the high-molecular-weight form of adiponectin. The molecular mechanisms underlying this specificity remain unclear. Here we used a combination of X-ray crystallography and protein engineering to define the details of adiponectin's interaction with T-cadherin. We found that T-cadherin binds to the globular domain of adiponectin, relying on structural stabilization of this domain by bound metal ions. Moreover, we show that the adiponectin globular domain can be engineered to enhance its binding affinity for T-cadherin. These results help to define the molecular basis for the interaction between adiponectin and T-cadherin, and our engineered globular domain variants may be useful tools for further investigating adiponectin's functions.
Keywords: adiponectin, crystallography, protein engineering, cadherin, adipokine
Introduction
Adiponectin is the most abundant adipokine produced exclusively by adipocytes, and it accounts for 0.01% of total plasma proteins, with a serum concentration ranging from 5–30 μg/ml (1). Mature adiponectin is composed of two major domains: an N-terminal collagen-like domain and a C-terminal globular domain (2). It is structurally related to the members of the complement factor C1q family (3, 4). Adiponectin is extensively posttranslationally modified, with modifications including hydroxylation at conserved proline and lysine residues on the collagen-like domain (4–7), glycosylation on hydroxylated lysine residues (5), and O-glycosylation on threonine residues (8). All of these modifications are important for subsequent multimerization of adiponectin to form trimers, hexamers, and high-molecular-weight (HMW)4 oligomers (1, 4, 9–11). No monomeric form of adiponectin has been found under native conditions, and the fundamental building block of the circulating hormone is the trimeric form, which is stabilized by extensive hydrophobic interactions within the globular domains as well as in the triple helix of the collagen-like domain (9). A cysteine residue in position 36 near the N terminus of human adiponectin allows formation of intermolecular disulfide bonds, mediating the association of two trimers to form hexamers. What drives the assembly to high-molecular-weight species is less clear but likely also involves posttranslational modifications and disulfide formation via Cys-36 (7, 12).
Multimerization of adiponectin has been shown to be important for its biological function (13, 14). The HMW form of adiponectin has the most pronounced role in improving insulin sensitivity and protecting against diabetes (9, 15, 16), and impaired multimerization is associated with different metabolic diseases, including obesity, type 2 diabetes, insulin resistance, and arteriosclerosis (1, 17–19).
The biological functions of adiponectin are thought to be carried out by at least three receptors: AdipoR1, AdipoR2, and T-cadherin. The first two are members of a class of seven-transmembrane domain receptors called the progesterone and AdipoQ receptor superfamily (20) and are structurally and functionally unrelated to G protein–coupled receptors (21). Their expression and therefore their biological activity vary among different tissues, with AdipoR1 being mostly expressed in skeletal muscle, heart, kidney, and liver, whereas AdipoR2 is expressed almost exclusively in the liver (22, 23).
The third receptor, T-cadherin, is a glycosyl phosphatidylinositol–anchored protein that lacks transmembrane and cytoplasmic regions (24). Hug et al. (25) identified T-cadherin as the receptor for hexameric and HMW adiponectin but not for the trimeric or globular form. T-cadherin is composed of a propeptide and five extracellular cadherin domains. T-cadherin is highly expressed in the cardiovascular system, in particular in smooth muscle cells and pericytes and on endothelial cells in all types of blood vessels (26), where it interacts with adiponectin; T-cadherin knockout mice show an increase in circulating adiponectin and reduced adiponectin bound to endothelial cells in vascular tissues (27–30). Recently, it has been shown that the prodomain of T-cadherin, together with its extracellular domains 1 and 2, is essential and sufficient for binding of adiponectin (31). More recently, native adiponectin from serum has been shown to bind to T-cadherin–expressing cells but, surprisingly, not to cells expressing AdipoRs (32). Despite its increasingly clear importance, the molecular details of T-cadherin's interactions with adiponectin are largely unknown and have received relatively less attention than proposed interactions with AdipoR1 and AdipoR2. Here we investigate adiponectin–T-cadherin interactions using a combination of biochemical approaches and protein engineering.
Results
Structural characterization of full-length adiponectin
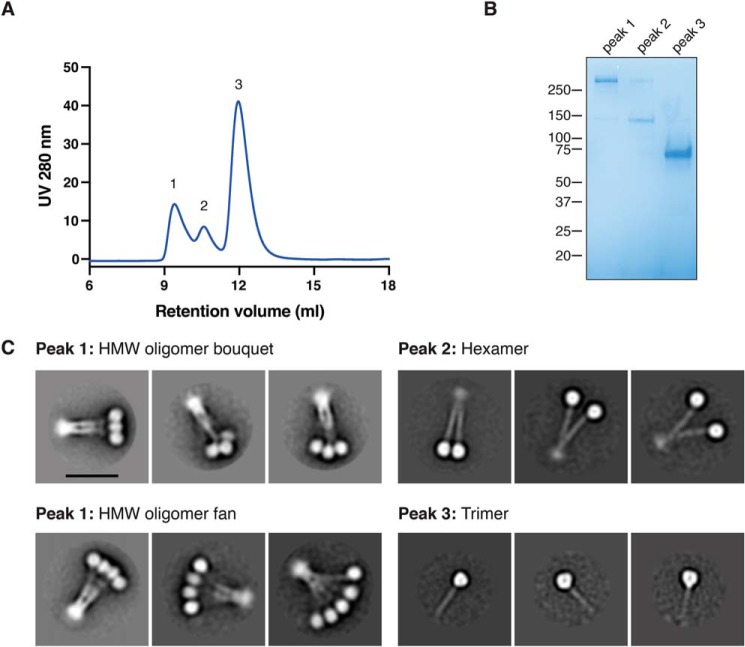
We initially sought to characterize the oligomerization states of adiponectin purified by size exclusion chromatography from HEK293 cells stably expressing FLAG-tagged adiponectin. The protein eluted in three peaks with differing size exclusion retention volumes and electrophoretic mobilities, consistent with 18-mer, hexamer, and trimer forms of the protein (Fig. 1, A and B). Negative-stain EM of each peak confirmed this assignment (Fig. 1C and Fig. S1). As reported previously (33), we found that the HMW 18-mer exists as a dynamic bundle resembling either a flower bouquet or fan in negative-stain EM, with the N terminus of the collagen-like domain held together at one end and the trimers of the globular domain interacting with each other. The adiponectin hexamer is also held together by the collagen-like domain but shows multiple conformations because of the presence or absence of interactions between the two globular domain trimers.
Figure 1.
Oligomerization states of full-length adiponectin. A, size exclusion chromatography of full-length adiponectin shows three peaks at three different retention volumes. B, the three peaks from the size exclusion chromatography were run on nonreducing SDS-PAGE and stained with Coomassie dye. C, two-dimensional negative-stain EM class averages show the adiponectin oligomerization state of each peak from size exclusion chromatography. Each image has the same scale, but different circular masks were used for classification according to the size of the particle. Scale bar = 20 nm.
