Abstract
Understanding the mechanisms by which viruses evade host cell immune defenses is important for developing improved antiviral therapies. In an unusual twist, human cytomegalovirus co-opts the antiviral radical SAM enzyme viperin (virus-inhibitory protein, endoplasmic reticulum–associated, interferon-inducible) to enhance viral infectivity. This process involves translocation of viperin to the mitochondrion, where it binds the β-subunit (HADHB) of the mitochondrial trifunctional enzyme complex that catalyzes thiolysis of β-ketoacyl–CoA esters as part of fatty acid β-oxidation. Here we investigated how the interaction between these two enzymes alters their activities and affects cellular ATP levels. Experiments with purified enzymes indicated that viperin inhibits the thiolase activity of HADHB, but, unexpectedly, HADHB activates viperin, leading to synthesis of the antiviral nucleotide 3′-deoxy-3′,4′-didehydro-CTP. Measurements of enzyme activities in lysates prepared from transfected HEK293T cells expressing these enzymes mirrored the findings obtained with purified enzymes. Thus, localizing viperin to mitochondria decreased thiolase activity, and coexpression of HADHB significantly increased viperin activity. Furthermore, targeting viperin to mitochondria also increased the rate at which HADHB is retrotranslocated out of mitochondria and degraded, providing an additional mechanism by which viperin reduces HADHB activity. Targeting viperin to mitochondria decreased cellular ATP levels by more than 50%, consistent with the enzyme disrupting fatty acid catabolism. These results provide biochemical insight into the mechanism by which human cytomegalovirus subverts viperin; they also provide a biochemical rationale for viperin's recently discovered role in regulating thermogenesis in adipose tissues.
Keywords: enzyme inhibitor, enzyme degradation, metabolic regulation, metalloenzyme, mitochondria, innate immunity, fatty acid oxidation, cytomegalovirus, HADHB thiolase, mitochondrial trifunctional protein, radical SAM enzyme, viperin
Introduction
Viruses and cells have coevolved so that the increasingly sophisticated antiviral defenses of host cells have been met with equally ingenious mechanisms by which viruses evade or neutralize them (1–5). The innate immune system provides the primary line of defense against viruses and other pathogens, with type 1 IFN mediating the induction of over 300 IFN-stimulated genes that collectively act to disrupt viral replication and transmission (6–9). Viruses have evolved multiple strategies to evade various components of the innate immune system, which often appear to be virus-specific; these include binding cellular signaling proteins to prevent the expression of interferon-stimulated genes, proteolytic degradation or deubiquitination of cellular proteins by virally encoded proteases and deubiquitinases, and sequestering viral nucleic acids within cellular membranes (1, 3, 5, 6, 10, 11). A better understanding of the diverse mechanisms by which viruses evade the immune system promises to lead to more effective antiviral drugs.
Here we investigated one of the more unusual examples of a virus co-opting the cellular antiviral machinery to enhance infectivity. This involves human cytomegalovirus (HCMV),2 which co-opts the antiviral protein viperin (virus-inhibitory protein, endoplasmic reticulum–associated, interferon-inducible; also called RSAD2 or CIG5 in humans), to facilitate release of the virus from the cell (12, 13).
Viperin appears to be conserved in all animals (12, 14, 15). Viperin-like enzymes have also been reported in fungi and archaea, indicating that it is an ancient component of the antiviral response (16, 17). Viperin is a radical SAM enzyme, (18) one of only eight annotated in the human genome (19). Radical SAM enzymes are predominately found in microbes, where they catalyze a remarkably wide range of reactions that are initiated by reductive cleavage of SAM to produce adenosyl radical (20–32). Viperin has been shown recently to catalyze the formation of 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP) through dehydration of CTP by a radical mechanism (33). This novel nucleotide acts as a chain-terminating inhibitor of viral RNA–dependent RNA polymerases that are essential for replication of flaviviruses (33).
Although the enzymatic activity of viperin explains its antiviral activity against dsRNA viruses such as flaviviruses, viperin has antiviral activity against a much wider range of RNA and DNA viruses (14). It is also involved in regulating innate immune signaling (34) and lipid metabolism (35–37). Intriguingly, very recent studies of mice in which viperin was knocked out have implicated the enzyme in regulating thermogenesis in adipose tissues through regulation of fatty acid β-oxidation (38).
These varied aspects of viperin's activity appear to result from interactions with a wide variety of other cellular and viral proteins. For example, we recently investigated viperin's role in innate immune signaling through the Toll-like receptor 7 and 9 (TLR7/9) signaling pathways (39). We found that viperin mediates formation of a complex between interleukin-1 receptor–associated kinase-1 (IRAK1) and the E3 ubiquitin ligase tumor necrosis factor receptor–associated factor 6 (TRAF6) and that this complex is necessary to observe Lys63-linked polyubiquitination of IRAK1 by TRAF6, a key step in the signaling pathway. However, although IRAK1 ubiquitination appeared to depend on structural changes induced by SAM binding to viperin, it did not require catalytically active viperin (39).
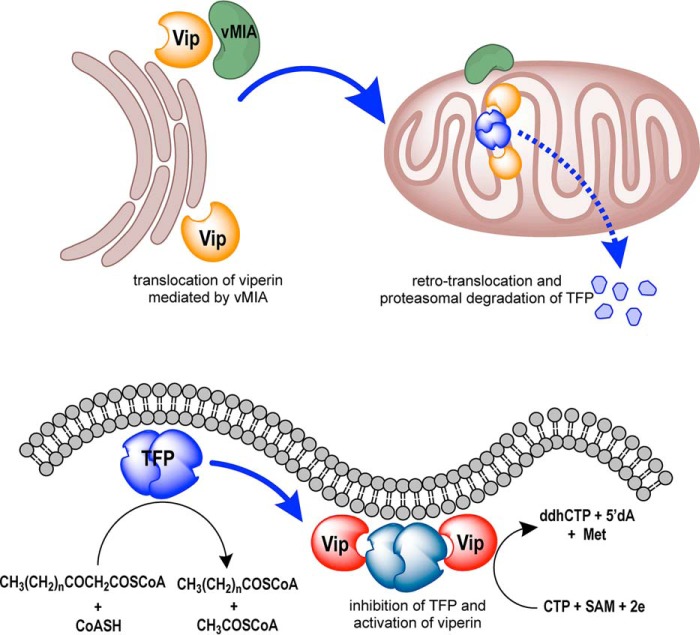
Previous studies have shown that, perhaps counterintuitively, HCMV directly induces expression of viperin upon infection (12, 13). Viperin is normally associated with the cytosolic face of the endoplasmic reticulum (ER), but the virally encoded protein mitochondrial inhibitor of apoptosis (vMIA) binds to viperin and causes it to be targeted to the mitochondrion (Fig. 1) (13, 40). In the mitochondrion, viperin binds to HADHB, the β-subunit of the mitochondrial trifunctional protein (41), a multienzyme complex that catalyzes the last three steps in the β-oxidation pathway of fatty acids. Targeting viperin to the mitochondrion disrupts the cellular pools of ATP and NADH, which, in turn, appears to disrupt the actin cytoskeleton (12, 13). This disruption is hypothesized to aid newly synthesized viruses in escaping from the cell.
Figure 1.
