Abstract
Aberrant Ras signaling drives 30% of cancers, and inhibition of the Rho family small GTPase signaling has been shown to combat Ras-driven cancers. Here, we present the discovery of a 16-mer cyclic peptide that binds to Cdc42 with nanomolar affinity. Affinity maturation of this sequence has produced a panel of derived candidates with increased affinity and modulated specificity for other closely-related small GTPases. The structure of the tightest binding peptide was solved by NMR, and its binding site on Cdc42 was determined. Addition of a cell-penetrating sequence allowed the peptides to access the cell interior and engage with their target(s), modulating signaling pathways. In Ras-driven cancer cell models, the peptides have an inhibitory effect on proliferation and show suppression of both invasion and motility. As such, they represent promising candidates for Rho-family small GTPase inhibitors and therapeutics targeting Ras-driven cancers. Our data add to the growing literature demonstrating that peptides are establishing their place in the biologics arm of drug discovery.
Keywords: cancer, CDC42, peptide conformation, nuclear magnetic resonance (NMR), GTPase Kras (KRAS), drug discovery, cyclic peptide, cell proliferation, cell signaling, cell migration, biologics, cancer therapeutics, drug discovery
Introduction
Aberrant signaling from the small G-protein Ras has been shown to drive approximately one-third of all human cancers making it a compelling therapeutic target (1). Targeting Ras isoforms directly has proven difficult historically, although some progress has been made recently (2–4). Cdc42, together with Rac1, RhoA, and other members of the Rho family small GTPases, is a key regulator of the actin cytoskeleton and therefore cell architecture and motility (5). Cdc42 is often found to signal downstream of the master regulator, Ras, and induces the formation of filopodia by initiating actin remodeling, which is key to cell motility. Deregulation of Cdc42 therefore results in the progression of several disease states, including tumor metastasis (6). Genetic deletion of Cdc42 in Ha-Ras G12V-transformed fibroblasts results in a significant block in cell cycle progression and therefore in cell proliferation, demonstrating an essential role of Cdc42 in Ras-induced transformation (7). Correspondingly, overexpression of Cdc42 is implicated in several human cancers where it correlates with poor prognosis (8–10). Although mutations in Cdc42 are rarely found in human cancers, oncogenic mutations have been extensively characterized in its regulators, e.g. the guanine-exchange factors Dbl (11) and Asef2 (12) and the GTPase-activating protein DLC1 (13). Cdc42 participates in both physiological and tumorigenic processes by interacting with its effector proteins. One such family of effectors, the CRIB7 proteins, includes activated Cdc42-associated kinase (ACK), the p21-activated kinases (PAKs), and Wiskott-Aldrich syndrome proteins (WASPs) (14).
Inhibition of Cdc42 has been demonstrated to reverse the oncogenic properties of v-Ha-Ras–transformed cells in experiments employing the 42-amino acid G-protein–binding region (GBD) of ACK, a previously mentioned Cdc42 effector (15). This peptide binds to Cdc42 at a binding surface that prevents it from interacting with its downstream effector proteins, all of which employ overlapping binding surfaces on Cdc42 (16). This inhibits Cdc42-driven signaling pathways, which must be necessary for transformation by Ras in this model.
This observation, together with our structural and biophysical analyses of the Cdc42–ACK GBD complex (17, 18), prompted us to undertake a peptide discovery campaign, using CIS display technology, to identify a Cdc42-binding peptide that would similarly inhibit effector interaction and Ras-driven oncogenesis but would possess properties more compatible with future use as a lead therapeutic. CIS display is a biological display technology that links the DNA-coding sequence of library members directly to the encoded peptide in a cell-free system by employing the binding properties of the R1 RepA protein to the origin of replication, i.e. the cis-binding activity of the protein to its coding DNA (19). The technology allows the generation and interrogation of the very large library sizes (routinely 1013 library members) associated with cell-free display and offers greater stability than many orthogonal methods (for example, ribosome or RNA display) by employing DNA and the robust CIS RepA system.
Despite the knowledge for over 30 years that targeting Ras and other small G proteins would have enormous therapeutic potential, multiple issues have been encountered in attempts to attack them (20). Small G proteins all contain a conserved G domain comprising five α helices surrounding a six-stranded β sheet (21) This architecture mediates binding to Mg2+ and either GTP or GDP, with GTP binding associated with activation. The picomolar affinity for guanine nucleotides together with the ubiquitous nature of guanine nucleotide usage and cellular abundance have prevented the development of small-molecule therapeutics based on inhibition of nucleotide binding (22). Likewise, the relatively smooth surface of the small G proteins has also confounded searches for small-molecule therapeutics due to the lack of other binding pockets, although some progress has been made in this regard in recent years (23).
Peptides and their orthologues can bind surfaces of proteins inhospitable to small molecules and as such can inhibit protein–protein interactions successfully, with the added property of exquisite specificity. Thus, peptides or peptidomimetics provide an opportunity to mimic the effect of ACK GBD overexpression, which was effective in inhibiting transformation in oncogenic NIH3T3 cells (15) by allowing targeting of the functional face of the Cdc42. Peptide therapeutic candidates have traditionally suffered from poor delivery and stability; however, progress on both fronts continues to be made, using cell-penetrating peptides (CPPs) (24) and delivery vehicles (25) and introducing unnatural modifications into peptide structures (26–29), allowing peptide biologics to join the current repertoire of therapeutics (30).
Here, we report the identification and maturation of a lead 16-mer cyclic peptide using CIS display (19) to produce a panel of cyclic peptides that bind Cdc42 with low nanomolar affinity. Binding has been characterized by biophysical methods, indicating the docking site on Cdc42 and demonstrating varying selectivity between peptides and related small GTPases. The peptides are shown to have promising phenotypic effects in cells when linked to a nona-arginine motif facilitating entry, reducing proliferation, and particularly inhibiting motility and invasion. These peptides provide a promising lead platform for development of a therapeutic peptidomimetic targeting Rho-family signaling and therefore preventing oncogenesis in Ras-driven cancers.
Results
CIS display biopanning
Selections were performed following the work plan detailed in Ref. 19. Ten naïve peptide libraries, encoding peptides 10–25 amino acids long, were separated into three pools, short linear (10- and 12-mer), long linear (16-, 20-, and 25-mer), and cyclic (14-, 15-, 16-, 17-, and 18-mer), and were selected against Cdc42·GMPPNP. The five libraries in the cyclic pool contained two constant cysteines (at positions N + 4 and C − 4) designed to allow production of disulfide-linked cyclic peptides under nonreducing conditions. Two methods of elution were used in parallel for each library pool: heating to 75 °C or competition with the ACK GBD peptide, generating six individual selections. Competition with the ACK GBD was designed to select for peptides that bind to an interface on Cdc42 that overlaps with that of ACK. After four rounds of biopanning, DNA recovery significantly increased (data not shown), indicating binding peptide enrichment. Next-generation sequence analysis of the peptide-coding DNA indicated three highly-enriched peptides (each of which represented over 20% of recovered sequences), two from the short linear and one from the cyclic library pools, as shown in Fig. 1A. The three selection pools showed different patterns of enrichment. In the long linear libraries, individual sequences are hydrophobic, suggestive of less specific binders, with no dominant motif. In contrast, the short linear pool produced a highly-dominant motif (HISWPXN) in both heat and competitive elution experiments, whereas in the cyclic libraries one sequence (C1) was strongly enriched, especially under competitive elution conditions where it comprised ∼70% of the recovered sequences. Several of the more frequently-occurring alternative sequences selected from the short linear and cyclic libraries were single-site variants stemming from the dominant sequence. No strong homology was shared across the cyclic and short linear consensus sequences.
Figure 1.
A, bar charts showing the relative enrichment of peptides in each library pool and the 10 most frequently selected sequences under each set of conditions. Sequences taken forward for further evaluation are highlighted in red. B, ELISA screening of phage presenting the three most enriched sequences. The relative absorbances from ELISA for binding against a panel of small GTPase targets are as indicated, and a BSA control is plotted as a modified box plot. n = 3–4. C, displacement of [3H]GTP·Cdc42 from GST–WT ACK GBD by C1 binding to Cdc42. Increasing concentrations of C1 or C1C4S, C13S were titrated into a fixed concentrations of [3H]GTP·Cdc42 and GST–WT ACK GBD in the presence or absence of 50 mm DTT, in competition SPAs. The Kd value for Cdc42 binding to GST–WT ACK GBD was fixed to the value (30 nm) obtained for this Kd in direct SPAs. The data were fitted to an isotherm describing a pure competition model (58) to give an apparent Kd (Ki) value for C1. The data and curve fits are displayed as a percentage of the maximal SPA signal in each assay.
Peptide validation
Phage display of the three most highly-selected sequences was used to validate peptide specificity for Cdc42 in an ELISA, as well as testing the nucleotide dependence of the interaction. Results for the three dominant sequences, C1, SL1, and SL2, are shown in Fig. 1B. SL2 bound strongly but nonspecifically to all the proteins tested, including the negative control, BSA. SL1 gave weaker signals, which were again nonspecific. C1, however, generated strong signals with Cdc42·GMPPNP and low signals with Cdc42·GDP, RalB·GMPPNP, and BSA. Some binding between C1 and Rac1·GMPPNP was also observed. These data suggested that, among these targets, C1 was specific for Cdc42·GMPPNP.
Quantitative peptide-binding affinities were next determined by competition with the Cdc42–ACK GBD complex in scintillation proximity assays (SPAs). Peptide solubility at pH 7.5 (<200 μm) tended to limit the concentration in the assays, such that the SL1 and SL2 peptides did not produce full competition data and could not be fitted (data not shown). C1, however, inhibited the Cdc42·GMPPNP–ACK GBD interaction with a Kd of 350 nm (Fig. 1C). The competition observed did not go to completion, i.e. the signal due to Cdc42–ACK GBD did not reach zero, suggesting that the binding site for C1 on Cdc42 partially overlaps with the ACK–Cdc42 interaction surface. Interestingly, the affinity of C1 for Cdc42·GMPPNP was also shown to be dependent on the disulfide bond in the cyclic peptide, because addition of 50 mm DTT or mutation of the cysteine residues to serine lowered the binding affinity significantly (Fig. 1C).
C1 validation in cells
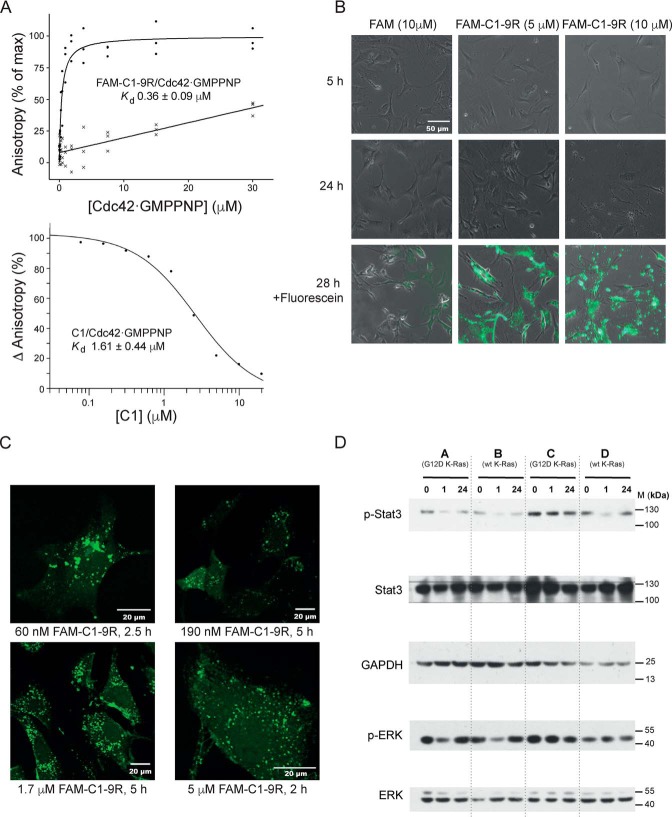
The C1 sequence was re-synthesized with the addition of a C-terminal nona-arginine motif (9R) to facilitate cell penetration and an N-terminally–linked carboxyfluorescein (FAM) group for visualization. The binding of this modified peptide to Cdc42 was confirmed by fluorescence polarization (FP), and the results are shown in Fig. 2A. Direct binding of Cdc42·GMPPNP to the FAM-C1–9R peptide gave a dissociation constant of 0.36 ± 0.09 μm (Fig. 2A, upper panel). The binding of FAM-9R was so low it could not be accurately determined. The Kd value of untagged C1 for Cdc42·GMPPNP in competition FP was determined to be 1.61 ± 0.44 μm (Fig. 2A, lower panel), indicating that the addition of 9R and FAM did not adversely affect Cdc42 binding in vitro. The very-low affinity of FAM-9R for Cdc42 suggests that the slight difference in affinity between FAM-C1–9R and C1 may just reflect technical differences in the experimental methodologies.