Structural metal ions in adiponectin are important for T-cadherin binding
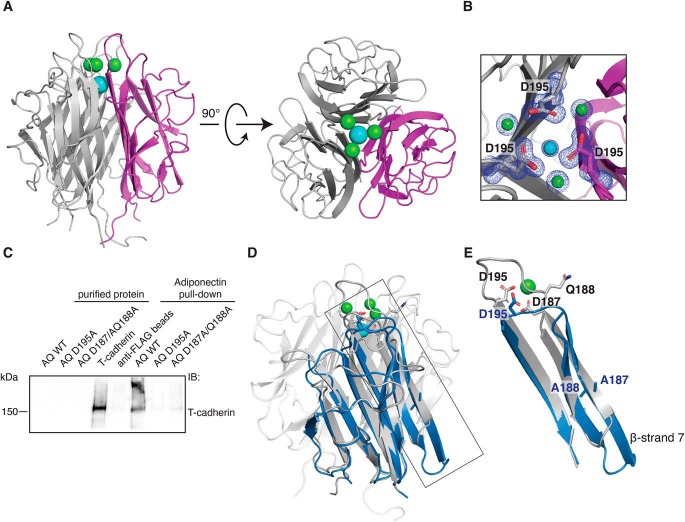
Full-length adiponectin is a large and heavily posttranslationally modified protein, features that do not favor crystallization; we thus decided to first focus our attention on the globular domain, which lacks these modifications. This domain could be readily expressed and purified from Escherichia coli. Crystallization was straightforward, and a 1.0 Å–resolution structure was obtained (Table S1). Our structure is similar overall to a previously reported structure of a single-chain adiponectin variant in which three globular domains are expressed as a single polypeptide (34). In our structure, each of the three protomers is a separate polypeptide, eliminating flexible linkers between protomers and potentially accounting for the superior crystallographic resolution. The asymmetric unit in our structure contains three copies of each globular domain, with the N and C termini located near one another at the base of the trimer (Fig. 2A). The overall structure of the adiponectin globular domain shows a tightly associated trimer, where each monomer is composed of eight β-sheets interconnected by well-ordered loops. The electron density is excellent throughout the molecule and clearly defines the structure of the metal ion coordination region (Fig. 2B and Fig. S2A). Four metal ions are resolved, and on the bases of interatomic distances, coordination geometry, and electron density, three of these were identified as calcium ions and the fourth as a sodium ion. Side chains of Asp-187, Asn-193, and Asp-195 interact with the calcium ions, whereas Gln-188 and Val-194 contribute to calcium coordination via their main-chain carbonyls. A single sodium ion is coordinated by the backbone carbonyl of Gln-196 from all three protomers together with three water molecules.
Figure 2.
Organization of the metal ion binding site is necessary for T-cadherin binding. A, crystal structure of the WT adiponectin globular domain. The protein self-assembles as a trimer; one monomer is colored magenta. Three calcium ions are depicted in green, and a single sodium ion is shown in cyan. B, close-up view of the metal ion coordination site. The 2Fo-Fc map is shown in blue and is contoured at 1 σ. C, FLAG-adiponectin pulldown assay showing that T-cadherin cannot be pulled down efficiently by mutant forms of adiponectin (abbreviated AQ for AdipoQ) in which the metal ion binding site is disrupted. D, superimposition of adiponectin D187A/Q188A (blue) on the WT structure (gray). The side chains of the mutated amino acids Asp-187 and Gln-188 are shown in both structures, together with Asp-195. E, close-up of the region affected by mutations. The superimposition shows a major change in the conformation of β-strand 7 and in loop 7.
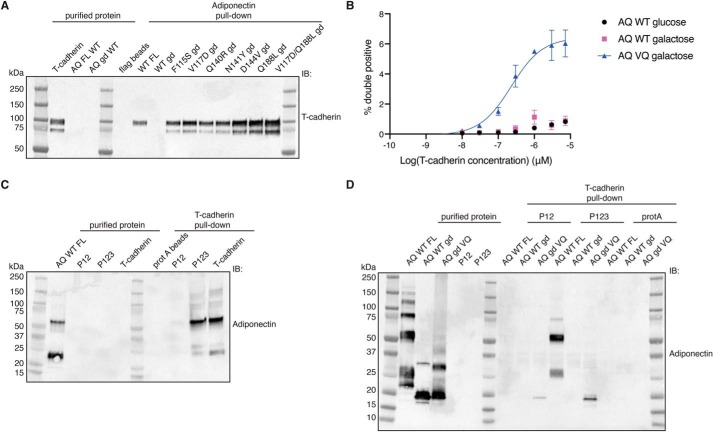
We decided to investigate the role of this metal coordination pocket in binding with T-cadherin. To this end, we created two point mutants in the context of full-length adiponectin: a D195A single mutant and a D187A/Q188A double mutant. We first assessed the binding of these mutants to T-cadherin by performing a pulldown assay in which we immobilized FLAG-tagged full-length adiponectin variants and tested binding of purified T-cadherin. T-cadherin failed to bind to both mutants, suggesting that proper folding of the metal coordination region is essential for the interaction between adiponectin and T-cadherin (Fig. 2C).
We then proceeded to structurally characterize the mutants to confirm disruption of the metal-binding site. Using the globular domain construct described above, we introduced the mutations, purified the resulting protein, and set crystallization trays. Although we were not able to obtain diffraction-quality crystals of the D195A mutant, we were able to obtain strongly diffracting crystals of the D187A/Q188A double mutant, giving a dataset of 2.15 Å resolution (Table S1). The asymmetric unit contains one copy of the globular domain with a three-fold crystallographic symmetry axis at the center of the biological trimer. The overall fold of the protein closely resembles that of WT adiponectin (Fig. 2D) but differs in β-strand 6, which is extended. The metal coordination site is completely eliminated with major rearrangements in this region, positioning the D187A/Q188A mutated residues in β-strand 7. Importantly, even Asp-195, located in loop 7, is shifted substantially, preventing metal coordination entirely (Fig. 2E). Together with the pulldown results, these data indicate that proper organization of the metal ion coordination site and adjacent regions is important for adiponectin to bind to T-cadherin.
The globular domain alone does not bind T-cadherin
Given the essentiality of the metal ion coordination site, we speculated that T-cadherin's interaction with adiponectin might be entirely mediated by the globular domain. To test the roles of different domains, we performed a pulldown assay. Purified FLAG-tagged adiponectin variants were immobilized and then incubated with T-cadherin (Fig. S3A). We assessed the binding through Western blotting, and, as expected, the WT protein binds the receptor, but neither the globular domain alone nor a trimeric C36S mutant (unable to multimerize because of its inability to form disulfide bonds) was able to pull down T-cadherin. This suggests that the globular domain is not sufficient to bind T-cadherin, perhaps because of a reliance on avidity conferred by high-molecular-weight forms.
To test this possibility, we decided to mimic the high-molecular-weight form of adiponectin by preparing site-specific, biotinylated, globular-domain adiponectin and incubating it with streptavidin. In this case, we used a 3× tandem repeat construct described previously (34), resulting in formation of 12-mers upon incubation with streptavidin tetramers. However, this dodecameric form also failed to bind T-cadherin in a pulldown assay (Fig. S3B), suggesting that not only avidity but also the geometry of the oligomeric form is important for interaction of adiponectin with T-cadherin.
