We designed two new quality assessment metrics namely “fraction of mis-identifications” (FMI) and “fraction of interprotein cross-links from known interactions” (FKI) for proteome-wide cross-link mass spectrometry data. We developed a new robust cross-link search engine named MaXLinker, with an “MS3-centric” approach that demonstrated high specificity and sensitivity. We performed a proteome-wide human XL-MS study and identified more than 9,300 cross-links. We further experimentally validated a large subset of novel interactions identified in our study using an orthogonal assay, thereby confirming the quality of our data and the robustness of our MaXLinker software.
Keywords: Protein cross-linking, systems biology, protein-protein interactions, computational biology, mass spectrometry
Graphical Abstract
Highlights
New quality assessment metrics to evaluate proteome-wide cross-linking mass spectrometry (XL-MS) data sets.
New “MS3-centric” cross-link search engine named MaXLinker with high sensitivity and specificity.
More than 9300 cross-links from our human proteome-wide XL-MS study.
Orthogonal experimental validation of novel interactions identified in our study.
Abstract
Protein-protein interactions play a vital role in nearly all cellular functions. Hence, understanding their interaction patterns and three-dimensional structural conformations can provide crucial insights about various biological processes and underlying molecular mechanisms for many disease phenotypes. Cross-linking mass spectrometry (XL-MS) has the unique capability to detect protein-protein interactions at a large scale along with spatial constraints between interaction partners. The inception of MS-cleavable cross-linkers enabled the MS2-MS3 XL-MS acquisition strategy that provides cross-link information from both MS2 and MS3 level. However, the current cross-link search algorithm available for MS2-MS3 strategy follows a “MS2-centric” approach and suffers from a high rate of mis-identified cross-links. We demonstrate the problem using two new quality assessment metrics [“fraction of mis-identifications” (FMI) and “fraction of interprotein cross-links from known interactions” (FKI)]. We then address this problem, by designing a novel “MS3-centric” approach for cross-link identification and implementing it as a search engine named MaXLinker. MaXLinker outperforms the currently popular search engine with a lower mis-identification rate, and higher sensitivity and specificity. Moreover, we performed human proteome-wide cross-linking mass spectrometry using K562 cells. Employing MaXLinker, we identified a comprehensive set of 9319 unique cross-links at 1% false discovery rate, comprising 8051 intraprotein and 1268 interprotein cross-links. Finally, we experimentally validated the quality of a large number of novel interactions identified in our study, providing a conclusive evidence for MaXLinker's robust performance.
In the post-genomic era, one of the main goals of systems biology is to determine the functions of all the proteins of various organisms. In the cell, most proteins function through interacting with other proteins. Therefore, generating interactome network models with high quality and coverage is a necessary step in the process of developing predictive models for protein functions at the scale of the whole cell (1). Furthermore, structural information for protein-protein interactions can serve as a crucial prerequisite for understanding the mechanism of protein function (2).
Rapid advancements in the fields of cross-linking and mass spectrometry lead to the development of a powerful technique known as cross-linking mass spectrometry (XL-MS)1 (3–5). XL-MS has been demonstrated to be an efficient technology to capture distance constraints, thereby providing crucial information to decipher the interaction partners and dynamics of protein-protein interactions (6). Efficient MS-cleavable chemical cross-linkers such as disuccinimidyl sulfoxide (DSSO) (7), disuccinimidyl dibutyric urea (DSBU) (8) and protein interaction reporters (PIRs) (9) have expanded the applications of XL-MS to discovering proteome-wide interactions along with their structural dynamics (10). Moreover, different cross-linkers exhibit distinct cleavage mechanisms and hence need specific and optimized fragmentation strategies. DSSO is currently one of the most popular commercially available MS-cleavable linkers. It yields signature peaks with ∼32 Dalton mass difference at MS2, facilitating the downstream MS3 fragmentation and analysis (7). Liu et al. (11) demonstrated the high-throughput capability of DSSO with a proteome-wide XL-MS study on HeLa cell lysate, using their XlinkX search engine. They adapted the traditional “target-decoy” approach for estimating false discovery rate (FDR) in peptide spectrum matches (PSMs) to estimate quality of the identified cross-links (each individual cross-link identification is also known as a Cross-link Spectrum Match (CSM).