Figure 2.
A, affinity of C1 and FAM-C1–9R for Cdc42. The top panel shows anisotropy data for direct binding of 60 nm FAM-C1–9R or FAM-9R to varying concentrations of Cdc42·GMPPNP with data fitted to a direct binding model containing a linear nonspecific-binding term. n = 3. The bottom panel shows the competition between 60 nm FAM-C1–9R and 500 nm Cdc42·GMPPNP with a range of concentrations of unmodified C1 fitted to a competition binding model to calculate binding affinity. n = 2. B, effect of higher concentrations of FAM-C1–9R on cells. MEFs treated with peptide or a FITC control for varying times are shown under ×20 phase contrast. After 28 h, cells were washed twice in PBS and imaged by phase contrast and fluorescence using an EVOS GFP light cube. These images are shown in overlay in the bottom panels. C, entry of FAM-C1–9R into cells. Peptide at five different concentrations was applied to MEFs, and the cells were monitored by fluorescence confocal microscopy at four different time points. Representative images are shown and labeled accordingly. n = 3. D, effects of the peptides on signaling pathways. MEFs were incubated with FAM-C1–9R for 1 and 24 h alongside untreated controls (0). Cell lines A and C expressed K-Ras G12D, and B and D were WT. Western blottings from whole-cell lysate were probed for Stat3 phospho-Tyr-705, total Stat3, ERK1/2 phospho-Thr-202/204, total ERK1/2, and GAPDH. Data are representative of three independent experiments.
To assess peptide entry into cells, mouse embryo fibroblasts (MEFs) were incubated in media containing FAM-C1–9R at concentrations from 60 nm to 10 μm for between 2 and 5 h at 37 °C before being washed and imaged by live-cell confocal microscopy. Fig. 2B shows images of cells exposed to 10 μm peptide compared with a 5 μm peptide treatment and a FITC control. At peptide concentrations of 10 μm, the MEFs showed signs of membrane disruption and cell death. Neither the 5 μm peptide nor the FITC-treated cells show such atypical cells at the time points observed. These data corroborate previous observations of a toxicity threshold for the nona-arginine CPP (31), and therefore 5 μm FAM-C1–9R was the maximum concentration used in subsequent experiments. Example images for treatments with ≤5 μm peptide in Fig. 2C show that the peptide can access the cell interior at a variety of concentrations and within 2 h of dosing. The cells in these images appear healthy with the majority of the peptide dispersed in a punctate pattern in the cytoplasm, suggesting an endosomal location, as is often seen with peptides (32, 33). The cytosol and nuclei of the cells, however, show fluorescence above background levels, suggesting that not all peptide is confined to endosomes and that endosomal exit has allowed a wider distribution in the cells.
Next, we investigated the effects of FAM-C1–9R on relevant cell-signaling pathways. As our ultimate aim was to engineer peptide inhibitors of Cdc42 to attack Ras-driven cancers, we chose to examine signaling pathways using MEFs derived from four mice, two of which carried a Cre-induced G12D oncogenic K-Ras mutant (34). Previous work in our laboratory8 and from others (35, 36) has shown Stat3 Tyr-705 is a target for phosphorylation downstream of ACK, so pTyr-705 Stat3 was chosen as a readout for an active Cdc42–ACK-signaling pathway. Cells were treated with 1 μm FAM-C1–9R for 1 and 24 h alongside the untreated controls. Four different MEF isolates were used from four littermates: A and C expressing G12D K-Ras and B and D expressing WT K-Ras. Fig. 2D shows pTyr-705 Stat3 levels demonstrating a marked decrease at 1 h in cell lines A, B, and D, returning to untreated levels by the 24-h time point, while total Stat3 remained relatively constant, demonstrating a reduction in signaling by Cdc42 on exposure to FAM-C1–9R. ERK phosphorylation was also probed, in order to assess activity through the MAPK pathway, and the results are shown in Fig. 2D. MEK induces phosphorylation of ERK1/2 at residues threonine 202 and tyrosine 204, so levels of pThr-202/pTyr-204 ERK1/2 were analyzed. Suppression of pThr-202/pTyr-204 ERK1/2 phosphorylation is visible at 1 h for cells lines A and B, but not in cell lines C and D, suggesting that the MAPK pathway, downstream of Ras, can be affected by FAM-C1–9R.
Taken together, these data demonstrate that FAM-C1-R9 enters cells, likely by an endosomal route, but can escape into other parts of the cell where it is able to elicit the signaling effects that it was designed to accomplish.
Maturation of C1
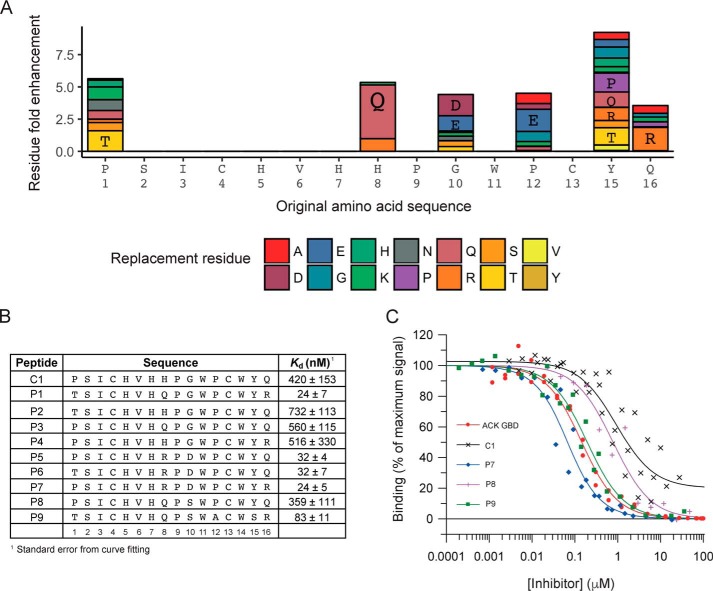
Although CIS display offers the potential to interrogate some of the largest naïve sequence space available, the naïve 16-mer library used in the original selections covers only ∼0.0125% of possible sequences, suggesting the likelihood that other potentially higher-affinity sequences could exist that had not been available for selection. Thus a maturation library designed to search the sample space around the C1 sequence was constructed and selected against Cdc42·GMPPNP. Sequence analysis of the selected peptides, after four rounds of biopanning, showed that residue replacement was favored at six positions, whereas three other positions saw no substitutions highlighting their possible key roles in the target engagement. The fold enhancement of individual amino acids at these positions compared with the initial library are shown in Fig. 3A. A panel of peptides was then synthesized, based on the enriched sequences observed in the NGS analysis of the selections, to represent the most favorable residue replacements, as shown in Fig. 3B. Binding affinities for this panel of peptides were determined by competition SPA, as described above, and dissociation constants are listed in Fig. 3B with representative data shown in Fig. 3C. For five of these second generation peptides, the affinity for Cdc42 was increased significantly (5–20-fold) and reached values close to the affinity of the complete ACK GBD for Cdc42 (20–30 nm), lending further promise for their ability to act as competitive inhibitors of Cdc42 effector proteins in cells.
Figure 3.
A, enrichment of replacement residues in second generation peptides after maturation by CIS display. At positions where the parent residue was proportionately decreased in abundance after four rounds of biopanning of the cyclic maturation library, the relative enrichment of replacement residues at each amino acid is shown in the bar chart. Amino acids are individually colored as shown in the key and labeled using the one-letter code where they have a greater than 2-fold enrichment. B, second generation peptides. The nine selected sequences from the maturation screen are presented. Their affinities for Cdc42·GTP were determined by competition SPA, and their Kd values are given together with the standard error form curve fitting. n ≥ 2. C, representative SPA data. Increasing concentrations of the ACK GBD or the peptides were titrated into a fixed concentration of [3H]GTP·Cdc42 and GST–WT ACK GBD in competition SPAs. The Kd for Cdc42 binding to GST–WT ACK GBD was fixed to the value (30 nm) obtained for this Kd in direct SPAs. The data were fitted to an isotherm describing a pure competition model (58) to give an apparent Kd (Ki) value for the peptides. The data and curve fits are displayed as a percentage of the maximal SPA signal in each assay.
The maturation data suggest that the combined H8R and G10D substitutions in peptide candidates are important for increasing binding affinity, as all peptides with these substitutions have an affinity of 32 nm or lower for Cdc42. Effects of other replacements are less clear; however, the combined effect of P1T, H8Q, and Q16R are favorable in P1 and P9.
Mode of peptide binding to Cdc42
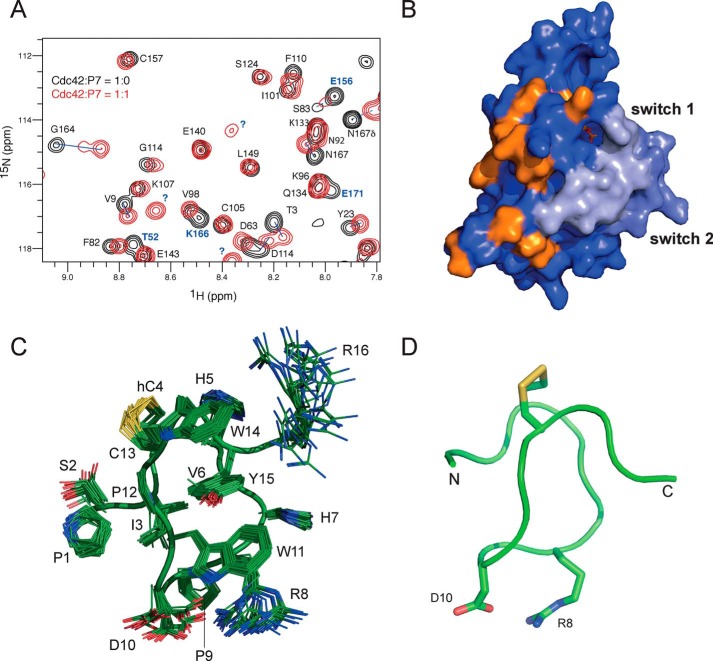
One of the second generation peptides with the tightest binding, P7, was chosen for structural analysis by NMR. 15N HSQC spectra of Cdc42 were recorded, and the chemical shifts of the backbone amides were monitored with increasing P7 peptide concentrations (Fig. 4A). Most of the peaks in the Cdc42 spectrum that experienced chemical shift changes were in slow exchange, so that peaks corresponding to both the free and bound species were observable in the intermediate time points of the titration. Residues, whose peaks in the complex spectrum could be assigned and whose chemical shift changes were higher than average, were identified (Fig. S1). To these were added those residues whose peaks had moved too far from their position in the free Cdc42 spectrum to be reliably assigned (labeled in blue in Fig. 4A). The residues with significant shift changes and that are solvent-accessible are shown mapped onto the Cdc42 structure in Fig. 4B. Chemical shift perturbations can indicate that a particular residue is involved in the interface with the binding partner, but may also be due to longer-range allosteric effects. Residues that are not accessible to the solvent are unlikely to be involved in direct interactions, and their removal reduces the number of indirect contributions to chemical shift mapping. The chemical shift mapping localizes the P7-binding site to a face of Cdc42 that is involved in binding to a critical section of the N termini of the GBDs of effector proteins (18, 37).