Overview illustrating how viperin affects the activity of the mitochondrial trifunctional protein when translocated to the mitochondrion by the cytomegalovirus protein vMIA. The HCMV protein vMIA binds to viperin (Vip) at the ER membrane and causes it to be translocated into the matrix of the mitochondrion. In the mitochondrion, viperin binds to the β-subunit of the mitochondrial trifunctional protein (TFP, HADHB). Viperin binding causes HADHB to be retrotranslocated from the mitochondrion and degraded by the proteasome. Binding also inhibits thiolase activity and prevents fatty acid oxidation from occurring. At the same time, binding HADHB activates viperin to synthesize ddhCTP.
However, the functional significance of viperin binding to HADHB remains unclear. Therefore, we investigated the interaction between HADHB and viperin using purified proteins in vitro and in transiently transfected HEK293T cell lines that express viperin tagged with a mitochondrial localization signal. Our results provide support for the hypothesis that HCMV co-opts viperin to inhibit HADHB and thereby facilitates escape of viral particles from the cell. They also shed light on the recent and intriguing observation that viperin is involved in regulating thermogenesis in murine adipose tissue (38).
Results
Viperin inhibits the thiolase activity of HADHB
The downstream effects on cellular physiology that result from vMIA translocating viperin to the mitochondrion have been attributed to inhibition of the fatty acid β-oxidation pathway (13). To test this hypothesis, we investigated the effect of viperin on the activity of HADHB in vitro. HADHB catalyzes the last step in β-oxidation, which is thiolytic cleavage of the β-ketoacyl–CoA ester by CoASH to produce acetyl-CoA and the corresponding N-2-acyl-CoA ester (41). This reaction may be conveniently followed by measuring the decrease in absorbance at 303 nm because of coordination of Mg2+ to the β-ketoacyl–CoA (42).
These experiments used a viperin construct lacking the first 50 residues corresponding to the N-terminal amphipathic helix, termed ΔN-viperin (35), as it has not proved possible to recombinantly express the full-length enzyme in active form. We also constructed an inactive mutant, ΔN-viperinC83A, in which one of the cysteine residues ligating the iron–sulfur cluster was mutated to alanine. The ΔN-viperin and ΔN-viperinC83A proteins were expressed and purified from Escherichia coli as described under “Experimental procedures.” Recombinantly expressed and purified HADHB was purchased from a commercial supplier (AdooQ Bioscience). Prior to experiments, the iron–sulfur cluster of ΔN-viperin was reconstituted under anaerobic conditions to provide active holoenzyme, as described previously (Fig. S1a) (35). As expected, the ΔN-viperinC83A was unable to bind an iron–sulfur cluster and was purified as the colorless apoenzyme (Fig. S1b).
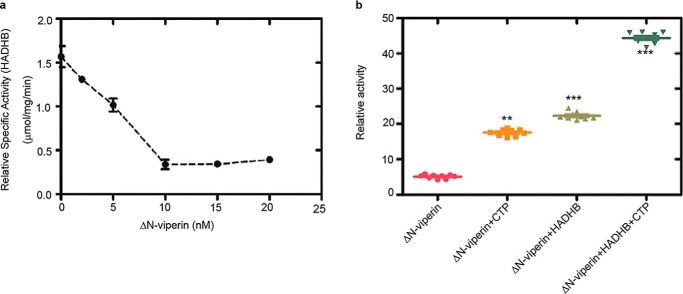
To investigate viperin's effect on HADHB thiolase activity, assays were conducted under anaerobic conditions because of the air sensitivity of viperin. Reactions were performed at 22 °C and contained 100 μm acetoacetyl-CoA, 100 μm CoASH, and 10 nm HADHB. Prior to the assay, HADHB was incubated with various concentrations of ΔN-viperin for 50 min. In the absence of viperin, the apparent kcat of HADHB was ∼ 2 s−1 under the conditions of the assay. As the concentration of viperin was increased in the assay, the thiolase activity of HADHB was progressively inhibited, with the activity of HADHB declining to a minimum of 15%–20% of the initial activity at a 1:1 ratio of HADHB to viperin (Fig. 2a and Fig. S2). Interestingly, increasing the concentration of viperin did not result in any further decrease in HADHB activity, suggesting that viperin does not act as a simple competitive inhibitor. In contrast, preincubation with ΔN-viperinC83A had no significant inhibitory effect on HADHB activity (Fig. S2), although, as discussed below, the mutant still bound HADHB.
Figure 2.
In vitro activity of HADHB and viperin. a, the thiolase activity of purified recombinant HADHB was assayed in the presence of increasing concentrations of ΔN-viperin. Maximal inhibition was observed at equimolar concentrations of the two enzymes. b, reductive SAM cleavage activity of purified recombinant ΔN-viperin, assayed in the absence or presence of HADHB and/or the cosubstrate CTP (100 μm). The uncoupled rate of SAM cleavage and the rate of SAM cleavage in the presence of CTP are significantly accelerated by HADHB. Values are presented as mean ± S.E. (n = 6). **, p < 0.005; ***, p < 0.001 (Student's t test for independent samples).
HADHB increases the activity of viperin
We have shown previously that the enzymatic activity of viperin increases significantly when it forms a complex with IRAK1and TRAF6 as part of the TLR7/9 innate immune signaling pathway (39). This observation prompted us to investigate whether HADHB might similarly activate viperin (Fig. 2b). Assays were conducted at 22 °C under anaerobic conditions as described under “Experimental procedures.” After 1 h, the reactions were quenched, and the amount of 5′-dA formed was measured by LC-MS. In the absence of HADHB, the apparent kcat of viperin was 2.2 ± 0.1 h−1, which is similar to values measured previously (33). However, in the presence of HADHB, the apparent kcat of viperin increased by about 2.5-fold to 4.4 ± 0.1 h−1 (Table S1). We also found that, in the absence of CTP, the presence of HADHB resulted in a substantial increase in the rate of uncoupled 5′-dA formation by viperin (Fig. 2b). The activation of viperin was specific to HADHB, as no increase in activity was observed for viperin in the presence of proteins such as BSA (Fig. S2c), which are often used to stabilize enzymes. It is interesting that, although IRAK1/TRAF6 and HADHB perform very different functions, both activate viperin to catalyze formation of ddhCTP, providing further support for the idea that protein–protein interactions play an important role in activating viperin.
Mitochondrially targeting viperin results in HADHB degradation by the proteasomal pathway
Having established that ΔN-viperin inhibits HADHB in vitro, we extended our studies to examine how relocalization of viperin to the mitochondrion, as occurs in HCMV infection, may affect HADHB activity. Viperin is normally targeted to the membrane of the ER through its N-terminal amphipathic domain. Therefore, to redirect viperin to the mitochondrion, this domain was replaced by a mitochondrial localization sequence (MLS). Residues 1–42, comprising the ER membrane-localizing domain of WT viperin, were replaced by the MLS of the human mitochondrial protein Tom70 (residues 35–68), with this construct being designated MLS-viperin, as described previously (13).