In a more recent study, multiple fragmentation schemes were comparatively evaluated using DSSO, that include CID-MS2, CID-MS2-ETD-MS2, CID-MS2-MS3 and CID-MS2-MS3-ETD-MS2 (a combination of CID-MS2-ETD-MS2 and CID-MS2-MS3 acquisition strategies) utilizing an updated version of XlinkX (12). Apart from CID-MS2, all other strategies combine spectra from multiple MS levels (MS2 and MS3) or from different types of energy fragmentations (CID and ETD) or both. Their analysis revealed that, the ensemble strategy (i.e. CID-MS2-MS3-ETD-MS2) resulted in the highest number of cross-links, followed by CID-MS2-MS3, CID-MS2-ETD-MS2 and CID-MS2. Moreover, utilizing sequence information only from MS3 spectra (a subset of CID-MS2-MS3) for cross-link identification (“MS3-Only”) resulted in the least number of crosslinks among all the strategies. Hence the study concluded CID-MS2-MS3-ETD-MS2 and MS3-Only to be the most and least informative strategies, respectively. However, the study did not assess quality of different strategies at the given FDR cut-off using a rigorous comparative analysis.
In this study, we perform systematic and rigorous quality assessment across different XL-MS acquisition strategies, inspired by approaches widely-used in machine learning (1, 13). Based on these analyses, we noted that MS3-level information is crucial for reliable XL identification and observed that XlinkX results in high fraction of mis-identifications. Therefore, we developed and validated a novel search algorithm named MaXLinker, which is based on an innovative “MS3-centric” approach for MS2-MS3 XL-MS strategy, designed to efficiently eliminate incorrect cross-link candidates. With MaXLinker in hand, we performed a large-scale proteome-wide XL-MS study on K562 cell lysate, yielding more than 9319 XLs. We further carried out an orthogonal systematic experimental validation of the novel interactions and thereby confirming the reliable quality interprotein cross-links identified in our study.
EXPERIMENTAL PROCEDURES
Cell Culture and Whole Cell Lysate Preparation
The K562 cells (ATCC® CCL-243™) were purchased from American Type Culture Collection (ATCC) and cultured in the Iscove's Modified Dulbecco's Medium (IMDM) (ATCC, Manassa, VA) supplemented with 10% fetal bovine serum (FBS) (ATCC) at 37 °C under humidified ambient atmosphere containing 5% CO2. For harvest, 1 × 107 cells were collected at 1000 × g for 3 mins and wash with cold PBS for three times. The cell pellet were resuspended in cold buffer composed of 50 mm HEPES, 150 mm NaCl, pH 7.5 and Protease Inhibitor Mixture (Roche, Mannheim, Germany). The cell lysis was carried out by sonication with the setting of 5-s on and 10-s off on Amplitude 10% for 6 cycles on ice, followed by centrifugation at 15,000 × g for 10 min at 4 °C. The cell lysate in the supernatant was collected. The protein concentration of the lysate was determined using Bio-Rad Protein Assay Dye (Bio-Rad, Hercules, CA).
Cross-linking of Bovine Glutamate Dehydrogenase (GDH) and Human Proteome
DSSO (Thermo Fisher Scientific, Rockford, IL) stock solution (50 mm) was freshly prepared by dissolving in anhydrous DMSO (Invitrogen). To perform cross-linking, 1 mm DSSO was mixed with 1 mg/ml purified bovine glutamate dehydrogenase (GDH) protein (Sigma-Aldrich, Gillingham, United Kingdom) in 50 mm HEPES, 150 mm NaCl, pH 7.5 and reacted for 30 min at room temperature. Similarly, the 1 mg/ml K562 cell lysate were incubated with 1 mm DSSO for 1 h at room temperature. Both cross-linking reactions were terminated by 50 mm Tris-Cl buffer, pH 7.5.