Figure 4.
Structural analysis of peptide P7 and its interactions with Cdc42. A, HSQC spectra of 15N-labeled Cdc42 in its free (black) and bound (red) forms. The peaks corresponding to the free form are labeled with their assignment in one-letter code and are connected by a blue line to the corresponding resonance in the bound form. Peaks that had shifted too far to be reliably assigned are labeled in blue. B, surface representation of Cdc42 with those residues that experienced a larger than average shift change and that were more than 50% solvent-exposed are colored orange. C, family of structures of peptide P7 is shown in green with the disulfide bond in yellow. Side chain nitrogen atoms are in blue, and oxygen atoms are in red (PDB accession code 6R28). The side chains are well-defined in the structures with the exception of the C-terminal Arg-16 and the two residues in the loop, Arg-8 and Asp-10, which are responsible for high-affinity binding of this peptide. D, backbone representation of the closest structure to the mean.
The structure of the P7 peptide in isolation was determined by NMR. The peptide was fully assigned using standard homonuclear experiments, and the NOESY was used to generate 440 unique distance restraints. The RMSD of the final water-refined structures is 0.37 Å over all backbone atoms, and the large number of distance restraints means that the structure is well-defined (Table 1). The disulfide bond, in combination with the presence of two proline residues within the middle of the sequence, which lend rigidity, means that the peptide adopts a tightly-constrained structure (Fig. 4, C and D). Stacking interactions between the three aromatic residues within the sequence (Trp-11, Trp-14, and Tyr-15) further stabilize the structure. There is no regular secondary structure in the peptide, which merely forms a series of turns. The overall structure forms a single turn of a loose, irregular, left-handed helix between residues 3 and 15, pinned by the disulfide bond between Cys-4 and Cys-13, with the N and C termini protruding out on opposite sides (Fig. 4C). Arg-8 and Asp-10, which drive higher binding affinities, are displayed on a loop opposite the disulfide bond, which presumably constitutes the Cdc42 binding face (Fig. 4D). The structure explains why the disulfide bond is necessary for high-affinity binding to Cdc42, because there is only one residue, Val-6, in its hydrophobic core (Fig. 4C). Therefore, without the disulfide this compact structure would not form stably and would be unable to display the Cdc42-binding face correctly.
Table 1.
Experimental restraints and structural statistics
PDB accession code is 6R28. Chemical shifts deposited at BMRB under accession code 34380.
| Experimental restraints | ||
| Distance restraints: | ||
| Total nondegenerate | 440 | |
| Unambiguous | 408 | |
| Ambiguous | 32 | |
| Intraresidue NOEs | 170 | |
| Sequential NOEs | 115 | |
| Medium and long-range NOEs | 155 | |
| 〈SA〉a | 〈SA〉b | |
| Coordinate precision for well-ordered regions | ||
| RMSD of all backbone atoms (Å) | 0.37 ± 0.13 | 0.15 |
| RMSD of all heavy atoms (Å) | 0.69 ± 0.09 | 0.63 |
| RMSD for all residues | ||
| From the experimental restraints: | ||
| NOE distances (Å) | 0.025 ± 0.0014 | 0.025 |
| From idealized geometry: | ||
| Bonds (Å) | 0.0045 ± 0.00023 | 0.0044 |
| Angles (°) | 0.83 ± 0.025 | 0.83 |
| Impropers (°) | 1.30 ± 0.17 | 1.18 |
| Ramachandran plot analysisc | ||
| Core region (%) | 59.5 | 50.0 |
| Allowed region (%) | 30.7 | 41.7 |
| Generously allowed region (%) | 9.0 | 8.3 |
| Disallowed region (%) | 0.7 | 0.0 |
a 〈SA〉 represents the average RMSD for the ensemble of 36 structures.
b 〈SA〉 represents values for the structure that is closest to the mean.
c Data were analyzed using Procheck.
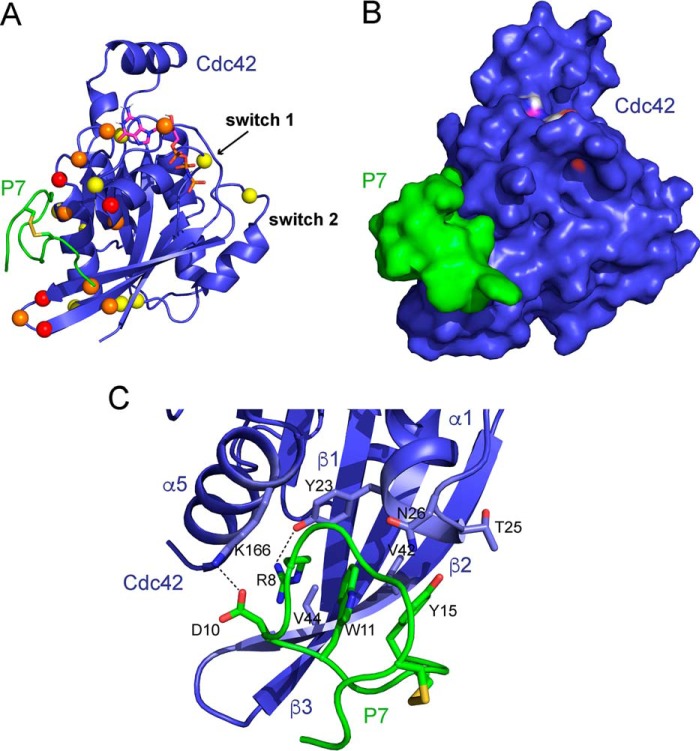
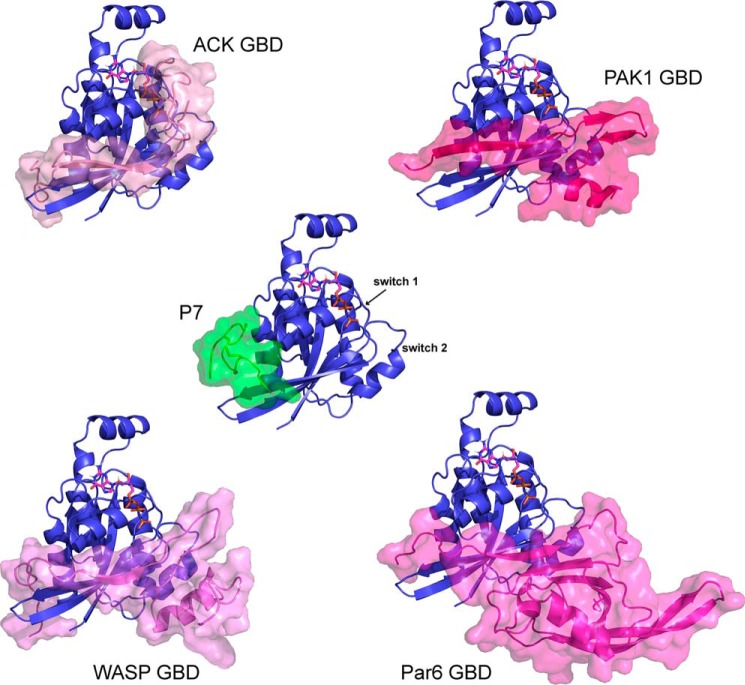
Data-driven docking (HADDOCK) was next used to predict the structure of the Cdc42–P7 complex (38, 39). The NMR chemical shift mapping was used to determine the active residues in Cdc42, and the matured peptide sequences and affinity measurements (Fig. 3B) were used to determine the active residues in the peptide (Arg-8 and Asp-10). The complete Cdc42–P7 docked model is shown in Fig. 5, A and B, and identifies both hydrophobic and polar interactions between Cdc42 and P7. In particular, the conserved peptide residues Trp-11 and Tyr-15 (found in all second generation peptides except P9) form the hydrophobic core of interactions with a Cdc42 pocket between strand β1 and helices α1 and α5. This pocket is lined by Cdc42 residues such as Val-42, Val-44, and Tyr-23. Tyr-15 is also close to Cdc42 residues Thr-25 and Asn-26, which could make polar interactions with the Tyr OH group. Arg-8 and Asp-10 are predicted to form a salt bridge with each other in the free peptides, but Asp-10 is within 3 Å of Lys-166Cdc42 in the model and hence may form a salt bridge. Arg-8 is within 3 Å of Tyr-23Cdc42 and potentially could form a hydrogen bond (Fig. 5C), although the backbone amide of Tyr-23Cdc42 did not show chemical shift changes when the peptide was bound. The Cdc42 pocket that binds to P7 also contacts the N terminus of the ACK GBD in the Cdc42–ACK complex (Fig. 6). This is consistent with the previous observation that P7 can compete with ACK GBD binding to Cdc42 (Fig. 1C), which implies overlapping binding sites. Importantly, this region of Cdc42 contributes significantly thermodynamically to the binding of both ACK and WASP (18, 37) and is also demonstrated to bind the other known CRIB effectors PAK1 and Par6 where structural information is available (Fig. 6), suggesting that P7 will disrupt binding to these proteins as well.
Figure 5.
A, data-driven model of the Cdc42–peptide P7 complex generated using HADDOCK. Peptide P7 is shown in green with the disulfide bond in yellow, and Cdc42 is blue with the nucleotide shown in a stick representation. Residues identified from the chemical shift mapping are depicted as colored spheres; those that had shifted the most (more than average + 1 S.D.) are colored red, and those that had shifted by more than average but less than average + 1 S.D. are colored yellow, and those that had shifted too far to reliably assign are colored orange. B, Cdc42–P7 model is shown in the same orientation as in A in a surface representation. P7 is shown in green and Cdc42 in blue. The peptide binds in a Cdc42 pocket between the β2–β3 hairpin and helix α5, which is lined by residues that showed chemical shift changes upon peptide binding. C, close-up of some of the interactions in the model indicates that Arg-8, Asp-10, Trp-11, and Tyr-15 of the peptide loop insert into the Cdc42 pocket and that salt bridges form between Asp-10P7 and Lys-166Cdc42 and Arg-8P7 and Tyr-23Cdc42.
Figure 6.
P7-binding site overlaps with the binding surface of the CRIB effectors on Cdc42. ACK, PAK1, WASP, and the Par6 semi-CRIB all form contacts with the same pocket so that the peptide binding will cause steric clashes with CRIB effectors. Cdc42 is shown as a blue cartoon, and peptide P7 (green) and the CRIB effectors (shades of pink) are shown as a semi-transparent surface overlaid on the cartoons.
Second generation peptide specificity and cellular target engagement
The two tightest-binding second generation peptides, P1 and P7, were next synthesized with FAM and 9R additions for use in cell assays. The tagged proteins were first tested for their ability to bind Cdc42. As the specificity of the interaction of these peptides is likely to be paramount for any future therapeutics, a survey of the binding affinities of these peptides across a wider panel of small GTPases was also undertaken. The panel of small G proteins tested included both Cdc42·GDP and Cdc42·GMPPNP along with the classic Rho-family exemplars, Rac1 and RhoA. We also included members of the Ras family, K-Ras and RalA, as examples of more diverse small G proteins. Binding of tagged C1, P1, and P7 with the selected small G proteins was assessed using FP. The results are shown in a radar plot in Fig. 7 and are tabulated in Table 2. All three peptides have relatively weak affinity for K-Ras and RalA as expected. The data demonstrate that overall P7 has higher specificity than C1 or P1, showing considerably tighter binding to Cdc42 and Rac1 than other small GTPases. P1 and C1 have more similar specificity profiles and bind RhoA with comparable affinity to Cdc42 and K-Ras to a lesser extent. All three peptides, however, show only marginally better binding to Cdc42·GMPPNP than Cdc42·GDP.