Affinity maturation of the adiponectin globular domain to find T-cadherin interactors
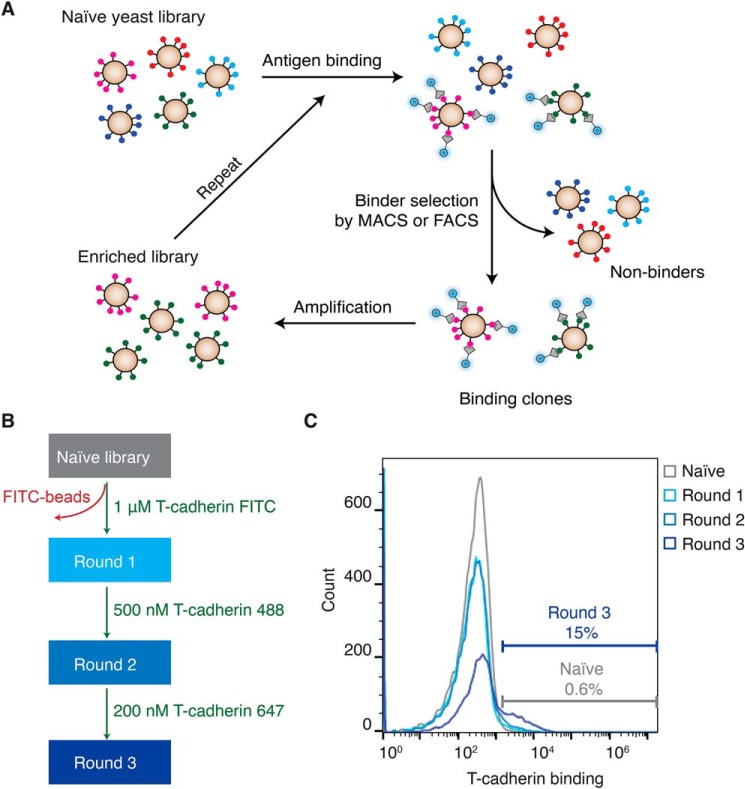
Because a properly folded globular domain of adiponectin appears to be necessary but not sufficient for T-cadherin binding, we hypothesized that it might be possible to enhance the affinity of adiponectin's globular domain for T-cadherin and bypass the requirement for oligomerization. To do this, we used a yeast surface display (35) with the display vector pYDS (36). This approach allows expression of a protein variant library on the surface of Saccharomyces cerevisiae yeast, which can then be sorted by FACS to identify binders to a target of interest (Fig. 3A). We first assessed whether we could express the globular domain of adiponectin on the yeast surface, confirming that the protein could be displayed efficiently (Fig. 5, top left panel). In this format, we observed weak but detectable binding to T-cadherin, which we then sought to optimize.
Figure 3.
Affinity maturation of the adiponectin globular domain to find T-cadherin interactors. A, selection procedure to identify T-cadherin binders. B, the approach used for selection involves one step of MACS using 1 μm T-cadherin FITC, followed by one round of FACS employing 500 nm T-cadherin 488 and a second round of FACS utilizing 200 nm T-cadherin 647. C, the different rounds of selection were plotted in a histogram. On the x axis, T-cadherin binding is shown by fluorescence intensity (arbitrary units), whereas on the y axis, the count of cells is shown.
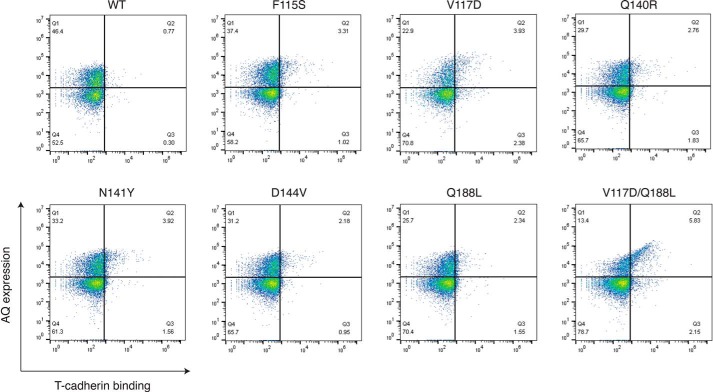
Figure 5.
Each mutation independently enhances T-cadherin binding to the adiponectin globular domain on yeast. Each adiponectin (AQ) mutant was tested for its binding to purified T-cadherin (100 nm) in a yeast surface display assay. On the x axis, T-cadherin binding is shown, whereas on the y axis, adiponectin expression is reported. The upper right quadrant indicates yeasts that express adiponectin and bind T-cadherin.
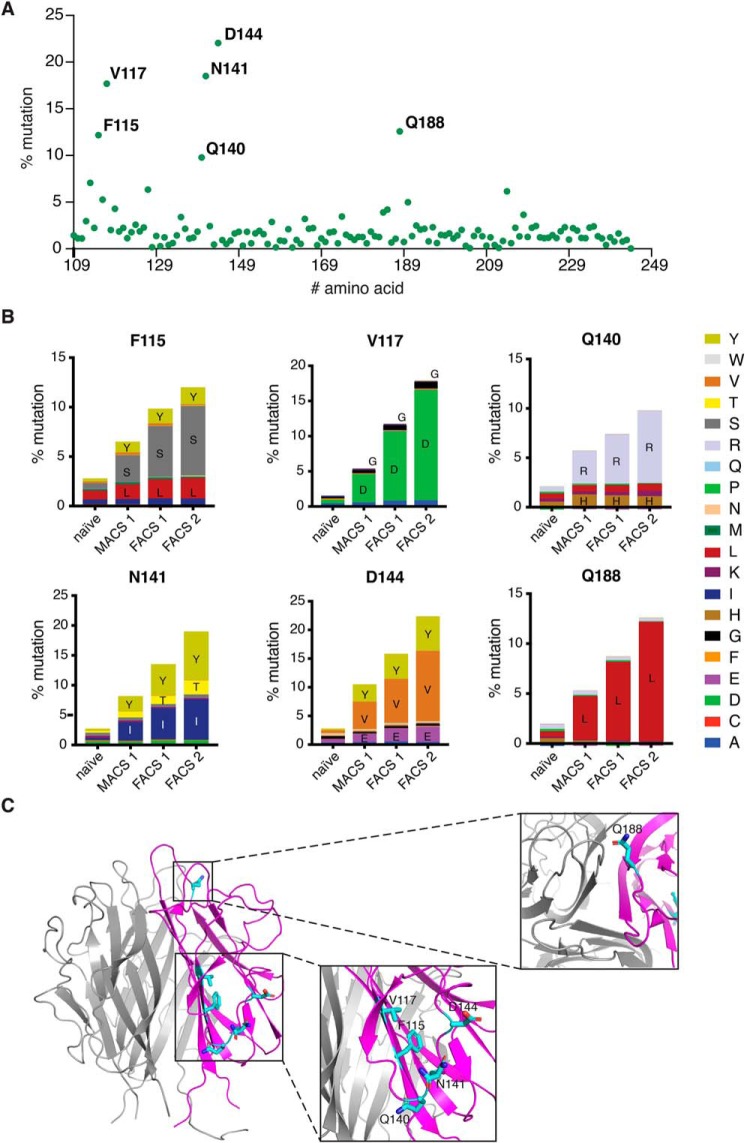
We created an error-prone PCR library of adiponectin globular domain variants with roughly two to three mutations per clone spread throughout the gene. Transformation into yeast yielded a library of 5 × 108 transformants. This library was then used to screen for T-cadherin binding by using progressively lower concentrations of T-cadherin coupled to fluorophores (Fig. 3B). Each round of selection increased the proportion of adiponectin variants binding to T-cadherin (Fig. 3C). We then analyzed all rounds of selection compared with the naïve library through amplicon deep sequencing to identify “hot spots” with high mutation frequency. Certain regions of the protein were preferentially enriched in mutations after selection (Fig. 4A), with each round showing an increase in the percentage of mutant at each position. We analyzed each residue that showed a mutant enrichment above 10% in the second FACS round (Fig. 4B). Interestingly, the amino acid substitution preference in each position was apparent from the first magnetic cell sorting round (MACS1) and was maintained throughout the selection process, nearing fixation in most positions after the final selection round. The mutations were localized in several regions of the protein (Fig. 4C). Gln-188 is located at the “top” of the trimer, opposite the N and C termini regions, and it is involved in calcium coordination through its backbone carbonyl. Phe-115 and Val-117 are in the first β-sheet, with their side chains pointing in the opposite direction of the core of the trimer. Gln-140, Asn-141, and Asp-144 are positioned in the second loop, with their side chains pointing toward the solvent.