DSSO-cross-linked Samples Processing for Analysis
The DSSO-treated protein samples were processed as previously described (14, 15). Briefly, the cross-linked GDH was denatured in 1% sodium dodecyl sulfate (SDS), reduced by dithiothreitol (DTT), and alkylated with iodoacetamide, followed by precipitated in cold acetone-ethanol solution (acetone/ethanol/acetic acid = 50:49.9:0.1, v/v/v). The precipitates were reconstituted in 50 mm Tris-Cl, 150 mm NaCl, 2 m urea, pH 8.0 and digested by Trypsin Gold (Promega, Madison, WI) at 37 °C overnight. The digested samples were then acidified by 2% trifluoroacetic acid-formic acid (TFA-FA) solution and desalted through Sep-Pak C18 cartridge (Waters, Dublin, Ireland). The eluents were dried using SpeedVacTM Concentrator (Thermo Fisher Scientific, Pittsburgh, PA). The samples were then reconstituted in 0.1% TFA and stored in −80 °C before mass spectrometry analysis. The DSSO-cross-linked human proteome was processed identically as described above except that the TPCK-treated trypsin (Worthington Biochemical Corporation, Lakewood, NJ) was used for digestion and the sample was further processed by fractionation after drying.
Sample Fractionation by Strong Cation Exchange (SCX)
The SCX fractionation was performed on a Dionex UltiMate 3000 Series instrument (Thermo Fisher Scientific, Sunnyvale, CA) using a PolySULFOETHYL A column (5 μm, 200 Å, 2.1 × 200 mm; PolyLC, Columbia, MD) with 10 mm potassium phosphate monobasic in 25% acetonitrile, pH 3.0 as Buffer A and 10 mm potassium phosphate monobasic/500 mm potassium chloride in 25% acetonitrile, pH 3.0 as Buffer B. All eluents were filtered through a 0.22 μm Durapore membrane (EMD Millipore Corporation, Billerica, MA) and stored at 4 °C until use. Prior to injection, the 1 mg of trypsin-digested sample was reconstituted in 25% acetonitrile/0.1% formic acid (v/v) and filtered through a Spin-X centrifuge tube filters (cellulose acetate membrane, 0.22 μm; Corning Incorporated, Corning, NY) by following manufacturer's recommended protocol. The fractionation was performed at a flow rate of 200 μl/min using a linear gradient from 5–60% of Buffer B in 40 min and 60–100% of Buffer B in an additional 10 min. A total of 60 fractions were collected using a 96-well plate at 1-min intervals monitored by the absorbance at 220 nm and 280 nm. The fractions collected from 23 to 60 min were desalted using SOLA HRP SPE cartridges (Thermo Scientific). The eluted peptides were dried by speed vacuum and stored at −20 °C until LC-MS analysis.
Fractionation of Cross-linked Peptides by Hydrophilic Interaction Liquid Chromatography (HILIC)
The DSSO-cross-linked human peptides in 70% acetonitrile and 1% formic acid were fractionated and enriched by hydrophilic interaction liquid chromatography (HILIC). The HILIC fractionation was performed on a Dionex UltiMate 3000 Series instrument (Thermo Fisher Scientific, Sunnyvale, CA) equipped with a TSKgel Amide-80 column (3 μm, 4.6 mm × 15 cm; Tosoh Bioscience, Griesheim, Germany). The three following solvents were used: 90% acetonitrile (solvent A), 80% acetonitrile and 0.005% trifluoroacetic acid (solvent B), 0.025% trifluoroacetic acid (solvent C). All the runs were performed at a flow rate of 600 μl/min using the following gradients: 0–5 min (0–98% B and 0–2% C); 5–55 min (98–75% B and 2–25% C); and 55–60 min (75–5% B and 25–95% C). The fractions were collected from 5–55 min per 30 s. Each fraction was dried and stored at −80 °C for further analysis.