Figure 7.

Affinities of peptides FAM-C1–9R, FAM-P1–9R, and FAM-P7–9R for a panel of small GTPases were measured by FP. The Kd values (see also Table 1), determined from n ≥ 3 assays, are plotted as reciprocals on a radar chart to demonstrate the relative affinity of the peptides for small GTPases related to Cdc42. Units of 1/Kd = μm−1.
Table 2.
Affinities of peptides for a panel of small G proteins measured by FP
|
Kd (nm) ± S.D. |
|||
|---|---|---|---|
| C1 | P1 | P7 | |
| Cdc42·GDP | 876.0 ± 170.0 | 98.4 ± 9.1 | 32.0 ± 4.3 |
| Cdc42·GMPPNP | 409.0 ± 44.2 | 24.5 ± 8.6 | 14.6 ± 3.4 |
| Rac1·GMPPNP | 355.0 ± 79.0 | 896.0 ± 123.0 | 436.0 ± 107.0 |
| RhoA·GMPPNP | 475.0 ± 152.0 | 166.0 ± 52.2 | NDa |
| K-Ras 4B·GMPPNP | 3418.0 ± 581.0 | 3970.0 ± 614.0 | 19,600.0 ± 5600.0 |
| RalA·GMPPNP | 44,000.0 ± 1251.0 | 37,040.0 ± 1340.0 | ND |
a ND (no binding) denotes data that could not be fitted to the binding isotherm.
Cellular entry of these second generation peptides was demonstrated by confocal microscopy using A549 cells. The A549 cell line is a human lung epithelial cell line, which is homozygous for K-Ras G12S. As such, it is a good representative of the type of cells in which these peptides would need to be active therapeutically. The images in Fig. 8A show representative examples of cells treated with C1, P1, and P7 at varying time points after addition. The peptides concentrate at the periphery of cells in vesicular structures at the 1-h time points but are able to access the interior of the cell and appear to be distributed in the cytosol rather than concentrated, potentially in endosomes, after 3–6 h, in contrast to the previous studies using MEFs.
Figure 8.
A, entry of FAM-9R–conjugated peptides into A549 cells. Peptides were applied to A549 cells at a final concentration of 1 μm, and the cells were monitored by confocal microscopy at a variety of time points. Representative images are shown and labeled accordingly. B, inhibition of Cdc42–effector interactions by C1 and second generation peptides (P1 and P7). The ability of Cdc42 to interact with its effector WASP in the A549 lysate was monitored by GST–WASP pulldown experiments. GST–WASP GBD immobilized on GSH-agarose beads was used to pull down endogenous Cdc42 from A549 lysates in the presence of various peptide concentrations. The levels of Cdc42 bound were analyzed by Western blotting. Representative Western blottings from unconjugated C1, P1, and P7 treatments are shown in the upper panels. Densitometry was performed on the results from replicate experiments (n ≥ 3), and the amount of Cdc42 present in pulldowns, expressed as a percentage of levels in the absence of peptides, are plotted in the lower panel. Data for FAM-9R–conjugated peptides are also included. C1, pink; FAM-C1–9R, red; P1, green; P7, cyan; FAM-P7–9R, purple. Approximate IC50 values were calculated from the curve fits: C1, 190 ± 62 nm; FAM-C1–9R, 2200 ± 740 nm; P1, 18 ± 3.5 nm; P7, 1.2 ± 0.28 nm; FAM-P7–9R, 65 ± 12 nm. C, target engagement by peptides. CETSA experiments were run in A549 lysate at 56 °C treated with varying concentrations of FAM-P7–9R peptide to assess target engagement. Cdc42 and actin (as a control) levels were quantified by Western blotting, and a representative blot is presented showing destabilization of Cdc42 with increasing peptide concentrations. n = 3. D, target engagement after cell penetration. Whole-cell CETSA experiments were run in A549 cells at 51 °C after 4 h of incubation with varying concentrations of FAM-P7–9R peptide to assess target engagement after external application of peptide. After lysis, Cdc42 and actin levels were quantified by Western blotting, and a representative blot is presented showing destabilization of Cdc42 with increasing peptide concentrations. n = 3.
The efficacy of the peptides in inhibiting the interaction of Cdc42 with its effector proteins was next tested using GST-effector domain pulldown inhibition assays in cell lysate. Increasing concentrations of unconjugated C1, P1, and P7 were introduced into the A549 cell lysate, and the ability of Cdc42 to interact with the effector binding domains was assessed. The data in Fig. 8B (upper panels) show the inhibition of the endogenous Cdc42 interaction with immobilized GST–WASP GBD by peptides C1, P1, and P7 (experiments were also performed with ACK GBD showing similar inhibition, data not shown). These data suggest that the binding of the peptides to Cdc42 is sufficiently stringent to inhibit Cdc42–effector interactions among the myriad of proteins in the cell lysate.
These experiments were repeated with FAM-9R–conjugated peptides to determine whether these additions affected targeting of Cdc42 in cell lysates. These data were quantified and are shown, together with the data from the unconjugated peptides, in the graph in Fig. 8B (lower panel). The data show that the FAM-9R–conjugated peptides inhibited the Cdc42–WASP interaction, but a considerably higher concentration was needed to achieve this than with the unconjugated peptides. The IC50 values calculated from these data suggest that, in fact, the addition of FAM-9R to the peptides results in a >10× decrease in efficacy in cell lysates. For example, IC50 C1 is 190 ± 62 nm but FAM-C1–9R is 2200 ± 740 nm. For P7, the effect is even greater with the IC50 P7 calculated as 1.2 ± 0.28 nm but FAM-P7–9R being 65 ± 12 nm. Importantly, these data suggest that any cellular effects seen with the peptides will be greatly enhanced when using an alternative delivery system and/or without the reporter group.
Peptide binding to Cdc42 in vitro was further demonstrated using cellular thermal shift assays (CETSAs) (40). These assays monitor protein stability during thermal denaturation in cells or in lysates, in the presence of potential binding partners or inhibitors. Changes in thermal stability are taken as an indication of protein engagement by the peptides. Cdc42 levels were monitored under appropriate conditions in A549 cell lysates, with the addition of increasing concentrations of peptide P7, and the results are shown in Fig. 8C. Cdc42 in lysates is destabilized in the presence of P7, indicating target engagement by the peptide. These experiments were repeated in A549 cells with peptide P7 added in to the cell growth media and allowed to enter the cells for 3.5 h (Fig. 8D). The results show a similar destabilization pattern of Cdc42 when the cells were exposed to peptide. Interestingly, at higher peptide concentrations (3–6 μm), which are toxic to the cells, a rise in Cdc42 levels was noticed, which then similarly decreased with concentration, potentially implying a different transport mechanism takes over that does not allow similar levels of target engagement.
Second generation peptides significantly inhibit transformation phenotypes in A549 cells
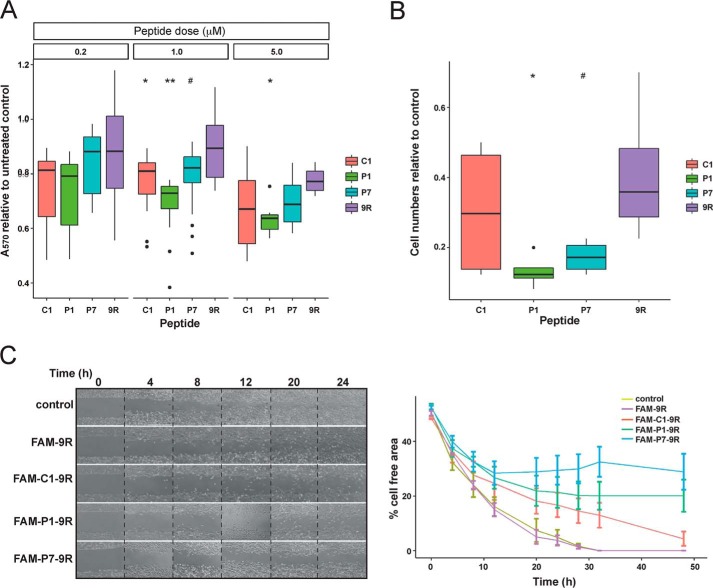
As the peptides were able to enter cells and engage with Cdc42, the effects of the peptides on cellular properties associated with transformation were assessed. First, proliferation was measured. The longevity of either the peptide sequences themselves or the disulfide bond was unknown in cells; however, as the experiments in MEFs (Fig. 2D) showed signaling restoration with time, the peptides were redosed at 24, and 48 h was used as an upper time limit in proliferation experiments. After 48 h of incubation, cells treated with a 1 μm dose of the peptides showed significantly lower proliferation levels compared with the 9R control, as shown in Fig. 9A. The most pronounced effect was seen for P1, whereas C1 and P7 show similar but slightly reduced potency. At 5 μm doses, greater inhibition of proliferation is seen, with a similar pattern of peptide efficacy. Overall, for all three peptides, C1, P1, and P7, statistically there is a significant linear trend both with increasing dose and increasing time of incubation (using general linear models in R, data not shown), although this is not seen for 9R, further demonstrating biological activity for these peptides.
Figure 9.
A, peptide effects on proliferation of A549 cells. The proliferation of A549 cells was analyzed over 48 h by MTT assay. Proliferation data are shown for 48-h time points from 6 to 12 replicate experiments at three different peptide concentrations. A570 values are shown as a proportion of the untreated control values and displayed as modified boxplots. Group significance testing at each dosing level was performed by t test (n = 12–34): #, p < 0.1; *, p < 0.05; **, p < 0.01. B, peptide effects on migration and invasion of A549 cells. Boyden chamber assays were used to assess cell migration and invasion. Cells were seeded in the top chamber in 0.1% serum, and migration through the membrane to 10% serum-containing media was measured. Cell numbers relative to untreated control cells are shown as modified boxplots, and a significant decrease in proliferation relative to 9R was assessed using the one-way t test (n = 4): #, p < 0.1; *, p < 0.05. C, peptide effects on proliferation and migration of A549 cells. Scratch assays were used to measure cell proliferation and coordinated migration. Representative images over the first 24 h of the assay are shown for each of the peptides in the left panel. The complete data set (n = 3) is shown in the graph (right panel) with the percentage area of the scratch left uncovered by cells for each condition plotted at each time point, with error bars representing the standard error of the mean.
Cdc42 and other Rho family small G proteins are traditionally associated with cytoskeletal rearrangements, which govern cell motility (41). High migration and motility are also associated with aggressive oncogenic phenotypes, and inhibition of this would therefore be highly desirable. It was notable from the proliferation assays that the 9R CPP alone begins to have a significant effect on cell proliferation at concentrations above 5 μm, and as such peptides were only dosed at 1 μm in subsequent experiments. Peptides were first assessed in Boyden chamber assays monitoring invasion and migration toward higher serum concentrations, and the results are shown in Fig. 9B. Potent effects for the peptides dosed at 1 μm were observed. The data also show significant inhibition of invasion and migration by the 9R control (∼65%); however, the peptide-9R conjugates showed greater inhibitory effects, in particular P1 and P7, which reduce invasion and migration by around 90 and 85%, respectively.