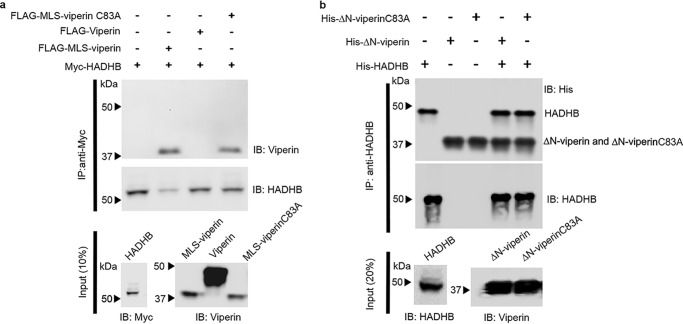
We first established that HADHB and viperin interact within the mitochondrion by conducting pulldown experiments using transiently expressed proteins cotransfected in HEK293T cells (Fig. 3a and Fig. S4, a–d). Myc-tagged HADHB, which is naturally translocated into the mitochondrion, was cotransfected with either WT viperin (which localizes to the ER membrane) or MLS-viperin. These experiments found that only MLS-viperin was coprecipitated with HADHB, consistent with viperin and HADHB forming a complex in the mitochondrial matrix. Fluorescence microscopy of fixed cells that had been transfected with MLS-viperin and stained with both MitoTracker-640, a mitochondrion-staining dye, and immunostained for viperin further confirmed that MLS-viperin colocalized with mitochondria (Fig. S4e). To further establish the interaction, we performed immunoprecipitation experiments with purified proteins. Both ΔN-viperin and ΔN-viperinC83A were efficiently pulled down by HADHB (Fig. 3b), confirming that the interaction does not depend on the Fe–S cluster of viperin.
Figure 3.
Viperin binds HADHB when targeted to the mitochondrion. a, immunotagged genes were transfected into HEK293T cells, and cell extracts were prepared 20 h after transfection. FLAG-MLS-viperin or FLAG-MLS-viperinC83A cell extracts were mixed with Myc-HADHB cell extracts at a ratio of 1:1. Proteins were immunoprecipitated with anti-Myc (HADHB) antibodies and analyzed by immunoblotting (IB) with the indicated antibodies. Control experiments confirmed the specificity of the antibodies used for immunoprecipitation (IP) (Fig. S4). Immunoprecipitation of MLS-viperin and MLS-viperinC83A indicates that they bind to HADHB, whereas cytosolically expressed viperin does not bind to HADHB; representative blots are shown from two independent experiments. Cytosolic extracts were also immunoblotted to confirm the expression levels of each individual protein of interest (10% input). b, in vitro pulldown experiments were performed with recombinant proteins to examine the binding of HADHB to ΔN-viperin and ΔN-viperinC83A. Recombinant proteins (ΔN-viperin and ΔN-viperinC83A) purified from E. coli were immunoprecipitated with anti-HADHB antibody, and the blots were probed with anti-His antibody.
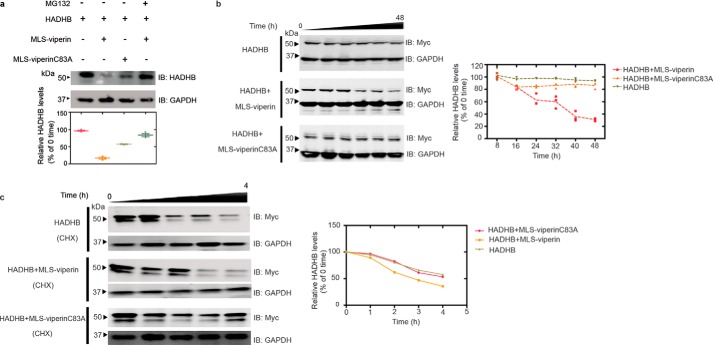
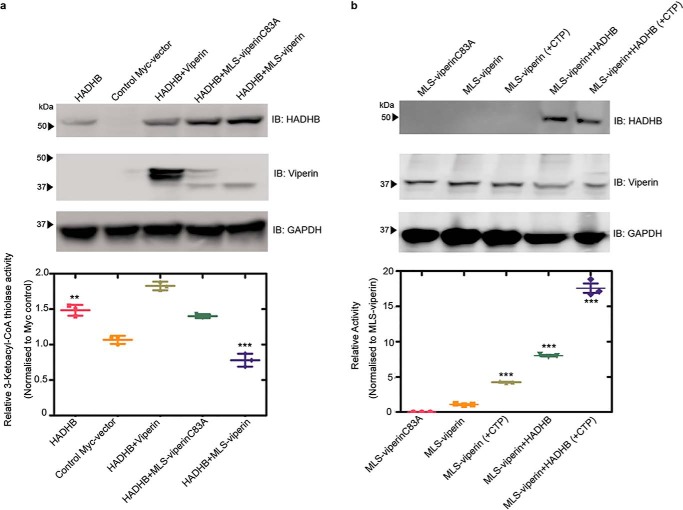
Viperin expression has been shown to be associated with degradation of several cellular and viral proteins (14, 35, 43). Therefore, we next examined the effect of targeting viperin to the mitochondrion on the cellular levels of HADHB. The expression levels of HADHB were compared at 48 h following transfection of HEK293T cells with Myc-tagged HADHB and either WT viperin, MLS-viperin, or MLS-viperin-C83A. In the latter viperin construct, one of the three cysteines that coordinate the Fe4S4 cluster is mutated, rendering the enzyme catalytically inactive.
In initial experiments, we found that coexpression of WT viperin had no significant effect on the cellular concentration of HADHB; however, targeting viperin to the mitochondrion resulted in a significant decrease in HADHB levels (Fig. 4a). In contrast, coexpression of the inactive MLS-viperin-C83A mutant caused a much smaller decrease in HADHB levels. To investigate this observation in more detail, we studied HADHB levels as a function of time after cotransfection with MLS-viperin. The results show that MLS-viperin causes a time-dependent decrease in HADHB levels. However, in this experiment, coexpression of MLS-viperin-C83A had no significant effect on HADHB levels (Fig. 4b). These results were confirmed when the experiment was repeated in the presence of cycloheximide (20 μm) to block de novo protein synthesis (Fig. 4c).
Figure 4.
Targeting viperin to mitochondria results in degradation of HADHB. a, representative immunoblot (IB) of HEK293T cells transfected with HADHB and either MLS-viperin or the inactive mutant MLS-viperinC83A in the presence and absence of the proteasomal inhibitor MG132. Bottom, quantification of HADHB levels determined by immunoblotting. b, 48-h time course showing HADHB levels in cells transfected with HADHB, HADHB and MLS-viperin, and HADHB and MLS-viperinC83A. c, cycloheximide (CHX) chase followed by immunoblotting indicates that MLS-viperin accelerates HADHB degradation. HEK293T cells were transfected with HADHB, HADHB and MLS-viperin, or HADHB and MLS-viperinC83A, and the amount of HADHB was followed for 4 h after cycloheximide addition. HADHB levels are plotted as a function of time for each condition (to = 100%). The data are averages of three experiments, with representative blots shown.