Figure 4.
Identification of hot spot mutations through amplicon deep sequencing. A, schematic of mutation frequency at each amino acid in the adiponectin globular domain (residues 109–244) after the final FACS2 selection round. B, residues showing more than a 10% mutation rate in FACS round 2 were then analyzed to identify preferred substitutions. C, hot spot mutations are represented as side chains in a ribbon representation of the structure.
To explore the contribution of each mutation to binding affinity, we measured T-cadherin binding to yeast cells displaying the adiponectin globular domain containing each single mutant. All single substitutions modestly enhanced T-cadherin binding when measured in this format (Fig. 5), but combination of two mutations (V117D and Q188L) significantly enhanced the interaction (Fig. 5, bottom right). When we assessed binding to purified adiponectin globular domain mutants through a pulldown assay, each mutant could bind the receptor, with the double mutant V117D/Q188L (VQ) showing more effective pulldown (Fig. 6A). This mutant was further analyzed in a dose–response binding assay in yeast, showing an apparent KD of 250 nm (Fig. 6B). To investigate whether this mutant could recapitulate the features of the full-length protein, we checked that it retained the ability to bind the same domains of T-cadherin. Fukuda et al. (31) discovered that the prodomain of T-cadherin together with extracellular domains 1 and 2 were necessary and sufficient for interaction with adiponectin. We confirmed these data with full-length adiponectin, finding also that addition of the third extracellular domain of T-cadherin significantly enhanced binding (Fig. 6C). We found that the adiponectin globular domain VQ mutant follows the same trend (Fig. 6D), indicating that the mutations likely enhance intrinsic affinity rather than conferring a fundamentally new binding mode.
Figure 6.
Mutations in the globular domain confer binding to T-cadherin in vitro. A, FLAG pulldown assay. Purified FLAG-adiponectin proteins were immobilized on FLAG resin and then incubated with T-cadherin. Binding of T-cadherin was assessed by Western blotting. IB, immunoblot. B, dose–response binding assay in yeast by flow cytometry. Test proteins (adiponectin globular domain and mutant) were expressed under the control of galactose induction. Data are shown as means ± S.D. from two independent biological replicates, each performed in triplicate. On the y axis, the percentage of cells positive for both the HA epitope (to confirm display) and T-cadherin binding is shown. C, protein A pulldown assay. Purified T-cadherin constructs fused with human IgG1 Fc were immobilized on protein A resin and then incubated with adiponectin. Binding of adiponectin was assessed by Western blotting. D, protein A pulldown assay. Purified T-cadherin constructs fused with Fc were immobilized on protein A resin and then incubated with different adiponectin proteins. Binding of adiponectin was assessed by Western blotting. All pulldown data are representative of experiments performed at least in duplicate.
Discussion
Data from a variety of studies indicate that the primary biologically active form of adiponectin is its high-molecular-weight oligomeric form (17, 37–39), which is the only form of adiponectin that binds with high affinity to T-cadherin (25). However, the mechanistic basis for this specificity has remained unclear. Here we showed that the adiponectin globular domain is necessary but not sufficient for this interaction. Consistent with prior data (34), we observed structural calcium and sodium ions within the trimer of the adiponectin globular domain, and we found that these are important for proper folding of the globular domain. A metal ion coordination mutant, D187A/Q188A, displayed significant rearrangement of the region and lacked T-cadherin binding activity. However, even with an intact metal coordination site, an artificial globular domain dodecamer failed to bind to T-cadherin. This highlights the importance not only of the oligomeric state of the protein but also the unique C1q-like geometry of full-length adiponectin.
Although the WT globular domain of adiponectin is not sufficient to bind T-cadherin, we speculated that it might be possible to restore binding activity to this isolated domain. We diversified this region through error-prone PCR and created a library of ∼500 million mutants displayed on the surface of yeast. After sorting for T-cadherin binding, we identified a variety of individual point mutations that confer T-cadherin binding to the isolated adiponectin globular domain trimer, overcoming the avidity and geometric requirements of the native protein. Although it is possible, in principle, that these mutations alter the binding mode or induce an artificial interaction. The diversity of mutations identified and the small number of changes (even just a single amino acid) required to confer binding suggests that these mutations more likely enhance a low intrinsic affinity. Consistent with this, the most effective mutant (VQ mutant) retains specificity for the same domains of T-cadherin as full-length adiponectin.
One of the most intriguing and puzzling features of adiponectin biology is its numerous proposed receptors. Although T-cadherin shows robust binding to adiponectin, at least in its high-molecular-weight form, the other putative receptors, AdipoR1 and AdipoR2, as well as calreticulin appeared to be unimportant for binding in a recent study (32), in contrast to previous reports (22, 23, 40). Adding to the complexity, other recent work showed that adiponectin can bind to certain negatively charged lipids (41), suggesting a possible opsonin or lipid transport role for the protein, perhaps in conjunction with signaling activity through other receptors. Accordingly, a key future challenge is to understand the relationship among these different binding partners and to definitively determine whether AdipoR1 and AdipoR2 are, in fact, direct receptors of adiponectin or whether they serve some other role. Our work here adds to an increasingly extensive body of literature validating T-cadherin binding to adiponectin, defining the site of interaction and showing that adiponectin's complex quaternary structure is important for T-cadherin binding. Remarkably, the requirement for adiponectin oligomerization to bind T-cadherin can be overcome by a variety of point mutations in adiponectin. These adiponectin globular domain mutations may also prove to be useful tools for investigating other aspects of adiponectin's functions in the future. Combinatorial biology approaches similar to those described here might also be applicable to studies of other proposed adiponectin binding partners, providing a path to a more complete mechanistic understanding of this system.
