Abstract
The calcineurin/nuclear factor of activated T cell (CN/NFAT) signaling pathway plays a critical role in the immune response. Therefore, inhibition of the CN/NFAT pathway is an important target for inflammatory disease. The conserved PXIXIT and LXVP motifs of CN substrates and targeting proteins have been recognized. Based on the affinity ability and inhibitory effect of these docking sequences on CN, we designed a bioactive peptide (named pep3) against the CN/NFAT interaction, which has two binding sites derived from the RCAN1-PXIXIT motif and the NFATc1-LXVP motif. The shortest linker between the two binding sites in pep3 is derived from A238L, a physiological binding partner of CN. Microscale thermophoresis revealed that pep3 has two docking sites on CN. Pep3 also has the most potent inhibitory effect on CN. It is suggested that pep3 contains an NFATc1-LXVP–substrate recognition motif and RCAN1-PXIXIT–mediated anchoring to CN. Expression of this peptide significantly suppresses CN/NFAT signaling. Cell-permeable 11-arginine–modified pep3 (11R-pep3) blocks the NFAT downstream signaling pathway. Intranasal administration of the 11R-pep3 peptide inhibits airway inflammation in an ovalbumin-induced asthma model. Our results suggest that pep3 is promising as an immunosuppressive agent and can be used in topical remedies.
Keywords: calcineurin, NFAT transcription factor, enzyme inhibitor, cytokine, cellular immune response
Introduction
Calcineurin (CN)5 is a serine/threonine phosphatase that is activated by calcium/calmodulin (Ca2+/CaM). CN is a heterodimer composed of a catalytic subunit called calcineurin A (CNA, ∼61 kDa) and a regulatory subunit (CNB, ∼19 kDa). CN plays important roles in the immune response, learning and memory, and cardiac hypertrophy (1). To carry out its biological functions, it interacts with multiple substrates and targeting proteins, such as nuclear factor of activated T cells (NFAT) (2), endogenous regulator of calcineurin 1 (RCAN1) (3), transcription factor EB (TFEB) (4), and dynamin-related protein 1 (Drp1) (5). NFAT has been the center of attention in studies on CN-substrate interactions. In resting cells, the hyperphosphorylated NFAT proteins reside in the cytoplasm (NFAT cytoplasmic 1–4, NFATc1-c4). Upon activation, CN dephosphorylates these proteins, which translocate to the nucleus. They regulate gene transcription via interaction with other transcription factors in the nucleus. The highly conserved PXIXIT motif (PALESPRIEITSCLGL, 16 aa, NFATc1-PXIXIT motif) was identified by Aramburu et al. (6). They developed a high-affinity PVIVIT peptide (MAGPHPVIVITGPHEE, PVIVIT peptide) based on the PXIXIT docking motif (7). This peptide potently inhibited NFAT activation (NFATc1-c3) and NFAT-dependent gene expression in T cells without affecting CN activity. Recently, the second motif (DQYLAVPQHPYQWAK, 15 aa, NFATc1-LXVP motif) in NFATc1 has attracted new attention (8–10). The LXVP motif binds to a hydrophobic pocket at the interface of the CNA and CNB subunits. LXVP-containing sequences have a similar mechanism of action as the two immunosuppressant-immunophilin complexes, cyclosporin A-cyclophilin (CsA-CyP) and FK506-FKBP (11). The DQYLAVPQHPYQWAK peptide derived from NFATc1-LXVP inhibits not only CN activity directly but also NFAT-dependent transcription regulation. It is well-known that the T cell receptor (TcR)/Ca2+/CN signaling pathway plays a critical role in T cell activation. The NFAT family of proteins are important transcription factors that promote the expression of a panel of genes required for activation. The above peptides blocking the interaction between CN and NFAT, similar to CsA, could be targeted to alter the transcriptional program, thereby modulating immune responses.
Subsequently, researchers found binding motifs in RCAN1 (12). RCAN1 also has the PXIXIT motif, and the LXVP motif closely matches the two motifs of NFAT. Our previous study demonstrated that the binding between CN and the RCAN1-LXVP motifs is weaker than that between CN and the NFATc1-LXVP motifs (13–15). However, the RCAN1-PXIXIT motif has a strong binding affinity for CN. It is worth noting that the RCAN1-PXIXIT motif in many studies refers to two independent and closely connected CN-interacting motifs: a PXIXIT-like site (208TPSVVVHVCES218, also called the E-motif), which resembles the PXIXIT motif from NFAT, located near the C terminus, and the more N-terminal CIC (calcineurin-inhibitor CALP1) motif (195PGEKYELHAATDT207, also called the V-motif). Replacement of the hydrophobic residues in the RCAN V-motif with the polar amino acid glutamine eliminated the interaction of the EV-motif with CN. Alanine substitution of the two conserved glutamic acid residues found in the E-motif reduced CN binding but did not abolish it. Therefore, it is suggested that the E-motif contributes to binding of the EV-motif to CN and that the major determinant lies within the hydrophobic PSVVVH motif. More importantly, only the synthetic peptide derived from the whole EV-motif (195PGEKYELHAATDTTPSVVVHVCES218) in RCAN1 could suppress CN-mediated NFAT-dependent cytokine gene expression in human T cells (16). Carme Mulero et al. (17) developed an optimized in vitro high-throughput fluorescence polarization assay based on the disruption of the interaction between the RCAN1-PXIXIT motif and CNA for identifying molecules with immunosuppressant potential.
The protein inhibitor A238L from African swine fever virus is a physiological binding partner of CN. Grigoriu et al. (18) used X-ray crystallography to investigate the structure of the A238L-CN complex and identified two binding motifs in A238L similar to those in NFAT. A238L (residues 200–239) binds CN in a largely extended conformation, stretching from the A238L-PXIXIT–binding site to the CNA/CNB interface and then looping back along the CNA/CNB interface to occupy the A238L-LXVP–binding pocket. They further summarized the features of the 10 CN substrates and regulators. The 17-aa GCEDNVYEKLPEQNSN sequence in A238L is the shortest linker in these CN-targeting proteins. The distance between the A238L-PXIXIT motif and the A238L-LXVP motif is 45 Å. By comparison, the linker between the two motifs in NFAT is 263 aa, as well as 54 aa in RCAN1.
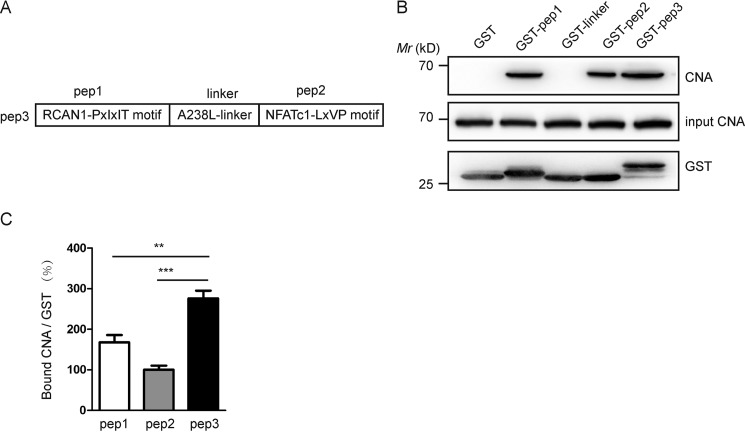
Based on our previous investigation of the two conserved motifs in CN substrates and targeting proteins, we designed a short polypeptide (Fig. 1A). The sequence of the peptide is constructed from three different substrates of CN: PGEKYELHAATDTTPSVVVH derived from the RCAN1-PXIXIT motif (pep1), GCEDNVYEKLPEQNSN derived from the A238L (linker), and YLAVPQHPYQWAK derived from the NFATc1-LXVP motif (pep2). We named the peptide pep3 and investigated the effects of pep3 in vitro and in vivo.