We also examined the effect of inhibiting the proteasomal degradation pathway on HADHB levels. Preliminary investigations established that HADHB levels are sensitive to the proteasomal inhibitor MG132, implying that it is degraded by retrotranslocation to the mitochondrial outer membrane followed proteolysis by the proteasomal pathway (Fig. 4a and Fig. S5). HEK 293T cells were cotransfected with HADHB and MLS-viperin and treated with MG132 (1 μm final concentration). The levels of HADHB in the cells were analyzed by immunoblotting. We observed that MG132 effectively counteracted the degradative effect of viperin on HADHB (Fig. 4a). This suggests that viperin reduces HADHB levels by increasing the rate of HADHB retrotranslocation from the mitochondrion.
Targeting viperin to the mitochondrion inhibits HADHB thiolase activity
To examine the effect of viperin on the activity of HADHB, cell lysates were prepared from HEK293T cells that had been transfected with HADHB and cotransfected with WT viperin, MLS-viperin, or the MLS-viperin-C83A mutant. Cells were harvested 12 h after transfection, at which point HADHB levels were not yet significantly depleted by the action of viperin; cells transfected with empty vector served as a control. The thiolase activity of the lysates was measured using acetoacetyl-CoA and CoASH as substrates (36), as described under “Experimental procedures.”
The thiolase activity of cells transfected with HADHB alone was significantly higher compared with the background activity observed in cells transfected with an empty vector (Fig. 5a). After subtracting the background, the additional thiolase activity because of transfected HADHB was ∼ 3.1 μm·min−1 mg−1 of total protein. Lysates from cells cotransfected with HADHB and WT viperin (which does not enter mitochondria) exhibited similar levels of thiolase activity, 3.8 μm·min−1 mg−1. However, in lysates prepared from cells cotransfected with HADHB and MLS-viperin, thiolase activity was significantly reduced to levels below that of the empty vector control. In contrast, lysates made from cells cotransfected with HADHB and MLS-viperinC83A exhibited levels of thiolase activity very similar to the HADHB-only lysates (Fig. 5a). These assays were carried out 12 h post-transfection because, at this early time, the HADHB levels did not vary greatly between the different conditions (Fig. S6). Therefore, differences in HADHB expression levels did not contribute significantly to the differences in thiolase activity.
Figure 5.
Effect of targeting viperin to mitochondria on the enzymatic activity of HADHB and viperin. a, cell lysates were prepared from HEK293T cells transfected with the indicated constructs, and the thiolase activity of the lysate was measured. Top panel, expression of enzymes, verified by immunoblotting (IB). Bottom panel, thiolase activity of the extracts relative to the control. b, cell lysates were prepared from HEK293T cells transfected with the indicated constructs. Lysates were assayed anaerobically and contained 100 μm SAM and either no additional CTP or 100 μm CTP. Top panel, expression of enzymes, verified by immunoblotting. Bottom panel, activity is plotted relative to the MLS-viperin only condition (activity = 1) after normalizing for the viperin concentration. For details, see the text. Values are presented as mean ± S.E. (n = 6). **, p < 0.005; ***, p < 0.001(Student's t test for independent samples).
HADHB activates the radical SAM activity of viperin
The unexpected observation that, with purified enzymes in vitro, HADHB activates viperin prompted us to investigate whether HADHB exerted a similar effect on viperin in the context of the mitochondrial matrix (Fig. 5b). HEK293T cells were cotransfected with either MLS-viperin or MLS-viperinC83A and HADHB. 12 h post-transfection, cell lysates were prepared under anaerobic conditions (because of the in vitro oxygen sensitivity of viperin). The enzymatic activity of viperin was assayed under reducing conditions as described under “Experimental procedures” (33). The specific activity was quantified by measuring the amount of 5′-dA formed in 1 h. The amount of viperin present in the cell extracts was quantified by immunoblotting, using methods described previously (39). Control experiments confirmed that substitution of the N-terminal of viperin with the MLS sequence had no effect on viperin's enzymatic activity (Fig. S3).
In cell lysates transfected with MLS-viperin alone, the specific activity of viperin, expressed as a turnover number, was 12.1 ± 0.5 h−1. Viperin activity increased significantly when HADHB was coexpressed in the cells by ∼2-fold to give a turnover number of 27 ± 1 h−1. It appears, therefore, that HADHB activates viperin in the mitochondrion. We note that these turnover numbers are considerably higher than those measured with purified enzymes but similar to those we reported previously for viperin activation by TRAF6/IRAK1 (39). This discrepancy may, in part, reflect differences in the methods used to determine viperin concentration but also suggests that, when expressed in eukaryotic cells, viperin is intrinsically more active. The availability of the endogenous iron–sulfur cluster–assembling machinery in eukaryotic cells may be one factor that contributes to the higher activity of viperin.
Interestingly, we also observed significant activation of viperin by HADHB in assays conducted in the absence of CTP. Under these conditions, lysates of cells transfected with only MLS-viperin exhibited a low level of activity, 1.6 ± 0.1 h−1, which may represent uncoupled reductive cleavage of SAM and/or reaction with endogenous CTP in the cell lysates (Fig. 5b). However, when coexpressed with HADHB, this “background” activity increased ∼4-fold to 6.7 ± 0.3 h−1, an activity substantially higher than in lysates to which CTP was added but HADHB was absent (Table S2).
To investigate whether HADHB only acts to uncouple the consumption of SAM from synthesis of ddhCTP, we undertook LC-MS analysis of ddhCTP formed in these reactions, using methodology described previously. The analysis indicated that coexpression of HADHB with viperin resulted in a significant increase in the amount of ddhCTP formed in the cell lysates (Fig. S7). This result implies that HADHB does not simply activate viperin toward uncoupled turnover but specifically activates viperin to increase the overall rate at which ddhCTP is synthesized.
Targeting viperin to the mitochondrion reduces cellular ATP levels
HADHB is a component of the multienzyme complex involved in β-oxidation of fatty acids, which is a major route for ATP production. Therefore, we examined the effect of targeting viperin to the mitochondrion on cellular ATP levels. We measured cellular ATP levels in transfected HEK293T cells expressing HADHB and/or MLS-viperin or MLS-viperinC83A, with an empty vector serving as a control. The results, summarized in Fig. 6, show that ATP levels were reduced significantly in cells expressing MLS-viperin, by over 50%. In contrast, ATP levels were only slightly reduced in cells expressing the inactive MLS-viperinC83A mutant. Interestingly, attempts to restore ATP levels by overexpressing HADHB with MLS-viperin did not result in an increase in ATP levels, suggesting that viperin is a potent inhibitor of HADHB. The reduction in cellular ATP levels is consistent with earlier studies (13) and with our observation that the interaction of mitochondrially targeted viperin with HADHB reduces β-thiolase activity and blocks β-oxidation.
Figure 6.
