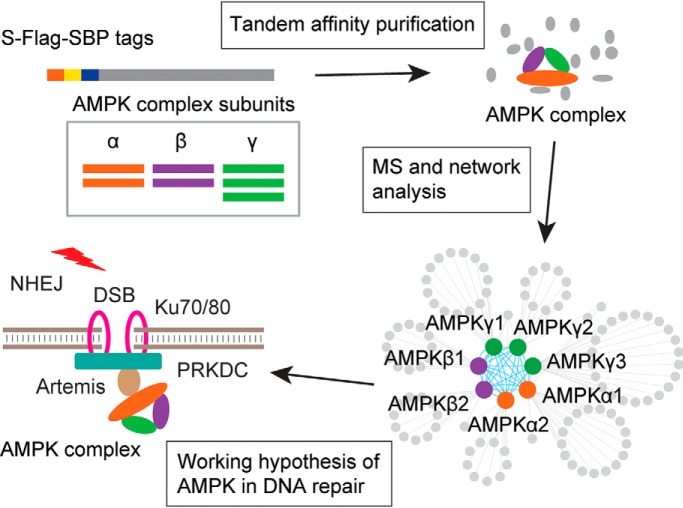
Adenosine monophosphate-activated protein kinase (AMPK) is an obligate heterotrimer that regulates cellular energy homeostasis. Chen et al. performed an interactome study of AMPK subunits using Tandem Affinity Purification coupled to mass spectrometry (TAP-MS). In addition to identifying many known and novel AMPK-interacting proteins, they uncovered a new function of AMPK in the NHEJ DNA repair pathway through its association with Artemis.
Keywords: Kinases, knockouts, mass spectrometry, non-covalent interaction MS, protein complex analysis, protein identification, protein-protein interactions
Graphical Abstract
Highlights
Each component of the AMPK trimeric complex was profiled by interaction proteomics.
The subunit composition of the AMPK complex directs interactions to distinct proteins.
AMPK interacts with Artemis and plays a role in Non-Homologous End Joining.
Abstract
Adenosine monophosphate-activated protein kinase (AMPK) is an obligate heterotrimer that consists of a catalytic subunit (α) and two regulatory subunits (β and γ). AMPK is a key enzyme in the regulation of cellular energy homeostasis. It has been well studied and is known to function in many cellular pathways. However, the interactome of AMPK has not yet been systematically established, although protein-protein interaction is critically important for protein function and regulation. Here, we used tandem-affinity purification, coupled with mass spectrometry (TAP-MS) analysis, to determine the interactome of AMPK and its functions. We conducted a TAP-MS analysis of all seven AMPK subunits. We identified 138 candidate high-confidence interacting proteins (HCIPs) of AMPK, which allowed us to build an interaction network of AMPK complexes. Five candidate AMPK-binding proteins were experimentally validated, underlining the reliability of our data set. Furthermore, we demonstrated that AMPK acts with a strong AMPK-binding protein, Artemis, in non-homologous end joining. Collectively, our study established the first AMPK interactome and uncovered a new function of AMPK in DNA repair.
Adenosine monophosphate-activated protein kinase (AMPK)1 has been identified as a key enzyme that regulates energy homeostasis, which is crucial for cell survival (1). When the ratios of AMP/ATP or ADP/ATP increase, AMPK is activated and regulates many downstream pathways, such as glucose metabolism, protein metabolism, fatty acid metabolism, autophagy, and mitochondrial homeostasis (2). As a result, AMPK positively regulates signaling pathways to generate more ATP and inhibits the anabolic pathways that consume ATP.
AMPK is an obligate heterotrimer consisting of a catalytic subunit (α) and two regulatory subunits (β and γ). In mammals, there are two α subunits (α1 and α2), two β subunits (β1 and β2), and three γ subunits (γ1, γ2, and γ3) (3). The tumor suppressor liver kinase B1 (LKB1) and calmodulin-dependent protein kinase kinase beta (CAMKKβ) are two upstream AMPK regulators that control AMPK activity (4, 5). In response to distinct stimuli, they activate AMPK by phosphorylating the activation loop T172 in the catalytic subunit α (6). AMPK is involved in different downstream pathways through the phosphorylation of its substrates. More and more AMPK substrates have been identified, which reveal a common conserved AMPK substrate motif (7). Screening for AMPK substrates has been attempted via different strategies, such as 14–3-3 binding and AMPK substrate motif searching (7, 8), a chemical genetic screen and peptide capture (9), a global phosphoproteomic analysis (10, 11), and an anti-AMPK motif antibody pulldown assay (12). Together, the functions of these newly identified AMPK substrates confirm that AMPK participates in many different biological processes.
Physical contacts between proteins in vivo are crucial for their regulation and function (13). Affinity purification combined with an MS-based analysis is highly efficient and has advantages in protein interactome research (14–17). To identify candidate binding partners of the AMPK complex, Moon et al. purified AMPKα1 and AMPKβ1 subunits using overexpressed Myc-tagged AMPK subunits and Myc-tag antibody, followed by an MS analysis (14). Pilot-Storch et al. used the yeast two-hybrid system to screen for the interactome of the PI3K-mTOR pathway genes that include one of the AMPK complex subunits, AMPKα1. They identified 27 interactors that potentially bind to AMPKα1 (18). However, a comprehensive analysis of the AMPK interactome, which includes all seven AMPK subunits, has yet to be conducted.
In this report, we used our modified tandem affinity purification and mass spectrometry (TAP-MS) analysis to characterize the interactomes of the seven AMPK subunits (AMPKα1 and α2, AMPKβ1 and β2, and AMPKγ1, γ2, and γ3). We generated stable cell lines that express these SFB-tagged (S-protein, FLAG, and streptavidin binding peptide) AMPK subunits in HEK293T cells. Interactome data filtration was performed using three different methods: upgraded significance analysis of interactome (SAINTexpress) (19), contaminant repository for affinity purification (CRAPome) (20), and enrichment analysis (21); we identified 138 candidate high-confidence interacting proteins (HCIPs) that may bind to AMPK subunits. With this list of HCIPs, we built an interaction network that included all seven AMPK subunits. We then selected five putative AMPK binding proteins and experimentally validated their interactions with AMPK. We revealed a new function of AMPK in non-homologous end joining (NHEJ) repair through its interaction with Artemis.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
TAP-MS analyses of seven AMPK subunits were performed using two biological replicates in HEK293T cells. The raw MS data were searched using Mascot, and the identified proteins and peptides were filtered by FDR<0.01 at the peptide level using the target-decoy method. The identified proteins were filtered for HCIPs using three strategies: SAINTexpress, CRAPome, and background enrichment. The negative control TAP-MS group in these data analyses included 46 experiments with baits that had no reported connection to the AMPK signaling pathway. The peptide-spectrum match (PSM) value of the identified protein was used in the HCIP analysis. The functional enrichment of the HCIPs was revealed by an Ingenuity Pathway Analysis (22) (Qiagen, Inc., Germantown, MD). The clonogenic survival assays were performed using at least three biological replicates, and a statistical analysis was performed using Student's t test.