Next, wound-healing assays were used to monitor coordinated cell motility and proliferation. A549 cells were treated with 1 μm FAM–peptides–9R and monitored for 48 h. Representative images and data plots are shown in Fig. 9C. All peptides showed significant inhibitory effects on wound closure; in the case of P1 and P7, the scratch area did not close within the duration of the assay. In contrast to the Boyden chamber assays, no inhibitory effect was noted for 9R alone.
Taken together, these data suggest that the high-affinity Cdc42-binding peptides developed here have the hallmarks of early lead molecules for therapeutic intervention in Ras-driven cancers.
Discussion
The isolation and maturation of a peptide, C1, capable of binding Cdc42 with mid-nanomolar affinity, from the naïve libraries used in the original display experiments described here, validates CIS display as a robust technique for isolating small G-protein–binding peptides. Molecular display methods all have to rely on the chance sampling of library members to some extent, as seldom, even with larger libraries, can all sequence space be covered for peptides beyond 10 residues in length. However, as a wholly acellular technology, CIS display, with larger, stable libraries, is likely to sample greater sequence space and thus allow the identification of tighter binding peptides. Subsequent maturation of C1 increased both the specificity and affinity of this peptide further, with the best second generation binders containing 2–4 mutations at six positions and possessing ∼17-fold increased binding to their primary target, Cdc42·GTP. Although the use of a randomly doped maturation library has identified six positions as being important for the improvement of binding efficiency, display of this library may still not have allowed sampling of all the most beneficial combinations of mutant residues. Further affinity maturation may be possible by using mutagenesis directed to these six positions particularly if harnessed with competitive regimes of selection. Alternatively, a rational design approach and the incorporation of unnatural amino acids may offer further scope for improved affinities.
Closer investigation of C1 and its derivatives has demonstrated that the specificity for active Cdc42 was not as high as initially indicated (see Fig. 1B versus Fig. 7) with the peptides binding closely-related small GTPases. Binding to Cdc42 is also insensitive to the nucleotide state, and thus the peptides may also interfere with Cdc42 interactions when inactive. Peptide maturation has, however, resulted in tighter Cdc42 binding and also modulation of Rho family specificity. P7, in particular, shows improved targeting of Rac1 and Cdc42 over other Rho family proteins and K-Ras.
In general, tighter binding to a target does correlate with increased specificity. This holds true for general off-target binding events; however, it does not necessarily follow for proteins very closely related to the target itself. For a potential therapeutic designed to target small G proteins, there are many very closely-related proteins that the molecule could encounter.
The data-driven model we generated of the Cdc42–P7 complex suggests some hypotheses as to how residue replacements in the second generation peptides have influenced binding. The key replacements at the Arg-8 and Asp-10 positions may allow a salt bridge to form between Asp-10 and Lys-166Cdc42, whereas Arg-8 lies close to Tyr-23 in Cdc42 helix α1 and could form a polar interaction here. The replacement of His-8 and Gln-10 in C1 and P1 for charged residues in P7 therefore increases the affinity of P7, by imparting the ability to form the ionic interaction involving Asp-10P7 and Arg-8P7.
The most marked difference between P7 and C1/P1 is the reduced binding to RhoA by P7, which is likely due to Arg-8 and Asp-10. Tyr23Cdc42, which, in the model, is close to Arg-8 in P7, is replaced by Phe in RhoA, which would remove any polar interaction between these side chains. Lys-166Cdc42, which forms a salt bridge with Asp-10P7 in the model, is still present in RhoA, but Met-45Cdc42, which is also close to Asp-10, is replaced by Glu in RhoA. This could potentially destabilize the binding of Asp-10 at this site. These interactions and substitutions between Rho family members potentially explain the specificity of P7 for Cdc42 and Rac1 and could be used to guide fine-tuning of specificity, although detailed structural data on the Cdc42–P7 complex would be necessary to underpin any rational design campaign.
Our structure of P7 also explains some important structural features of the peptide. Our binding data indicate the importance of the cyclized nature of the peptides in engaging their target. In fact, cyclizing libraries were deliberately utilized in the initial stages of this project to exploit the utility of cyclic peptides as therapeutics. As it is difficult to introduce stable macrocycle linkages into a biological display strategy, the nonvariant incorporation of cysteine residues was exploited in these libraries to allow disulfide formation and provide a useful platform to screen cyclic peptides. However, the potential instability of a disulfide bond in cells was always acknowledged.
Disulfide bonds are regulated inside the cell by an array of enzymes and redox molecules such as thioredoxin, GSH, and NADPH that increase the naturally slow turnover of disulfides (42). The major redox couples that govern cellular redox potential are not necessarily in equilibrium with each other, and indeed, the equilibria vary between intracellular compartments, generally being defined by the reduced/oxidized GSH ratio (42). Endosomes have an oxidizing potential, whereas the cytosol is mainly reducing (43), and it is thought that a disulfide could survive intact through endocytosis (44), but upon endosomal escape, the peptide enters a reducing environment. How long the disulfide would last under these conditions is not easy to define as data are confounded by uptake and release rates from endosomes (45). Available data suggest that a disulfide bond in cyclized peptides may last in the extracellular environment but could start to come under attack from the point of interaction with a cell; however, they are likely to pass through the endocytic pathway in an oxidized form. Upon release into the cytosol, disulfide-bridged cyclic peptides should be quickly reduced, diminishing their binding affinity and proteolytic stability. Peptides, however, will be taken up and released into the cell as a gradual process, meaning release into the cytosol will be continual.
In light of the possible instability of our lead peptides in cells, we have trialed various strategies to produce stable cyclized versions of these peptides. The replacement of the disulfide with a lanthionine bridge by alkali-catalyzed desulfurization (46) rendered this linkage nonlabile in reducing environments, and the resulting peptides retained the same binding affinity as disulfide-linked forms in vitro and in CETSA lysate experiments (data not shown), suggesting that, with the appropriate chemistry, stable cyclized versions of these peptides can be successfully engineered if necessary. However, the biological data presented here were all obtained using disulfide-linked peptides. Our data suggest that these disulfide-linked peptides are actually relatively stable in a cellular environment, with biological effects seen up to 24 h after exposure to peptides. The structure of the P7 peptide reveals that it is remarkably rigid, and it is possible that the highly-compact nature of P7 protects the disulfide bond to some extent, so that it is less labile in a cellular environment than a more flexible peptide might be. The rigid P7 structure contributes to its high affinity, because the binding loop containing Arg-8 and Asp-10 is pre-formed, and there will be no entropic penalty when it binds to the G protein. The peptide-binding site, determined by experimental chemical shift mapping, is within a pocket formed by the β2 strand and helices α1 and α5 in Cdc42. This site is away from the nucleotide-binding site and the nucleotide-sensitive switch regions of Cdc42, which explains the ability of P7 to bind Cdc42 without nucleotide specificity. Nevertheless, peptide P7 can still compete with ACK for binding to Cdc42 (Fig. 3) and is presumably an orthosteric inhibitor, given the overlap between the ACK and P7-binding sites. However, Cdc42 is, like all small GTPases, an allosteric protein (47), and it is possible that P7 will disrupt binding of other proteins, such as Guanine nucleotide dissociation inhibitors (GDIs) and exchange factors, that do not directly engage with the P7-binding pocket.
The model of the Cdc42–P7 complex may also explain the observation that Cdc42 engagement by P7 leads to destabilization of the small G protein (Fig. 8, C and D). This was potentially unexpected, given that the binding of the CRIB effectors is likely to stabilize both binding partners as two disordered regions coalesce into a more rigid complex (48). Other systems using CETSAs have reported stabilization, destabilization, and no change with target engagement, and the effect appears difficult to predict (49, 50). However, the distinct binding mode of P7, interacting with regions that are already structured in the free Cdc42, would not necessarily stabilize the protein and could potentially inhibit interaction with binding partners that confer thermal stability.
The addition of a CPP motif allowed the peptides access to the cell interior in both MEF and A549 cells. The general mechanism of CPP entry has been a subject of some debate but appears most likely to be through endosomal uptake and escape (51). Certainly, this seems to be the case with the peptides described here. The kinetics of cellular distribution appear to be different in MEF and A549 cells; however, in both cell types the effects observed suggest sufficient peptide delivery to the target to obtain measurable biological effects.
Pulldown inhibition in lysates has demonstrated that the peptides are specific and soluble enough among the cell contents to bind and inhibit Cdc42–effector interactions, but, interestingly, the addition of N-terminal FAM and C-terminal 9R considerably reduces the potency of Cdc42 interaction inhibition. The most likely reasons for this ∼10–50-fold reduction in potency are that the 9R motif binds nonspecifically to other moieties or causes aggregation in the lysate. These results suggest that the cellular potency of the peptides is reduced by this CPP motif, an effect that is likely to be general to peptides using a 9R CPP. No therapeutics have made it to clinical trials using such a motif, and aside from the toxicity issues that are confirmed in Fig. 2C, this reduction in target specificity could be a contributory reason in some cases. An alternative delivery system therefore should dramatically improve potency in this case. Methods to improve delivery could either employ a cleavable linker or another cell-penetration strategy.
The early promise of C1 in decreasing signaling to STAT3 and ERK1/2 in MEFs has been consolidated by potent second generation peptide effects on proliferation and especially migration in the more aggressive A549 cell line. The fact that these peptides were selected to target the Rho-family small GTPases, which are known to control cytoskeletal structure and migration, makes it encouraging that the most potent effects of the matured peptides are on cell motility. Data presented here have demonstrated effective target engagement in the cellular environment; however, it is uncertain how important inhibition of off-target small GTPases is to overall peptide efficacy. P1 shows more potency than P7 in the Boyden chamber and proliferation assays despite having a lower affinity for Cdc42. P1, however, has relatively high affinity for RhoA, and it may be that inhibition of RhoA, or indeed another related Rho GTPase, is important for these effects. P7 performs best in wound-healing assays, where, presumably, high affinity for Cdc42 is most efficacious. Our data, therefore, suggest that simultaneous targeting of several Rho family GTPases could result in improved anti-oncogenic effects; indeed, arguments for true polypharmacology are presently redefining drug discovery paradigms (52). This opens up interesting future studies into how inhibition by different peptide derivatives from C1 could tune various signaling pathways and phenotypic effects.
The effect we see on ERK1/2 signaling is an interesting example of the type of effects we were hoping to see in these peptides. Traditionally, ERK1/2 phosphorylation is monitored as a readout for Ras signaling. We see down-regulation of pERK1/2. However, in this case we assume that this is due to inhibition of the Cdc42–PAK complex. PAK is another CRIB effector of Cdc42 (and indeed of Rac1), which is known to be involved in regulation of ERK activity as it regulates phosphorylation of both Raf and MEK, contributing to activation of the Raf–MEK–ERK pathway (53, 54). Thus, the peptides appear to inhibit the interaction of Cdc42 (and possibly other Rho-family GTPases) with multiple effectors due to the similarity in the effector engagement sites and act as blockades of Cdc42 signaling. The similarity in the CRIB-effector interfaces and their overlap with the P7-binding site (Fig. 6) provide a rational explanation for the ability of P7 to inhibit multiple effector pathways.
In summary, the peptides isolated in this study show promise as inhibitors of oncogenic phenotypes in Ras-driven cancers, a hugely important cohort of cancers that have proved difficult to treat to date. Their activity depends on their cyclic nature, and although they appear to contain a relatively stable disulfide bond, which confers activity in cells for several hours, introduction of a nonlabile cyclization was also possible without compromising activity. Other strategies to increase stability of the peptides using unnatural bonds and residues are currently under investigation. Utilization of a 9R CPP motif for experimental purposes was successful, but a 9R control peptide did show significant toxic effects on cells above a threshold concentration. More importantly, the 9R CPP, when conjugated to the peptides, reduced target engagement by ∼10–50-fold in cell lysate. Alternative peptide entry strategies are also currently under investigation and should dramatically improve efficacy of these peptides in vivo. Slight changes in the C1 peptide sequence were also shown to modulate specificity to small GTPases and to change phenotypic effects suggesting that the target selection of these peptides can be adapted. A degree of polypharmacology is also likely to be desirable in future anti-cancer therapeutics, and tuning alternative-target binding should improve therapeutic properties further.