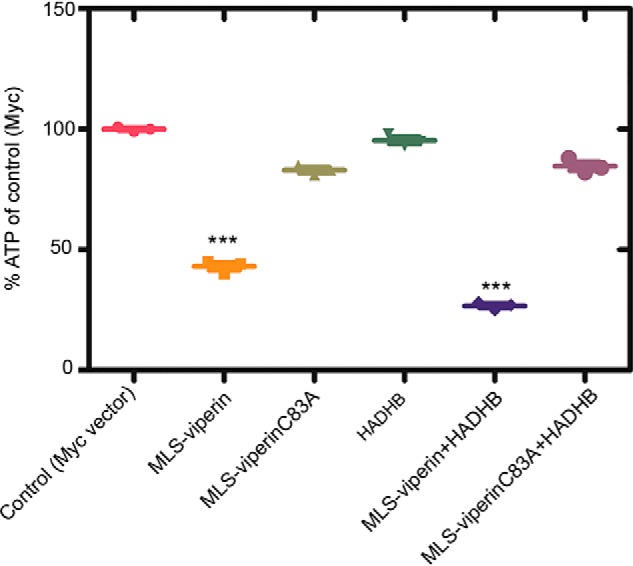
Targeting viperin to mitochondria results in a decrease in cellular ATP levels. HEK293T cells were transfected with the indicated constructs. 24 h after transfection, the cells were harvested, and ATP concentrations were determined. Results are plotted relative to the empty vector control (100%). Values are presented as mean ± S.E. (n = 6). ***, p < 0.001 (Student's t test for independent samples).
Discussion
There is considerable interest in the mechanisms by which viruses evade cellular defenses because a detailed understanding of such mechanisms is a prerequisite for developing new antiviral therapies. Co-opting of viperin by HCMV, which leads to increased infectivity, was first described by Cresswell and co-workers (13). They established that the viral protein vMIA is responsible for relocating viperin to the mitochondrion and that this results in a significant decrease in cellular ATP levels. Their investigations also identified HADHB as a target of viperin, and on this basis, they hypothesized that viperin inhibits fatty acid β-oxidation. They further hypothesized that the reduction in cellular ATP levels contributed to the observed disruption of the actin cytoskeleton, facilitating release of the virus from the cell. However, at the time of these studies, the biochemical function of viperin was unknown, which limited the conclusions that could be drawn regarding its effects on mitochondrial metabolism.
Through experiments with purified enzymes, we directly demonstrated that ΔN-viperin tightly binds to HADHB and inhibits its enzymatic activity, indicating that the two enzymes form a stoichiometric complex. This is consistent with the original observation that targeting viperin to the mitochondrion inhibits fatty acid oxidation and causes downstream effects on ATP levels and cytoskeleton integrity (13). Our results further suggest that vMIA is only responsible for transporting viperin into the mitochondrion but does not play a role in mediating inhibition of HADHB by viperin. Most interestingly, HADHB activity was not completely inhibited by ΔN-viperin, even when the enzyme was in 2-fold excess (Fig. 2a). This suggests that viperin may regulate HADHB activity by an allosteric mechanism rather than simply preventing the substrate from binding. It is also interesting that, although the ΔN-viperinC83A mutant still bound to HADHB, it had no effect on activity. It therefore appears that the Fe–S cluster may play an important role in viperin's regulation of HADHB.
Previous studies did not examine the effect of targeting viperin to the mitochondrion on the thiolase activity of HADHB. Here we show that coexpression of MLS-viperin with HADHB results in a significant decrease in thiolase activity in cell extracts that do not occur with WT viperin. This decrease could be due to viperin reducing the cellular levels of HADHB, as appears to occur with other protein targets of the enzyme (35, 36, 43), or viperin could inhibit HADHB directly by binding to it. We found that coexpression of MLS-viperin with HADHB does result in a significant decrease in the cellular levels of HADHB, which could be reversed by blocking the proteasomal degradation pathway. This observation suggests that viperin causes HADHB to be retrotranslocated to the outer mitochondrial membrane, where it is subject to ubiquitination and subsequently degraded by the proteasome in a process known as outer mitochondrial membrane–associated degradation (44, 45). However, details regarding the mechanism by which viperin binding to HADHB leads to its retrotranslocation out of the mitochondrion remain to be elucidated. Overall, our data, summarized in Figs. 2, 4, and 5, indicate that viperin may regulate HADHB by directly inhibiting it and through proteostasis by increasing its rate of degradation.
The original discovery that HCMV up-regulates viperin to enhance viral infectivity (13) was surprising because it appeared to rely on a seemingly adventitious association between viperin and HADHB. However, the recent finding that viperin is intrinsically expressed in mouse adipose tissue, where it appears to regulate thermogenesis (38), suggests to us that viperin binding to HADHB likely serves a pre-existing regulatory function the virus has evolved to exploit. In particular, the observation that mice lacking viperin exhibited increased heat production when fed a high-fat diet is consistent with viperin either inhibiting HADHB and/or reducing HADHB levels in the mitochondrion, as we have demonstrated, and regulating β-oxidation. These observations suggest that the interaction between viperin and HADHB may provide a new target for anti-HCMV therapeutic agents and regulation of thermogenesis in metabolic diseases.
The activating effect of HADHB on viperin is an interesting and unanticipated observation. We observed, in HEK293T cell lysates and with purified proteins, that HADHB activates the radical SAM activity of viperin by severalfold. Whether the production of ddhCTP in the mitochondrion also contributes to enhancing the infectivity of HCMV or plays a role in regulating thermogenesis is at this point unclear. ddhCTP has been shown to inhibit replication of some RNA viruses by acting as a chain terminator during replication of the genome by viral RNA–dependent RNA polymerases (33). Although ddhCTP does not appear to be misincorporated by nuclear RNA polymerases such as Pol II, which are complex multisubunit proteins, the mitochondrial polymerase comprises only a single subunit and is structurally most closely related to bacteriophage T7 polymerase (46, 47). The mitochondrial RNA polymerase may therefore be more susceptible to misincorporating ddhCTP. In this regard, we note that development of antiviral compounds based on synthetic ribonucleotide analogs to treat HCMV infection, among other diseases, has been hampered because of their cytotoxicity. One reason for this toxicity is misincorporation of these ribonucleotides by the mitochondrial RNA polymerase (48), which, in contrast to Pol II, lacks a proofreading function.
These observations raise the intriguing possibility that ddhCTP might act as a regulator of mitochondrial RNA transcription under nonpathological conditions by causing low levels of chain termination. Under pathological conditions, such as HCMV infection, high levels of ddhCTP, resulting from viperin's translocation to the mitochondrion, might further enhance virus infectivity by extensively disrupting mitochondrial transcription though chain termination.
Experimental procedures
Cell lines
The HEK293T cell line was obtained from the ATCC.
Antibodies
Rabbit polyclonal RSAD2/viperin antibody (11833-1-AP) was obtained from ProteinTech. The mouse monoclonal viperin antibody clone MaP (MABF106) was obtained from EMD Millipore. The rabbit polyclonal Myc tag antibody (16286-1-AP) was obtained from ProteinTech. Mouse monoclonal HADHB(E-1) (sc-271495) was obtained from Santa Cruz Biotechnology. Goat anti-rabbit (170-6515) and anti-mouse (626520) Ig secondary antibodies were purchased from Bio-Rad and Life Technologies, respectively. Rabbit polyclonal GAPDH (TAB1001) was purchased from Thermo Scientific, and mouse monoclonal GAPDH antibody (6C5) was obtained from EMD Millipore.
Plasmids
The HADHB (Myc-DDK–tagged) human hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein) plasmid (RC209853) was purchased from Origene (Rockville, MD). The MLS-viperin plasmid was a kind gift from Dr. Peter Cresswell (Yale School of Medicine). The MLS-viperinC83A mutant was constructed by site-directed mutations of the first cysteine residue to alanine (C83A) using the QuikChange site-directed mutagenesis kit (Agilent). The following primer pair was used for introducing the change: 5′-CACTTCACTCGCCAGTGCAACTACAAATGCGGC-3′ and 5′-GCCGCATTTGTAGTTGCACTGGCGAGTGAAGTG-3′. Synthetic genes encoding human viperin (GenBank accession number AAL50053.1) were purchased from GenScript. The pCMV-Myc plasmid (empty vector, 631604) was purchased from Takara Biosciences.