Experimental procedures
Error-prone PCR library
The DNA library for the adiponectin globular was constructed through error-prone PCR as described previously (42). The protocol takes advantage of a polymerase that does not possess 3′ to 5′ exonuclease proofreading activity, resulting in low replication fidelity, which is further decreased by an unbalanced ratio of nucleotides and addition of Mn2+ and increased Mg2+ concentration. The protocol for PCR consisted of a step at 94 °C for 30 s and 10 cycles at 94 °C for 1 min and 72 °C for 4 min. The polymerase used was Thermus aquaticus polymerase.
The DNA library was amplified for yeast transformation with lib_fwd (ATCGCTGCTAAGGAAGAAGGTGTTCAATTGGACAAGAGAGAAGCT) and lib_rev (GGGTGAGGATGTTTGAGCGTAATCTGGAACATCGTATGGGTAGGATCC) primers. 500 ml of BJ5465 yeast (MATa ura352 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL) was grown to A600 = 1.5, transformed with 245 μg of nanobody insert DNA and 50 μg of pYDS2.0 plasmid, and digested with NheI-HF and BamHI-HF (New England Biolabs) using an ECM 830 Electroporator (BTX-Harvard Apparatus) as described previously (36). Dilutions of transformed yeast were then plated on -Trp dropout medium, and colonies were counted to obtain an estimate of library diversity.
Library selection
T-cadherin binders were isolated after one round of magnetic activated cell sorting (MACS) by staining the yeast library with FITC-labeled T-cadherin and anti-FITC microbeads (Miltenyi). This was followed by two rounds of FACS, staining the yeast library with Alexa Fluor 488– or Alexa Fluor 647–labeled T-cadherin at decreasing concentrations (1 μm, 500 nm, and 200 nm). MACS selection was performed as described in Ref. 36. Cells from each round of selection were then analyzed by amplicon deep sequencing as described below.
Next-generation sequencing amplicon deep sequencing
The yeast DNA library of each round was extracted through Zymoprep Yeast Plasmid Miniprep II, PCR-amplified using primers GTTCAATTGGACAAGAGAGAAGCT and GAACATCGTATGGGTAGGATCC, gel-purified, and submitted for amplicon deep sequencing at the Massachusetts General Hospital Center for Computational and Integrative Biology DNA Core. Data were analyzed using Geneious Prime 2019 and a custom Python 3 script. In brief, reads were aligned to the reference sequence using Bowtie 2 (43). Mutation percentages were compiled using pysam's pileup method as the number of nucleotides (or amino acids) different from the reference nucleotide over the total number of nucleotides sequenced at this position.
Protein expression and purification
The full-length adiponectin gene (both WT and mutants) was cloned and purified as described in Ref. 36. In brief, the adiponectin gene was cloned into the pTARGET vector with an HA signal peptide and a FLAG tag. An Expi293 (Thermo) stable cell line was created to express FLAG-adiponectin in the medium and was maintained in a bioreactor (CELLine flasks, Wheaton). Cells were harvested by centrifugation at 4000 × g for 10 min, the supernatant was diluted 1:1 with HEPES-buffered saline, and 2 mm CaCl2 was added before loading onto M1 anti-FLAG affinity resin. This was first washed with 300 mm NaCl, 20 mm HEPES (pH 7.5), and 2 mm CaCl2 and then with 150 mm NaCl, 20 mm HEPES (pH 7.5), and 2 mm CaCl2; elution was performed with 150 mm NaCl, 20 mm HEPES (pH 7.5), 0.2 mg/ml FLAG peptide, and 5 mm EDTA. Adiponectin was dialyzed in HEPES-buffered saline overnight.
For crystallography studies, adiponectin globular domain WT (from residue 108 to the end) was cloned into a pMAL vector containing an N-terminal His tag, followed by maltose-binding protein and a 3C protease cleavage site. E. coli BL21 (DE3) cells were grown in Terrific broth supplemented with 0.2% (w/v) glucose, 2 mm MgCl2, and 100 μg/ml ampicillin at 37 °C until an A600 of 0.8 was reached, at which point the temperature was shifted to 18 °C, and protein expression was induced by addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. After 16 h of induction, cells were collected and resuspended in 150 mm NaCl and 20 mm HEPES (pH 7.5). Cells were lysed by sonication and then centrifuged at 20,000 × g for 20 min. The supernatant was recovered, filtered with a 0.45-μm filter, and applied to nickel-nitrilotriacetic acid resin. The nickel-nitrilotriacetic acid resin was first washed with 500 mm NaCl, 20 mm HEPES (pH 7.5), and 50 mm imidazole and then with 150 mm NaCl, 20 mm HEPES (pH 7.5), and 50 mm imidazole; elution was performed with 150 mm NaCl, 20 mm HEPES, and 250 mm imidazole (pH 8). The protein was further purified by size exclusion chromatography.
For all other studies, adiponectin globular domain, both WT and mutants, (from residue 108 to the end) was cloned in the vector pMS211 (44) containing an N-terminal small ubiquitin-like modifier fusion followed by a FLAG epitope tag and a 3C protease cleavage site. E. coli C43 (DE3) harboring an arabinose-inducible Ulp1 protease plasmid (pAM174) was transformed with the pAM205 vector containing the protein of interest. Bacteria were grown in Terrific broth supplemented with 0.2% glucose, 2 mm MgCl2, 100 μg/ml ampicillin, and 35 μg/ml chloramphenicol at 37 °C until an A600 of 0.8 was reached, at which point the temperature was reduced to 18 °C, and protein expression was induced by addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside and 0.2% arabinose for the adiponectin globular domain and Ulp1, respectively. After 16 h of induction, cells were collected and resuspended in 150 mm NaCl and 20 mm HEPES (pH 7.5). Cells were lysed by sonication and then centrifuged at 20,000 × g for 20 min. The supernatant was recovered, filtered with a 0.45-μm filter, and applied to M1 anti-FLAG affinity resin. The resin was first washed with 500 mm NaCl, 20 mm HEPES (pH 7.5), and 2 mm CaCl2 and then with 150 mm NaCl, 20 mm HEPES (pH 7.5), and 2 mm CaCl2; elution was performed with 150 mm NaCl, 20 mm HEPES (pH 7.5), 0.2 mg/ml FLAG peptide, and 5 mm EDTA. The protein was further purified by size exclusion chromatography using a Superdex 200 increase column.
For biotinylation experiments, an adiponectin globular domain single-chain trimer (34) was cloned with an N-terminal hemagglutinin signal peptide and a FLAG epitope tag; at the C terminus, it was fused to an AVI tag (45) to allow biotinylation by the BirA enzyme. This construct was expressed in Spodoptera frugiperda Sf9 insect cells (Expression Systems, Davis, CA) using the FastBac baculovirus system (Thermo Fisher) according to the manufacturer's instructions. Protein was harvested by centrifugation to remove cells, and the supernatant was treated with 50 mm Tris (pH 8), 1 mm NiCl2, and 5 mm CaCl2. The medium was centrifuged again and then filtered with a 0.45-μm filter to remove precipitated material. Clarified supernatant was loaded by gravity flow over anti-FLAG resin. The column was first washed with 300 mm NaCl, 20 mm HEPES (pH 7.5), and 2 mm CaCl2 and then with 150 mm NaCl, 20 mm HEPES (pH 7.5), and 2 mm CaCl2; protein was then eluted in 150 mm NaCl, 20 mm HEPES (pH 7.5), 0.2 mg/ml FLAG peptide, and 5 mm EDTA.