The peptides we describe here are examples of peptide macrocycle technology applied to the highly-intractable target class of small G proteins and represent a promising new modality for future therapeutics. Our data also add to the growing literature demonstrating that peptides are establishing their place in the biologics arm of drug discovery. Importantly, here we create early lead therapeutic biologics for one of the most urgent clinical needs in cancer presented today.
Experimental procedures
Protein preparation
Target proteins were expressed in pGEX-2T (GE Healthcare) as GST fusion proteins. Constructs expressing the ACK and WASP GBDs, Cdc42 Δ7 Q61L, Cdc42 Δ7, Rac1 Δ7 Q61L, RhoA Δ12 Q63L, RalA Δ21 Q72L, and RalB Δ21 Q72L have been previously described (55–57). Human K-Ras 4B (residues 1–169) was amplified by PCR and cloned into pGEX-6P (GE Healthcare) using BamHI and EcoRI sites that had been incorporated into the PCR primers. A Q61L mutation was introduced using QuikChange Lighting mutagenesis (Agilent), following the manufacturer's instructions. Recombinant proteins were expressed in Escherichia coli, affinity-purified on GSH-agarose resin (Sigma), and cleaved (except for GST ACK/WASP) from GST using thrombin (Novagen) or C3 protease. Cleaved proteins were purified by gel filtration (S75 16/60, GE Healthcare), and GST ACK/WASP was purified by ion exchange as described previously (18, 37). Nucleotide exchange reactions were performed for small G proteins to add the nonhydrolyzable GTP analogue, GMPPNP, or tritiated GTP as described previously (18). Cdc42 was biotinylated for use in CIS display. EZ-link Sulfo-NHS–biotin (Thermo Fisher Scientific) was incubated for 30 min at room temperature with Cdc42 at a molar ratio of 20:1. Reactions were performed in PBS according to manufacturer's instructions, and the protein was recovered from the reaction mix using PD10 desalting columns (GE Healthcare).
CIS display
Three naïve library populations were created by mixing 10 random peptide encoding libraries generated from oligonucleotide primers purchased from Ella Biotech GmbH. The variant regions of these primers were built from DNA trimers designed to give equal amino acid frequencies (with the exclusion of cysteine) and optimized for expression in E. coli. Linear libraries comprised varying lengths of contiguous random amino acids, whereas cyclic libraries followed a sequence pattern of X3CXnCX3, where X denotes random amino acid representation (excluding Cys), and n was defined by the library length. The C1 maturation library conserved the cysteine residues for cyclic linkage but introduced biased mutagenesis at all other nucleotide positions, with 79% parental conservation and 7% incorporation of each nonparental nucleotide. CIS display construct synthesis, and biopanning reactions were performed as described previously (19). For heat elutions, DNA was eluted from streptavidin beads by heating to 75 °C for 10 min, whereas in competitive elutions, beads were incubated for 1 h in 100 μl of PBS + 2% BSA + 50 μm ACK GBD, and the supernatant was recovered. Four rounds of biopanning were performed for each selection and for the maturation campaign, before library analysis by next generation sequencing using the MiSeq Desktop Sequencer (Illumina).
Phage ELISAs
Library DNA from selections and pHEN-1 phagemid vector were digested with NcoI and NotI, ligated together, and transformed into TG1 E. coli cells (Lucigen). Transformants were picked at random, and 30 μl of overnight culture derived from individual colonies was used to seed cultures in 300 μl of 2TY, 2% glucose, 100 μg/ml ampicillin, which were incubated at 37 °C with shaking to OD600 = 0.3. 108 pfu of M13K07 helper phage in culture medium were added to each well, and the cultures were incubated for 1 h at 37 °C without shaking. Cells were centrifuged, the supernatant discarded, and bacteria resuspended in 400 μl of 2TY, 100 μg/ml ampicillin, 25 μg/ml kanamycin per well, and incubated overnight at 37 °C with shaking to allow phage expression. The recombinant phage were recovered in the supernatant after centrifugation for screening.
NUNC Maxisorp plates were coated overnight with streptavidin (Pierce) at 250 ng per well for assays using biotinylated Cdc42. Assay plates for specificity tests were coated directly with 500 ng per well, recombinant small GTPases, or BSA. Wells were washed in PBS and blocked in 1% BSA in PBS for 2 h at 20 °C. The streptavidin plates were then incubated with biotinylated Cdc42·GMPPNP (100 ng per well for 30 min), and the wells were subsequently washed in PBS.
In parallel, the phage were blocked in PBS containing 1% BSA for 2 h and then added to target-coated ELISA plate and shaken for 1–2 h at room temperature. Plates were washed (three times in PBS/Tween 0.1%, two times in PBS) to remove unbound phage and then incubated with anti-M13/HRP antibody conjugate (B62-FE2, Abcam: 1:5000 dilution in PBS containing 2% BSA). Plates were washed as before, and 3,3',5,5'-Tetramethylbenzidine (eBioscience) (50 μl per well) was added. Wells were quenched with 50 μl of 0.5 m H2SO4 after blue coloration was visible, and A450 was measured using a Multiskan EX (Thermo Fisher Scientific). DNA from phage clones was amplified by PCR and sequenced (Beckman Coulter).
Peptide synthesis
Peptides with an amidated C terminus were synthesized on a 10-μmol scale by standard Fmoc solid-phase chemistry on an automated peptide synthesizer (MultiPep RS, Intavis, Germany). Following the final Fmoc deprotection, the peptide N terminus was acetylated (4% acetic anhydride, 16% N-methylpyrrolidone in dimethyl fluoride (DMF) for 1 h), biotinylated (12 eq of biotin, 12 eq of hexafluorophosphate benzotriazole tetramethyl uronium (HBTU), 5% N-methylmorpholine (NMM) in DMF overnight), or 5(6)-FAM–labeled (5 eq of 5(6)-FAM, 5 eq of HBTU, 10 eq of NMM in DMF overnight). Resin was extensively washed with dichloromethane and methanol and dried overnight. After deprotection and cleavage from the resin (TentaGel, Intavis, Germany) with 5% water, 5% phenol, 5% thioanisole, 2.5% ethanedithiol in TFA for 3 h, the peptides were precipitated in diethyl ether at −20 °C. The crude peptides were purified by reverse-phase HPLC (Shimadzu) using a C18 column (50 × 250 mm, SunFire, Waters) and a linear gradient elution of 20–50% acetonitrile in 0.1% TFA. The molecular masses were determined by MALDI-TOF-TOF mass spectrometer (AXIMA Assurance, Shimadzu). Nona-arginine–linked peptides were purchased from Cambridge Research Biochemicals.
SPAs
30 nm GST WT-ACK GBD was immobilized on protein A SPA fluoromicrospheres (PerkinElmer Life Sciences) via an anti-GST antibody (Sigma). 30 nm [3H]GTP·Cdc42 was added, and the effect of competition was measured by measuring the signal in the presence of increasing concentrations of the peptide under test. Scintillation counts were measured at each concentration and fitted to the appropriate binding isotherm as described previously (58).
Fluorescence polarization
FP experiments were measured on a BMG Labtech Pherastar fluorimeter at 298 K with excitation 485 nm and emission 520 nm. Solutions were made up in black flat-bottom 384-well plates (Corning) to a final volume of 30 μl. Anisotropy was calculated from polarization measurements in MARS analysis software and data fitted to binding isotherms in R (59). Direct binding data were fitted using Equation 1, whereas the competition data were fitted to a previously described equation (58).
| (Eq. 1) |
where y equals anisotropy; Smax equals the maximum binding signal; Smin equals the minimum binding signal; C is the target concentration, and NSB is the nonspecific binding component. For direct binding assays, a concentration range of small GTPase was added to FAM-labeled peptides (60 nm), and in competition assays dilutions of unlabeled C1 peptide was added to 60 nm FAM-C1 and 500 nm Cdc42·GMPPNP.
NMR spectroscopy and structure calculations
15N HSQC experiments were recorded on a Bruker DRX500 at 298 K in 50 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 5 mm DTT, 150 mm NaCl, 10% D2O. For the titration experiments, 0.25, 0.5, 1, and 1.25 eq of peptide were added to 15N-labeled Cdc42, and spectra were recorded after each peptide addition. Overall chemical shift perturbations, δ, were calculated using Eq. 2,
| (Eq. 2) |
where δ1H and δ15N are the chemical shift changes for the 1H and 15N dimensions, respectively.
2D NOESY (250-ms mixing time), TOCSY (65-ms mixing), and DQF-COSY experiments were recorded on peptide P7 on a Bruker AV800 at 298 K in 50 mm sodium phosphate, pH 7, 10% D2O.
All NMR data were processed using the AZARA package (Wayne Boucher, University of Cambridge) and analyzed using CCPN ANALYSIS (60). Peptide structures were calculated using ARIA 1.2 (61) interfaced to CNS (62), where the ambiguity of the restraints was decreased during eight iterations. Cdc42 residues whose backbone amides had shifted by more than 0.043 (the average shift change) and that were more than 40% solvent-exposed in either of two structures (according to NACCESS (63)) were used as active residues in HADDOCK (38, 39). HADDOCK was also run including only residues whose backbone amides had shifted by more than 0.090 (the average shift change plus 1 S.D.). In all cases, residues whose peaks had shifted too far to be reliably assigned were included as active residues. The Cdc42 structures used were those from complexes with Par6 (PDB code 1nf3) or IRSp53 (PDB code 4js0), where the effector was removed in each case. HADDOCK was run twice for each set of active residues, once with each of these structures as a starting point. In all HADDOCK runs, the top cluster showed the peptide binding in the same orientation on the same surface of Cdc42. The active Cdc42 residues were as follows (after filtering for solvent exposure): shifted by more than average +1 S.D., 25, 46, 49, 164; shifted by between average and average +1 S.D., 3, 12, 26, 62, 76, 116, 153, 167, 177, 178; and shifted too far for unambiguous assignment to be possible, 24, 30, 43, 48, 52, 160, 162, 166. Passive residues were defined automatically by HADDOCK. Active residues for the peptide were defined as 8 and 10, with all other peptide residues being passive.
Immunoblotting
Lysates were taken from 2 × 6-cm dishes in 100 μl of lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm sodium orthovanadate, 1 mm β-glycerophosphate disodium salt hydrate, 1× protease inhibitor mixture (Mammalian, Sigma), 1% Triton X-100) and centrifuged at 17,000 × g for 20 min. Cell samples were resolved by SDS-PAGE and immunoblotted using the following primary antibodies: α-Stat3 (F-2) sc-1019; α-Stat3 (Y705) sc-7993 (Santa Cruz Biotechnology); α-GAPDH–HRP ab9482 (Abcam), α-Erk1/2 9107S, α-pErk1/2 4376S (Cell Signaling Technology).