Recombinant human HADHB
Purified, recombinantly expressed HADHB (AP4092) was purchased from AdooQ Bioscience LLC.
Reagents
The sources of all other reagents have been described previously (35, 39)
Transfection
HEK293T cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 100 units/ml penicillin, 100 g/ml streptomycin, and 2 mm l-glutamine. Transient transfections were carried out either using FuGENE® HD (Promega) or PEI MAX transfection-grade linear polyethyleneimine hydrochloride (Polysciences Inc.) following the manufacturer's instructions.
Expression and purification of recombinant truncated viperin (ΔN-viperin/ΔN-viperinC83A)
For in vitro assays of viperin, a truncated version of the enzyme (pET28b-ΔN-viperin) that lacked the N-terminal amphipathic helix was used. This enzyme, ΔN-viperin, was overexpressed and purified from E. coli as described previously (35) The pET28b-ΔN-viperinC83A mutant was constructed by site-directed mutations of the first cysteine residue to alanine (C83A) using the QuikChange site-directed mutagenesis kit (Agilent). The following primer pair was used for introducing the change: 5′-CACTTCACTCGCCAGTGCAACTACAAATGCGGC-3′ and 5′-GCCGCATTTGTAGTTGCACTGGCGAGTGAAGTG-3′. The expression, purification, and reconstitution conditions were similar as for the WT construct, as described above.
Assays of viperin activity using purified recombinant ΔN-viperin
Enzyme assay reactions were performed under anaerobic conditions in 10 mm Tris-Cl buffer (pH 8.0), 300 mm NaCl, 10% glycerol, and 5 mm sodium dithionite. Assays contained, in a total volume of 100 μl, 10 nm ΔN-viperin, 100 μm SAM, and 100 μm CTP. The assay was incubated for 60 min at room temperature, and then the reaction was stopped by heating at 95 °C for 10 min. The solution was chilled to 4 °C, and the precipitated proteins were removed by centrifugation at 14,000 rpm for 25 min. For assays with HADHB, HADHB was added at a concentration of 10 nm, and the other conditions remained the same. The 5′-dA generated during the reaction was then extracted with acetonitrile, as described previously (35, 39). Samples were analyzed in triplicate by Ultra-performance liquid chromatography (UPLC) tandem MS as described previously (35, 49).
Assay of MLS-viperin activity in HEK293T cell lysates
HEK293T cells transfected with MLS-viperin, MLS-viperinC83A, and/or HADHB were harvested from one 10-cm diameter tissue culture plate each, resuspended in 400 μl of anoxic Tris-buffered saline (50 mm Tris-Cl (pH 7.6) and 150 mm NaCl) containing 1% Triton X-100, sonicated within an anaerobic glove box (Coy Chamber), and centrifuged at 14,000 × g for 20 min. DTT (5 mm) and dithionite (5 mm) were added to the cell lysate together with CTP (300 μm). The assay mixture was incubated at room temperature for 30 min prior to starting the reaction by addition of SAM (200 μm). The assay was incubated for 50 min at room temperature, and then the reaction was stopped by heating at 95 °C for 10 min. The solution was chilled to 4 °C, and the precipitated proteins were removed by centrifugation at 14,000 rpm for 25 min. The supernatant was then extracted with acetonitrile, and the samples were derivatized with benzoyl chloride as described previously (49) and analyzed in triplicate by UPLC tandem MS, as detailed in the supporting information (Table S3). For details regarding standard curve construction and calculations, refer to Ref. 39. The viperin activity measurements reported here represent the average of at least three independent biological replicates.
Immunoblotting and immunoprecipitation assays
Immunoprecipitation using HEK293T cell lysates was performed with anti-Myc–tagged magnetic beads according to the manufacturers' protocol, as detailed previously (32). Immunoprecipitation for E. coli purified proteins using Protein A beads was performed according to the manufacturer's protocol. For immunoprecipitation, the ratio of suspension to packed gel volume was 2:1. Resin pre-equilibration was done according to the manufacturer's protocol. The purified proteins, HADHB:ΔN-viperin/ΔN-viperinC83A, were mixed at a 1:1 ratio and incubated for 2 h at 4 °C with gentle rotation. Beads (attached to HADHB antibody) were pelleted by centrifugation at 5000 × g for 30 s at 4 °C and washed three times with washing buffer (50 mm Tris (pH 7.4),150 mm NaCl, 10% glycerol, 1 mm EDTA, 10 mm NaF, and 0.2 mm phenylmethylsulphonyl fluoride with protease inhibitor mixture from Sigma). Immunocomplexes were eluted by boiling in SDS-PAGE sample buffer, separated by SDS-PAGE, and transferred to a PVDF membrane. Immunoblotting was performed using standard methods as described previously (39), with total protein concentrations determined by the BCA method and sample loading concentrations normalized accordingly. Blots were visualized and band intensities quantified using a Bio-Rad ChemiDoc Touch imaging system, with GAPDH used as a control to compare relative expression levels. The quantitative measurements of protein expression levels reported here represent the average of at least three independent biological replicates.
Assay of HADHB thiolase activity using purified recombinant protein
Thiolase activity of HADHB was measured using acetoacetyl-CoA and CoASH as substrates and following the decrease in absorbance at 303 nm as described by Zhou et al. (42). A typical assay contained 100 mm Tris-Cl (pH 8.3), 25 mm MgCl2, 1 mm DTT, 100 μm CoASH, 100 μm acetoacetyl-CoA, and 10 nm HADHB complex in a 1-ml reaction volume. The reaction was started by adding HADHB, and after 5 min of incubation at room temperature, the decrease in absorbance at 303 nm was monitored; under these conditions, the reaction rate remained linear for at least 30 min. The rate of reaction was calculated assuming an extinction coefficient of 16,900 cm−1 m−1 for the complex of acetoacetyl-CoA with Mg2+.
For thiolase activity measurements in the presence of viperin, the assay was performed with the following modifications. All assay buffers were made anaerobic prior to the assay, and the assay components were assembled in anaerobic cuvettes in an anaerobic glove box (Coy Laboratory). Prior to addition to the assay mixture, HADHB (10 nm) and ΔN-viperin were mixed together at varying concentrations of ΔN-viperin (0, 2, 5, 10, 15, and 20 nm) under anaerobic conditions and incubated with gentle agitation at room temperature for 50 min. After adding the enzyme, the cuvettes were sealed to the atmosphere, removed from the anaerobic chamber, and introduced into the spectrophotometer, with reaction rates being recorded 5 min after the addition of enzyme. For follow-up assays, ΔN-viperin (10 nm) and HADHB (10 nm) were mixed together in equimolar concentrations and assayed as described above. For ΔN-viperinC83A assays, similar conditions were followed. The thiolase activity reported here represents the average of at least three independent measurements.
