Abstract
Blood test is a kind of liquid biopsy that checks cancer cells or cancer nucleic acids circulating freely from cells in the blood. A liquid biopsy may be used to distinguish cancer at early stages and it could be a game-changer for both cancer diagnosis and prognosis strategies. Liquid biopsy tests consider several tumor components, such as DNA, RNA, proteins, and the tiny vesicles originating from tumor cells. Actually, liquid biopsy signifies the genetic alterations of tumors through nucleic acids or cells in various body fluids, including blood, urine, cerebrospinal fluid, or saliva in a noninvasive manner. In this review, we present an overall description of liquid biopsy in which circulating tumor cells, cell-free nucleic acids, exosomes, and extrachromosomal circular DNA are included.
Keywords: Liquid biopsy, cell-free nucleic acids (cfNAs), circulating tumor cells (CTCs), exosomes
Introduction
Cancer is a major worldwide health problem that originates from abnormal and uncontrolled cell division and can eventually spread into other tissues.1 In the past, it was wrongly thought that tumor genes and cells are only present in the exact tumor site. In 1896, the suggestion that circulating tumor cells (CTCs) are a major requirement to metastasis was first proposed by an Australian pathologist, Thomas Ashworth.2 The presence of cell-free DNA (cfDNA) was reported in human plasma by Mandel and Metals in 1948.3 Unfortunately, the concept of liquid biopsy was totally ignored till 1977, when researchers made the novel discovery that cancer patients carried cell-free nucleic acids (cfNAs) and cells in their peripheral blood.4
The exact word of “liquid biopsy” is classically applied for cfDNA and CTCs, but its component can be more. In fact it is a diagnostic molecular test to run on a sample of plasma to discover cancer cells that are shedding from primary or metastatic tumors that are circulating in the blood or DNA fragments of tumor cells.5 Tissue biopsy is a snapshot of tumor and cannot cover tumor heterogeneity.6 Liquid biopsy can be beneficial for early diagnosis of cancer or to examine the treatment efficacy, and because of its noninvasive nature, several samplings of blood over time are possible.7-9
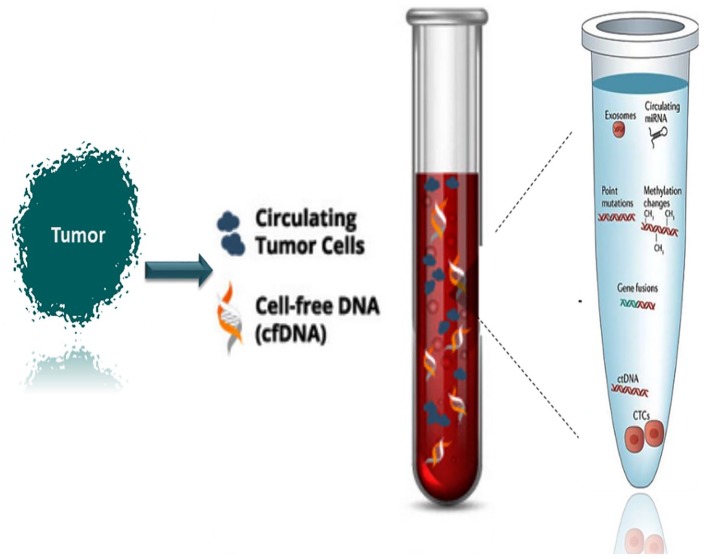
In this review, we explain the molecular importance of the CTCs, cfNAs, and exosomes as the main component of liquid biopsy, in addition to the extrachromosomal circular DNA (eccDNA) as a possible component of liquid biopsy (Figure 1).
Figure 1.
Schematic figure of the liquid biopsy and the way that its genetic components are presented in the blood.
Advantages of Liquid Biopsy
In the field of cancer management, there are some problematic issues concerning the early diagnosis, prognosis, and prediction of treatment resistance.10-12 Tumor heterogeneity brings a lot of difficulties for clinicians and stand in the way of cancer treatment.13 The molecular genetics profile of a tumor changes over time and standard tissue biopsies are able to represent an overall view of the tumor.14-16 Moreover, tissue biopsy is an invasive procedure and so is not easily repeatable considering the cost and the risk factors.17 Traditionally, routine histological evaluation and immunohistochemical study have an essential role in several aspects of diagnosis in both neoplastic and non-neoplastic disorders.