CETSA
For CETSA lysate experiments, A549 cells were trypsinized, washed in PBS, and resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 5 mm MgCl2, 1× protease inhibitor mixture (Mammalian, Sigma)) at 60,000 cells/50 μl. Cells were lysed by three freeze-thaw cycles in liquid nitrogen and centrifuged. Supernatant was divided into 50-μl aliquots, which were exposed to varying concentrations of peptide or temperatures. Cell lysates were first heated over a wide range of temperatures to determine the melting temperature of the protein under investigation. Samples were heated for 3 min in a Veriti 96-well thermal cycler (Applied Biosystems), cooled for 3 min, and then centrifuged. Supernatants were analyzed by Western blotting. Lysates were then tested over a narrower range of temperatures below the melting temperature, +/− a fixed concentration of the peptide under test, and a temperature was identified where a peptide effect was apparent; this temperature was set as the experimental temperature. Finally, lysates were tested at the experimental temperature with a range of peptide concentrations. For whole-cell experiments, 600,000 A549 cells were seeded in 12-well culture plates and incubated for 3.5 h. Cells were trypsinized, collected, and washed in PBS. Samples were heated for 3 min as above, and 30 μl of lysis buffer were added (25 mm Tris-HCl, pH 7.5, 5 mm β-glycerophosphate, 2 mm DTT, 0.1 mm sodium vanadate, 10 mm MgCl2) and then subjected to two freeze-thaw cycles before centrifugation. The supernatant was analyzed by Western blotting. An experimental temperature was determined as above. Cells were then plated and incubated with a range of peptide concentrations for 3.5 h and analyzed as above.
GST pulldown assays
GSH-Sepharose (GE Healthcare) in PBS was incubated for 15 min at 4 °C with GST–WASP and then washed three times in PBS, 0.1% Tween. 25 μg of GST–WASP attached to beads was incubated with 100 μl of A549 lysate for 45 min in the presence of varying peptide concentrations. Beads were washed twice in PBS, 0.1% Tween. Samples were resolved by SDS-PAGE and immunoblotted using an anti-Cdc42 antibody (610928, BD Biosciences). Western blottings were quantified using ImageJ, and the data were fitted to Equation 3 to calculate the IC50 values.
| (Eq. 3) |
Proliferation assays
100 μl of A549 cells were seeded in 96-well culture plates (CellStar) at 50,000 cells/ml and allowed to settle overnight. Peptides were dosed in 4 μl of PBS every 24 h. After 48 h, 10 μl of MTT (Invitrogen, 5 mg/ml in PBS) was added, and the plates were incubated for 2–3 h. Medium was removed, and 100 μl of DMSO was added to solubilize the contents. After 4 h, the A570 absorbance was measured for each well in a SoftMax Pro 5 (Molecular Devices).
Boyden chamber assays
Upper chambers of Boyden chambers (BRAND) were seeded with 50,000 A549 cells in 100 μl of RPMI medium, 0.1% FCS with 1 μm peptide doses. The bottom chamber was filled with RPMI medium, 10% FCS, and the plates incubated for 5.5 h at 37 °C. Cells and media were removed from the top chamber using cotton buds, and the media in the bottom well was replaced with trypsin and incubated for 10 min at 37 °C. The cells were removed and centrifuged, and the cell pellets were resuspended in 20 μl of PBS. Cell suspensions were counted on the Countess automated cell counter (Invitrogen), and data were analyzed and plotted in R.
Wound-healing assays
Three-well microdishes (Ibidi) were seeded with 20,000 A549 cells in 100 μl of RPMI medium/well and allowed to settle overnight. Wells were dosed with 1 μm peptide and incubated for 3 h. 0.5 ml of medium with 1 μm peptide was added to the outside of the dishes, and the inserts were removed. Images were taken at various time points using an EVOS fl digital inverted microscope (AMG) at ×4 magnification. Images were cropped to 680 by 512 pixels (width ∼1 mm), and the scratch area was analyzed using the MiToBo package in Fiji (64, 65). Data were processed in R.
Confocal microscopy
Cells were seeded at 50,000/ml in micro-insert μ-dishes (Ibidi) and incubated overnight. Peptides were added in fresh medium, and the cells were incubated for the respective time points. Cells were washed four times with PBS and imaged in imaging medium (H0887, Sigma) in a stage top heated chamber (Okolabs) at 37 °C. Confocal images were acquired on an inverted Ti-E microscope (Nikon) with a spinning-disk (X-light Nikpow, Crest), 250-μm Lunencor Spectra X LED illumination, and a piezo-driven Z stage controller (NanoScanZ) using a ×100 1.40 oil objective lens (Plan Apo VC, Nikon). Images were collected with an EMCDD camera (Evolve Delta, Photometrics) with Metamorph software (version 7.8.2.0). 5(6)-FAM samples were visualized using a 470/40 excitation and 525/50 emission filters, and images were processed in Fiji (64, 65).
Author contributions
G. J. N. T., N. P. M., S. B., G. I.-B., H. R. M., R. N. C., and D. O. formal analysis; G. J. N. T., N. P. M., S. B., G. I.-B., H. R. M., and R. N. C. investigation; G. J. N. T., H. R. M., R. N. C., and D. O. writing-original draft; G. J. N. T., N. P. M., S. B., G. I.-B., J. R., H. R. M., R. N. C., and D. O. writing-review and editing; J. R., H. R. M., R. N. C., and D. O. supervision; H. R. M., R. N. C., and D. O. conceptualization; H. R. M., R. N. C., and D. O. funding acquisition; D. O. project administration.
Supplementary Material
Acknowledgments
We are extremely grateful to Dr. Chris Ullman and Dr. Kevin Matthews for their belief in peptide therapeutics and their support of this project. We thank Dr. Cathy Wilson for expert assistance and advice in producing MEFs for this work. We are also indebted to Dr. Jenny Gallop and Dr. Jonathan Gadsby for help in performing confocal experiments.
This work was supported by MRC CASE Studentship MR/K017101/1 (to D. O. and R. N. C.), BBSRC DTP iCASE Studentship Grant BB/M011194/1 (to D. O. and J. R.), and a short-term Glover Research Fund Fellowship (to G. J. N. T.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
The atomic coordinates and structure factors (code 6R28) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The NMR chemical shift data of this paper are available from the Biological Magnetic Resonance Data Bank under BMRB accession number 34380.
D. Owen, unpublished results.
- CRIB
- Cdc42/Rac-interactive binding domain
- ACK
- activated Cdc42 kinase
- CETSA
- cellular thermal shift assay
- CPP
- cell-penetrating peptide
- FAM
- carboxyfluorescein
- FP
- fluorescence polarization
- GBD
- G protein-binding domain
- FCS
- fetal calf serum
- GST
- glutathione S-transferase
- MEF
- mouse embryonic fibroblast
- PAK
- p21-activated kinase
- SPA
- scintillation proximity assay
- WASP
- Wiskott-Aldrich syndrome protein
- 9R
- nona-arginine
- GMPPNP
- guanosine-5′-[(β,γ)-imido]triphosphate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- HSQC
- heteronuclear single quantum coherence
- DMF
- dimethyl fluoride
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- NMM
- N-methylmorpholine
- HBTU
- hexafluorophosphate benzotriazole tetramethyl uronium
- RMSD
- root mean square deviation
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- MAPK/ERK kinase.
References
- 1. Li S., Balmain A., and Counter C. M. (2018) A model for RAS mutation patterns in cancers: finding the sweet spot. Nat. Rev. Cancer 18, 767–777 10.1038/s41568-018-0076-6 [DOI] [PubMed] [Google Scholar]
- 2. Ostrem J. M., and Shokat K. M. (2016) Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15, 771–785 10.1038/nrd.2016.139 [DOI] [PubMed] [Google Scholar]
- 3. Spencer-Smith R., and O'Bryan J. P. (2019) Direct inhibition of RAS: Quest for the Holy Grail? Semin. Cancer Biol. 54, 138–148 10.1016/j.semcancer.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mott H. R., and Owen D. (2019) Bioblockades join the assault on small G protein signaling. Semin. Cancer Biol. 54, 149–161 10.1016/j.semcancer.2018.01.001 [DOI] [PubMed] [Google Scholar]
- 5. Ridley A. J. (2015) Rho GTPase signaling in cell migration. Curr. Opin. Cell Biol. 36, 103–112 10.1016/j.ceb.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arias-Romero L. E., and Chernoff J. (2013) Targeting Cdc42 in cancer. Expert Opin. Ther. Targets 17, 1263–1273 10.1517/14728222.2013.828037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stengel K. R., and Zheng Y. (2012) Essential role of Cdc42 in Ras-induced transformation revealed by gene targeting. PLoS ONE 7, e37317 10.1371/journal.pone.0037317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fritz G., Brachetti C., Bahlmann F., Schmidt M., and Kaina B. (2002) Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br. J. Cancer 87, 635–644 10.1038/sj.bjc.6600510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tucci M. G., Lucarini G., Brancorsini D., Zizzi A., Pugnaloni A., Giacchetti A., Ricotti G., and Biagini G. (2007) Involvement of E-cadherin, β-catenin, Cdc42 and CXCR4 in the progression and prognosis of cutaneous melanoma. Br. J. Dermatol. 157, 1212–1216 10.1111/j.1365-2133.2007.08246.x [DOI] [PubMed] [Google Scholar]
- 10. Liu Y., Wang Y., Zhang Y., Miao Y., Zhao Y., Zhang P. X., Jiang G. Y., Zhang J. Y., Han Y., Lin X. Y., Yang L. H., Li Q. C., Zhao C., and Wang E. H. (2009) Abnormal expression of p120-catenin, E-cadherin, and small GTPases is significantly associated with malignant phenotype of human lung cancer. Lung Cancer 63, 375–382 10.1016/j.lungcan.2008.12.012 [DOI] [PubMed] [Google Scholar]
- 11. Lin R., Cerione R. A., and Manor D. (1999) Specific contributions of the small GTPases Rho, Rac, and Cdc42 to Dbl transformation. J. Biol. Chem. 274, 23633–23641 10.1074/jbc.274.33.23633 [DOI] [PubMed] [Google Scholar]
- 12. Hamann M. J., Lubking C. M., Luchini D. N., and Billadeau D. D. (2007) Asef2 functions as a Cdc42 exchange factor and is stimulated by the release of an autoinhibitory module from a concealed C-terminal activation element. Mol. Cell. Biol. 27, 1380–1393 10.1128/MCB.01608-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Durkin M. E., Yuan B. Z., Zhou X., Zimonjic D. B., Lowy D. R., Thorgeirsson S. S., and Popescu N. C. (2007) DLC-1: a Rho GTPase-activating protein and tumour suppressor. J. Cell Mol. Med. 11, 1185–1207 10.1111/j.1582-4934.2007.00098.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pirone D. M., Carter D. E., and Burbelo P. D. (2001) Evolutionary expansion of CRIB-containing Cdc42 effector proteins. Trends Genet. 17, 370–373 10.1016/S0168-9525(01)02311-3 [DOI] [PubMed] [Google Scholar]
- 15. Nur-E-Kamal M. S., Kamal J. M., Qureshi M. M., and Maruta H. (1999) The CDC42-specific inhibitor derived from ACK-1 blocks v-Ha-Ras–induced transformation. Oncogene 18, 7787–7793 10.1038/sj.onc.1203215 [DOI] [PubMed] [Google Scholar]
- 16. Mott H. R., and Owen D. (2015) Structures of Ras superfamily effector complexes: what have we learnt in two decades? Crit. Rev. Biochem. Mol. Biol. 50, 85–133 10.3109/10409238.2014.999191 [DOI] [PubMed] [Google Scholar]