Cell Culture and Transfection
HEK293T and HEK293A cells were purchased from the American Type Culture Collection and maintained in Dulbecco modified essential medium supplemented with 10% fetal bovine serum at 37 °C in 5% CO2 (v/v). The culture media contained 1% penicillin and streptomycin. Plasmid transfection was performed with polyethyleneimine reagent, as reported previously (23).
Plasmids and Antibody
The plasmids were purchased from Harvard Plasmids Resource, addgene, or Open Biosystems. All expression constructs were generated by polymerase chain reaction and subcloned into pDONOR201 vector as the entry clones using Gateway Technology (Invitrogen, Carlsbad CA). All the entry clones were subsequently recombined into a lentiviral-gateway-compatible destination vector to determine the expression of N-terminal triple-tagged (S protein, Flag epitope, and streptavidin-binding peptide) fusion proteins (24).
Anti-AMPKα1 (2795S), anti-AMPKα2 (2757S), anti-AMPKβ1 (4178S), anti-AMPKβ2 (4148S), anti-AMPKγ1 (4187S), anti-AMPKγ2 (2536S), and anti-Artemis (13381S) were purchased from Cell Signaling Technology (Danvers, MA) and used at 1:1000 dilution. Anti-(E3-independent) E2 ubiquitin-conjugating enzyme (UBE2O) antibody was purchased from Bethyl Laboratories (Montgomery, TX) and used at 1:2000 dilution. Anti-α-tubulin (T6199–200UL) and anti-Flag (M2) (F3165–5MG) monoclonal antibodies were purchased from Sigma-Aldrich (St. Louis, MO) and used at 1:5000 dilution. Anti-endosome-associated-trafficking regulator 1 (ENTR1) (sc-398909) was purchased from Santa Cruz Biotechnology (Dallas, TX) and used at 1:200 dilution in the immunofluorescence analysis.
Tandem Affinity Purification of SFB-tagged Proteins
HEK293T cells were transfected with plasmids encoding SFB-tagged AMPK family subunits or control proteins. Stable cell lines were selected with media containing 2 μg/ml puromycin and confirmed by immunostaining and Western blot analysis.
For tandem affinity purification, HEK293T cells were subjected to lysis with NETN buffer (100 mm NaCl; 1 mm EDTA; 20 mm Tris HCl; and 0.5% Nonidet P-40) and protease inhibitors at 4 °C for 20 min. Crude lysates were subjected to centrifugation at 14,000 rpm for 20 min at 4 °C. The supernatant was incubated with streptavidin-conjugated beads (GE Healthcare, Chicago, IL) for 2 h at 4 °C. The beads were washed three times with NETN buffer, and bounded proteins were eluted with NETN buffer containing 2 mg/ml biotin (Sigma-Aldrich) for 1 h at 4 °C. The elutes were incubated with S-protein agarose (Sigma-Aldrich) for 2 h, followed by three washes using NETN buffer. The beads were subjected to SDS-PAGE, and the gel was fixed and stained with Coomassie brilliant blue. The whole lane of the sample in the gel was excised and subjected to MS analysis. To generate a profiling background control for the HCIP analysis, we fractionated the HEK293T cell lysates into 18 fractions by SDS-PAGE gel and then analyzed them by MS using the same method that was used in the tandem affinity purification sample analysis.
MS Analysis
The excised gel bands were de-stained completely and washed with H2O three times before being dehydrated with 75% acetonitrile and subjected to trypsin (V5280, Promega Corporation, Madison, WI) digestion in 50 mm NH4HCO3 at 37 °C overnight. The peptides were extracted with acetonitrile and vacuum dried.
The samples were reconstituted in the MS loading solution (2% acetonitrile and 0.1% formic acid), delivered onto a nano reverse-phase high-performance liquid chromatography system, and eluted with 5–35% acetonitrile gradient containing 0.1% formic acid for 75 min at 700 nL/min. The eluate was electrosprayed into LTQ Orbitrap Velos Pro MS (Thermo Fisher Scientific, Waltham, MA) under positive ion mode and in a data-dependent manner, with a full MS scan of 350–1200 m/z, resolution of 60,000, minimum signal threshold of 1000, and isolation width of 2 Da.
The MS/MS spectra from the raw data were treated with Proteome Discoverer 1.4 (Thermo Fisher Scientific) and searched against the database using Mascot 2.4 (Matrix Science, Boston, MA), with proteins in Homo sapiens downloaded from Uniprot (March 2015, 20,203 entries). An automatic decoy database search was performed. Several parameters in Mascot were set for peptide searching, including oxidation for methionine and carboxyamidomethyl for cysteine as variable modifications, tolerance of two missed cleavages of trypsin, and a precursor mass tolerance of 10 ppm; the product ion tolerance was 0.5 Da. The identified peptides were filtered by Percolator, using a “Strict” Target FDR level of 0.01 based on the target-decoy method. The peptides were grouped into proteins by Proteome Discoverer, which removed the proteins that did not have unique peptides. Common contaminants were excluded.
AMPK Interactome Data Analysis
To evaluate potential protein-protein interactions, we assessed the identified peptides using the SAINTexpress method. The PSMs from AMPK proteins and the control group were assembled as a matrix for all of the bait and prey proteins. The interactions with a probability score > 0.8 from the SAINTexpress analysis were kept for the following analysis. We also used CRAPome to remove the high-count background contaminants. The interactions with a FC-B score < 2 were removed from the list. We compared the TAP-MS analysis results to the background using the HEK293T proteome profiling results, following a previously reported strategy (21). Proteins with a fold of enrichment > 2 were kept as candidate binding proteins. Using these three different analyses, we only chose the prey proteins that passed all three cut-off values as our HCIPs. The interactome network of AMPK family genes was generated by Cytoscape (25) based on these HCIPs. We used the Ingenuity Pathway Analysis to reveal the potential functional pathways involving these AMPK HCIPs against the background data set of all human proteins.
Pulldown and Western Blot Analysis
For the pulldown assay, 1 × 107 cells were lysed with NETN buffer containing protease inhibitors on ice for 20 min. The cell lysates were collected after centrifugation and incubated with 20 μl of S-beads for 2 h at 4 °C. The beads were washed with NETN buffer three times and boiled in 2× Laemmli buffer. The samples were resolved using SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane; immunoblotting was carried out with antibodies as indicated in the figures.
CRISPR/Cas9-mediated Gene Knockout
The guide RNAs that targeted Artemis were designed using an online tool, CHOPCHOP (26), and ligated into the LentiCRISPR plasmid according to a previously described protocol (27). The guide RNAs used to generate Artemis knockout clones were CTTCGATAGGGAGAACCTGA GGG and CTCCATAGACCGCTTCGATA GGG. They were co-transfected with Cas9 expression construct into HEK293A cells. Twenty-four hours later, we treated the cells with puromycin for 2 days and then placed them in 96-well plates. After 12 days of cell incubation, single clones were analyzed by Western blot analysis to screen for Artemis knockout cell clones.