T-cadherin was produced in Tni insect cells. In brief, the gene from residues 23–690 was cloned into expression vector pAcGP67a, followed by a His6 tag. Baculovirus was produced by cotransfection of the transfer vector and linearized “BestBac” viral DNA in S. frugiperda Sf9 cells (Expression Systems). Protein was harvested by centrifugation to remove cells, and the supernatant was treated with 50 mm Tris (pH 8), 1 mm NiCl2, and 5 mm CaCl2. The medium was centrifuged again and then filtered with a 0.45-μm filter to remove precipitated material. Clarified supernatant was loaded by gravity flow over nickel-bound chelating Sepharose resin (GE Healthcare). The column was first washed with 300 mm NaCl, 20 mm HEPES (pH 7.5), and 20 mm imidazole and then with 150 mm NaCl, 20 mm HEPES (pH 7.5), and 20 mm imidazole; protein was then eluted in the same buffer supplemented with 250 mm imidazole. For the pulldown shown in Fig. 6, C and D, and Fig. S3B, another construct was used. T-cadherin was cloned in the pFUSE vector, where, at the N terminus, it was preceded by the IL2 signal sequence, and at the C terminus, it was fused to the heavy chain CH2-CH3 fragment of human IgG1. Construct P12 comprised T-cadherin residues 23–359, construct P123 comprised residues 23–473, and the full-length construct comprised residues 23–690, as described in Ref. 31.
Crystallization
Proteins for crystallographic studies were produced in E. coli BL21 (DE3). The WT adiponectin globular domain and mutants were expressed as above and then digested with 3C protease (1:1000 (w/w)) to remove the maltose-binding protein tag (for the WT construct) or the FLAG tag (for the mutant). The proteins were further purified by size exclusion chromatography on a Superdex S200 increase column (GE Healthcare) in a buffer containing 150 mm NaCl and 20 mm HEPES.
The WT adiponectin globular domain was concentrated to 6.3 mg/ml prior to crystallization and was crystallized by vapor diffusion using a Gryphon LCP crystallization robot (Art Robbins Instruments) to mix 100 nl of protein with 100 nl of a precipitant solution consisting of 0.2 mm Li2SO4, 0.1 m sodium acetate (pH 4.5), and 50% PEG 400. Crystals grew in a few minutes and continued to grow over the course of several days. The crystals were transferred to a solution of mother liquor supplemented with 20% (v/v) glycerol for cryoprotection prior to freezing in liquid nitrogen.
Adiponectin globular domain D187A/Q188A was concentrated to 6.3 mg/ml prior to crystallization and was crystallized by vapor diffusion by mixing 100 nl of protein with 100 nl of a precipitant solution consisting of 0.2 m sodium nitrate, 0.1 m MES (pH 5.5), and 20% PEG 3350. Crystals grew in 2 days. The crystals were transferred to a solution of mother liquor supplemented with 20% (v/v) glycerol for cryoprotection prior to freezing in liquid nitrogen.
Data collection was performed at Advanced Photon Source beamline 23ID-D for the WT adiponectin globular domain and at beamline 23ID-B for the adiponectin globular domain D187A/Q188A mutant (NE-CAT) using a single crystal for each dataset. Data reduction and scaling were carried out using XDS and XSCALE, respectively (46), and the structure was solved by molecular replacement in Phaser (47) using a single-chain trimer of human adiponectin globular domain (PDB code 4DOU) as the search model. Refinement was conducted using Phenix 1.16 (48) with manual building in COOT (49). Because of its high resolution, refinement for the WT adiponectin structure was conducted with riding hydrogens and anisotropic B-factors for all nonhydrogen atoms. Structure quality was assessed using MolProbity (50). The refined structures were deposited in the Protein Data Bank under accession codes 6U66 for the adiponectin globular domain WT and 6U6N for the adiponectin globular domain D187A/Q188A mutant.
EM
Purified FLAG-adiponectin full-length was loaded onto a Superdex S200 column (GE Healthcare) equilibrated with a buffer containing 150 mm NaCl and 20 mm HEPES to separate the three oligomerization states of adiponectin by size exclusion chromatography. Fractions from each peak were diluted to a concentration of 0.01 mg/ml, and 2.5 μl was applied to a glow-discharged, carbon-coated copper grid (Electron Microscopy Sciences). The protein solution was allowed to adsorb for 30 s, and then grids were stained with three drops of 1.5% (w/v) uranyl formate. Filter paper (Whatman #1) was used to absorb excess solution between each staining step, and the grids were allowed to air-dry.
Negative-stain micrographs were collected at room temperature with a Philips Tecnai T12 electron microscope with a LaB6 filament operated at 120 kV. Micrographs were collected on a Gatan 4K charge-coupled device camera at a magnification of ×52,000, corresponding to a pixel size of 2.13 Å, and a defocus value of −1.5 μm. Particles were manually picked in RELION (51) to generate initial two-dimensional class averages that were used as references for autopicking the remaining micrographs. Several iterations of two-dimensional classification were carried out in RELION to generate the final class averages.
Biochemical assays
For FLAG pulldown, 1 μm of FLAG-adiponectin was immobilized on M1 anti-FLAG resin and then incubated for 1 h at 4 °C with 3 μm T-cadherin. Beads were washed four times in 150 mm NaCl, 20 mm HEPES, 5 mm CaCl2, and 0.05% Triton X-100, and then the reaction was eluted in Laemmli buffer. For protein A pulldown, 3 μm T-cadherin–Fc was immobilized on the resin and then incubated with 1 μm FLAG-adiponectin for 1 h at 4 °C. Beads were washed four times in 150 mm NaCl, 20 mm HEPES, 5 mm CaCl2, and 0.05% Triton X-100, and then the reaction was eluted in Laemmli buffer. The biotinylation reaction was performed using a BirA-500 kit (Avidity). The protein was dialyzed in 10 mm Tris (pH 8) and 50 mm NaCl for the reaction and incubated according to manufacturer's instructions. The biotinylated protein was then dialyzed in 150 mm NaCl and 20 mm HEPES (pH 7.5) to remove any excess of free biotin before performing the pulldown assay.
Author contributions
R. P. and A. C. K. conceptualization; R. P. validation; R. P., S. C. E., D. J. B., S. Z., and A. C. K. investigation; R. P. writing-original draft; S. C. E. and A. C. K. writing-review and editing; D. J. B. formal analysis; A. C. K. supervision; A. C. K. funding acquisition.
Supplementary Material
This work was supported by National Institutes of Health Grant DP5OD021345 (to A. C. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (codes 6U6N and 6U66) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HMW
- high-molecular-weight
- MACS
- magnetic activated cell sorting.