Currently, liquid biopsy, as a real-time representative of tumor, is a minimally invasive biopsy method which has been studied by scientists and oncologists over the past decades.9,18,19 The purpose of liquid biopsy is to identify and examine the biological material circulating in body fluid, originating within and from the tumor.20,21 Several research works have defined certain benefits of molecular information about primary tumors by using liquid biopsies and compared the result of liquid biopsy with tumor biopsy.22-30 There are controversies that liquid biopsy can take the place of tissue biopsy or will support “gold standard” tissue biopsies. According to Miro Venturi, Roche’s global head of diagnostics biomarkers, “Liquid biopsies could be a game-changer in cancer testing.” In fact, the minimally invasive nature of liquid biopsy brings a new insight for malignancy checking without delay, expenses, and risks, possibly at a microscopic stage, before the radiological tests.31 Liquid biopsy promises early detection of cancer because of its potential for detecting a classically incurable malignancy at an earlier, more curable, and even curable stage.32 The use of circulating tumor DNA (ctDNA) genetic changes within blood test has extraordinary applications for the period of cancer treatment, with dynamic monitoring of therapy response, quick detection of resistance, and foretelling the tumor recurrence ahead of clinical relapse.33-35
Circulating Tumor Cells
Circulating tumor cells are tumor cells that are driven from the primary tumor and are released into the blood vessels or lymphatic vessels.36 The existence of CTCs in the blood of metastatic prostate cancer patients was reported by Thomas Ashworth, an Australian physician, in 1869.2 Metastasis is the most challenging process of tumor and the key cause of cancer-associated death (9 of 10 deaths). The “seed and soil” hypothesis described that tumor invasion—the intrinsic characteristics of the tumor cells (seeds) and host microenvironment (soil)—are the central elements of the place of tumor development.37 Watanabe in 1954 by injecting bronchogenic carcinoma cells of mice showed that the CTCs can be the key role players in metastasis.38 It was shown that the metastases development, to some extent, depends on the size and quantity of the CTC clusters.39,40 The CTCs are highly heterogenic, and several types of CTCs are suggested, which are given below:
Traditional CTCs: The hallmark of these cells is having large and irregular shapes, with an intact, viable nucleus; they express cytokeratins with epithelial origin and not hematopoietic origin (they do not have CD45).41,42
Cytokeratin-negative (CK−) CTCs: These types of CTCs are cancer stem cells passing through the epithelial-mesenchymal transition steps and forming the cytokeratin-negative CTCs. They can be opposed to treatment and is the most potential one to invade and form metastasis because they do not have cytokeratins and CD45.43-45
Apoptotic CTCs: These are the traditional CTCs that undergo apoptosis. They are distinguishable with nuclear fragmentation or cytoplasmic blabbing associated with cell death. The proportion of apoptotic CTCs compared with the traditional CTCs can be a predictor of treatment efficacy.46,47
Small CTCs: These are recognized as the cytokeratin-positive and CD45-negative cells, similar in size to leukocytes. Notably, the small CTCs have been concerned with progressive disease and differentiation into small cell carcinomas, which often need special therapeutic strategies.48,49
However, the new classification system suggested that CTCs should be characterized by their size and the expression of several markers such as EpCAM.50 The principle of CTC isolation, quantification, and characterization is based on the different physical characteristics (magnitude, electric charges) and genetic properties of the CTCs compared with the nontumor cells.51,52 Isolation by size of the epithelial tumor cell (ISET) platform is used for CTC cluster isolation based on tumor and nontumor cell differences.53 Interestingly, physical property can also support micro-fluidic technology, which results in making a new flexible micro-spring array device.54-57 The CellSearch platform, Menarini Silicon Biosystems (https://www.cellsearchctc.com/) system, which is based on EpCAM- and cytokeratin-positive CTC selection, is the only Food and Drug Administration (FDA)–approved one and is used in numerous clinical studies.58-61 In breast cancer, it was found that metastatic breast cancer patients who had CTCs ⩾5, contrary to patients with CTCs <5, in 7.5 mL of blood had failed first-line treatment.62-64 The cutoff value (CTCs ⩾5/7.5 mL of blood) in prostate cancer highlighted the fact that the higher CTC numbers resulted in poorer overall survival.65,66 The new cutoff is a CTC threshold of 5 cells per 7.5 mL, which means patient with CTCs ⩾5 were classified as Stage IVaggressive and those with CTCs <5 as Stage IVindolent.67 In colorectal cancer, the comparable predictive value of the CTC counts was optional for the selection of first-line treatment.68,69 Higher quantity of CTC number was linked to lower survival in lung cancer as well.
Cell-Free Nucleic Acids
“Cell-free nucleic acids” is a general term and comprises cfDNA, cell-free RNA (cfRNA), and cell-free mitochondrial DNA (cf-mtDNA). Cell-free DNA are nonencapsulated DNA, free from cells in the bloodstream. They originate from a tumor clone.70 In eukaryotic nucleus, the DNA exists in a structured form termed nucleosomes which consist of around 170 base pairs of DNA wrapped around a core histone octamer (H2A, H2B, H3, and H4), linked by 10 to 100 base pairs of naked DNA as a linker DNA.71,72 The fragmented DNA, in the form of nucleosomes, can be released by various tumor cells, and then these enter the bloodstream during apoptosis or necrosis and normally are removed with macrophages.73,74 In cancerous condition, these cfDNA fragments are overproduced by the tumor cells, which are left behind that the macrophages cannot clean up completely.75 The first connection between cancer and elevated cfDNA was shown in 1977 by Leon et al.76 Then, Stroun et al77 established this link by separating DNA gained from the plasma of cancer patients and analyzing them. Patients with malignant epithelial gastrointestinal tumors have more cfDNA in comparison with patients with benign disease.78 The cfDNA is typically present in plasma as a double-stranded form (dsDNA), even though the single-stranded form can also been recognized.79,80
In 1987, the circulating cfRNA, in the form of RNA proteo-lipid complex, was isolated from the serum of malignant ones.70,81 A cfRNA scan of messenger RNA and microRNA (miRNA) is made. In 1999, the presence of cfRNAs was established in the plasma or serum of patients with nasopharyngeal carcinoma and malignant melanoma.82-84 Moreover, the presence of cfRNA was proven in patients with breast cancer, colorectal cancer, follicular lymphoma, and hepatocellular carcinoma.85-87 The circulating cell-free circulating microRNAs are the new course of biomarkers as they possess all the crucial features such as sensitivity, predictability, specificity, robustness, translatability, and noninvasiveness. MicroRNAs are small noncoding RNAs that act as suppressors of protein translation and disturb the protein expression panel at posttranscriptional mechanisms.88
Thanks to the latest advances in molecular genetics techniques, taking the genetic and epigenetic alterations of cfDNA and cfRNA into consideration is not very hard.89,90 Several studies on “Catalogue of Somatic Mutations in Cancer” (COSMIC) mutations, using next-generation sequencing (NGS), verified that circulating cfDNA analysis can be an excellent prognostic and diagnostic tool.91-93 One of the most promising applications of ctDNA is treatment response monitoring.94-96 It is also suggested that ctDNA-based analysis possibly will develop the controlling of patients with possibly treatable or metastatic disease.97 Recently, Newman et al. and Murtaza M et al.98,99 have suggested an ultrasensitive and cost-effective method entitled cancer-personalized profiling by deep sequencing for cfDNA quantification.