- 17. Mott H. R., Owen D., Nietlispach D., Lowe P. N., Manser E., Lim L., and Laue E. D. (1999) Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK. Nature 399, 384–388 10.1038/20732 [DOI] [PubMed] [Google Scholar]
- 18. Tetley G. J. N., Mott H. R., Cooley R. N., and Owen D. (2017) A dock and coalesce mechanism driven by hydrophobic interactions governs Cdc42 binding with its effector protein ACK. J. Biol. Chem. 292, 11361–11373 10.1074/jbc.M117.789883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Odegrip R., Coomber D., Eldridge B., Hederer R., Kuhlman P. A., Ullman C., FitzGerald K., and McGregor D. (2004) CIS display: in vitro selection of peptides from libraries of protein-DNA complexes. Proc. Natl. Acad. Sci. U.S.A. 101, 2806–2810 10.1073/pnas.0400219101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCormick F. (2018) Targeting KRAS directly. Annu. Rev. Cancer Biol. 2, 81–90 10.1146/annurev-cancerbio-050216-122010 [DOI] [Google Scholar]
- 21. Vetter I. R., and Wittinghofer A. (2001) Signal transduction–the guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- 22. Stephen A. G., Esposito D., Bagni R. K., and McCormick F. (2014) Dragging Ras back in the ring. Cancer Cell 25, 272–281 10.1016/j.ccr.2014.02.017 [DOI] [PubMed] [Google Scholar]
- 23. Chakraborty A., Linnane E., and Ross S. (2018) Ras proteins as therapeutic targets. Biochem. Soc. Trans. 46, 1303–1311 10.1042/BST20170529 [DOI] [PubMed] [Google Scholar]
- 24. Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., and Sugiura Y. (2001) Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 276, 5836–5840 10.1074/jbc.M007540200 [DOI] [PubMed] [Google Scholar]
- 25. Wang K., Qi J., Weng T., Tian Z., Lu Y., Hu K., Yin Z., and Wu W. (2014) Enhancement of oral bioavailability of cyclosporine A: comparison of various nanoscale drug-delivery systems. Int. J. Nanomedicine 9, 4991–4999 10.2147/IJN.S72560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heard K. R., Wu W., Li Y., Zhao P., Woznica I., Lai J. H., Beinborn M., Sanford D. G., Dimare M. T., Chiluwal A. K., Peters D. E., Whicher D., Sudmeier J. L., and Bachovchin W. W. (2013) A general method for making peptide therapeutics resistant to serine protease degradation: application to dipeptidyl peptidase IV substrates. J. Med. Chem. 56, 8339–8351 10.1021/jm400423p [DOI] [PubMed] [Google Scholar]
- 27. Werle M., and Bernkop-Schnürch A. (2006) Strategies to improve plasma half-life time of peptide and protein drugs. Amino Acids 30, 351–367 10.1007/s00726-005-0289-3 [DOI] [PubMed] [Google Scholar]
- 28. Martinek T. A., and Fülöp F. (2012) Peptidic foldamers: ramping up diversity. Chem. Soc. Rev. 41, 687–702 10.1039/C1CS15097A [DOI] [PubMed] [Google Scholar]
- 29. Walensky L. D., and Bird G. H. (2014) Hydrocarbon-stapled peptides: principles, practice, and progress. J. Med. Chem. 57, 6275–6288 10.1021/jm4011675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Henninot A., Collins J. C., and Nuss J. M. (2018) The current state of peptide drug discovery: back to the future? J. Med. Chem. 61, 1382–1414 10.1021/acs.jmedchem.7b00318 [DOI] [PubMed] [Google Scholar]
- 31. Tünnemann G., Ter-Avetisyan G., Martin R. M., Stöckl M., Herrmann A., and Cardoso M. C. (2008) Live-cell analysis of cell penetration ability and toxicity of oligo-arginines. J. Pept. Sci. 14, 469–476 10.1002/psc.968 [DOI] [PubMed] [Google Scholar]
- 32. Räägel H., Säälik P., and Pooga M. (2010) Peptide-mediated protein delivery–which pathways are penetrable? Biochim. Biophys. Acta 1798, 2240–2248 10.1016/j.bbamem.2010.02.013 [DOI] [PubMed] [Google Scholar]
- 33. LeCher J. C., Nowak S. J., and McMurry J. L. (2017) Breaking in and busting out: cell-penetrating peptides and the endosomal escape problem. Biomol. Concepts 8, 131–141 10.1515/bmc-2017-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tuveson D. A., Shaw A. T., Willis N. A., Silver D. P., Jackson E. L., Chang S., Mercer K. L., Grochow R., Hock H., Crowley D., Hingorani S. R., Zaks T., King C., Jacobetz M. A., Wang L., et al. (2004) Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 5, 375–387 10.1016/S1535-6108(04)00085-6 [DOI] [PubMed] [Google Scholar]
- 35. Mahendrarajah N., Paulus R., and Krämer O. H. (2016) Histone deacetylase inhibitors induce proteolysis of activated CDC42-associated kinase-1 in leukemic cells. J. Cancer Res. Clin. Oncol. 142, 2263–2273 10.1007/s00432-016-2229-x [DOI] [PubMed] [Google Scholar]
- 36. Mahendrarajah N., Borisova M. E., Reichardt S., Godmann M., Sellmer A., Mahboobi S., Haitel A., Schmid K., Kenner L., Heinzel T., Beli P., and Krämer O. H. (2017) HSP90 is necessary for the ACK1-dependent phosphorylation of STAT1 and STAT3. Cell Signal. 39, 9–17 10.1016/j.cellsig.2017.07.014 [DOI] [PubMed] [Google Scholar]
- 37. Tetley G. J. N., Szeto A., Fountain A. J., Mott H. R., and Owen D. (2018) Bond swapping from a charge cloud allows flexible co-ordination of upstream signals through WASP: multiple regulatory roles for the WASP basic region. J. Biol. Chem. 293, 15136–15151 10.1074/jbc.RA118.003290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dominguez C., Boelens R., and Bonvin A. (2003) HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 10.1021/ja026939x [DOI] [PubMed] [Google Scholar]
- 39. van Zundert G. C. P., Rodrigues J. P. G. L. M., Trellet M., Schmitz C., Kastritis P. L., Karaca E., Melquiond A. S. J., van Dijk M., de Vries S. J., and Bonvin A. M. J. J. (2016) The HADDOCK2.2 Web Server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 10.1016/j.jmb.2015.09.014 [DOI] [PubMed] [Google Scholar]
- 40. Martinez Molina D., and Nordlund P. (2016) The cellular thermal shift assay: a novel biophysical assay for in situ drug target engagement and mechanistic biomarker studies. Annu. Rev. Pharmacol. Toxicol. 56, 141–161 10.1146/annurev-pharmtox-010715-103715 [DOI] [PubMed] [Google Scholar]
- 41. Lawson C. D., and Ridley A. J. (2018) Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 217, 447–457 10.1083/jcb.201612069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. López-Mirabal H. R., and Winther J. R. (2008) Redox characteristics of the eukaryotic cytosol. Biochim. Biophys. Acta 1783, 629–640 10.1016/j.bbamcr.2007.10.013 [DOI] [PubMed] [Google Scholar]
- 43. Gilbert H. F. (1995) in Biothiols, Pt. A: Monothiols and Dithiols, Protein Thiols, and Thiyl Radicals (Packer L., ed) pp. 8–28, Academic Press, San Diego [Google Scholar]
- 44. Austin C. D., Wen X., Gazzard L., Nelson C., Scheller R. H., and Scales S. J. (2005) Oxidizing potential of endosomes and lysosomes limits intracellular cleavage of disulfide-based antibody-drug conjugates. Proc. Natl. Acad. Sci. U.S.A. 102, 17987–17992 10.1073/pnas.0509035102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Santra S., Kaittanis C., Santiesteban O. J., and Perez J. M. (2011) Cell-specific, activatable, and theranostic prodrug for dual-targeted cancer imaging and therapy. J. Am. Chem. Soc. 133, 16680–16688 10.1021/ja207463b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Galande A. K., and Spatola A. F. (2001) A facile method for the direct synthesis of lanthionine containing cyclic peptides. Letters in Peptide Science 8, 247–251 10.1023/A:1016285331403 [DOI] [Google Scholar]
- 47. Owen D., and Mott H. R. (2018) CRIB effector disorder: exquisite function from chaos. Biochem. Soc. Trans. 46, 1289–1302 10.1042/BST20170570 [DOI] [PubMed] [Google Scholar]
- 48. Buck M., Xu W., and Rosen M. K. (2004) A two-state allosteric model for autoinhibition rationalizes WASP signal integration and targeting. J. Mol. Biol. 338, 271–285 10.1016/j.jmb.2004.02.036 [DOI] [PubMed] [Google Scholar]
- 49. Cimmperman P., Baranauskiene L., Jachimoviciüte S., Jachno J., Torresan J., Michailoviene V., Matuliene J., Sereikaite J., Bumelis V., and Matulis D. (2008) A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys. J. 95, 3222–3231 10.1529/biophysj.108.134973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lim Y. T., Prabhu N., Dai L., Go K. D., Chen D., Sreekumar L., Egeblad L., Eriksson S., Chen L., Veerappan S., Teo H. L., Tan C. S. H., Lengqvist J., Larsson A., Sobota R. M., and Nordlund P. (2018) An efficient proteome-wide strategy for discovery and characterization of cellular nucleotide-protein interactions. PLoS ONE 13, e0208273 10.1371/journal.pone.0208273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qian Z., Martyna A., Hard R. L., Wang J., Appiah-Kubi G., Coss C., Phelps M. A., Rossman J. S., and Pei D. (2016) Discovery and mechanism of highly efficient cyclic cell-penetrating peptides. Biochemistry 55, 2601–2612 10.1021/acs.biochem.6b00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hopkins A. L. (2008) Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 4, 682–690 10.1038/nchembio.118 [DOI] [PubMed] [Google Scholar]
- 53. Zang M., Hayne C., and Luo Z. (2002) Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J. Biol. Chem. 277, 4395–4405 10.1074/jbc.M110000200 [DOI] [PubMed] [Google Scholar]
- 54. Park E. R., Eblen S. T., and Catling A. D. (2007) MEK1 activation by PAK: a novel mechanism. Cell Signal. 19, 1488–1496 10.1016/j.cellsig.2007.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Owen D., Mott H. R., Laue E. D., and Lowe P. N. (2000) Residues in Cdc42 that specify binding to individual CRIB effector proteins. Biochemistry 39, 1243–1250 10.1021/bi991567z [DOI] [PubMed] [Google Scholar]
- 56. Owen D., Lowe P. N., Nietlispach D., Brosnan C. E., Chirgadze D. Y., Parker P. J., Blundell T. L., and Mott H. R. (2003) Molecular dissection of the interaction between the small G proteins Rac1 and RhoA and protein kinase C-related kinase 1 (PRK1). J. Biol. Chem. 278, 50578–50587 10.1074/jbc.M304313200 [DOI] [PubMed] [Google Scholar]
- 57. Campbell L. J., Peppa M., Crabtree M. D., Shafiq A., McGough N. F. ott H. R., and Owen D. (2015) Thermodynamic mapping of effector protein interfaces with RalA and RalB. Biochemistry 54, 1380–1389 10.1021/bi501530u [DOI] [PubMed] [Google Scholar]
- 58. Elliot-Smith A. E., Owen D., Mott H. R., and Lowe P. N. (2007) Double mutant cycle thermodynamic analysis of the hydrophobic Cdc42–ACK protein–protein interaction. Biochemistry 46, 14087–14099 10.1021/bi701539x [DOI] [PubMed] [Google Scholar]
- 59. R Core Team (2018) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 60. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M, Ulrich E. L., Markley J. L., Ionides J., and Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- 61. Linge J. P., O'Donoghue S. I., and Nilges M. (2001) Automated assignment of ambiguous NOEs with ARIA. Methods Enzymol. 339, 71–90 10.1016/S0076-6879(01)39310-2 [DOI] [PubMed] [Google Scholar]
- 62. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography and NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- 63. Hubbard S. J., and Thornton J. M. (1993) NACCESS. Department of Biochemistry and Molecular Biology, University College London [Google Scholar]
- 64. Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T., and Eliceiri K. W. (2017) ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.