Figure 1.
The binding capacity between CNA and pep3 is more potent in mouse brain lysates. A, schematic representation of pep1, pep2, linker, and pep3. B, pep3 binds CNA in mouse brain lysates in GST pulldown assays. Bound CNA was visualized by Western blotting via monoclonal anti-CNA antibody, shown in the upper blot; the input is shown in the middle panel. The GST fusion proteins were shown in the bottom blot. C, CNA bound by pep1, pep2, and pep3 was densitometrically quantified, and histograms illustrate the relative intensity units of bound CN. Data are presented as the mean ± S.E. (n = 3), **, p < 0.01; ***, p < 0.001 compared with the pep3 group.
Results
The binding ability of pep3 and CNA is stronger than that of each independent fragment with CNA
We constructed plasmids encoding peptides fused to GST and compared the binding ability of pep1, pep2, linker, and pep3 with CNA in lysates of the mouse brain as a source of CN. CN normally consists of one catalytic subunit of CNA and a tightly bound regulatory subunit of CNB. This anti-CNA antibody used in our experiments could detect endogenous levels of CNA. We used an equivalent amount of input CNA in the GST pulldown assays. Anti-CNA immunoblotting showed that a GST control did not interact with CNA and that GST-pep3 pulled down more CNA than GST-pep1, GST-pep2, and GST-linker (Fig. 1B). Our pulldown data shows that A238L-linker and CNA have no obvious combination. Accordingly, the ratios of bound CNA to GST proteins were calculated (Fig. 1C). Compared with pep1 or pep2, pep3 has a stronger binding capacity with CNA.
Pep3 presents two binding phases with CN and potently inhibits CN activity
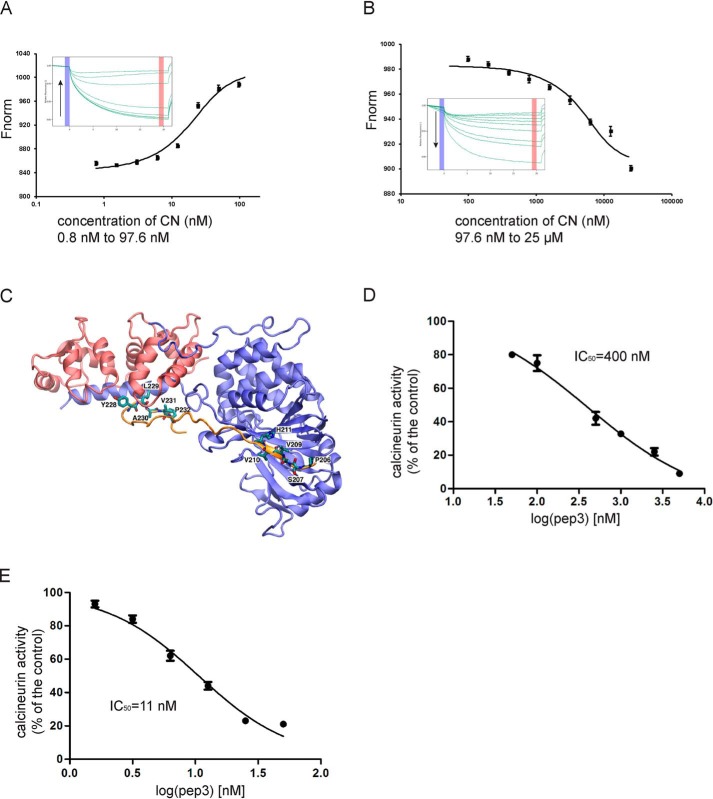
To quantitatively define the interaction between CN and pep3, we resorted to MST. MST is a sensitive and versatile biophysical technique for protein-peptide interaction studies. We mixed 31.3 nm fluorescently labeled pep3 with serial dilutions of CN (25 μm to 0.8 nm). The experiment was conducted in PBS. The result of the MST is shown in Fig. 2, A and B. The titration of CN yielded a biphasic MST signal. We performed an SDS denaturation test (SD-Test) to analyze the ligand-induced fluorescence changes. This test helps to distinguish between fluorescence changes caused by an interaction and changes caused by nonspecific effects, such as loss of protein due to aggregation or adsorption to labware. Our SD-Test results showed that the specific binding of ligand induced the fluorescence change. Then, we quantified the high-affinity binding from the first half of the data points, which yielded a dissociation constant Kd = 8.6 ± 0.3 nm. Fitting the low-affinity binding yielded a dissociation constant Kd = 1.6 ± 0.2 μm. The MST results demonstrate two binding phases between pep3 and CN. Pep3 binds CN with high affinity. In a previous study, Seidel et al. (19) extensively characterized the binding between a short peptide and a protein. Their study shows that MST can yield Kd data that are very comparable with those obtained by other well-established biophysical methods, including fluorescence polarization (FP) as a solution-based and surface plasmon resonance as a solid-phase technique. A more specific aspect of this system is the observation of a second binding phase that was identified in both the MST and surface plasmon resonance techniques.
Figure 2.
Pep3 binds to purified recombinant CN and inhibits its activity. A and B, titrating CN to a constant FAM-labeled pep3 system yielded a biphasic MST signal. Fitting the high affinity phase (A) reveals a Kd = 8.6 ± 0.3 nm, and fitting the low affinity phase (B) yields a Kd = 1.6 ± 0.2 μm. The inset panels in A and B show the isotherm derived from the raw data, fitted to a sigmoidal dose-response curve, and the raw thermophoresis data were recorded at 20 °C using 20% LED power and 20% MST power. C, the modeled structure of the pep3-CN complex. CNA (blue), CNB (red), and pep3 (orange) are shown in cartoon presentation, and the two critical CN-binding motifs YLAVP and PSVVVH are shown as sticks. The complex structure is stable during the 100-ns MD simulation. D, inhibition of purified recombinant CN activity by pep3. E, inhibition of CN activity in the lysates of Jurkat cells. All data are expressed as the mean ± S.E. of phosphatase activity in three duplicable experiments.
Based on our experimental data, we propose that pep3 binds CN at two major binding sites, a similar binding pattern to that between A238L and CN. Thus, the pep3-CN complex structure was modeled based on the crystal structure of the A238L-CN complex (PDB ID 4F0Z). A molecular dynamics (MD) simulation initiated from the model structure suggests that the two CN-binding motifs YLAVP and PSVVVH dock at the binding sites (Fig. 2C), which are highly similar to the LXVP- and PXIXIT-binding motifs in A238L and other peptide inhibitors.
We also assayed the inhibition of the purified CN by pep3. Pep3 had a potent inhibitory effect on CN activity (Fig. 2D). The IC50 value for pep3 was 400 nm. We further determined the CN activity in lysates of Jurkat T cells in the presence of various concentrations of pep3. The IC50 value of pep3 for CN was 11 nm (Fig. 2E). Our results show that pep3 is a very strong inhibitor of CN.