References
- 1. Arita Y., Kihara S., Ouchi N., Takahashi M., Maeda K., Miyagawa J., Hotta K., Shimomura I., Nakamura T., Miyaoka K., Kuriyama H., Nishida M., Yamashita S., Okubo K., Matsubara K., et al. (1999) Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 257, 79–83 10.1006/bbrc.1999.0255 [DOI] [PubMed] [Google Scholar]
- 2. Shapiro L., and Scherer P. E. (1998) The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8, 335–338 10.1016/S0960-9822(98)70133-2 [DOI] [PubMed] [Google Scholar]
- 3. Reid K. B., Gagnon J., and Frampton J. (1982) Completion of the amino acid sequences of the A and B chains of subcomponent C1q of the first component of human complement. Biochem. J. 203, 559–569 10.1042/bj2030559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scherer P. E., Williams S., Fogliano M., Baldini G., and Lodish H. F. (1995) A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 270, 26746–26749 10.1074/jbc.270.45.26746 [DOI] [PubMed] [Google Scholar]
- 5. Wang Y., Xu A., Knight C., Xu L. Y., and Cooper G. J. (2002) Hydroxylation and glycosylation of the four conserved lysine residues in the collagenous domain of adiponectin: potential role in the modulation of its insulin-sensitizing activity. J. Biol. Chem. 277, 19521–19529 10.1074/jbc.M200601200 [DOI] [PubMed] [Google Scholar]
- 6. Wang Y., Lu G., Wong W. P., Vliegenthart J. F., Gerwig G. J., Lam K. S., Cooper G. J., and Xu A. (2004) Proteomic and functional characterization of endogenous adiponectin purified from fetal bovine serum. Proteomics 4, 3933–3942 10.1002/pmic.200400826 [DOI] [PubMed] [Google Scholar]
- 7. Richards A. A., Stephens T., Charlton H. K., Jones A., Macdonald G. A., Prins J. B., and Whitehead J. P. (2006) Adiponectin multimerization is dependent on conserved lysines in the collagenous domain: evidence for regulation of multimerization by alterations in posttranslational modifications. Mol. Endocrinol. 20, 1673–1687 10.1210/me.2005-0390 [DOI] [PubMed] [Google Scholar]
- 8. Richards A. A., Colgrave M. L., Zhang J., Webster J., Simpson F., Preston E., Wilks D., Hoehn K. L., Stephenson M., Macdonald G. A., Prins J. B., Cooney G. J., Xu A., and Whitehead J. P. (2010) Sialic acid modification of adiponectin is not required for multimerization or secretion but determines half-life in circulation. Mol. Endocrinol. 24, 229–239 10.1210/me.2009-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsao T. S., Tomas E., Murrey H. E., Hug C., Lee D. H., Ruderman N. B., Heuser J. E., and Lodish H. F. (2003) Role of disulfide bonds in Acrp30/adiponectin structure and signaling specificity: different oligomers activate different signal transduction pathways. J. Biol. Chem. 278, 50810–50817 10.1074/jbc.M309469200 [DOI] [PubMed] [Google Scholar]
- 10. Pajvani U. B., Du X., Combs T. P., Berg A. H., Rajala M. W., Schulthess T., Engel J., Brownlee M., and Scherer P. E. (2003) Structure-function studies of the adipocyte-secreted hormone Acrp30/adiponectin: implications for metabolic regulation and bioactivity. J. Biol. Chem. 278, 9073–9085 10.1074/jbc.M207198200 [DOI] [PubMed] [Google Scholar]
- 11. Waki H., Yamauchi T., Kamon J., Ito Y., Uchida S., Kita S., Hara K., Hada Y., Vasseur F., Froguel P., Kimura S., Nagai R., and Kadowaki T. (2003) Impaired multimerization of human adiponectin mutants associated with diabetes: molecular structure and multimer formation of adiponectin. J. Biol. Chem. 278, 40352–40363 10.1074/jbc.M300365200 [DOI] [PubMed] [Google Scholar]
- 12. Wang Y., Lam K. S., Chan L., Chan K. W., Lam J. B., Lam M. C., Hoo R. C., Mak W. W., Cooper G. J., and Xu A. (2006) Post-translational modifications of the four conserved lysine residues within the collagenous domain of adiponectin are required for the formation of its high molecular weight oligomeric complex. J. Biol. Chem. 281, 16391–16400 10.1074/jbc.M513907200 [DOI] [PubMed] [Google Scholar]
- 13. Tsao T. S., Murrey H. E., Hug C., Lee D. H., and Lodish H. F. (2002) Oligomerization state-dependent activation of NF-κB signaling pathway by adipocyte complement-related protein of 30 kDa (Acrp30). J. Biol. Chem. 277, 29359–29362 10.1074/jbc.C200312200 [DOI] [PubMed] [Google Scholar]
- 14. Schraw T., Wang Z. V., Halberg N., Hawkins M., and Scherer P. E. (2008) Plasma adiponectin complexes have distinct biochemical characteristics. Endocrinology 149, 2270–2282 10.1210/en.2007-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomas E., Tsao T. S., Saha A. K., Murrey H. E., Zhang C. C., Itani S. I., Lodish H. F., and Ruderman N. B. (2002) Enhanced muscle fat oxidation and glucose transport by ACRP30 globular domain: acetyl-CoA carboxylase inhibition and AMP-activated protein kinase activation. Proc. Natl. Acad. Sci. U.S.A. 99, 16309–16313 10.1073/pnas.222657499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamauchi T., Kamon J., Minokoshi Y., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S., Ueki K., Eto K., Akanuma Y., Froguel P., Foufelle F., Ferre P., et al. (2002) Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 8, 1288–1295 10.1038/nm788 [DOI] [PubMed] [Google Scholar]
- 17. Basu R., Pajvani U. B., Rizza R. A., and Scherer P. E. (2007) Selective downregulation of the high molecular weight form of adiponectin in hyperinsulinemia and in type 2 diabetes: differential regulation from nondiabetic subjects. Diabetes 56, 2174–2177 10.2337/db07-0185 [DOI] [PubMed] [Google Scholar]
- 18. Koenen T. B., van Tits L. J., Holewijn S., Lemmers H. L., den Heijer M., Stalenhoef A. F., and de Graaf J. (2008) Adiponectin multimer distribution in patients with familial combined hyperlipidemia. Biochem. Biophys. Res. Commun. 376, 164–168 10.1016/j.bbrc.2008.08.111 [DOI] [PubMed] [Google Scholar]
- 19. Pajvani U. B., Hawkins M., Combs T. P., Rajala M. W., Doebber T., Berger J. P., Wagner J. A., Wu M., Knopps A., Xiang A. H., Utzschneider K. M., Kahn S. E., Olefsky J. M., Buchanan T. A., and Scherer P. E. (2004) Complex distribution, not absolute amount of adiponectin, correlates with thiazolidinedione-mediated improvement in insulin sensitivity. J. Biol. Chem. 279, 12152–12162 10.1074/jbc.M311113200 [DOI] [PubMed] [Google Scholar]
- 20. Tang Y. T., Hu T., Arterburn M., Boyle B., Bright J. M., Emtage P. C., and Funk W. D. (2005) PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J. Mol. Evol. 61, 372–380 10.1007/s00239-004-0375-2 [DOI] [PubMed] [Google Scholar]
- 21. Thomas P., Pang Y., Dong J., Groenen P., Kelder J., de Vlieg J., Zhu Y., and Tubbs C. (2007) Steroid and G protein binding characteristics of the seatrout and human progestin membrane receptor α subtypes and their evolutionary origins. Endocrinology 148, 705–718 10.1210/en.2006-0974 [DOI] [PubMed] [Google Scholar]