Assay of HADHB thiolase activity in HEK293T cell lysates
Cell pellets were thawed on ice and suspended in 300 μl of lysis buffer (100 mm Tris-Cl (pH 8.3), 2 mm β-mercaptoethanol, 200 mm NaCl, 0.5 mm EDTA, and 0.5% Tween 20) containing Halt protease inhibitor mixture. Cells were lysed by brief sonication. Cell debris was removed by centrifugation of the lysate at 14,000 rpm for 25 min at 4 °C. The clarified lysates were used in thiolase assays, with the total protein concentration of the lysate determined using the BCA assay (Pierce BCA protein assay kit, Thermo Scientific). A typical assay contained 10 μl of cell lysate, 10 mm Tris-Cl, 25 mm MgCl2, 100 μm CoA, 1 mm DTT, and 100 μm acetoacetyl-CoA in a 1-ml reaction volume. Thiolase activity was measured spectrophotometrically using the protocol described above. HADHB concentrations were determined by immunoblotting as described above. Thiolase activities were normalized to Myc-only control cell lysates. The measurements of thiolase activities reported here represent the average of at least three independent biological replicates.
ATP assays
HEK293T cells were transfected with the indicated constructs, and cellular ATP levels were measured with the ATP bioluminescence assay kit HS II (Roche) following the manufacturer's instructions. To normalize the measured ATP levels, the total protein content of transfected cells was measured using the Bio-Rad protein assay kit. Serial dilutions of BSA were used as standards. Finally, ATP in cells transfected with the indicated constructs was normalized to those in control cells transfected with vector alone.
Statistical analyses
Results from all studies were compared with unpaired two-tailed Student's t test using GraphPad Prism 5 software. p values of less than 0.05 were considered significant.
Author contributions
A. B. D., K. A. Z., and E. N. G. M. conceptualization; A. B. D., K. S., R. T. K., and E. N. G. M. resources; A. B. D., K. A. Z., and E. N. G. M. formal analysis; A. B. D., K. S., R. T. K., and E. N. G. M. supervision; A. B. D. and E. N. G. M. funding acquisition; A. B. D., K. A. Z., K. S., R. T. K., and E. N. G. M. investigation; A. B. D., K. A. Z., K. S., R. T. K., and E. N. G. M. methodology; A. B. D. and K. A. Z. writing-original draft; A. B. D., K. S., R. T. K., and E. N. G. M. project administration; A. B. D., K. A. Z., K. S., R. T. K., and E. N. G. M. writing-review and editing.
Supplementary Material
Acknowledgment
We thank Prof. Peter Cresswell (Yale University) for the kind gift of the MLS-viperin expression vector.
This work was supported in part by National Institutes of Health Grants GM 093088 (to A. B. D., K. S., and E. N. G. M.) and DK 046960 (to K. A. Z. and R. T. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7 and Tables S1–S3.
- HCMV
- human cytomegalovirus
- ddhCTP
- 3′-deoxy-3′,4′-didehydro-CTP
- ER
- endoplasmic reticulum
- MLS
- mitochondrial localization sequence.
References
- 1. Horner S. M. (2014) Activation and evasion of antiviral innate immunity by hepatitis C virus. J. Mol. Biol. 426, 1198–1209 10.1016/j.jmb.2013.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Horst D., Verweij M. C., Davison A. J., Ressing M. E., and Wiertz E. (2011) Viral evasion of T cell immunity: ancient mechanisms offering new applications. Curr. Opin. Immunol. 23, 96–103 10.1016/j.coi.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 3. Iannello A., Debbeche O., Martin E., Attalah L. H., Samarani S., and Ahmad A. (2006) Viral strategies for evading antiviral cellular immune responses of the host. J. Leukoc. Biol. 79, 16–35 10.1189/jlb.0705397 [DOI] [PubMed] [Google Scholar]
- 4. Jiang X., and Chen Z. J. (2012) The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 12, 35–48 10.1038/nri3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor K. E., and Mossman K. L. (2013) Recent advances in understanding viral evasion of type I interferon. Immunology 138, 190–197 10.1111/imm.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhoj V. G., and Chen Z. J. (2009) Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437 10.1038/nature07959 [DOI] [PubMed] [Google Scholar]
- 7. Jo E. K., Yang C. S., Choi C. H., and Harding C. V. (2007) Intracellular signalling cascades regulating innate immune responses to Mycobacteria: branching out from Toll-like receptors. Cell Microbiol. 9, 1087–1098 10.1111/j.1462-5822.2007.00914.x [DOI] [PubMed] [Google Scholar]
- 8. O'Neill L. A., and Bowie A. G. (2010) Sensing and signaling in antiviral innate immunity. Curr. Biol. 20, R328–R333 10.1016/j.cub.2010.01.044 [DOI] [PubMed] [Google Scholar]
- 9. Schneider W. M., Chevillotte M. D., and Rice C. M. (2014) Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morrison J., Aguirre S., and Fernandez-Sesma A. (2012) Innate immunity evasion by dengue virus. Viruses 4, 397–413 10.3390/v4030397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orange J. S., Fassett M. S., Koopman L. A., Boyson J. E., and Strominger J. L. (2002) Viral evasion of natural killer cells. Nat. Immunol. 3, 1006–1012 10.1038/ni1102-1006 [DOI] [PubMed] [Google Scholar]
- 12. Chin K. C., and Cresswell P. (2001) Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U.S.A. 98, 15125–15130 10.1073/pnas.011593298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seo J. Y., Yaneva R., Hinson E. R., and Cresswell P. (2011) Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332, 1093–1097 10.1126/science.1202007 [DOI] [PubMed] [Google Scholar]
- 14. Helbig K. J., and Beard M. R. (2014) The role of viperin in the innate antiviral response. J. Mol. Biol. 426, 1210–1219 10.1016/j.jmb.2013.10.019 [DOI] [PubMed] [Google Scholar]
- 15. Mattijssen S., and Pruijn G. J. (2012) Viperin, a key player in the antiviral response. Microbes Infect. 14, 419–426 10.1016/j.micinf.2011.11.015 [DOI] [PubMed] [Google Scholar]
- 16. Chakravarti A., Selvadurai K., Shahoei R., Lee H., Fatma S., Tajkhorshid E., and Huang R. H. (2018) Reconstitution and substrate specificity for isopentenyl pyrophosphate of the antiviral radical SAM enzyme viperin. J. Biol. Chem. 293, 14122–14133 10.1074/jbc.RA118.003998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Honarmand Ebrahimi K., Carr S. B., McCullagh J., Wickens J., Rees N. H., Cantley J., and Armstrong F. A. (2017) The radical-SAM enzyme viperin catalyzes reductive addition of a 5′-deoxyadenosyl radical to UDP-glucose in vitro. FEBS Lett. 591, 2394–2405 10.1002/1873-3468.12769 [DOI] [PubMed] [Google Scholar]
- 18. Duschene K. S., and Broderick J. B. (2010) The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 584, 1263–1267 10.1016/j.febslet.2010.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landgraf B. J., McCarthy E. L., and Booker S. J. (2016) Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 85, 485–514 10.1146/annurev-biochem-060713-035504 [DOI] [PubMed] [Google Scholar]