Expression of pep3 suppresses the CN/NFAT signaling pathway
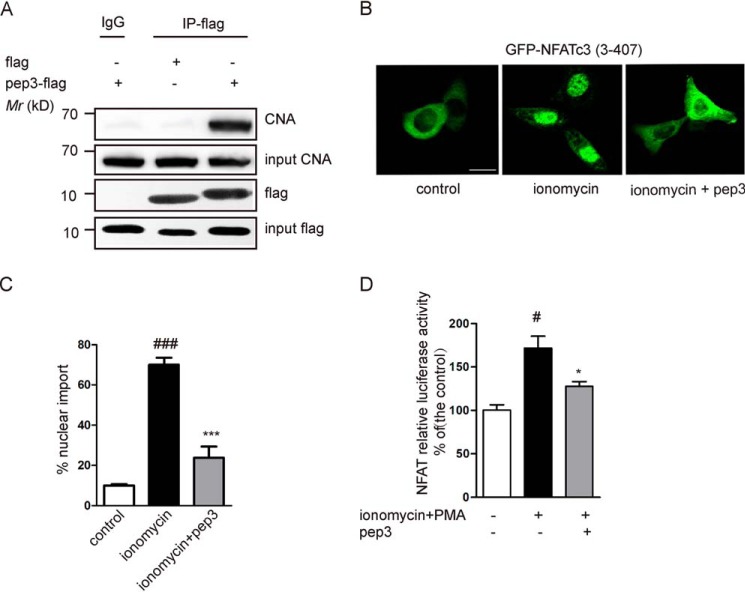
We also tested the CN-pep3 interaction in live cells in coprecipitation experiments in HEK293 cells. Compared with the DMSO control group, CNA was clearly pulled down after transfection of 2 μg of pep3 (Fig. 3A).
Figure 3.
Expression of pep3 suppresses CN-NFAT signaling pathway. A, Western blotting was utilized to detect CNA pulled down by an anti-DDK (flag) antibody in pep3-flag–transfected HEK293 cells. B, GFP-NFATc3 (3–407) plasmids were transfected into HeLa cells. Representative images shows that transfection of pep3-flag blocks ionomycin-induced GFP-NFATc3 translocation. Scale bar, 10 μm. C, bar graph shows the percentages of NFAT nuclear translocation in HeLa cells. Data were presented as mean ± S.E. of four independent experiments with n = 100 cells per experiment, ###, p < 0.001 compared with the control group; ***, p < 0.001 compared with the ionomycin group. D, pep3 inhibits NFAT-driven gene expression. HEK293 cells were transiently transfected with pep3-flag, together with pGL3-NFAT luciferase. pRL-null-Renilla-luc (7.5 ng) was used as an internal transfection control. Cells were stimulated with ionomycin + PMA for 1.5 h. Control values were taken as 100%. All data are expressed as the mean ± S.E. of three separate experiments. #, p < 0.001 compared with the control group; *, p < 0.05 compared with the ionomycin + PMA group.
We also examined the in vivo effect of pep3 on the CN/NFAT signaling pathway. Fig. 3, B and C, show that transfection of pep3 could prevent the ionomycin-triggered nuclear import of GFP-NFATc3 in HeLa cells. Using luciferase reporter gene assays, we found that pep3 also inhibited NFAT-driven promoters (Fig. 3D). Our experimental results show that expression of pep3 effectively inhibits the CN/NFAT signaling pathway.
A cell-permeable 11R-pep3 blocks NFAT dephosphorylation and downstream signaling
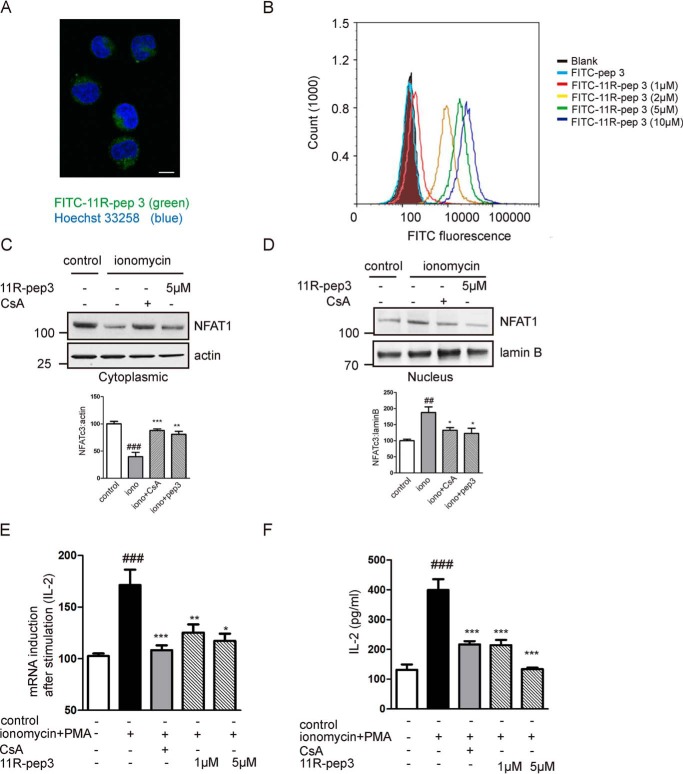
11-Arginine has been developed for the delivery of bioactive peptides into eukaryotic cells (20–22). We synthesized an 11R-modified pep3 and investigated the polypeptide transduction efficiency. We used FITC-pep3 as a control peptide. Jurkat cells were treated with FITC-conjugated 11R-pep3. To examine the intracellular localization of 11R-pep3, Jurkat cells were incubated with 1, 2, 5, and 10 μm FITC-conjugated 11R-pep3 for 20 min, and the FITC fluorescence was analyzed by fluorescence microscopy. Twenty minutes after transduction, only FITC-11R-pep3 was observed as a fluorescent signal in all living Jurkat cells. FITC fluorescence was visualized in the cytoplasm (Fig. 4A). FITC-11R-pep3 was transduced in a concentration-dependent manner (Fig. 4B). We also examined the effect of 11R-pep3 on the phosphorylation of endogenous NFAT1. Compared with the ionomycin group, incubation with 11R-pep3 resulted in a significant increase of NFAT1 in cytoplasm (Fig. 4C) and a decrease in the nuclear fraction (Fig. 4D). Our data suggests that the cell-permeable peptide prevented the dephosphorylation of NFAT induced by ionomycin stimulation.
Figure 4.
A cell-permeable 11R-pep3 blocks NFAT dephosphorylation and downstream signaling. A, FITC-11R-pep3 was visualized in the cytoplasm. Jurkat cells were prepared for fluorescence microscopy. Scale bar, 10 μm. B, FITC-pep3 or FITC-11R-pep3 was incubated with Jurkat cells at different concentrations. Intracellular fluorescent levels were analyzed by flow cytometry. C and D, the preincubated cells were stimulated with ionomycin (1 μm). CsA was used as a positive control. Western blotting detection of NFAT1 in the cytoplasmic and nuclear fractions in Jurkat cells. The β-actin (cytoplasmic) and lamin B (nuclear) expression levels were used as the loading control. The histograms represent the quantification of cytoplasmic and nuclear NFAT1 corrected by the loading control. All histograms in C and D represent values as a percentage compared with the control group. Data are presented as mean ± S.E. (n = 4). ##, p < 0.01; ###, p < 0.001 compared with the control group, *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with the ionomycin (iono) group. E, 11R-pep3 inhibits NFAT-driven gene expression in Jurkat cells. Total mRNA was extracted 6 h after stimulation with ionomycin (1 μm) + PMA (50 ng/ml). The mRNA levels of IL-2 were quantified by quantitative RT-PCR. The mRNA levels of the control group were set at 100%. The data are shown as the mean ± S.E. of three independent experiments. ###, p < 0.001 compared with the control group; *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with the ionomycin + PMA group. F, 11R-pep3 inhibits IL-2 protein expression induced by ionomycin + PMA. IL-2 protein expression (pg/ml) in Jurkat cells was determined by ELISA experiments. The data are shown as the mean ± S.E. of three independent experiments. ###, p < 0.001 compared with the control group; ***, p < 0.001 compared with the ionomycin + PMA group.