Mainly, DNA methylation in CpG islands as the epigenetic event happens nearly at the beginning of cancer development, and so it has the potential of being a biomarker for early diagnosis.100-102 So, DNA methylation of cfDNA can be checked and several markers can be proposed, including both global genomic hypomethylation (Alu elements) and gene-specific methylation such as GSTP1 methylation in prostate cancer, methylation of cfDNA in SEPT9 promoter region in colorectal cancer, and RASSF1A in different cancer types.103,104 It is shown that genome-wide cfDNA methylation profiles are extremely counterpart with detected methylation in corresponding tumor tissues.105 The methylated cfDNA biomarkers are a comprehensive noninvasive monitoring tool of treatment response in metastatic colorectal cancer.106,107 SOX17 promoter methylation in CTCs and matched cfDNA isolated from plasma of patients with breast cancer indicated a direct connection between the presence of CTCs and cfDNA in patients with operable breast cancer, after surgical removal of the primary tumor.108
Extrachromosomal Circular DNA
In the 1980s, the presence of endogenous DNA circles originating from canonical linear chromosomal loci, identified as eccDNA, was described in nuclear fractions of plant cells (wheat and tobacco).109 In fact, the main machinery of oncogenes to aggregate their copy number occurs by eccDNA.110 It was shown that eccDNA can be seen in approximately half of human cancers, while its frequency is different by tumor type.111,112 The presence of tumor eccDNA in blood as a liquid biopsy component has been suggested very recently.113
Exosomes
Microvesicles and exosomes, collectively referred to as extracellular vesicles (EVs), are lipid bilayer structure vesicles that are released from all eukaryotic cells and play an important role in the instruction of extracellular communication, cellular differentiation, cell migration, and maintenance of normal tissue condition.114 The size of the exosomes varies from 30 to 100 nm, and they are secreted through the interior budding of the plasma cell membrane.115 The exosomes can be released through both normal (epithelial, mesenchymal, and immune) and cancerous cells in different settings such as blood, urine, and sputum.116 They were first described by Pan and Johnstone in 1983 at McGill University.117 It was suggested that there is a connection among the existence of cfNAs in plasma and exosomes because one possible mechanism for the release of the cfNAs into blood is by exosomes.118,119 The transferring of genetic information from the exosomes to the host cells (receiver of exosomes) is possibly involved in the metastatic conversion of the host/receiver cells.120 Exosomes of diverse cell types have unlike proteins that can be potentially used as biomarkers in clinical experiments.121
Exosomes contain dsDNA of the parent cell, so they could be released from a specific tissue or from a specific tumor via the exosomal surface biomarkers.122,123 Using sensitive detection technologies such as nano-particle tracking analysis (ZetaView), Western blotting techniques, transmission electron microscopy, the Agilent Bioanalyzer system, and modern droplet digital polymerase chain reaction techniques, we are able to assess the exosomal nucleic acids.124,125
Exosome-based liquid biopsy in comparison with the CTCs and cfNAs are more homogeneous in terms of size.126 Many isolation and characterization protocols are established to prepare the exosomes for the diagnosis of cancer and its therapy.126,127
Clinical Applications of Liquid Biopsy
In fact, CTC, ctDNA, and exosomes have broad biomarker potential because they can timely and dynamically represent the tumor’s genetic status both for diagnosis and for prognosis applications. It was suggested that liquid biopsy has a better sensitivity and is extra convenient as a cancer diagnosis tool in comparison with the traditional tissue biopsy methods.128 In the middle of 2016, the first liquid biopsy test was approved by the FDA.129,130 The point mutation of exon 19 deletion or exon 21 [L858R] in the epidermal growth factor receptor (EGFR) gene of ctDNA was approved as a good predictor of the response to the EGFR tyrosine kinase inhibitors in non–small-cell lung cancer patients.131,132 When the first test of ctDNA was approved, several studies had already shown the impact of liquid biopsy in the field of cancer management. Several studies have stated that breast and ovarian cancers with ctDNA microsatellite instability have deprived prognosis.133-135 The presence of apoptotic CTCs or fragmented CTCs in the peripheral blood can be an indicator of cancer.24,136 Although imaging and tissue biopsy are still the gold standards in solid tumor screening and monitoring, prospective studies have suggested that CTC detection with imaging combination is the greatest choice.137,138 The quick diagnostic value of CTCs in primary stages of cancer has been considered several times, and in an animal model, it was shown that CTCs were established very quickly in the “carcinoma in situ” stage, implying the fact that the tumor cells had spread prior to diagnosis.139,140 The exceeding indication showed that the CTCs may be a valuable tool for very early cancer diagnosis. The detection of CTCs in different cancer types including colorectal cancer, lung cancer, and prostate cancer correlates with different pathological stages, clinical outcome, and patient’s survival.141-146 The first and only actionable test for detecting CTCs in cancer patients is the CellSearch system, which is applicable as a self-determining forecaster of overall and progression-free survival in metastatic breast, prostate, and colorectal cancer.147,148 In addition to CTC detection, ctDNA is a gifted approach for primary cancer diagnosis. Many researchers have indicated a positive connection of ctDNA quantity and tumor stage to predict the severity of malignancies and efficacy of treatment.90,149,150 The use of exosomes as a predictive biomarker and indicator of treatment response completely relies on its protein and miRNA expression profiles. It was shown that the downregulation of exosomal miR-92a in hepatocellular carcinoma was connected to cancer development and a high risk of relapse, and the overexpression of exosomal miR-21-3p is a sign of cisplatin resistance in ovarian cancer.151,152