Clonogenic Survival Assays
The clonogenic survival assays were performed as described in a previous paper (28). In brief, 250 cells were seeded onto 6-well plates, including HEK293A WT, AMPKα1/α2 double knockout, and Artemis knockout cells. Twenty-four hours later, the cells were exposed to different doses of ionizing radiation (IR). After IR treatment, the cells were incubated for 12 days. The colonies were stained with crystal violet and counted manually. The results were the averages of data from three independent experiments, and the statistical analysis was performed using Student's t test.
RESULTS
Overview of TAP-MS Proteomics Analysis of AMPK Complex
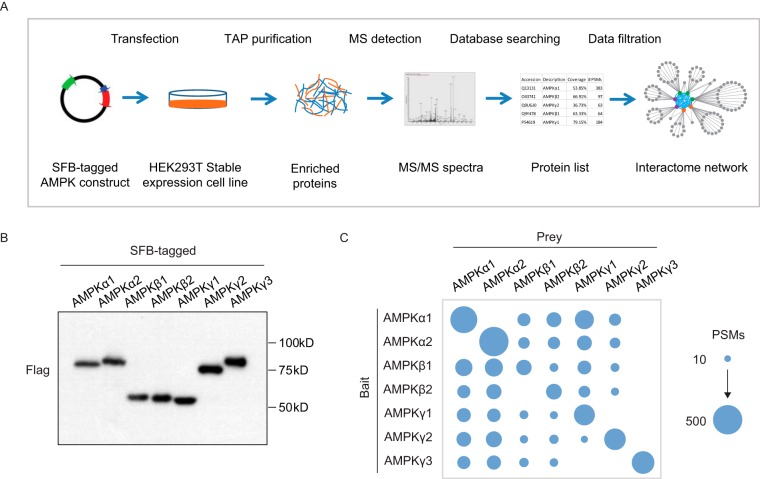
To determine the interaction network of the AMPK complex, we used the seven AMPK subunits (AMPKα1 and α2, β1 and β2, and γ1, γ2, and γ3) to perform a TAP-MS analysis following the protocol shown in Fig. 1A. We first generated HEK293T derivative cell lines that stably express the respective SFB-tagged AMPK subunits and verified these stable clones by Western blotting analyses (Fig. 1B). Cell lysates were extracted, followed by TAP purification. The enriched proteins were digested by Trypsin and analyzed by LTQ-Orbitrap Velos Pro MS (Thermo Fisher Scientific); the results were searched against the Homo sapiens database using Mascot. A biological repeat was conducted for each AMPK subunit. The protein and peptide identification list for each bait protein can be found in supplemental Tables S1 and S2.
Fig. 1.
Characterize AMPK interactome using TAP-MS analysis. A, AMPK interactome study workflow using a TAP-MS approach. Constructs encoding SFB-tagged AMPK subunits were expressed in HEK293T cells. Cell lysates underwent two steps of affinity purification using streptavidin beads and S-protein agarose beads. The samples were analyzed by LTQ-Orbitrap Velos Pro, and the identified peptides and proteins were further filtered using HCIP analysis, as described in the text. B, Validation of cell lines that stably expressed each of the seven AMPK subunits. C, The identified PSMs for AMPK subunits in each TAP-MS experiment are depicted. The size of the blue cycle reflects the number of identified PSMs.
AMPK functions as a protein complex that consists of three AMPK subunits. We confirmed the identification of each subunit from the TAP-MS results. As shown in Fig. 1C and Supplemental Fig. S1A–S1C, the subunits were captured with relatively high PSMs. However, AMPKγ3 was not identified in any of the TAP-MS analyses using other AMPK subunits. We checked the protein expression level using the whole proteome profiling data of HEK293T cells and the protein abundance database (supplemental Fig. S1D and S1E); the abundance of AMPKγ3 was much lower than that of any other AMPK subunits. However, when we overexpressed AMPKγ3 and performed the TAP-MS analysis, the two AMPKα subunits and two AMPKβ subunits were found to have high PSMs, indicating that they were able to bind strongly to the AMPKγ3 subunit. The TAP-MS results revealed that there was no interaction between AMPKα1 and AMPKα2. A similar situation appeared to be true for the other isoforms of the same AMPK subunit, except that we identified one unique peptide of AMPKβ2 in the AMPKβ1 purification and two unique peptides of AMPKγ1 in the AMPKγ2 purification.
The protein sequence similarity between AMPK subunits is high (e.g. 75% between AMPKα1 and AMPKα2); therefore, we determined whether this was the reason for the weak or absent interactions among AMPK isoforms. We performed an in-silico prediction of peptides obtained following trypsin digestion for all AMPK subunits. In this analysis, any peptides with fewer than seven amino acids were removed. In total, we obtained 35 peptides for AMPKα1 and 33 peptides for AMPKα2. There are only four shared peptides between AMPKα1 and AMPKα2. For the other AMPK subunits, there is one shared peptide between AMPKβ1 (16 peptides) and AMPKβ2 (15 peptides); three between AMPKγ1 (21 peptides) and AMPKγ2 (33 peptides); and one between AMPKγ3 (23 peptides) and AMPKγ1 or AMPKγ2 subunits. There is no common peptide among the three AMPKγ subunits. Therefore, the shared peptides between AMPK subunits is not as high as one may think based on the identities of their protein sequences. In addition, we checked the identified peptides and found that we failed to detect AMPKα1 and AMPKα2 in each other's IP samples, because there was no unique peptide identified in these IPs. Based on the results of these analyses, we believe that different isoforms of the same subunits show very little or no interaction.
Establishing the AMPK Interactome with HCIPs
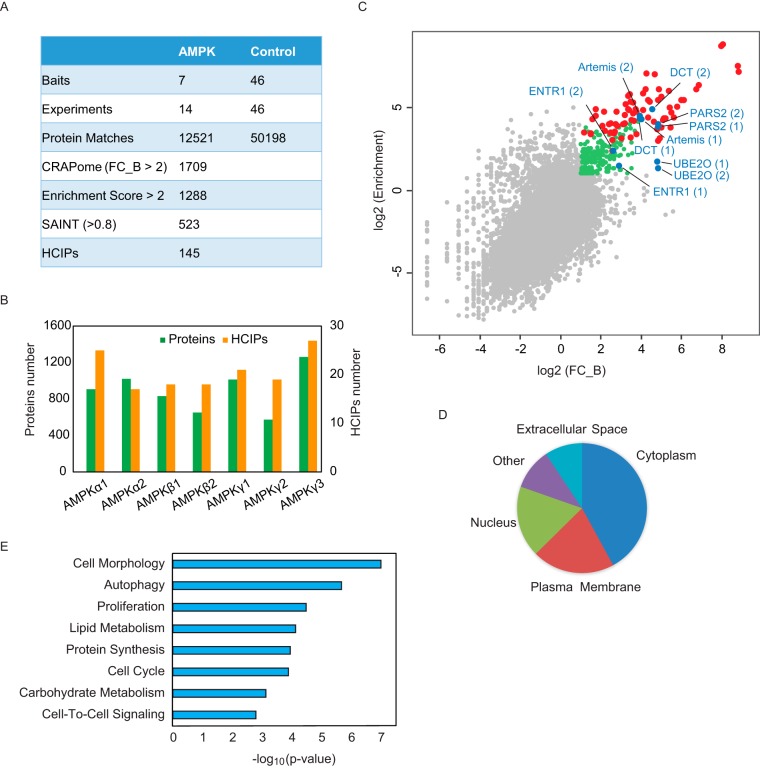
We filtered the TAP-MS results following the strategy we published previously (21). We used CRAPome and SAINTexpress to compare the AMPK subunit TAP-MS results to those of negative controls, which included 46 TAP-MS results with baits that are not functionally related to AMPK. We obtained 1709 proteins that scored FC_B > 2 CRAPome and 523 proteins that scored > 0.8 with SAINTexpress.