To test the effect of 11R-pep3 on endogenous interleukin-2 transcription, we treated Jurkat cells with 11R-pep3 or CsA for 1 h and then treated them with PMA + ionomycin for an additional 6 h. We examined the IL-2 mRNA expression. Compared with the ionomycin group, IL-2 transcription in the cells treated with 11R-pep3 was significantly inhibited. As a positive control, CsA inhibited the expression of IL-2 mRNA (Fig. 4D). ELISA experiments further confirmed the effects of 11R-pep3 at the IL-2 protein level (Fig. 4E).
Intranasal administration of 11R-pep3 inhibited airway inflammation and hyperresponsiveness
To evaluate the functional effects of pep3 in vivo, we intranasally administered 11R-pep3 in an animal model of asthma. CsA was used as a positive control. Some articles have reported the inhibitory effects of T cells in OVA-induced asthma murine model by CsA (23, 24). We used the OVA-induced experimental asthma model. Before the OVA challenge, 200 μg of 11R-pep3 or CsA was administered intranasally.
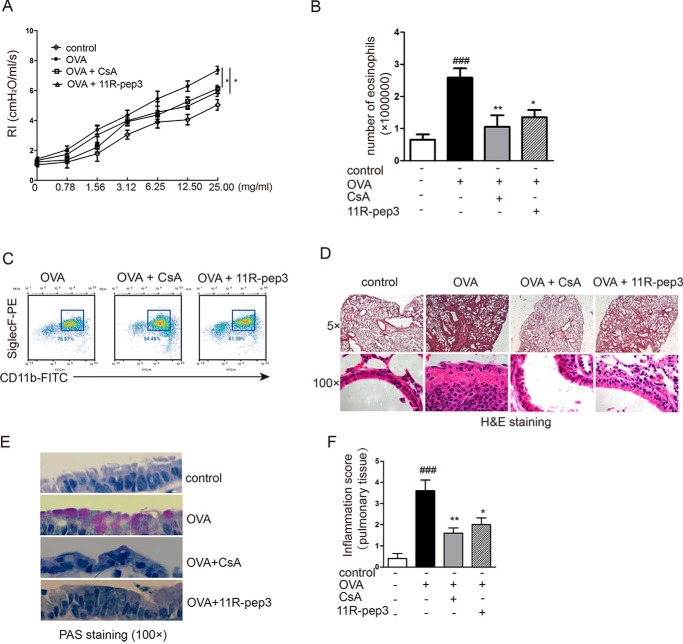
As expected, the OVA group of mice showed obvious airway hyperresponsiveness. Inhalation of methacholine (MCh) increased the values of RI in mice. Compared with animals with experimental allergic asthma, 11R-pep3–treated animals revealed a significant improvement in reactivity toward inhaled MCh (Fig. 5A). Furthermore, 11R-pep3 treatment inhibited allergic airway inflammation with significantly decreased numbers of eosinophils in bronchoalveolar lavage fluid (Fig. 5, B and C). The histological examination by H&E staining showed that the OVA challenge induced obvious infiltration of inflammatory cells around the trachea and interstitium, thickening of the tracheal wall, and exuberant secretion of tracheal mucosal epithelium and eosinophil cells (Fig. 5D). Remarkably, in the 11R-pep3–treated group, a small amount of inflammatory cells infiltrated, and no significant tracheal wall thickening or eosinophils were observed. Our PAS staining shows that OVA treatment also resulted in goblet cell metaplasia. The treatment of 11R-pep3 can significantly improve the histopathological phenomenon induced by OVA (Fig. 5E). Subsequently, inflammatory changes were graded by histopathological assessment. A relatively high grade was observed in the OVA challenge group. The grade was decreased significantly in the treatment of 11R-pep3 (Fig. 5F). Our results suggest that the intranasal administration of 11R-pep3 strongly suppressed allergic airway inflammation as an efficient immunosuppressant.
Figure 5.
Intranasal administration of 11R-pep3 inhibits airway inflammation and hyperresponsiveness. A, airway reactivity was assessed by MCh provocation testing. The mean ± S.E. is presented for each group (n = 6). *, p < 0.05 compared with the OVA group. B, total numbers of eosinophils in leukocytes of BAL fluids. Bar graph depicts the numbers of eosinophils in leukocytes of BAL fluids for each group. Data are presented as the mean ± S.E. for each group (n = 6). ###, p < 0.001 compared with the control group; *, p < 0.05; **, p < 0.01 compared with the OVA group. C, representative results of flow cytometry methods. We defined eosinophils in leukocytes as CD45+CD11b+SiglecF+. D, lung tissue was prepared for histological analysis, including morphology and the infiltration of inflammatory cells by H&E staining. Representative results of H&E staining in four groups are shown. Scale bars, 200 μm (top) and 10 μm (bottom). E, lung tissue was prepared for histological analysis. PAS-stained airway cross-sections of each group are shown. Scale bar, 10 μm. F, inflammatory changes were graded by histopathological assessment using a semiquantitative scale of 0–5. The results are presented as the mean ± S.E. for each group (n = 6).
Discussion
Protein-protein interactions play important regulatory roles in pathological and physiological processes and are closely related to human diseases. NFAT has been the focus of studies of CN-substrate interactions (25, 26). Recently, the PXIXIT-type motif and the LXVP-type motif in other CN substrates and targeting proteins have been the subject of in-depth study (18).
We previously reported that two LXVP-type peptides derived from RCAN1 and TFEB could bind to CN and inhibit its phosphatase activity (13–15). However, among the studied LXVP-type peptides in our experiments, the DQYLAVPQHPYQWAK peptide derived from NFATc1 shows a very strong binding ability with CN and potent inhibition of CN activity. The hydrophobic Leu and Val residues for targeting substrates to CN are very important, and the aromatic residue Tyr acts as a binding strength enhancer. Substitutions of the Tyr, Leu, and Val residues weakened the interactions between CN and LXVP-type motifs, such as YLAVP to HLAPP in the RCAN1-LXVP motif. We confirmed the results by experiments with GST pulldown and CN activity assays. We also found that the proline (YLAVP or HLAPP) residues in the LXVP motif were very important. Amino acids vary in their ability to form the various secondary structure elements. In natural proteins and peptides, proline is the most efficient helix breaking residue because it conflicts sterically with the helical conformation and, additionally, because its nitrogen cannot engage in hydrogen bonding. For example, Lys232 of A238L, which is a noncanonical residue in the LXVP motif because it has a “Lys” instead of the expected “Pro,” does not interact with CN and is thus not important for CN recognition by A238L. Therefore, the NFATc1-LXVP sequence was adopted in our design.
Mulero et al. (27) reported that expression of the peptide derived from the RCAN1-PXIXIT motif (195PGEKYELHAATDTTPSVVVHVCES218) in COS-7 cells could inhibit CN/NFATc2 signaling in vivo. The synthesized RCAN1-PXIXIT peptide binds to CN with high affinity. The Kd value between the carboxyfluorescein (FAM)-labeled RCAN1-PXIXIT peptide and CN was ∼1.25 μm in the FP experiment. We repeated the FP experiment and obtained similar results in our laboratory (data not shown). The FAM-labeled RCAN1-PXIXIT peptide is very suitable for binding experiments. Martinez-Martinez et al. (16) discussed the RCAN1-PXIXIT motif in detail. The full-length RCAN1-PXIXIT sequence includes two motifs, one containing the ELHA sequence (E-motif) and another the valine-rich sequence PSVVVH (V-motif). The integrity of the motif is necessary for targeting RCAN1 to CN because peptides of the individual E-motif or V-motif could not pull CN down in GST pulldown experiments. The E-motif contributes to the binding of the RCAN1-PXIXIT motif with CN, and the major deciding factor lies within the hydrophobic PSVVVH motif. Therefore, the conserved and important residues in the RCAN1-PXIXIT motif were fully incorporated into the sequence of pep3.