At MD Anderson Cancer Center, the role of liquid biopsy as a medical tool for patients with papillary thyroid carcinoma (PTC), medullary thyroid carcinoma (MTC), and anaplastic thyroid carcinoma is analyzed. In patients with PTC and MTC, it is shown that the concordance between tissue and liquid biopsy was more than 80%.153
Thanks to the latest advances in molecular technology detection, in methods such as NGS, whole-genome shotgun bisulfate sequencing, and tagged-amplicon deep sequencing, day by day, the importance of liquid biopsy is highlighted more and more. Most scientists believe that its use in cancer treatment will support the conventional tissue biopsy method instead of replacing it. Liquid biopsies may not replace tissue biopsies at the present or in the future; but it will let many people to be tested. Furthermore, using a blood test along with tissue will be a breakthrough because, more often than not, the traditional biopsy does not pick enough tissue.27,154,155
Conclusion
Taking everything into consideration, liquid biopsy is a new noninvasive sampling tool which brings a new insight into the cancer diagnosis and prognosis perspectives. It can use several components such as cfDNA, CTCs, and exosomes as the real representative of tumor to evaluate the tumor genetic alterations and status. Mostly, for evaluation of the cancer treatment efficacy, the number of CTCs with cutoff 5 per 7.5 mL of blood works. For real-time tracking, the genetic alterations of tumor cfDNA and exosomes can be used to select the best treatment in the way of personalized medicine. In the near future, liquid biopsy will take the place of tissue biopsy or will support the suspicious and nondetermining result of it in an excellent way.
Acknowledgments
Special thanks to Urology Research Center (URC), Tehran University of Medical Sciences, Tehran, Iran.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration Of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: SMKA: Principal investigator, supervision of the team, and editing the manuscript, RH and ME running the project and data acquisition, FKH wrote the manuscript.
Availability of Data and Material: The datasets used and/or analyzed during the current study are available from the corresponding author.
Consent for Publication: This review article does not contain data from any individual person; consequently, the consent for publication is “Not Applicable” in this section.
Ethics Approval and Consent to Participate: This manuscript does not report on or involve the use of any animal or human data or tissue, so ethical approval is not applicable in this study.
ORCID iD: Fatemeh Khatami  https://orcid.org/0000-0002-6311-1336
https://orcid.org/0000-0002-6311-1336
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [DOI] [PubMed] [Google Scholar]
- 2. Ashworth T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med J Australia. 1869;14:146-147. [Google Scholar]
- 3. Mandel P. Les acides nucleiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil. 1948;142:241-243. [PubMed] [Google Scholar]
- 4. Leon S, Shapiro B, Sklaroff D, Yaros M. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-650. [PubMed] [Google Scholar]
- 5. Kidess E, Jeffrey SS. Circulating tumor cells versus tumor-derived cell-free DNA: rivals or partners in cancer care in the era of single-cell analysis? Genome Med. 2013;5:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castro-Giner F, Gkountela S, Donato C, et al. Cancer diagnosis using a liquid biopsy: challenges and expectations. Diagnostics (Basel). 2018;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472-484. [DOI] [PubMed] [Google Scholar]
- 8. Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013;73:6384-6388. [DOI] [PubMed] [Google Scholar]
- 9. Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med. 2017;15:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17:507. [DOI] [PubMed] [Google Scholar]
- 12. Prasad V, Fojo T, Brada M. Precision oncology: origins, optimism, and potential. Lancet Oncol. 2016;17:e81-e86. [DOI] [PubMed] [Google Scholar]
- 13. McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15-26. [DOI] [PubMed] [Google Scholar]
- 14. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arnaud A. Costs and outcomes comparison of tissue and blood based biopsies for the purpose of biomarker testing. Value Health. 2016;19:A143-A144. [Google Scholar]
- 18. Karachaliou N, Mayo-de-Las-Casas C, Molina-Vila MA, Rosell R. Real-time liquid biopsies become a reality in cancer treatment. Ann Transl Med. 2015;3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rossi G, Mu Z, Rademaker AW, et al. Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin Cancer Res. 2018;24:560-568. [DOI] [PubMed] [Google Scholar]