LC-MSn Analysis
The SCX-fractionated samples were analyzed using UltiMate3000 RSLCnano (Dionex, Sunnyvale, CA) coupled to an Orbitrap Fusion (Thermo Fisher Scientific, San Jose, CA) mass spectrometer. Each sample was loaded onto an Acclaim PepMap 100 C18 trap column (5 μm, 100 μm × 20 mm, 100 Å, Thermo Fisher Scientific) and separated on an Acclaim PepMap C18 nano column (3 μm, 75 μm × 25 cm, Thermo Fisher Scientific) by 5–35% B at 300 nL/min in 120 min. For MS data acquisition, the CID-MS2-HCD-MS3 workflow was used. The Orbitrap Fusion was operating in positive ion mode and the MS1 precursors were detected in Orbitrap mass analyzer (375–1575 m/z and resolution = 60,000). The precursor ions with the charge of 4+ to 8+ were selected for CID-MS2 acquisition in Orbitrap mass analyzer (resolution = 30,000, AGC target = 5 × 104, precursor isolation width = 1.6 m/z, and maximum injection time = 100 ms) with the collision energy of CID at 25%. The peaks with a mass difference (Δ = 31.9721) in the CID-MS2 spectrum triggered acquisition of HCD-MS3 spectra in Ion Trap with HCD collision energy of 35% and AGC target of 1 × 104. All spectra were recorded by Xcalibur 3.0 software and Orbitrap Fusion Tune Application v. 2.1 (Thermo Fisher Scientific).
The HILIC fractions were reconstituted in 0.1% trifluoroacetic acid. The samples were then analyzed using an EASY-nLC 1200 system (Thermo Fisher Scientific) equipped with an 125-μm × 25-cm capillary column in-house packed with 3-μm C18 resin (Michrom BioResources, Auburn, CA) and coupled online to an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). The LC analysis was performed using solvent A composed of 0.1% formic acid and solvent B composed of 80% acetonitrile and 0.1% formic acid and run 10–40% B for 180 min at 300 nl/min. For MSn data acquisition, the CID-MS2-HCD-MS3 method was used. The MS1 precursors were detected in Orbitrap mass analyzer (375–1500 m/z, resolution of 60,000). The precursor ions with the charge of 4+ to 8+ were selected for MS2 analysis in Orbitrap mass analyzer (resolution = 30,000, AGC target = 1 × 105, precursor isolation width = 1.6 m/z, and maximum injection time = 105 ms) with the collision energy of CID at 25%. The peaks with a mass difference of 31.9721 Da in CID-MS2 spectra were selected for further MS3 analysis. The selected ions were fragmented in Ion Trap using HCD with the collision energy at 35% and AGC target of 2 × 104. All spectra were recorded by Xcalibur 4.1 software and Orbitrap Fusion Lumos Tune Application v. 3.0 (Thermo Fisher Scientific). In total, we performed two biological replicates using SCX fractionation, and one of them has two technical replicates. For the HILIC fractionated samples, we performed four biological replicates. We analyzed two biological replicates from SCX and HILIC fractionation in terms of unique cross-linked peptides and observed an overlap of ∼58 and ∼63%, respectively (supplemental Fig. S1). Furthermore, we observed a higher overlap of ∼71% between two SCX technical replicates (supplemental Fig. S1). Additionally, given the stochasticity of mass spectrometric identifications from complex proteome-wide XL-MS samples, we ran some of the later fractions multiple times in anticipation of extracting maximum possible data from such complex and cross-link rich fractions (resulting in 344 raw files in total).
Experimental Design and Statistical Rationale
Validation of Newly Identified Protein-Protein Interactions by Protein Complementation Assay (PCA)
The ORFs of a total of 49 protein pairs in pDONR223 plasmid were picked from hORFeome v8.1 library (16). The bait and prey protein of each protein pair was cloned into the expression plasmids containing the complementation fragments of a fluorescent protein Venus using Gateway LR reactions. The success of the LR reactions with desired ORF was confirmed by PCR using the plasmid-specific primers. To perform PCA, HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (ATCC) in black 96-well flat-bottom plates (Corning, Kennebunk, ME) with 5% CO2 at 37 °C. At 60–70% confluency, the cells were co-transfected with the plasmids containing the bait and prey ORF (100 ng for each) pre-mixed with polyethylenimine (PEI) (Polysciences, Inc., Warrington, PA) and OptiMEM (Gibco, Grand Island, NY). A total of 49 bait and prey ORF pairs along with previously published 45 positive reference pairs and 45 negative reference pairs were examined and distributed across different plates (17, 18). After 68 h, the fluorescence of the transfected cells was measured using Infinite M1000 microplate reader (Tecan) (excitation = 514 ± 5 nm/emission = 527 ± 5 nm). The PCA experiments were performed and analyzed in triplicate. The p values were calculated using a paired one-tailed t test. In order to have a high confidence in our validation, we used a stringent fluorescence cutoff during postprocessing of the PCA raw data. We noted that the detection rate of our positive reference set is comparable to the detection rate achieved by previous studies utilizing PCA and other high-throughput methodologies such as Y2H, wNAPPA, and LUMIER to study protein interactions in various model organisms (18–21).