Abrams et al. (28) reported that the purified A238L fragment (residues 157–238, 82 aa) acted as a potent inhibitor of CN in vitro with an IC50 of ∼70 nm. The protein derived from A238L was tagged with His. The CN phosphatase activity was measured by release of [32P]phosphate from the 32P-labeled substrate peptide. It is well-known that the radioactive isotope method is more sensitive. Grigoriu et al. (18) used the ITC method to determine the thermodynamic parameters and dissociation (Kd) between CN and A238L (residues 200–239). The Kd is ∼4 ± 1 nm. In our MST experiment, the high affinity of pep3 with CN is ∼8 nm. The method of measuring CN activity that we used is a colorimetric experiment. The IC50 value for pep3 on purified CN or lysates of the Jurkat T cell line was 400 or 11 nm. Pep3 contains an NFATc1-LXVP substrate recognition motif and requires RCAN1-PXIXIT–mediated anchoring to CN. We have reported that the IC50 value for the NFATc1-LXVP peptide is 9 μm (15). Therefore, addition of the RCAN1-PXIXIT sequence in pep3 enhanced its inhibitory effect on CN. Therefore, the two sequences contribute to reducing CN-mediated dephosphorylation of the RII phosphopeptide.
Polypeptides are not able to easily enter cells. Many studies have focused on the delivery of bioactive peptides into cells. Based on the NFAT-PXIXIT motif, Aramburu et al. (7) developed a MAGPHPVIVITGPHEE peptide that selectively interferes with the CN/NFAT interaction without affecting CN phosphatase activity and may be useful as a therapeutic agent that is less toxic than CsA. Subsequently, Noguchi et al. (20) modified the MAGPHPVIVITGPHEE peptide at the N terminus with an 11-arginine transduction domain and a 3-glycine linker sequence. The cell-permeable 11R-PVIVIT peptide (RRRRRRRRRRR-GGG-MAGPHPVIVITGPHEE) could prevent the activation and proliferation of T cells both in vitro and in vivo. The 11R-PVIVIT peptide is effective at providing immunosuppression in a murine islet xenograft model without CN inhibition. However, 11R-PVIVIT affected cell viability in vitro when used at higher concentrations. Noguchi et al. (21) developed the RCAN1–11R peptide, which could selectively interfere with the CN/NFAT interaction without affecting cell viability even when it was used at a higher concentration. We modified pep3 with an 11-arginine transduction domain. FITC-11R-pep3 could enter the Jurkat T cell line and block the CN/NFAT signaling pathway.
The immunosuppressant CsA, used clinically to prevent transplant rejection, inhibits the CN activity toward all its substrates and targeting proteins, including NFAT. Although CsA has been crucial for the success of organ transplantation, its side effects are associated with progressive loss of renal function, hypertension, and neurotoxicity. The development of better, less toxic immunosuppressive agents is still a long and arduous task (29, 30).
Based on the amino acid sequences identified in the interaction between CN and its substrates or its targeting proteins, it is of particular interest that specific disruption of the CN/NFAT pathway has been obtained through the expression of peptides based on some of these sequences. The novel pep3 is a potent CN peptide inhibitor and can be used in topical remedy, such as intranasal administration in asthma animal models. Further studies could be helpful in furthering immunosuppressive therapy and identifying promising immunosuppressive agents.
Materials and methods
CN phosphatase assay
The RII phosphopeptide (DLDVPIPGRFDRRVpSVAAE) derived from the cAMP-dependent protein kinase regulatory subunit, Type II, was synthesized by Scilight-Peptide Inc. (Beijing, China). Other peptides used in the experiments were also synthesized by Scilight-Peptide Inc. and are shown in Table 1. Cyclosporine A (CsA) and okadaic acid were purchased from Sigma-Aldrich. CNA, CNB, and CaM were expressed in Escherichia coli and purified in our laboratory (31, 32). A colorimetric assay was used to determine the inhibitory effects of pep3 on purified CN activity with RII phosphopeptide (33) as a substrate and using the improved method based on the Calcineurin Colorimetric Drug Discovery Kit (AK-804, Enzo Life Sciences). We used another improved method to determine the effects of pep3 on cell lysates based on the Calcineurin Cellular Assay Kit (AK-816, Enzo Life Sciences).
Table 1.
Abbreviations and sequences of the peptides used
| Abbreviation | Sequence |
|---|---|
| Pep1 | PGEKYELHAATDTTPSVVVH |
| Pep2 | YLAVPQHPYQWAK |
| A238L-linker | GCEDNVYEKLPEQNSN |
| Pep3 | PGEKYELHAATDTTPSVVVHGCEDNVYEKLPEQNSNYLAVPQHPYQWAK |
| FAM-labeled pep3 | 5 (6)-Carboxyfluorescein-labeled pep3 |
| 11R-pep3 | RRRRRRRRRRR-GGG-pep3 |
| FITC-pep3 | Fluorescein isothiocyanate-labeled pep3 |
| FITC-11R-pep3 | Fluorescein isothiocyanate-labeled 11R-pep3 |
MST
FAM-labeled pep3 was synthesized by Scilight-Peptide Co. The MST method has been described in detail elsewhere (19). The Kd values for the binding of FAM-labeled pep3 to CN were measured using a Monolith NT.115 from NanoTemper Technologies. FAM-labeled pep3 (10 μl) was added to serial dilutions of CN (CNA:CNB:CaM, 1:1:2) in PBS (10 μl). The samples were incubated at 25 °C for 20 min and loaded into silica capillaries (Polymicro Technologies). Measurements were performed at 20 °C with 20% LED power and 20% IR-laser power. The data were analyzed with NanoTemper Analysis software, version 1.2.101.
Cell culture and cell transfection
HeLa cells were cultured in Dulbecco's modified Eagle's medium (supplemented with 4.5 g/liter of d-glucose and 5 μm l-glutamine, Gibco) with 10% fetal calf serum. Human embryonic kidney 293 (HEK293) cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Human T lymphocyte (Jurkat, RIKEN) cells were cultivated in RPMI 1640 medium with 10% fetal calf serum. The cells were washed with PBS buffer (Gibco, pH 7.2) and digested with trypsin and 0.25% EDTA. All the cells were cultivated overnight (12 h) before any treatment and harvesting.
Plasmids encoding the pep3 peptide were fused to FLAG through direct cloning of the overhang–double-stranded annealed oligonucleotides into the suitable plasmid vector. The cells were transfected with plasmid DNA (∼1 μg/106) via utilization of the Lipofectamine 2000 Transfection Reagent (Invitrogen). Ionomycin (1 μm) was used alone to stimulate the Ca2+/CN signaling pathway. Ionomycin is commonly used in conjunction with phorbol myristate acetate (PMA) to activate T cells. The combination of PMA (50 ng/ml) and ionomycin activates the transcription factors NF-κB and NFAT, leading to the production of cytokines, such as IL-2. CsA (1 μm) was added as a positive inhibitor of stimulation.