- 20. Wang J., Woldring R. P., Román-Meléndez G. D., McClain A. M., Alzua B. R., and Marsh E. N. G. (2014) Recent advances in radical SAM enzymology: new structures and mechanisms. ACS Chem. Biol. 9, 1929–1938 10.1021/cb5004674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duschene K. S., and Broderick J. B. (2012) Viperin: a radical response to viral infection. Biomol. Concepts 3, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vey J. L., and Drennan C. L. (2011) Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 111, 2487–2506 10.1021/cr9002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marsh E. N., Patterson D. P., and Li L. (2010) Adenosyl radical: reagent and catalyst in enzyme reactions. ChemBioChem 11, 604–621 10.1002/cbic.200900777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frey P. A., Hegeman A. D., and Ruzicka F. J. (2008) The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 10.1080/10409230701829169 [DOI] [PubMed] [Google Scholar]
- 25. Marsh E. N., Patwardhan A., and Huhta M. S. (2004) S-adenosylmethionine radical enzymes. Bioorg. Chem. 32, 326–340 10.1016/j.bioorg.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 26. Cheek J., and Broderick J. B. (2001) Adenosylmethionine-dependent iron-sulfur enzymes: versatile clusters in a radical new role. J. Biol. Inorg. Chem. 6, 209–226 10.1007/s007750100210 [DOI] [PubMed] [Google Scholar]
- 27. Broderick W. E., and Broderick J. B. (2019) Radical SAM enzymes: surprises along the path to understanding mechanism. J. Biol. Inorg. Chem. 24, 769–776 10.1007/s00775-019-01706-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yokoyama K., and Lilla E. A. (2018) C-C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products. Nat. Prod. Rep. 35, 660–694 10.1039/C8NP00006A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y., Schnell B., Müller R., and Begley T. P. (2018) Iterative methylations resulting in the biosynthesis of the t-butyl group catalyzed by a B12-dependent radical SAM enzyme in cystobactamid biosynthesis. Methods Enzymol. 606, 199–216 10.1016/bs.mie.2018.05.018 [DOI] [PubMed] [Google Scholar]
- 30. Wang S. C. (2018) Cobalamin-dependent radical S-adenosyl-l-methionine enzymes in natural product biosynthesis. Nat. Prod. Rep. 35, 707–720 10.1039/C7NP00059F [DOI] [PubMed] [Google Scholar]
- 31. Rettberg L., Tanifuji K., Jasniewski A., Ribbe M. W., and Hu Y. (2018) Radical S-adenosyl-l-methionine (SAM) enzyme involved in the maturation of the nitrogenase cluster. Methods Enzymol. 606, 341–361 10.1016/bs.mie.2018.04.009 [DOI] [PubMed] [Google Scholar]
- 32. Pang H., and Yokoyama K. (2018) Lessons from the studies of a C-C bond forming radical SAM enzyme in molybdenum cofactor biosynthesis. Methods Enzymol. 606, 485–522 10.1016/bs.mie.2018.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gizzi A. S., Grove T. L., Arnold J. J., Jose J., Jangra R. K., Garforth S. J., Du Q., Cahill S. M., Dulyaninova N. G., Love J. D., Chandran K., Bresnick A. R., Cameron C. E., and Almo S. C. (2018) A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614 10.1038/s41586-018-0238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saitoh T., Satoh T., Yamamoto N., Uematsu S., Takeuchi O., Kawai T., and Akira S. (2011) Antiviral protein viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity 34, 352–363 10.1016/j.immuni.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 35. Makins C., Ghosh S., Román-Meléndez G. D., Malec P. A., Kennedy R. T., and Marsh E. N. (2016) Does viperin function as a radical S-adenosyl-l-methionine-dependent enzyme in regulating farnesylpyrophosphate synthase expression and activity? J. Biol. Chem. 291, 26806–26815 10.1074/jbc.M116.751040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seo J. Y., and Cresswell P. (2013) Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 9, e1003497 10.1371/journal.ppat.1003497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X., Hinson E. R., and Cresswell P. (2007) The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 10.1016/j.chom.2007.06.009 [DOI] [PubMed] [Google Scholar]
- 38. Eom J., Kim J. J., Yoon S. G., Jeong H., Son S., Lee J. B., Yoo J., Seo H. J., Cho Y., Kim K. S., Choi K. M., Kim I. Y., Lee H.-Y., Nam K. T., Cresswell P., et al. (2019) Intrinsic expression of viperin regulates thermogenesis in adipose tissues. Proc. Natl. Acad. Sci. U.S.A. 116, 17419–17428 10.1073/pnas.1904480116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dumbrepatil A. B., Ghosh S., Zegalia K. A., Malec P. A., Hoff J. D., Kennedy R. T., and Marsh E. N. G. (2019) Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol. Chem. 294, 6888–6898 10.1074/jbc.RA119.007719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hee J. S., and Cresswell P. (2017) Viperin interaction with mitochondrial antiviral signaling protein (MAVS) limits viperin-mediated inhibition of the interferon response in macrophages. PLoS ONE 12, e0172236 10.1371/journal.pone.0172236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bartlett K., and Eaton S. (2004) Mitochondrial beta-oxidation. Eur. J. Biochem. 271, 462–469 10.1046/j.1432-1033.2003.03947.x [DOI] [PubMed] [Google Scholar]
- 42. Zhou Z., Zhou J., and Du Y. (2012) Estrogen receptor β interacts and colocalizes with HADHB in mitochondria. Biochem. Biophys. Res. Comm. 427, 305–308 10.1016/j.bbrc.2012.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Panayiotou C., Lindqvist R., Kurhade C., Vonderstein K., Pasto J., Edlund K., Upadhyay A. S., and Överby A. K. (2018) Viperin restricts Zika virus and tick-borne encephalitis virus replication by targeting NS3 for proteasomal degradation. J. Virol. 92, e02054–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pickles S., Vigié P., and Youle R. J. (2018) Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor E. B., and Rutter J. (2011) Mitochondrial quality control by the ubiquitin-proteasome system. Biochem. Soc. Trans. 39, 1509–1513 10.1042/BST0391509 [DOI] [PubMed] [Google Scholar]
- 46. Schwinghammer K., Cheung A. C., Morozov Y. I., Agaronyan K., Temiakov D., and Cramer P. (2013) Structure of human mitochondrial RNA polymerase elongation complex. Nat. Struct. Mol. Biol. 20, 1298–1303 10.1038/nsmb.2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arnold J. J., Smidansky E. D., Moustafa I. M., and Cameron C. E. (2012) Human mitochondrial RNA polymerase: structure-function, mechanism and inhibition. Biochim. Biophys. Acta 1819, 948–960 10.1016/j.bbagrm.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 48. Arnold J. J., Sharma S. D., Feng J. Y., Ray A. S., Smidansky E. D., Kireeva M. L., Cho A., Perry J., Vela J. E., Park Y., Xu Y., Tian Y., Babusis D., Barauskus O., Peterson B. R., et al. (2012) Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 8, e1003030 10.1371/journal.ppat.1003030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wong J. M., Malec P. A., Mabrouk O. S., Ro J., Dus M., and Kennedy R. T. (2016) Benzoyl chloride derivatization with liquid chromatography-mass spectrometry for targeted metabolomics of neurochemicals in biological samples. J. Chromatogr. A 1446, 78–90 10.1016/j.chroma.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.