We compared the TAP-MS results with the proteome profiling data of HEK293T whole cell lysis. We uncovered 9481 proteins, as shown in supplemental Table S3. By comparison, we identified 1288 proteins with an enrichment score > 2 using the published method (21). We overlapped the lists from these three filtration analyses and identified 138 proteins as HCIPs (Fig. 2A, 2B, supplemental Table S4). The CRAPome and enrichment analysis results compared with the profiling data are plotted in Fig. 2C. The red dots are AMPK subunits that had high scores in both analyses, which confirmed the strong binding among the AMPK subunits. The blue and green dots are other proteins on the HCIP list. The blue dots are the AMPK binding candidates that we selected for further validation. We calculated the correlation of the HCIPs in the biological replicates and found high correlation, as shown in supplemental Fig. S1F.
Fig. 2.
Overview of the TAP-MS results for AMPK complexes. A, Summary of TAP-MS identification and data filtration. We performed two biological repeats for each AMPK subunit. Forty-six TAP-MS results underwent HCIP analysis using non-related baits as controls. The final HCIPs are the common genes obtained from all three HCIP filtration strategies. B, Total identified proteins and corresponding HCIPs for each AMPK subunit. C, The correlation between two different HCIP analysis strategies, CRAPome and enrichment analysis. The red dots are the identified AMPK subunits; the blue dots are the subunits selected for further validation; the green dots are the genes that passed both CRAPome and enrichment filtrations; and the gray dots are nonspecific binding proteins. D, Localization analysis for the HCIPs of AMPK subunits. E, Annotation of HCIPs using Ingenuity Pathway Analysis software.
To understand the function of these AMPK HCIPs, we used an Ingenuity Pathway Analysis. As shown in Fig. 2D, 42% of the proteins were cytoplasm proteins, 22% were plasma membrane proteins, and 19% were nuclear proteins. This broad distribution of HCIPs agrees with the results of numerous reports suggesting that AMPK functions at different locations and participates in a variety of biological processes. The functional characterization of these HCIPs, shown in Fig. 2E, further demonstrates that AMPK complexes participate in multiple cellular functions, including cell morphology, metabolism, and autophagy and the cell cycle.
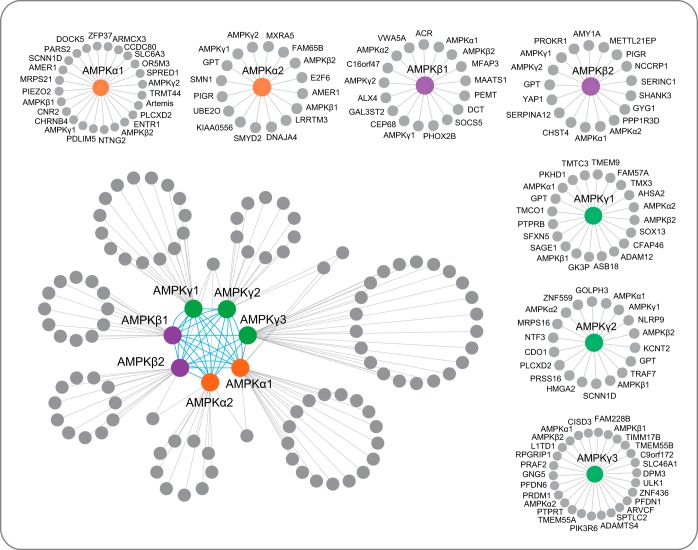
Using the AMPK HCIP list, we built a comprehensive interaction network using Cytoscape, as shown in Fig. 3. The interacting proteins for each AMPK subunit are also shown in supplemental Table S4. We identified several known AMPK-interacting proteins in the HCIPs of the AMPK complex. For example, in TAP-MS experiments using AMPKα2 as the bait, we identified UBE2O with 65 and 84 PSMs in two biological repeats. The results of the HCIP analysis also indicated that UBE2O is a strong AMPKα2 binding protein, and the results of our TAP-MS experiments suggest that it is a major binding partner of AMPKα2, but not AMPKα1. In agreement with our data, UBE2O was reported recently, because it specifically targets AMPKα2 for ubiquitination and degradation and promotes the activation of the mTOR-HIFα pathway (29). Another protein, PPP1R3D, was repeatedly identified in AMPKβ2 subunit TAP-MS experiments and passed all three of our HCIP analyses. This protein was found to interact directly with AMPKβ1/β2 subunits and may function in the regulation of glucose-induced AMPK dephosphorylation (30, 31). The identification of these known AMPK interaction proteins in our TAP-MS experiments led us to further validate AMPK HCIPs.
Fig. 3.
AMPK complex interaction network. Interaction network of the AMPK complex conducted by Cytoscape. The protein list was integrated from the two biological replicates. We removed the proteins identified as HCIPs in only one replicate, which also has fewer than three PSMs. The interactome has two parts, the integrated network and the individual interaction for each subunit. In the integrated interaction network, orange cycles are AMPKα subunits; purple cycles are AMPKβ subunits; green cycles are AMPKγ subunits; and gray cycles are HCIPs.
Validation of AMPK HCIPs
To demonstrate that our AMPK interactome is reliable, we selected five genes from the HCIP list for further validation, which included three AMPKα1 interacting proteins, Artemis, probable proline-tRNA ligase (PARS2), and ENTR1; one AMPKα2 interacting protein, UBE2O; and one AMPKβ1 binding protein, l-dopachrome tautomerase (DCT). These five genes were identified with relatively high scores in our HCIP analysis, as shown in Fig. 2C and Table I.