GST pulldown assay, co-immunoprecipitation, and Western blotting
Plasmids encoding peptides fused to GST were obtained by cloning overhang–double-stranded annealed oligonucleotides into EcoRI + XhoI-digested pGEX-4T-1 plasmid. The GST fusion proteins were expressed in E. coli and proteins were quantified by the Bradford procedure. Unless otherwise specified, all pulldown experiments were performed in 50 mm Tris-HCl, pH 7.5, 1.5 mm CaCl2, 2 μm CaM, 1.0 mm DTT, and 0.5 mm MnCl2. GSH-agarose beads coated with GST or GST peptide were incubated with brain lysates for 1 h at 4 °C with end-over shaking. CN was detected by immunoblotting with anti-CNA antibody, designated pan-calcineurin A antibody, at a 1:1000 dilution, or anti-GST antibody.
Plasmids encoding the pep3 peptide were fused to FLAG through direct cloning of the overhang–double-stranded annealed oligonucleotides into the pIRES2-EGFP vector. The plasmid was transiently transfected into cells and expressed EGFP and pep3-FLAG separately. This plasmid was kindly supplied by Prof. Hailong Wang of Capital Normal University of China. HEK293 cells transfected with the pep3-FLAG plasmid were lysed in lysis buffer (pH 7.4, 50 mm Tris-HCl, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, 2 mm DTT, 0.1% β-mercaptoethanol, and 1× protease inhibitor). For immunoprecipitation, 50 μl of protein A/G plus agarose beads (GE Healthcare) was incubated with 1:200 anti-DDDDK (anti-FLAG) tag mouse mAb (Abbkine, Wuhan, China)/anti-mouse IgG (Abbkine) at 4 °C overnight. After antibody additions were made to the cell lysates, the tubes were rotated at 4 °C for 1 h. After centrifugation, the beads were washed with PBS five times and boiled in 1× SDS Protein Loading Buffer for Western blotting. The protein mixture was loaded into the SDS-PAGE, fractionated at 80 volts for 30 min, and then at 120 volts for 1.5 h and transferred onto polyvinylidene difluoride membranes. Expressed CNA and pep3 were detected by immunoblotting with anti-CNA antibody at a 1:1000 dilution and anti-FLAG antibody at a 1:5000 dilution. The images were generated with a Tanon 5200 Imager.
Immunofluorescence of NFAT
HeLa cells were cultivated (37 °C, 0.05% CO2) on coverslips clinging to the bottom of 12-well-plates over 12 h and cotransfected with pep3-FLAG (p3×FLAG-CMV-10 vector) and HA-NFATc3(3–407)–GFP plasmids. After cultivation for ∼18–24 h, the cells were washed and fixed with pre-prepared 4% paraformaldehyde (Applygen Technologies Inc. Beijing, China) at room temperature for 10 min and then permeated with 0.3% Triton X-100 in PBS for 10 min. Next, 5% BSA was diluted in PBS and incubated with cells at room temperature for 1 h. The anti-GFP mouse mAb (Abbkine, anti-GFP tag) was diluted in 3% BSA and probed with the fusionally expressed NFATc3-GFP at 4 °C overnight. The Alexa Fluor® 488-conjugated anti-mouse IgG (H+L) was applied at room temperature for 1 h. The cell nucleus was stained with 5 mg/ml of Hoechst 33258 at room temperature for 10 min and incubated with an antifluorescence quencher. The coverslips were observed via a ZEISS LSM700 microscope system. The FITC image in this study was obtained via the same procedure.
Flow cytometry
The incubated Jurkat cells were resuspended in 500 μl of PBS and filtered using a flow cytometer tube to obtain a single cell suspension. Then, the cells were collected, and the cells with fluorescein (FITC)-conjugated peptide were counted by the ACEA Flow Cytometry System (NovoCyte).
ELISA
The secreted IL-2 protein was detected by the IL-2 ELISA kit (GeneTex Inc., San Antonio, TX). After centrifugation, 100 μl of the cell medium and standard IL-2 were incubated in an IL-2 antibody-conjugated ELISA-specific plate at 37 °C for 90 min. Then, 100 μl of biotin antibody, enzyme-conjugation, and chromogenic substrate (tetramethylbenzidine) was diluted and incubated at 37 °C, separately for 60, 30, and 15 min. The stop solution was added, and the absorbance (OD450) of each well was read by the Fluorescence Microplate System (Omega).
Complex structure modeling and simulation
The pep3-CN complex structure was built based on the published crystal structure of the A238L-CN complex obtained from the RCSB Protein Data Bank (PDB 4F0Z) (18). Pep3 was built based on the structure of A238L by mutating the CN-binding motif (residues 207 to 211) to “SVVVH” and the other motif (residues 228 to 234) to “YLAVPQH.” Two additional residues were included with the neutralized terminals at the N and C terminals of the original structures.
A molecular dynamics simulation was performed to test the structural stability of the CN-pep3 complex. For simulation, the titration states of the ionizable residues (aspartate, glutamate, lysine, arginine, histidine, and tyrosine) were assigned based on the pKa predictions performed with PROPKA (14, 15). Almost all the residues were in their default titration states, but only Asp121 had to be changed to a protonation state. Using the SOLVATE program, the system was dissolved in a box of water, with at least 10 Å between the protein and the border of the box. Then, 150 mm NaCl was used to neutralize the net charge of the system with the AUTOIONIZE plugin of Visual Molecular Dynamics.
The simulation was performed using NAMD2, with the CHARMM36 force field for proteins and ions, and the TIP3P model for explicit water, using periodic boundary conditions to perform all of the simulations with a time step of 2 fs. The SHAKE algorithm was applied to fix the bond distances involving hydrogen atoms in the simulations. After an initial 10,000 steps of energy minimization with all the Cα atoms fixed, the system was equilibrated in an NVT ensemble (constant number, volume, and temperature) at 310 K for 500 ps. During the process, all the protein Cα atoms were restricted (k = 1 kcal/mol/Å2) to allow the mitigation of the side chains and water. All the following equilibrium simulations were conducted in an NVT ensemble for 100 ns, restricting the active site to its configuration in the crystal structure. The Langevin dynamics were used to hold a constant temperature with a damping coefficient γ of 0.5 ps−1. We employed a cutoff distance of 12 Å to calculate the short-range, nonbonded interactions, and the long-range electrostatic forces were described by the particle mesh Ewald (PME) method.
OVA-induced experimental asthma model
Female BALB/c mice at 6–8 weeks of age were maintained under pathogen-free conditions at Beijing Normal University. All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines. All experimental procedures were approved by the Animal Ethics Committee of College of Life Sciences, Beijing Normal University.
On days 7 and 14 of the experiment, the mice were sensitized by an intraperitoneal injection of 20 μg of OVA (Sigma-Aldrich) absorbed in 2 mg of aluminum hydroxide (Thermo) in normal saline. The mice were subsequently challenged four times by intranasal administration of 1% OVA in saline every day beginning on day 21. On each day of OVA challenge, the mice were given 200 μg of pep3 (20 μl) or positive control CsA (2.8 mg/ml, 20 μl) 2 h before the challenge.