- 20. Huang W-L, Chen Y-L, Yang S-C, et al. Liquid biopsy genotyping in lung cancer: ready for clinical utility? Oncotarget. 2017;8:18590-18608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeffrey SS, Toner M. Liquid biopsy: a perspective for probing blood for cancer. Lab Chip. 2019;19:548-549. [DOI] [PubMed] [Google Scholar]
- 22. Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clinical Chemistry. 2013;59:110-118. [DOI] [PubMed] [Google Scholar]
- 23. Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623-631. [DOI] [PubMed] [Google Scholar]
- 24. Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479-491. [DOI] [PubMed] [Google Scholar]
- 25. Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pantel K, Speicher M. The biology of circulating tumor cells. Oncogene. 2016; 35:1216-1224. [DOI] [PubMed] [Google Scholar]
- 27. Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17: 223-238. [DOI] [PubMed] [Google Scholar]
- 28. Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017;141:220-230. [DOI] [PubMed] [Google Scholar]
- 29. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30: c836-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moore C, Kosgodage U, Lange S, Inal JM. The emerging role of exosome and microvesicle- (EMV-) based cancer therapeutics and immunotherapy. Int J Cancer. 2017;141:428-436. [DOI] [PubMed] [Google Scholar]
- 31. Krishnamurthy N, Spencer E, Torkamani A, Nicholson L. Liquid biopsies for cancer: coming to a patient near you. J Clin Med. 2017;6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaiser J. “Liquid Biopsy” for Cancer Promises Early Detection. Washington, DC: American Association for the Advancement of Science; 2018. [DOI] [PubMed] [Google Scholar]
- 33. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112-123. [DOI] [PubMed] [Google Scholar]
- 34. Macias M, Alegre E, Diaz-Lagares A, et al. Liquid biopsy: from basic research to clinical practice. Adv Clin Chem. 2018;83:73-119. [DOI] [PubMed] [Google Scholar]
- 35. Neoh KH, Hassan AA, Chen A, et al. Rethinking liquid biopsy: microfluidic assays for mobile tumor cells in human body fluids. Biomaterials. 2018;150:112-124. [DOI] [PubMed] [Google Scholar]
- 36. Riquet M, Rivera C, Gibault L, et al. Lymphatic spread of lung cancer: anatomical lymph node chains unchained in zones. Rev Pneumol Clin. 2014;70:16-25. [DOI] [PubMed] [Google Scholar]
- 37. Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;133:571-573. [PubMed] [Google Scholar]
- 38. Watanabe S. The metastasizability of tumor cells. Cancer. 1954;7:215-223. [DOI] [PubMed] [Google Scholar]
- 39. Liotta LA, Kleinerman J, Saidel GM. Quantitative relationships of intravascular tumor cells, tumor vessels, and pulmonary metastases following tumor implantation. Cancer Res. 1974;34:997-1004. [PubMed] [Google Scholar]
- 40. Knisely WH, Mahaley MS., Jr. Relationship between size and distribution of “spontaneous” metastases and three sizes of intravenously injected particles of VX2 carcinoma. Cancer Res. 1958;18:900-905. [PubMed] [Google Scholar]
- 41. Racila E, Euhus D, Weiss AJ, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci U S A. 1998;95:4589-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu X, Taftaf R, Kawaguchi M, et al. Homophilic CD44 interactions mediate tumor cell aggregation and polyclonal metastasis in patient-derived breast cancer models. Cancer Discov. 2019;9:96-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marrinucci D, Bethel K, Kolatkar A, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9:016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gkountela S, Castro-Giner F, Szczerba BM, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176:98-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCarthy JB, El-Ashry D, Turley E. Hyaluronan, cancer-associated fibroblasts and the tumor microenvironment in malignant progression. Front Cell Dev Biol. 2018;6:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Allen JE, Saroya BS, Kunkel M, et al. Apoptotic circulating tumor cells (CTCs) in the peripheral blood of metastatic colorectal cancer patients are associated with liver metastasis but not CTCs. Oncotarget. 2014;5:1753-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ren C, Han C, Zhang J, et al. Detection of apoptotic circulating tumor cells in advanced pancreatic cancer following 5-fluorouracil chemotherapy. Cancer Biol Ther. 2011;12:700-706. [DOI] [PubMed] [Google Scholar]
- 48. Wang L, Li Y, Xu J, et al. Quantified postsurgical small cell size CTCs and EpCAM+ circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett. 2018;412:99-107. [DOI] [PubMed] [Google Scholar]
- 49. Shaw Bagnall J, Byun S, Begum S, et al. Deformability of tumor cells versus blood cells. Sci Rep. 2015;5:18542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vallorani R, Bartolini G, Betti G, et al. Circulation type classifications for temperature and precipitation stratification in Italy. Int J Climatol. 2018;38:915-931. [Google Scholar]
- 51. Balasubramanian P, Lang JC, Jatana KR, et al. Multiparameter analysis, including EMT markers, on negatively enriched blood samples from patients with squamous cell carcinoma of the head and neck. PLoS ONE. 2012;7:e42048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stott SL, Hsu C-H, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392-18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krebs MG, Hou J-M, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and-independent approaches. J Thorac Oncol. 2012;7:306-315. [DOI] [PubMed] [Google Scholar]