Table I. Summary of the five candidate genes selected for validation.
| Experiment | Bait | Prey | # PSMs | FC_A | FC_B | SAINT | Enrichment score |
|---|---|---|---|---|---|---|---|
| AMPKα1_1 | AMPKα1 | ARTEMIS | 10 | 15.14 | 15.14 | 1 | 20.79 |
| AMPKα1_2 | AMPKα1 | ARTEMIS | 10 | 15.28 | 15.28 | 1 | 23.24 |
| AMPKα1_1 | AMPKα1 | PARS2 | 35 | 46.12 | 28.19 | 1 | 14.55 |
| AMPKα1_2 | AMPKα1 | PARS2 | 35 | 46.57 | 28.47 | 1 | 16.26 |
| AMPKα1_1 | AMPKα1 | ENTR1 | 5 | 7.75 | 6.02 | 1 | 5.2 |
| AMPKα1_2 | AMPKα1 | ENTR1 | 7 | 10.57 | 8.2 | 1 | 8.13 |
| AMPKα2_1 | AMPKα2 | UBE2O | 84 | 71.95 | 27.66 | 1 | 3.33 |
| AMPKα2_2 | AMPKα2 | UBE2O | 65 | 74.09 | 28.48 | 1 | 2.52 |
| AMPKβ1_1 | AMPKβ1 | DCT | 9 | 16.18 | 16.18 | 1 | 20.1 |
| AMPKβ1_2 | AMPKβ1 | DCT | 12 | 23.72 | 23.72 | 1 | 30.51 |
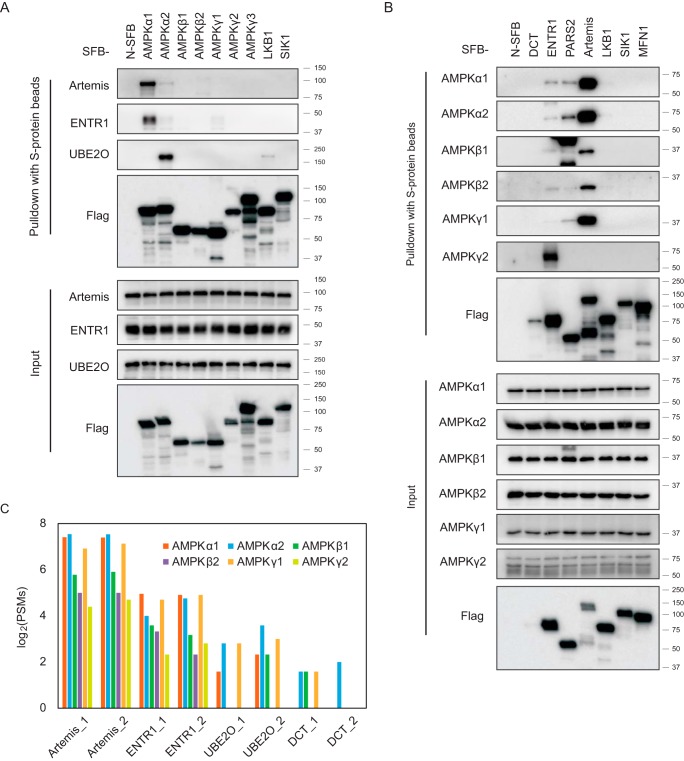
First, we overexpressed SFB-tagged AMPK complex subunits in HEK293T cells. Twenty-four hours after transfection, we performed a pulldown assay, incubating cell lysis with S-protein agarose in NETN buffer. The enriched protein lysis was analyzed by Western blot analysis. Antibodies against Artemis, ENTR1, and UBE2O were used to verify the interaction (Fig. 4A). Endogenous Artemis and ENTR1 showed a strong interaction with AMPKα1 but only a weak interaction with AMPKα2. On the other hand, UBE2O preferentially bound to the AMPKα2 subunit but not the AMPKα1 subunit. Next, constructs encoding SFB-tagged DCT, SDCCAGE, probable proline-tRNA ligase, or Artemis were transfected into HEK293T cells, and pulldown experiments were carried out with S-protein agarose beads. The samples were analyzed by Western blot analysis with the indicated AMPK antibodies that recognize different AMPK subunits (Fig. 4B). The results confirmed the interaction between AMPK subunits and these candidates, except DCT. We generated SFB-tagged cell lines with stable expression of Artemis, ENTR1, UBE2O, and DCT in HEK293T cells and performed a reverse TAP-MS analysis. The identified AMPK subunits are summarized with PSMs in Fig. 4C and supplemental Table S5. All AMPK subunits were identified with high PSMs when Artemis or ENTR1 was used as the bait protein. In the TAP-MS of UBE2O, we identified AMPKα1, α2, β1, and γ1. For the DCT results, only AMPKα2 was identified in the two biological repeats; AMPKβ1 and γ1 were identified in only one experiment with low PSMs, indicating that the interaction between DCT and AMPK was weak or transient.
Fig. 4.
Validation of the binding between AMPK complex and the candidate interaction proteins. A, Constructs encoding seven SFB-tagged AMPK subunits; the control genes LKB1, SIK1, or MFN1; or empty SFB construct were transfected into HEK293T cells. After 24 h, the cells were harvested, and the cell lysates were incubated with S-protein agarose and analyzed by Western blot analysis using the indicated antibodies. B, Constructs encoding the SFB-tagged AMPK complex binding candidates DCT, ENTR1, or Artemis; the control genes LKB1, SIK1, and MFN1; or empty SFB construct were transiently expressed in HEK293T cells. Cell lysates were subjected to pulldown assays with S-protein agarose and analyzed by Western blot analysis using the indicated antibodies. C, A TAP-MS analysis was performed using the candidate proteins interacting with the AMPK complex as the bait. The constructs encoding SFB-tagged Artemis, ENTR1, UBE2O, or DCT were transfected into HEK293T cells and selected with media containing 2 μg/ml puromycin. After TAP-MS, as shown in Fig. 1A, we identified the PSMs for each AMPK subunit. The number 1 or 2 on the x axis labels indicates two biological repeats for that gene.
AMPK May Participate in NHEJ DNA Repair Through Its Association with Artemis
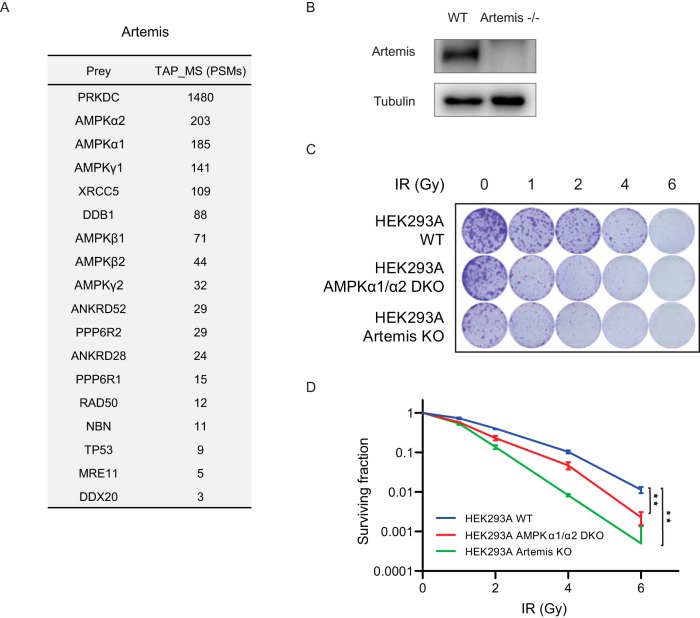
Based on the AMPK TAP-MS and validation experiment results shown above, we identified Artemis as a strong AMPK interacting protein. The TAP-MS results using Artemis as the bait protein are shown in Fig. 5A. AMPKα1, α2, β1, β2, γ1, and γ2 were all identified as top-ranked Artemis interacting proteins. DNA-dependent protein kinase catalytic subunit (PRKDC) was identified as an Artemis interacting protein; it has been reported that Artemis and PRKDC form a complex and play a major role in NHEJ DNA repair (32). Another known Artemis interacting protein, X-ray repair cross-complementing protein 5 (XRCC5), was also uncovered in our TAP-MS results and is a key component of the NHEJ repair pathway (33). We identified several other proteins with known functions in DNA repair, such as DDB1, RAD50, NBN, and MRE11. The proteins identified in these TAP-MS analyses are listed in supplemental Table S6.