Airway reactivity was assessed by MCh provocation testing (34), whereas respiratory mechanics were recorded using invasive lung function assessment (Buxco® FinePointeTM Resistance and Compliance units; Data Sciences International, New Brighton, MN). Mice were anesthetized with sodium pentobarbital (30 mg/kg; Sinopharm Chemical Reagent Co., Ltd.), tracheotomized with a cannula, and mechanically ventilated with 140 breaths/min. Mice were allowed to stabilize for 3 min before measurements started. MCh (acetyl-β-methylcholine chloride; Sigma-Aldrich) provocation testing started with PBS, followed by MCh aerosols with increasing concentrations (0, 0.78, 1.56, 3.12, 6.25, 12.50, and 25.00 mg/ml). Each aerosol provocation lasted 30 s, followed by a 180-s incubation time. Airflow and trans-pulmonary pressure in response to MCh inhalation were recorded and analyzed with FinePointe Review software (version 2.3.1.0; Data Sciences International), which calculated airway resistance (cm H2O/ml/s). The animals were sacrificed on day 25. The tissues were subjected to H&E staining (hematoxylin and eosin) and PAS staining (periodic acid-Schiff) for morphometric analysis. Histopathological assessment was performed in a blinded manner on randomized sections. Inflammatory changes were graded using a semiquantitative scale of 0–5 for perivascular eosinophilia and bronchiolar eosinophilia (35).
We repeated the experiment and next quantified the infiltrating leukocytes in the bronchoalveolar lavage fluid (BALF) by flow cytometry methods. The animals were sacrificed on day 25, and the lungs were repeatedly washed with PBS through a tracheal tube. Cells were stained with fluorescently labeled antibodies against CD45-PE/Cy7, CD11b-FITC, and SiglecF-PE (BD Biosciences) (36). Analysis was performed using a BD Flow Cytometer (FACSVantage SE). We defined eosinophils in leukocytes as CD45+CD11b+SiglecF+.
Statistical analysis
To quantitatively assess marker protein distributions, we used identical settings and exported them to ImageJ (NIH) to take images for imaging analyses. The data were analyzed with GraphPad Prism 5 using the mean ± S.E. Statistical significance was determined by comparing the means of different groups and conditions using one-way analysis of variance (ANOVA), followed by Tukey's test; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Author contributions
A. J. data curation; W. L., N. Z., Y. Y., and L. T. methodology; Y. Y., Q. W., and Z. L. funding acquisition; L. T. and G. L. writing-review and editing; L. W. first author; N. C. animal experiment; P. W. second author; J. Li complex structure modeling and simulation; W. L. for Western blotting; N. Z. for animal experiments; G. L. guide part of immunology experiment; Z. L. contribute clinical experience; J. Luo corresponding author.
Acknowledgments
We acknowledge Prof. Xiaoyan Gao and Dr. Yanling Wang, Beijing University of Traditional Chinese Medicine, for help with the use of Buxco® instruments, and Prof. Hailong Wang, Capital Normal University of China, for kindly providing the plasmid.
This work was supported by National Nature Science Foundation of China Project Grants 81373389 (to J. L.) and 31700695 (to Y. Y.) and the research fund for China Association Against Epilepsy-UCB program 2017010. The authors declare that they have no conflicts of interest with the contents of this article.
- CN
- calcineurin
- NFAT
- nuclear factor of activated T cells
- RCAN1
- regulator of calcineurin 1
- FP
- fluorescence polarization
- GST
- glutathione S-transferase
- aa
- amino acid(s)
- MST
- microscale thermophoresis
- FITC
- fluorescein isothiocyanate
- OVA
- ovalbumin
- CsA
- cyclosporine A
- CaM
- calmodulin
- HEK
- human embryonic kidney
- MCh
- methacholine
- PAS
- periodic acid-Schiff
- PDB
- Protein Data Bank
- PMA
- phorbol myristate acetate
- IL
- interleukin.
References
- 1. Rusnak F., and Mertz P. (2000) Calcineurin: form and function. Physiol. Rev. 80, 1483–1521 10.1152/physrev.2000.80.4.1483 [DOI] [PubMed] [Google Scholar]
- 2. Rao A. (2009) Signaling to gene expression: calcium, calcineurin and NFAT. Nat. Immunol. 10, 3–5 10.1038/ni0109-3 [DOI] [PubMed] [Google Scholar]
- 3. Mehta S., Li H., Hogan P. G., and Cunningham K. W. (2009) Domain architecture of the regulators of calcineurin (RCANs) and identification of a divergent RCAN in yeast. Mol. Cell Biol. 29, 2777–2793 10.1128/MCB.01197-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Medina D. L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., Settembre C., Wang W., Gao Q., Xu H., Sandri M., et al. (2015) Lysosomal calcium signaling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288–299 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slupe A. M., Merrill R. A., Flippo K. H., Lobas M. A., Houtman J. C., and Strack S. (2013) A calcineurin docking motif (LXVP) in dynamin-related protein 1 contributes to mitochondrial fragmentation and ischemic neuronal injury. J. Biol. Chem. 288, 12353–12365 10.1074/jbc.M113.459677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aramburu J., Garcia-Cózar F., Raghavan A., Okamura H., Rao A., and Hogan P. G. (1998) Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell 1, 627–637 10.1016/S1097-2765(00)80063-5 [DOI] [PubMed] [Google Scholar]
- 7. Aramburu J., Yaffe M. B., López-Rodríguez C., Cantley L. C., Hogan P. G., and Rao A. (1999) Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285, 2129–2133 10.1126/science.285.5436.2129 [DOI] [PubMed] [Google Scholar]
- 8. Garcia-Cozar F. J., Okamura H., Aramburu J. F., Shaw K. T., Pelletier L., Showalter R., Villafranca E., and Rao A. (1998) Two-site interaction of nuclear factor of activated T cells with activated calcineurin. J. Biol. Chem. 273, 23877–23883 10.1074/jbc.273.37.23877 [DOI] [PubMed] [Google Scholar]
- 9. Liu J., Arai K., and Arai N. (2001) Inhibition of NFATx activation by an oligopeptide: disrupting the interaction of NFATx with calcineurin. J. Immunol. 167, 2677–2687 10.4049/jimmunol.167.5.2677 [DOI] [PubMed] [Google Scholar]
- 10. Martínez-Martínez S., Rodríguez A., López-Maderuelo M. D., Ortega-Pérez I., Vázquez J., and Redondo J. M. (2006) Blockade of NFAT activation by the second calcineurin binding site. J. Biol. Chem. 281, 6227–6235 10.1074/jbc.M513885200 [DOI] [PubMed] [Google Scholar]
- 11. Rodríguez A., Roy J., Martinez-Martínez S., López-Maderuelo D., Niño-Moreno P., Ortí L., Pantoja-Uceda D., Pineda-Lucena A., Cyert M. S., and Redondo J. M. (2009) A conserved docking surface on calcineurin mediates interaction with substrates and immunosuppressants. Mol. Cell 33, 616–626 10.1016/j.molcel.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aubareda A., Mulero M. C., and Pérez-Riba M. (2006) Functional characterization of the calcipressin 1 motif that suppresses calcineurin-mediated NFAT-dependent cytokine gene expression in human T cells. Cell Signal. 18, 1430–1438 10.1016/j.cellsig.2005.11.006 [DOI] [PubMed] [Google Scholar]