- 22. Yamauchi T., Kamon J., Ito Y., Tsuchida A., Yokomizo T., Kita S., Sugiyama T., Miyagishi M., Hara K., Tsunoda M., Murakami K., Ohteki T., Uchida S., Takekawa S., Waki H., et al. (2003) Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423, 762–769 10.1038/nature01705 [DOI] [PubMed] [Google Scholar]
- 23. Zhou Y., Sun X., Jin L., Stringfield T., Lin L., and Chen Y. (2005) Expression profiles of adiponectin receptors in mouse embryos. Gene. Expr. Patterns 5, 711–715 10.1016/j.modgep.2005.02.002 [DOI] [PubMed] [Google Scholar]
- 24. Ranscht B., and Dours-Zimmermann M. T. (1991) T-cadherin, a novel cadherin cell adhesion molecule in the nervous system lacks the conserved cytoplasmic region. Neuron 7, 391–402 10.1016/0896-6273(91)90291-7 [DOI] [PubMed] [Google Scholar]
- 25. Hug C., Wang J., Ahmad N. S., Bogan J. S., Tsao T. S., and Lodish H. F. (2004) T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc. Natl. Acad. Sci. U.S.A. 101, 10308–10313 10.1073/pnas.0403382101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ivanov D., Philippova M., Antropova J., Gubaeva F., Iljinskaya O., Tararak E., Bochkov V., Erne P., Resink T., and Tkachuk V. (2001) Expression of cell adhesion molecule T-cadherin in the human vasculature. Histochem. Cell Biol. 115, 231–242 10.1007/s004180100252 [DOI] [PubMed] [Google Scholar]
- 27. Hebbard L. W., Garlatti M., Young L. J., Cardiff R. D., Oshima R. G., and Ranscht B. (2008) T-cadherin supports angiogenesis and adiponectin association with the vasculature in a mouse mammary tumor model. Cancer Res. 68, 1407–1416 10.1158/0008-5472.CAN-07-2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Denzel M. S., Scimia M. C., Zumstein P. M., Walsh K., Ruiz-Lozano P., and Ranscht B. (2010) T-cadherin is critical for adiponectin-mediated cardioprotection in mice. J. Clin. Invest. 120, 4342–4352 10.1172/JCI43464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parker-Duffen J. L., Nakamura K., Silver M., Kikuchi R., Tigges U., Yoshida S., Denzel M. S., Ranscht B., and Walsh K. (2013) T-cadherin is essential for adiponectin-mediated revascularization. J. Biol. Chem. 288, 24886–24897 10.1074/jbc.M113.454835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuda K., Fujishima Y., Maeda N., Mori T., Hirata A., Sekimoto R., Tsushima Y., Masuda S., Yamaoka M., Inoue K., Nishizawa H., Kita S., Ranscht B., Funahashi T., and Shimomura I. (2015) Positive feedback regulation between adiponectin and T-cadherin impacts adiponectin levels in tissue and plasma of male mice. Endocrinology 156, 934–946 10.1210/en.2014-1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fukuda S., Kita S., Obata Y., Fujishima Y., Nagao H., Masuda S., Tanaka Y., Nishizawa H., Funahashi T., Takagi J., Maeda N., and Shimomura I. (2017) The unique prodomain of T-cadherin plays a key role in adiponectin binding with the essential extracellular cadherin repeats 1 and 2. J. Biol. Chem. 292, 7840–7849 10.1074/jbc.M117.780734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kita S., Fukuda S., Maeda N., and Shimomura I. (2019) Native adiponectin in serum binds to mammalian cells expressing T-cadherin, but not AdipoRs or calreticulin. Elife 8, e48675 10.7554/eLife.48675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Radjainia M., Wang Y., and Mitra A. K. (2008) Structural polymorphism of oligomeric adiponectin visualized by electron microscopy. J. Mol. Biol. 381, 419–430 10.1016/j.jmb.2008.06.015 [DOI] [PubMed] [Google Scholar]
- 34. Min X., Lemon B., Tang J., Liu Q., Zhang R., Walker N., Li Y., and Wang Z. (2012) Crystal structure of a single-chain trimer of human adiponectin globular domain. FEBS Lett. 586, 912–917 10.1016/j.febslet.2012.02.024 [DOI] [PubMed] [Google Scholar]
- 35. Boder E. T., and Wittrup K. D. (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 10.1038/nbt0697-553 [DOI] [PubMed] [Google Scholar]
- 36. McMahon C., Baier A. S., Pascolutti R., Wegrecki M., Zheng S., Ong J. X., Erlandson S. C., Hilger D., Rasmussen S. G. F., Ring A. M., Manglik A., and Kruse A. C. (2018) Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 25, 289–296 10.1038/s41594-018-0028-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hara K., Horikoshi M., Yamauchi T., Yago H., Miyazaki O., Ebinuma H., Imai Y., Nagai R., and Kadowaki T. (2006) Measurement of the high-molecular weight form of adiponectin in plasma is useful for the prediction of insulin resistance and metabolic syndrome. Diabetes Care 29, 1357–1362 10.2337/dc05-1801 [DOI] [PubMed] [Google Scholar]
- 38. Inoue T., Kotooka N., Morooka T., Komoda H., Uchida T., Aso Y., Inukai T., Okuno T., and Node K. (2007) High molecular weight adiponectin as a predictor of long-term clinical outcome in patients with coronary artery disease. Am. J. Cardiol. 100, 569–574 10.1016/j.amjcard.2007.03.062 [DOI] [PubMed] [Google Scholar]
- 39. Hirose H., Yamamoto Y., Seino-Yoshihara Y., Kawabe H., and Saito I. (2010) Serum high-molecular-weight adiponectin as a marker for the evaluation and care of subjects with metabolic syndrome and related disorders. J. Atheroscler. Thromb. 17, 1201–1211 10.5551/jat.6106 [DOI] [PubMed] [Google Scholar]
- 40. Takemura Y., Ouchi N., Shibata R., Aprahamian T., Kirber M. T., Summer R. S., Kihara S., and Walsh K. (2007) Adiponectin modulates inflammatory reactions via calreticulin receptor-dependent clearance of early apoptotic bodies. J. Clin. Invest. 117, 375–386 10.1172/JCI29709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ye J., Bian X., Lim J., and Medzhitov R. (2019) Adiponectin and related C1q/TNF-related proteins bind selectively to anionic phospholipids and sphingolipids. bioRxiv 10.1101/776690 [DOI] [PMC free article] [PubMed]
- 42. Wilson D. S., and Keefe A. D. (2001) Random mutagenesis by PCR. Curr. Protoc. Mol. Biol. Chapter 8, Unit 8.3 10.1002/0471142727.mb0803s51 [DOI] [PubMed] [Google Scholar]
- 43. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sjodt M., Brock K., Dobihal G., Rohs P. D. A., Green A. G., Hopf T. A., Meeske A. J., Srisuknimit V., Kahne D., Walker S., Marks D. S., Bernhardt T. G., Rudner D. Z., and Kruse A. C. (2018) Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature 556, 118–121 10.1038/nature25985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fairhead M., and Howarth M. (2015) Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 1266, 171–184 10.1007/978-1-4939-2272-7_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 50. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scheres S. H. (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.