Fig. 5.
AMPK may be involved in DNA repair via the NHEJ pathway. A, TAP-MS analysis of Artemis. The list of Artemis-interacting proteins is shown, which was identified by TAP-MS using Artemis as the bait. The PSM numbers are the average of the two repeats. B, HEK293A Artemis knockout cells were validated using Western blot analysis. C, D, Clonogenic survival assays. C, Images of the clonogenic survival assay results. D, Quantification and statistical analysis of clonogenic survival assay results. Clonogenic survival assays were performed using at least three biological replicates, and a statistical analysis was performed using Student's t test.
The binding between AMPK and Artemis may indicate that AMPK functions in NHEJ repair. We generated Artemis knockout HEK293A cells, as shown in Fig. 5B. AMPKα1/α2 double-knockout cells were reported in our previous study (11). We performed clonogenic survival assays to compare these knockout cells with parental HEK293A cells after treating them with different doses of IR (Fig. 5C and 5D). As expected, Artemis knockout cells had higher IR sensitivity. Similarly, AMPKα1/α2 double-knockout cells also had higher sensitivity to IR than did parental HEK293A cells. These results suggest that AMPK is involved in NHEJ DNA repair.
DISCUSSION
In this study, we comprehensively analyzed the interactome of the AMPK complexes using CRAPome, SAINTexpress, and an enrichment analysis and identified 138 HCIPs that may function with AMPK. We further established an interaction network for the AMPK complexes using these HCIPs. We identified several reported AMPK interacting proteins, such as UBE2O and PPP1R3D.
We then compared our list to previously reported interactomes of specific AMPK subunits. For AMPKα1, we uncovered 75% (i.e. 130 of 173) of the identified genes and proteins reported by the Giulio Superti-Furga group, who performed TAP purification using Twin-Strep-tag and hemagglutinin epitope tag (17). We obtained 53% (269 of 511) of the AMPKα1-interacting proteins and 55% (354 of 645) of the AMPKβ1-interacting proteins reported by Moon et al. (14) using myc-tag purification. Storch et al. applied a yeast two-hybrid screen for 33 components of the PI3K-mTOR pathway, including AMPKα1 (18); however, only seven of the 27 genes reported in that study were identified in our AMPKα1 purification. This discrepancy may be caused by the difference between these two methods.
Next, we compared our AMPK complex purification results with the data available in the BioGRID 3.5 (34). Of the 276 unique AMPK complex-interacting proteins in the database, 111 were identified in our TAP-MS results. However, only 11 remained in our HCIP list. After checking these results carefully, we noticed that several well-studied AMPK complex interacting genes and proteins, such as UBE2O and PPP1R3D, were not deposited in the BioGRID database. Overall, these comparisons confirm the high quality of our AMPK complex interactome, in which we eliminated many potential nonspecific interacting proteins. Nevertheless, we discovered many novel AMPK complex interacting proteins that warrant further functional validation.
Because AMPK's major function results from its ability to phosphorylate its substrates, we also identified some known AMPK substrates. YAP1 was identified as a strong AMPK binding protein that was reported as an AMPK phosphorylation substrate by several research groups, including ours (35–37). However, many other well-known AMPK substrates were not significantly enriched in our TAP-MS study and were removed during the HCIP analysis. For example, although ACC1 was found to have high PSMs, it was removed because it is a highly abundant protein and has nonspecific binding to streptavidin beads. Many other known AMPK substrates showed low signals in the TAP-MS results, including HDAC5 (four PSMs), ULK1 (three PSMs), TSC2 (two PSMs), and Raptor (one PSM). These low signals may be caused by the weak or transient binding nature of any given kinase and its phosphorylation substrates, which may dissociate during the two steps of affinity purification and therefore be difficult to recover in our TAP-MS analysis.
To screen for AMPK substrates using large-scale approaches, several research strategies have been employed. Using 14–3-3 binding and AMPK substrate motif searching, Reubun Shaw and colleagues identified ULK1, Raptor, and MFF as AMPK substrates (7, 8, 38). Chemical genetic screen and peptide capture technique were used by Anne Brunet and colleagues to identify new direct AMPK phosphorylation sites (9). In addition, global phosphoproteomic analysis, described by James Burchfield and colleagues (10) and our group (11), is a proven method for identifying AMPK substrates. However, these strategies rely on specific knowledge about the AMPK substrate motif or suffer from low coverage, which led to the discovery of limited AMPK substrates. Better and more efficient strategies are needed to further define kinase-substrate relationships.
The AMPK complex is composed of three subunits (AMPKα, β, and γ). For each subunit, there are several different isoforms: AMPKα1 and α2, β1 and β2, and γ1, γ2, and γ3. Our TAP-MS results revealed that the number of overlapping HCIPs among AMPK isoforms was not very high. For example, UBE2O was only identified as an AMPKα2 interacting protein, not as an AMPKα1 interacting protein. These results indicate that each isoform has its own unique function and regulation. As previously reported, there are differences in the cellular or tissue localization of different AMPK subunit isoforms. For example, the AMPKα1 and α2 isoforms have different functions in the osteogenesis, osteoblast-associated induction of osteoclastogenesis and adipogenesis (39). The expression of the two isoforms in tissue is also different. The AMPKα1 isoform is broadly expressed in most tissues, but AMPKα2 is highly expressed in the skeleton, cardiac muscle, and liver (3). The AMPKβ1 subunit is enriched in the nucleus of neurons and neuron stem cells, whereas AMPKβ2 is predominantly cytoplasmic (40, 41). A previous study also reported that the regulation of AMPK complexes by AMP and ADP is different when they contain different γ subunits (42). Because the AMPK complex has three subunits, it has 12 different types of AMPK complex that contain different AMPK subunits. The localization or function of these different AMPK complexes may differ because of tissue-specific expression, among other reasons (43, 44). In our study, we identified different HCIPs with distinct AMPK subunits, which is the first step to uncovering the complex regulation and functions of AMPK complexes that warrant further investigation.