- 54. Adams AA, Okagbare PI, Feng J, et al. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J Am Chem Soc. 2008;130:8633-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Harouaka RA, Zhou M-D, Yeh Y-T, et al. Flexible micro spring array device for high-throughput enrichment of viable circulating tumor cells. Clin Chem. 2014;60:323-333. [DOI] [PubMed] [Google Scholar]
- 56. Zheng S, Lin H, Liu J-Q, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162:154-161. [DOI] [PubMed] [Google Scholar]
- 57. Sarioglu AF, Aceto N, Kojic N, et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12:685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Swennenhuis JF, van Dalum G, Zeune LL, Terstappen LWMM. Improving the CellSearch® system. Expert Rev Mol Diagn. 2016;16:1291-1305. [DOI] [PubMed] [Google Scholar]
- 59. Andreopoulou E, Yang LY, Rangel K, et al. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. Int J Cancer. 2012;130:1590-1597. [DOI] [PubMed] [Google Scholar]
- 60. Beije N, Jager A, Sleijfer S. Circulating tumor cell enumeration by the CellSearch system: the clinician’s guide to breast cancer treatment? Cancer Treat Rev. 2015;41:144-150. [DOI] [PubMed] [Google Scholar]
- 61. Khatami F, Aghayan HR, Sanaei M, Heshmat R, Tavangar SM, Larijani B. The potential of circulating tumor cells in personalized management of breast cancer: a systematic review. Acta Med Iran. 2017;55:175-193. [PubMed] [Google Scholar]
- 62. Hayes DF, Cristofanilli M, Budd GT, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218-4224. [DOI] [PubMed] [Google Scholar]
- 63. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302-6309. [DOI] [PubMed] [Google Scholar]
- 64. Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199-1209. [DOI] [PubMed] [Google Scholar]
- 65. Olmos D, Arkenau H-T, Ang J, et al. Circulating tumour cell (CTC) counts as intermediate end points in castration-resistant prostate cancer (CRPC): a single-centre experience. Ann Oncol. 2008;20:27-33. [DOI] [PubMed] [Google Scholar]
- 66. Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cristofanilli M, Pierga J-Y, Reuben J, et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): international expert consensus paper. Crit Rev Oncol Hematol. 2019;134:39-45. [DOI] [PubMed] [Google Scholar]
- 68. Tol J, Koopman M, Miller M, et al. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann Oncol. 2009;21:1006-1012. [DOI] [PubMed] [Google Scholar]
- 69. Cohen S, Punt C, Iannotti N, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol. 2009; 20:1223-1229. [DOI] [PubMed] [Google Scholar]
- 70. Garcia-Casas A, Garcia-Olmo DC, Garcia-Olmo D. Further the liquid biopsy: gathering pieces of the puzzle of genometastasis theory. World J Clin Oncol. 2017;8:378-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145-150. [DOI] [PubMed] [Google Scholar]
- 72. Holdenrieder S, Stieber P, Bodenmuller H, et al. Nucleosomes in serum of patients with benign and malignant diseases. Int J Cancer. 2001;95:114-120. [DOI] [PubMed] [Google Scholar]
- 73. Choi JJ, Reich CF, 3rd, Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology. 2005;115:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kajbafzadeh A-M, Payabvash S, Salmasi AH, Monajemzadeh M, Tavangar SM. Smooth muscle cell apoptosis and defective neural development in congenital ureteropelvic junction obstruction. J Urol. 2006;176:718-723; discussion 723. [DOI] [PubMed] [Google Scholar]
- 75. Gahan P, Stroun M. The biology of circulating nucleic acids in plasma and serum (CNAPS). In: Rykova EY, Kikuchi Y. eds. Extracellular Nucleic Acids. Berlin, Germany: Springer; 2010:167-89. [Google Scholar]
- 76. Leon S, Ehrlich G, Shapiro B, Labbate V. Free DNA in the serum of rheumatoid arthritis patients. J Rheumatol. 1977;4:139-143. [PubMed] [Google Scholar]
- 77. Stroun M, Anker P, Lyautey J, Lederrey C, Maurice PA. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol. 1987;23:707-712. [DOI] [PubMed] [Google Scholar]
- 78. Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116-2120. [DOI] [PubMed] [Google Scholar]
- 79. Tan E, Schur P, Carr R, Kunkel H. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Invest. 1966;45:1732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Koffler D, Agnello V, Winchester R, Kunkel HG. The occurrence of single-stranded DNA in the serum of patients with systemic lupus erythematosus and other diseases. J Clin Invest. 1973;52:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wieczorek AJ, Sitaramam V, Machleidt W, Rhyner K, Perruchoud AP, Block LH. Diagnostic and prognostic value of RNA-proteolipid in sera of patients with malignant disorders following therapy: first clinical evaluation of a novel tumor marker. Cancer Res. 1987;47:6407-6412. [PubMed] [Google Scholar]
- 82. Lo K-W, Lo YD, Leung S-F, et al. Analysis of cell-free Epstein-Barr virus-associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin Chem. 1999;45:1292-1294. [PubMed] [Google Scholar]
- 83. Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5:1961-1965. [PubMed] [Google Scholar]
- 84. Sarmadi S, Izadi-Mood N, Sotoudeh K, Tavangar SM. Altered PTEN expression; a diagnostic marker for differentiating normal, hyperplastic and neoplastic endometrium. Diagn Pathol. 2009;4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen XQ, Bonnefoi H, Pelte M-F, et al. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6: 3823-3826. [PubMed] [Google Scholar]
- 86. Dasi F, Lledo S, Garcia-Granero E, et al. Real-time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA: a simple blood test to monitor disease in cancer patients. Lab Invest. 2001;81:767-769. [DOI] [PubMed] [Google Scholar]