Fraction of Mis-identifications (FMI)
FMI is the fraction of cross-link identifications from a false search space (from an unrelated organism) among all the identified cross-links. It can be calculated using the following equation:
| (1) |
In this study, we searched the E. coli proteome-wide XL-MS fractions (12) against E. coli + S. cerevisiae database for FMI calculations. Where, PeptideE. coli − PeptideS. cerevisiae, PeptideS. cerevisiae − PeptideE. coli and PeptideS. cerevisiae − PeptideS. cerevisiae were considered as mis-identifications and everything else as true identifications (including pairs with shared peptides between homologous proteins from E. coli and S. cerevisiae). Additionally, we also utilized the following equation adapted from Fischer and Rappsilber (22) to account for the significantly larger size of the S. cerevisiae database compared with that of E. coli database.
| (2) |
where, TT is the number of target-target matches, DD is the number of decoy-decoy matches, and TD is number of target-decoy and decoy-target matches. Here, PeptideE. coli − PeptideE. coli were considered as TT (pairs with shared peptides between homologous proteins from E. coli and S. cerevisiae were categorized as TT); PeptideE. coli − PeptideS. cerevisiae, PeptideS. cerevisiae − PeptideE. coli were considered as TD; PeptideS. cerevisiae − PeptideS. cerevisiae were considered as DD. TDDB is the number of all possible unique target-decoy and decoy-target peptide pairs, and DDDB is the number of all possible unique decoy-decoy peptide pairs. We repeated the analysis shown in Fig. 1A using equation 2 to verify if the drastic difference in database sizes between E. coli and S. cerevisiae has any potential implications on the results. As observed in supplemental Fig. S2, we found no considerable effect of the database sizes on our conclusions from Fig. 1A.
Fig. 1.
Comparative quality assessment between various acquisition methods for Cross-linking Mass-Spectrometry on six E. coli fractions from Liu et al. (12) A, Comparison between different acquisition methods based on fraction of mis-identifications (FMI). (The search was performed using a database consisting amino acid sequences of E. coli and S. cerevisiae proteomes. Any CSM with either of the peptides exclusively from S. cerevisiae proteome was considered as a mis-identification). B, Quality comparison across multiple acquisition methods using the fraction of XLs from known interactions (FKI). A separate search was performed for panel “B” using only the E. coli database in order to avoid underestimation of FKI. (Significance was determined by a two-sided Z-test; The error bars represent the estimated standard error of mean).
Fraction of Interprotein Cross-links from Known Interactions (FKI)
FKI for proteome-wide XL-MS studies can be defined as the fraction of the identified interprotein cross-links from previously known protein-protein interactions. It can be derived using the following equation:
| (3) |
where, “positives” refer to all the identified interprotein cross-links, and “true positives” refer to cross-links from known protein-protein interactions. We compiled the known protein-protein interactions for E. coli (24,745) and H. sapiens (336,033) from seven primary interaction databases. These databases include IMEx (23) partners IntAct (24), MINT (25), and DIP (26); IMEx observer BioGRID (27); and additional sources HPRD (28), MIPS (29), and iRefWeb (30). Furthermore, iRefWeb combines interaction data from CORUM (31), BIND (32), MPPI (29) and OPHID (33). We converted all gene identifiers in each database to Entrez gene IDs and then mapped to uniport IDs. Furthermore, to ensure the reliable quality of the compiled set of interactions, we compared the FKI calculated for the datasets shown in Fig. 1 using these interactions with the FKI calculated using interactions from STRING database (34) filtered at a stringent probability score cutoff (≥0.7). We observed a great agreement between FKI calculated using both the interaction sets, confirming the utility of our set of compiled known interactions (supplemental Table S1).