- 13. Ma Y., Jiang G., Wang Q., Sun Y., Zhao Y., Tong L., and Luo J. (2015) Enzymatic and thermodynamic analysis of calcineurin inhibition by RCAN1. Int. J. Biol. Macromol. 72, 254–260 10.1016/j.ijbiomac.2014.08.042 [DOI] [PubMed] [Google Scholar]
- 14. Zhao Y., Zhang J., Shi X., Li J., Wang R., Song R., Wei Q., Cai H., and Luo J. (2016) Quercetin targets the interaction of calcineurin with LxVP-type motifs in immunosuppression. Biochimie (Paris) 127, 50–58 10.1016/j.biochi.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song R., Li J., Zhang J., Wang L., Tong L., Wang P., Yang H., Wei Q., Cai H., and Luo J. (2017) Peptides derived from transcription factor EB bind to calcineurin at a similar region as the NFAT-type motif. Biochimie (Paris) 142, 158–167 10.1016/j.biochi.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martínez-Martínez S., Genescà L., Rodríguez A., Raya A., Salichs E., Were F., López-Maderuelo M. D., Redondo J. M., and de la Luna S. (2009) The RCAN carboxyl end mediates calcineurin docking-dependent inhibition via a site that dictates binding to substrates and regulators. Proc. Natl. Acad. Sci. U.S.A. 106, 6117–6122 10.1073/pnas.0812544106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carme Mulero M., Orzáez M., Messeguer J., Messeguer A., Pérez-Payá E., and Pérez-Riba M. (2010) A fluorescent polarization-based assay for the identification of disruptors of the RCAN1-calcineurin protein complex. Anal. Biochem. 398, 99–103 10.1016/j.ab.2009.10.045 [DOI] [PubMed] [Google Scholar]
- 18. Grigoriu S., Bond R., Cossio P., Chen J. A., Ly N., Hummer G., Page R., Cyert M. S., and Peti W. (2013) The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin. Plos Biol. 11, e1001492 10.1371/journal.pbio.1001492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seidel S. A., Dijkman P. M., Lea W. A., van den Bogaart G., Jerabek-Willemsen M., Lazic A., Joseph J. S., Srinivasan P., Baaske P., Simeonov A., Katritch I., Melo F. A., Ladbury J. E., Schreiber G., Watts A., Braun D., and Duhr S. (2013) Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 10.1016/j.ymeth.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noguchi H., Matsushita M., Okitsu T., Moriwaki A., Tomizawa K., Kang S., Li S. T., Kobayashi N., Matsumoto S., Tanaka K., Tanaka N., and Matsui H. (2004) A new cell-permeable peptide allows successful allogeneic islet transplantation in mice. Nat. Med. 10, 305–309 10.1038/nm994 [DOI] [PubMed] [Google Scholar]
- 21. Noguchi H., Sugimoto K., Miyagi-Shiohira C., Nakashima Y., Kobayashi N., Saitoh I., Watanabe M., and Noguchi Y. (2017) RCAN-11R peptide provides immunosuppression for fully mismatched islet allografts in mice. Sci. Rep. 7, 3043 10.1038/s41598-017-02934-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi J. M., Sohn J. H., Park T. Y., Park J. W., and Lee S. K. (2012) Cell permeable NFAT inhibitory peptide Sim-2-VIVIT inhibits T-cell activation and alleviates allergic inflammation and hyper-responsiveness. Immunol. Lett. 143, 170–176 10.1016/j.imlet.2012.01.016 [DOI] [PubMed] [Google Scholar]
- 23. Roh S. S., Kim S. H., Lee Y. C., and Seo Y. B. (2008) Effects of radix adenophorae and cyclosporine A on an OVA-induced murine model of asthma by suppressing to T cells activity, eosinophilia, and bronchial hyperresponsiveness. Mediators Inflamm. 10.1155/2008/781425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fukuda T., Asakawa J., Motojima S., and Makino S. (1995) Cyclosporine A reduces T lymphocyte activity and improves airway hyperresponsiveness in corticosteroid-dependent chronic severe asthma. Ann. Allergy. Asthma. Immunol. 75, 65–72 [PubMed] [Google Scholar]
- 25. Ma B., Yu J., Xie C., Sun L., Lin S., Ding J., Luo J., and Cai H. (2015) Toll-Like receptors promote mitochondrial translocation of nuclear transcription factor of activated T-cells in prolonged microglial activation. J. Neurosci. 35, 10799–10814 10.1523/JNEUROSCI.2455-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo J., Sun L., Lin X., Liu G., Yu J., Parisiadou L., Xie C., Ding J., and Cai H. (2014) A calcineurin- and NFAT-dependent pathway is involved in α-synuclein-induced degeneration of midbrain dopaminergic neurons. Hum. Mol. Genet. 23, 6567–6574 10.1093/hmg/ddu377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mulero M. C., Aubareda A., Orzáez M., Messeguer J., Serrano-Candelas E., Martínez-Hoyer S., Messeguer A., Pérez-Payá E., and Pérez-Riba M. (2009) Inhibiting the calcineurin-NFAT (nuclear factor of activated T cells) signaling pathway with a regulator of calcineurin-derived peptide without affecting general calcineurin phosphatase activity. J. Biol. Chem. 284, 9394–9401 10.1074/jbc.M805889200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abrams C. C., Chapman D. A., Silk R., Liverani E., and Dixon L. K. (2008) Domains involved in calcineurin phosphatase inhibition and nuclear localisation in the African swine fever virus A238L protein. Virology 374, 477–486 10.1016/j.virol.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 29. Liu J., Farmer J. D. Jr, Lane W. S., Friedman J., Weissman I., and Schreiber S. L. (1991) Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66, 807–815 10.1016/0092-8674(91)90124-H [DOI] [PubMed] [Google Scholar]
- 30. Luo J., Ma J., Yu D. Y., Bu F., Zhang W., Tu L. H., and Wei Q. (2008) Infusion of FK506, a specific inhibitor of calcineurin, induces potent Tau hyperphosphorylation in mouse brain. Brain Res. Bull. 76, 464–468 10.1016/j.brainresbull.2007.12.005 [DOI] [PubMed] [Google Scholar]
- 31. Wei Q., Pervaiz S., Jackson A. E., Puett D., Carraway K. L. 3rd, Lee E. Y., and Brew K. (1988) Effects of interaction with calcineurin on the reactivities of calmodulin lysines. J. Biol. Chem. 263, 19541–19544 [PubMed] [Google Scholar]
- 32. Wei Q., and Lee E. Y. (1997) Expression and reconstitution of calcineurin A and B subunits. Biochem. Mol. Biol. Int. 41, 169–177 [DOI] [PubMed] [Google Scholar]
- 33. Blumenthal D., Takio K., Hansen R. S., and Krebs E. G. (1986) Dephosphorylation of cAMP-dependent protein kinase regulatory subunit (type II) by calmodulin-dependent protein phosphatase. J. Biol. Chem. 262, 8140–8145 [PubMed] [Google Scholar]
- 34. Lunding L. P., Webering S., Vock C., Behrends J., Wagner C., Hölscher C., Fehrenbach H., and Wegmann M. (2015) Poly(inosinic-cytidylic) acid-triggered exacerbation of experimental asthma depends on IL-17A produced by NK cells. J. Immunol. 194, 5615–5625 10.4049/jimmunol.1402529 [DOI] [PubMed] [Google Scholar]
- 35. Underwood S., Foster M., Raeburn D., Bottoms S., and Karlsson J. A. (1995) Time-course of antigen-induced airway inflammation in the guinea-pig and its relationship to airway hyperresponsiveness. Eur. Respir. J. 8, 2104–2113 10.1183/109031936.95.08122104 [DOI] [PubMed] [Google Scholar]
- 36. Li C., Bi Y., Li Y., Yang H., Yu Q., Wang J., Wang Y., Su H., Jia A., Hu Y., Han L., Zhang J., Li S., Tao W., and Liu G. (2017) Dendritic cell MST1 inhibits Th17 differentiation. Nat. Commun. 8, 14275 10.1038/ncomms14275 [DOI] [PMC free article] [PubMed] [Google Scholar]