AMPK's functions have been studied extensively. AMPK is known to participate in metabolic regulation, apoptosis, autophagy, cytoskeletal signaling, and transcriptional metabolism control (2, 45, 46). These biological functions mainly occur in cytoplasm. However, the AMPK complex has been shown to localize in both the nucleus and cytosol and may translocate from one to the other (47). More recent studies have reported that AMPK functions in the nucleus. For example, AMPK plays a role in UVB-induced DNA damage repair, and its activators prevent UVB-induced skin tumorigenesis (48). In another study, both AMPK and CHK1 contributed to S746 phosphorylation of EXO1 to promote fork protection in response to replication stress; in this case, AMPK was activated by CaMKKβ, not by LKB1 (49). In our study, we discovered a strong interaction between AMPK and Artemis, which is known to be critically important for the end-processing step during DNA repair via the NHEJ pathway (50). The results of our limited functional analysis indicate that AMPK is involved in NHEJ. Precisely whether and how this newly identified function of AMPK in DNA repair is mediated by its interaction with Artemis needs further mechanistic exploration.
In conclusion, we established the comprehensive interaction network of the AMPK complex using the TAP-MS approach. This AMPK interactome contains 138 HCIPs. We validated five HCIPs from the list and studied the potential functional link between AMPK and Artemis-dependent NHEJ. Our results suggest a newly identified function of AMPK in DNA repair, which further expands our understanding of the diverse functions of AMPK.
DATA AVAILABILITY
The MS proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository, with the data set identifier PXD015466.
Supplementary Material
Acknowledgments
We thank Drs. Yi Wang and Jong Min Choi for their kind help. We also thank the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for editing the manuscript.
Footnotes
* This work was supported by internal MD Anderson research support to J.C., who also received support from the Pamela and Wayne Garrison Distinguished Chair in Cancer Research. The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- AMPK
- adenosine monophosphate-activated protein kinase
- CAMKKβ
- calmodulin-dependent protein kinase kinase beta
- CRAPome
- contaminant repository for affinity purification
- DCT
- L-dopachrome tautomerase
- ENTR1
- endosome-associated-trafficking regulator 1
- HCIP
- high-confidence candidate interacting protein
- HPLC
- high-performance liquid chromatography
- IR
- ionizing radiation
- LKB1
- tumor suppressor liver kinase B1
- NHEJ
- non-homologous end joining
- PARS2
- probable proline–tRNA ligase
- PCR
- polymerase chain reaction
- PRKDC
- DNA-dependent protein kinase catalytic subunit
- PSM
- peptide-spectrum match
- SAINT
- significance analysis of interactome
- TAP-MS
- tandem affinity purification and mass spectrometry
- UBE2O
- (E3-independent) E2 ubiquitin-conjugating enzyme
- XRCC5/6
- x-ray repair cross-complementing protein 5/6.
REFERENCES
- 1. Hardie D. G., Ross F. A., and Hawley S. A. (2012) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mihaylova M. M., and Shaw R. J. (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stapleton D., Mitchelhill K. I., Gao G., Widmer J., Michell B. J., Teh T., House C. M., Fernandez C. S., Cox T., Witters L. A., and Kemp B. E. (1996) Mammalian AMP-activated protein kinase subfamily. J. Biol. Chem. 271, 611–614 [DOI] [PubMed] [Google Scholar]
- 4. Hurley R. L., Anderson K. A., Franzone J. M., Kemp B. E., Means A. R., and Witters L. A. (2005) The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 280, 29060–29066 [DOI] [PubMed] [Google Scholar]
- 5. Hong S. P., Leiper F. C., Woods A., Carling D., and Carlson M. (2003) Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. U.S.A. 100, 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woods A., Vertommen D., Neumann D., Turk R., Bayliss J., Schlattner U., Wallimann T., Carling D., and Rider M. H. (2003) Identification of phosphorylation sites in AMP-activated protein kinase (AMPK) for upstream AMPK kinases and study of their roles by site-directed mutagenesis. J. Biol. Chem. 278, 28434–28442 [DOI] [PubMed] [Google Scholar]
- 7. Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., and Shaw R. J. (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., Asara J. M., Fitzpatrick J., Dillin A., Viollet B., Kundu M., Hansen M., and Shaw R. J. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schaffer B. E., Levin R. S., Hertz N. T., Maures T. J., Schoof M. L., Hollstein P. E., Benayoun B. A., Banko M. R., Shaw R. J., Shokat K. M., and Brunet A. (2015) Identification of AMPK Phosphorylation Sites Reveals a Network of Proteins Involved in Cell Invasion and Facilitates Large-Scale Substrate Prediction. Cell Metab. 22, 907–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoffman N. J., Parker B. L., Chaudhuri R., Fisher-Wellman K. H., Kleinert M., Humphrey S. J., Yang P., Holliday M., Trefely S., Fazakerley D. J., Stockli J., Burchfield J. G., Jensen T. E., Jothi R., Kiens B., Wojtaszewski J. F., Richter E. A., and James D. E. (2015) Global Phosphoproteomic Analysis of Human Skeletal Muscle Reveals a Network of Exercise-Regulated Kinases and AMPK Substrates. Cell Metab. 22, 922–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Z., Lei C., Wang C., Li N., Srivastava M., Tang M., Zhang H., Choi J. M., Jung S. Y., Qin J., and Chen J. (2019) Global phosphoproteomic analysis reveals ARMC10 as an AMPK substrate that regulates mitochondrial dynamics. Nat. Commun. 10, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ducommun S., Deak M., Sumpton D., Ford R. J., Nunez Galindo A., Kussmann M., Viollet B., Steinberg G. R., Foretz M., Dayon L., Morrice N. A., and Sakamoto K. (2015) Motif affinity and mass spectrometry proteomic approach for the discovery of cellular AMPK targets: identification of mitochondrial fission factor as a new AMPK substrate. Cell Signal 27, 978–988 [DOI] [PubMed] [Google Scholar]
- 13. De Las Rivas J., and Fontanillo C. (2010) Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 6, e1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moon S., Han D., Kim Y., Jin J., Ho W. K., and Kim Y. (2014) Interactome analysis of AMP-activated protein kinase (AMPK)-alpha1 and -beta1 in INS-1 pancreatic beta-cells by affinity purification-mass spectrometry. Sci. Rep. 4, 4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Behrends C., Sowa M. E., Gygi S. P., and Harper J. W. (2010) Network organization of the human autophagy system. Nature 466, U68–U84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Varjosalo M., Sacco R., Stukalov A., van Drogen A., Planyavsky M., Hauri S., Aebersold R., Bennett K. L., Colinge J., Gstaiger M., and Superti-Furga G. (2013) Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MSMS. Nat. Methods 10, 307. [DOI] [PubMed] [Google Scholar]
- 17. Varjosalo M., Sacco R., Stukalov A., van Drogen A., Planyavsky M., Hauri S., Aebersold R., Bennett K. L., Colinge J., Gstaiger M., and Superti-Furga G. (2013) Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat. Methods 10, 307–314 [DOI] [PubMed] [Google Scholar]