- 87. Miura N, Shiota G, Nakagawa T, et al. Sensitive detection of human telomerase reverse transcriptase mRNA in the serum of patients with hepatocellular carcinoma. Oncology. 2003;64:430-434. [DOI] [PubMed] [Google Scholar]
- 88. Faraldi M, Gomarasca M, Banfi G, Lombardi G. Free circulating miRNAs measurement in clinical settings: the still unsolved Issue of the normalization. Adv Clin Chem. 2018;87:113-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Salvi S, Gurioli G, De Giorgi U, et al. Cell-free DNA as a diagnostic marker for cancer: current insights. Onco Targets Ther. 2016;9:6549-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Khatami F, Larijani B, Tavangar SM. Circulating tumor BRAF mutation and personalized thyroid cancer treatment. Asian Pac J Cancer Prev. 2017;18:293-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rothe F, Laes J-F, Lambrechts D, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol. 2014;25:1959-1965. [DOI] [PubMed] [Google Scholar]
- 92. Lebofsky R, Decraene C, Bernard V, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol. 2015;9: 783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Komatsu S, Kiuchi J, Imamura T, Ichikawa D, Otsuji E. Circulating microRNAs as a liquid biopsy: a next-generation clinical biomarker for diagnosis of gastric cancer. J Cancer Metastasis Treat. 2018;4:36. [Google Scholar]
- 94. O’Leary B, Cutts RJ, Liu Y, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8:1390-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. O’Leary B, Hrebien S, Morden JP, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zheng D, Ye X, Zhang M, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lianos GD, Mangano A, Cho WC, Dionigi G, Roukos DH. Circulating tumor DNA: new horizons for improving cancer treatment. Future Oncol. 2015;11:545-548. [DOI] [PubMed] [Google Scholar]
- 98. Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20: 548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Murtaza M, Dawson S-J, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013; 497:108-112. [DOI] [PubMed] [Google Scholar]
- 100. Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457-463. [DOI] [PubMed] [Google Scholar]
- 101. Sanii S, Saffar H, Tabriz HM, Qorbani M, Haghpanah V, Tavangar SM. Expression of matrix metalloproteinase-2, but not caspase-3, facilitates distinction between benign and malignant thyroid follicular neoplasms. Asian Pac J Cancer Prev. 2012;13:2175-2178. [DOI] [PubMed] [Google Scholar]
- 102. Lehmann-Werman R, Neiman D, Zemmour H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016;113:E1826-E1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mastoraki S, Strati A, Tzanikou E, et al. ESR1 methylation: a liquid biopsy-based epigenetic assay for the follow-up of patients with metastatic breast cancer receiving endocrine treatment. Clin Cancer Res. 2018;24:1500-1510. [DOI] [PubMed] [Google Scholar]
- 104. Gale D, Lawson AR, Howarth K, et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE. 2018;13:e0194630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhai R, Zhao Y, Su L, Cassidy L, Liu G, Christiani DC. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia. 2012;14:29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Barault L, Amatu A, Siravegna G, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2018;67:1995-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cheuk IWY, Shin VY, Kwong A. Detection of methylated circulating DNA as noninvasive biomarkers for breast cancer diagnosis. J Breast Cancer. 2017;20: 12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Chimonidou M, Strati A, Malamos N, Georgoulias V, Lianidou ES. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin Chem. 2013;59:270-279. [DOI] [PubMed] [Google Scholar]
- 109. Kinoshita Y, Ohnishi N, Yamada Y, Kunisada T, Yamagishi H. Extrachromosomal circular DNA from nuclear fraction of higher plants. Plant Cell Physiol. 1985;26:1401-1409. [Google Scholar]
- 110. Turner KM, Deshpande V, Beyter D, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Fan Y, Mao R, Lv H, et al. Frequency of double minute chromosomes and combined cytogenetic abnormalities and their characteristics. J Appl Genet. 2011;52:53-59. [DOI] [PubMed] [Google Scholar]
- 113. Khatami F, Larijani B, Tavangar SM. The presence of tumor extrachomosomal circular DNA (ecDNA) as a component of liquid biopsy in blood. Med Hypotheses. 2018;114:5-7. [DOI] [PubMed] [Google Scholar]
- 114. Johnstone RM, Adam M, Hammond J, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412-9420. [PubMed] [Google Scholar]
- 115. Kastelowitz N, Yin H. Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. Chembiochem. 2014;15:923-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sánchez-Vela P, Garcia NA, Campos-Segura M, Forteza-Vila J. Liquid biopsy and tumor derived exosomes in clinical practice. Rev Esp Patol. 2016;49:106-111. [Google Scholar]
- 117. Pan B-T, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967-978. [DOI] [PubMed] [Google Scholar]
- 118. Lasser C, Alikhani VS, Ekstrom K, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Silva J, Garcia V, Rodriguez M, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51: 409-418. [DOI] [PubMed] [Google Scholar]
- 120. Kawamura Y, Yamamoto Y, Sato TA, Ochiya T. Extracellular vesicles as trans-genomic agents: emerging roles in disease and evolution. Cancer Sci. 2017;108: 824-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lin J, Li J, Huang B, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal. 2015;2015:657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014; 289:3869-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Bellingham SA, Shambrook M, Hill AF. Quantitative analysis of exosomal miRNA via qPCR and digital PCR. Methods Mol Biol. 2017;1545:55-70. [DOI] [PubMed] [Google Scholar]
- 125. Helwa I, Cai J, Drewry MD, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS ONE. 2017;12:e0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907-1920. [DOI] [PubMed] [Google Scholar]
- 127. Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med. 2013;7:769-778. [DOI] [PubMed] [Google Scholar]
- 128. Zhang W, Xia W, Lv Z, Xin Y, Ni C, Yang L. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem. 2017;41:755-768. [DOI] [PubMed] [Google Scholar]
- 129. Brown P. The Cobas® EGFR Mutation Test v2 assay. Future Oncol. 2016;12:451-452. [DOI] [PubMed] [Google Scholar]
- 130. Greig SL. Osimertinib: first global approval. Drugs. 2016;76:263-273. [DOI] [PubMed] [Google Scholar]
- 131. Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Khozin S, Blumenthal GM, Zhang L, et al. FDA approval: ceritinib for the treatment of metastatic anaplastic lymphoma kinase-positive non-small cell lung cancer. Clin Cancer Res. 2015;21:2436-2439. [DOI] [PubMed] [Google Scholar]
- 133. Schwarzenbach H, Eichelser C, Kropidlowski J, Janni W, Rack B, Pantel K. Loss of heterozygosity at tumor suppressor genes detectable on fractionated circulating cell-free tumor DNA as indicator of breast cancer progression. Clin Cancer Res. 2012;18:5719-5730. [DOI] [PubMed] [Google Scholar]
- 134. Deng A, Yang J, Lang J, et al. Monitoring microsatellite instability (MSI) in circulating tumor DNA by next-generation DNA-seq. J Clin Oncol. 2018;36:12025. [Google Scholar]
- 135. Kasi PM. Mutational burden on circulating cell-free tumor-DNA testing as a surrogate marker of mismatch repair deficiency or microsatellite instability in patients with colorectal cancers. J Gastrointest Oncol. 2017;8:747-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Larson CJ, Moreno JG, Pienta KJ, et al. Apoptosis of circulating tumor cells in prostate cancer patients. Cytometry A. 2004;62:46-53. [DOI] [PubMed] [Google Scholar]
- 137. Collins LC, Tamimi RM, Baer HJ, Connolly JL, Colditz GA, Schnitt SJ. Outcome of patients with ductal carcinoma in situ untreated after diagnostic biopsy. Cancer. 2005;103:1778-1784. [DOI] [PubMed] [Google Scholar]
- 138. Sabetkish S, Kajbafzadeh AM, Sabetkish N, et al. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix liver scaffolds. J Biomed Mater Res A. 2015;103:1498-1508. [DOI] [PubMed] [Google Scholar]
- 139. Rhim AD, Mirek ET, Aiello NM, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Michaelson J, Cheongsiatmoy J, Dewey F, et al. Spread of human cancer cells occurs with probabilities indicative of a nongenetic mechanism. Br J Cancer. 2005;93:1244-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Di Costanzo F, Pinzani P, Orlando C, et al. Circulating tumour cells in colorectal cancer. Eur J Cancer Suppl. 2008;6:52-59. [Google Scholar]
- 142. Ilie M, Hofman V, Long-Mira E, et al. “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS ONE. 2014;9:e111597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang Y, Zhou Y, Hu Z. The functions of circulating tumor cells in early diagnosis and surveillance during cancer advancement. J Transl Int Med. 2017;5: 135-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Chen C-J, Sung W-W, Chen H-C, et al. Early assessment of colorectal cancer by quantifying circulating tumor cells in peripheral blood: ECT2 in diagnosis of colorectal cancer. Int J Mol Sci. 2017;18:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Chen JF, Lu YT, Cheng S, Tseng HR, Figlin RA, Posadas EM. Circulating tumor cells in prostate cancer: beyond enumeration. Clin Adv Hematol Oncol. 2017;15:63-73. [PubMed] [Google Scholar]
- 146. Hu B, Rochefort H, Goldkorn A. Circulating tumor cells in prostate cancer. Cancers. 2013;5:1676-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Miller MC, Doyle GV, Terstappen LW. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol. 2010;2010:617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Veridex L. CellSearch circulating tumor cell kit premarket notification—expanded indications for use—metastatic prostate cancer. https://www.accessdata.fda.gov/cdrh_docs/pdf7/K073338.pdf. Updated 2008. Accessed December 16, 2008.
- 149. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early-and late-stage human malignancies. Sci Transl Med. 2014;6: 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Perrone F, Lampis A, Bertan C, et al. Circulating free DNA in a screening program for early colorectal cancer detection. Tumori. 2014;100:115-121. [DOI] [PubMed] [Google Scholar]
- 151. Masyuk AI, Masyuk TV, Larusso NF. Exosomes in the pathogenesis, diagnostics and therapeutics of liver diseases. J Hepatol. 2013;59:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Pink RC, Samuel P, Massa D, Caley DP, Brooks SA, Carter DRF. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecol Oncol. 2015;137:143-151. [DOI] [PubMed] [Google Scholar]
- 153. Bernicker EH. Liquid biopsies. In: Dacic S, Allen TC, Cagle PT, et al. , eds. Precision Molecular Pathology of Lung Cancer. Cham, Switzerland: Springer; 2018:275-286. [Google Scholar]
- 154. Mino-Kenudson M. Cons: can liquid biopsy replace tissue biopsy?—the US experience. Transl Lung Cancer Res. 2016;5:424-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. The Lancet Oncology. Liquid cancer biopsy: the future of cancer detection. Lancet Oncol. 2016;17:123. [DOI] [PubMed] [Google Scholar]