- 18. Pilot-Storck F., Chopin E., Rual J. F., Baudot A., Dobrokhotov P., Robinson-Rechavi M., Brun C., Cusick M. E., Hill D. E., Schaeffer L., Vidal M., and Goillot E. (2010) Interactome mapping of the phosphatidylinositol 3-kinase-mammalian target of rapamycin pathway identifies deformed epidermal autoregulatory factor-1 as a new glycogen synthase kinase-3 interactor. Mol. Cell Proteomics 9, 1578–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teo G., Liu G., Zhang J., Nesvizhskii A. I., Gingras A. C., and Choi H. (2014) SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 100, 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., and Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Z., Tran M., Tang M., Wang W., Gong Z., and Chen J. (2016) Proteomic Analysis Reveals a Novel Mutator S (MutS) Partner Involved in Mismatch Repair Pathway. Mol. Cell Proteomics 15, 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kramer A., Green J., Pollard J. Jr, and Tugendreich S. (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Longo P. A., Kavran J. M., Kim M. S., and Leahy D. J. (2013) Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Srivastava M., Chen Z., Zhang H., Tang M., Wang C., Jung S. Y., and Chen J. (2018) Replisome dynamics and their functional relevance upon DNA damage through the PCNA interactome. Cell Rep. 25, 3869–3883 e3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Montague T. G., Cruz J. M., Gagnon J. A., Church G. M., and Valen E. (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelson T., Heckl D., Ebert B. L., Root D. E., Doench J. G., and Zhang F. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang C., Wang G., Feng X., Shepherd P., Zhang J., Tang M., Chen Z., Srivastava M., McLaughlin M. E., Navone N. M., Hart G. T., and Chen J. (2019) Genome-wide CRISPR screens reveal synthetic lethality of RNASEH2 deficiency and ATR inhibition. Oncogene 38, 2451–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vila I. K., Yao Y., Kim G., Xia W., Kim H., Kim S. J., Park M. K., Hwang J. P., Gonzalez-Billalabeitia E., Hung M. C., Song S. J., and Song M. S. (2017) A UBE2O-AMPKalpha2 axis that promotes tumor initiation and progression offers opportunities for therapy. Cancer Cell 31, 208–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garcia-Haro L., Garcia-Gimeno M. A., Neumann D., Beullens M., Bollen M., and Sanz P. (2010) The PP1-R6 protein phosphatase holoenzyme is involved in the glucose-induced dephosphorylation and inactivation of AMP-activated protein kinase, a key regulator of insulin secretion, in MIN6 beta cells. FASEB J. 24, 5080–5091 [DOI] [PubMed] [Google Scholar]
- 31. Oligschlaeger Y., Miglianico M., Dahlmans V., Rubio-Villena C., Chanda D., Garcia-Gimeno M. A., Coumans W. A., Liu Y., Voncken J. W., Luiken J. J., Glatz J. F., Sanz P., and Neumann D. (2016) The interaction between AMPKbeta2 and the PP1-targeting subunit R6 is dynamically regulated by intracellular glycogen content. Biochem. J. 473, 937–947 [DOI] [PubMed] [Google Scholar]
- 32. Ma Y., Pannicke U., Schwarz K., and Lieber M. R. (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108, 781–794 [DOI] [PubMed] [Google Scholar]
- 33. Roberts S. A., Strande N., Burkhalter M. D., Strom C., Havener J. M., Hasty P., and Ramsden D. A. (2010) Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature 464, 1214–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oughtred R., Stark C., Breitkreutz B. J., Rust J., Boucher L., Chang C., Kolas N., O'Donnell L., Leung G., McAdam R., Zhang F., Dolma S., Willems A., Coulombe-Huntington J., Chatr-Aryamontri A., Dolinski K., and Tyers M. (2019) The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mo J. S., Meng Z., Kim Y. C., Park H. W., Hansen C. G., Kim S., Lim D. S., and Guan K. L. (2015) Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 17, 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DeRan M., Yang J., Shen C. H., Peters E. C., Fitamant J., Chan P., Hsieh M., Zhu S., Asara J. M., Zheng B., Bardeesy N., Liu J., and Wu X. (2014) Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 9, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang W., Xiao Z. D., Li X., Aziz K. E., Gan B., Johnson R. L., and Chen J. (2015) AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 17, 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toyama E. Q., Herzig S., Courchet J., Lewis T. L. Jr, Loson O. C., Hellberg K., Young N. P., Chen H., Polleux F., Chan D. C., and Shaw R. J. (2016) Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y. G., Han X. G., Yang Y., Qiao H., Dai K. R., Fan Q. M., and Tang T. T. (2016) Functional differences between AMPK alpha1 and alpha2 subunits in osteogenesis, osteoblast-associated induction of osteoclastogenesis, and adipogenesis. Sci. Rep. 6, 32771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turnley A. M., Stapleton D., Mann R. J., Witters L. A., Kemp B. E., and Bartlett P. F. (1999) Cellular distribution and developmental expression of AMP-activated protein kinase isoforms in mouse central nervous system. J. Neurochem. 72, 1707–1716 [DOI] [PubMed] [Google Scholar]
- 41. Dasgupta B., and Milbrandt J. (2009) AMP-activated protein kinase phosphorylates retinoblastoma protein to control mammalian brain development. Dev. Cell 16, 256–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ross F. A., Jensen T. E., and Hardie D. G. (2016) Differential regulation by AMP and ADP of AMPK complexes containing different gamma subunit isoforms. Biochem. J. 473, 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dasgupta B., Ju J. S., Sasaki Y., Liu X., Jung S. R., Higashida K., Lindquist D., and Milbrandt J. (2012) The AMPK beta2 subunit is required for energy homeostasis during metabolic stress. Mol. Cell Biol. 32, 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quentin T., Kitz J., Steinmetz M., Poppe A., Bar K., and Kratzner R. (2011) Different expression of the catalytic alpha subunits of the AMP activated protein kinase–an immunohistochemical study in human tissue. Histol. Histopathol. 26, 589–596 [DOI] [PubMed] [Google Scholar]
- 45. Carling D., Mayer F. V., Sanders M. J., and Gamblin S. J. (2011) AMP-activated protein kinase: nature's energy sensor. Nat. Chem. Biol. 7, 512–518 [DOI] [PubMed] [Google Scholar]
- 46. Hardie D. G. (2011) AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miyamoto T., Rho E., Sample V., Akano H., Magari M., Ueno T., Gorshkov K., Chen M., Tokumitsu H., Zhang J., and Inoue T. (2015) Compartmentalized AMPK signaling illuminated by genetically encoded molecular sensors and actuators. Cell Rep. 11, 657–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu C. L., Qiang L., Han W., Ming M., Viollet B., and He Y. Y. (2013) Role of AMPK in UVB-induced DNA damage repair and growth control. Oncogene 32, 2682–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li S., Lavagnino Z., Lemacon D., Kong L., Ustione A., Ng X., Zhang Y., Wang Y., Zheng B., Piwnica-Worms H., Vindigni A., Piston D. W., and You Z. (2019) Ca(2+)-stimulated AMPK-dependent phosphorylation of Exo1 protects stressed replication forks from aberrant resection. Mol. Cell 74, 1123–1137 e1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lieber M. R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository, with the data set identifier PXD015466.