Data Processing
The raw data files were converted, and the spectra were exported as “.mgf” (MS1 spectra as “.dta”) files using Proteome Discoverer 2.1 software (PD 2.1). SEQUEST (35) searches were performed using PD 2.1 with the following settings: precursor mass tolerance: 20 ppm (10 ppm for MS2 rescue module); MS3 fragment ion mass tolerance: 0.6 Da (0.05 Da for MS2 rescue module); fixed modification: Cys carbamidomethylation; variable modifications: Met oxidation, Long arm of DSSO, Short arm of DSSO; max. equal modification per peptide: 3; Enzyme: Trypsin (full); max. missed cleavages: 3, minimum peptide length: 5. Concatenated target-decoy databases are used for various PSM searches performed during the study. Target sequences were downloaded from Uniprot database (36) (with filter “reviewed”) and a corresponding decoy database was generated by randomizing the sequences using an in-house python script. ((1) Escherichia coli: 5268 sequences; downloaded on 28th October 2017, (2) Saccharomyces cerevisiae: 7904 sequences; downloaded on 28th September 2017, and (3) Homo Sapiens: 42202 sequences; downloaded on 23rd June 2017).
For XlinkX searches, all the raw files were processed using XlinkX v2.0 implemented in Proteome Discoverer software version 2.2 (PD 2.2). PD templates for different XlinkX search methodologies were obtained from Rosa Viner (Thermo fisher Scientific). We understand that XlinkX available in PD 2.2 estimates FDR at CSM level on MS2-based identification and MS3-based identifications separately for MS2-MS3 acquisition strategy and then merges the information to infer a unique list of cross-links (https://assets.thermofisher.com/TFS-Assets/CMD/Reference-Materials/pp-structural-biology-cross-linking-studies-msum2017-en.pdf). All the searches were performed at 1% FDR cut-off (at redundant CSM level) and the CSMs were exported (after applying filter “Is Decoy: False”). For “MS3-Only” category, results from “CID-MS2-MS3” were reprocessed with option “Reprocess: Last Consensus Step” with “Ignore reporter scan: True” in “Xlinkx Crosslink Grouping” node. This set contained a list of all CSMs (includes multiple identifications representing a cross-linked peptide pair i.e. redundant). This set of data was used for comparisons shown in supplemental Fig. S1. Next, Those CSMs for were further processed to obtain a list of unique CSMs (In case of multiple CSMs with different cross-link positions, only one of them was retained to avoid potential biases because of over-representation of certain peptide pairs). The resulting set of CSMs were used for comparisons shown in Fig. 1, Fig. 3A, 3B, 3C, 3D, 3E, 3F, supplemental Fig. S4, supplemental Fig. S5, supplemental Fig. S8. Same procedure was followed to obtain the unique CSMs for GLUD1 analysis shown in Fig. 4B, 4C, and 4D. For obtaining CSMs that were exclusively identified using MS2 spectrum by XlinkX from MS2-MS3 acquisition strategy, the raw files were first processed using XlinkX's “MS2-MS3 workflow”. Then they were re-processed separately using “MS2-MS3 workflow with Ignore reporter scan: True”, and “MS2-only workflow”. Then, the CSMs from the initial MS2-MS3 workflow that were non-overlapping with that of MS2-MS3 workflow with Ignore reporter scan: True and overlapping with that of “MS2-only workflow” were labeled as exclusive identifications from MS2 spectra.
Fig. 3.
Comparison of MaXLinker's performance on proteome-wide XL-MS with that of XlinkX. A, Comparison of the fraction of mis-identifications from MaXLinker and XlinkX at 1% FDR using six E. coli MS2-MS3 XL-MS fractions from Liu et al. (12). B, Overlap between CSMs from MaXLinker and XlinkX at 1% FDR showing the respective fraction of mis-identifications in the parentheses. C, Overlap between CSMs at 1% FDR from MaXLinker and additional filtering on 1% FDR with “ΔXlinkX score” ≥ 50 for XlinkX, showing the respective fraction of mis-identifications in the parentheses. D, Comparison between MaXLinker and XlinkX in terms of fraction of XLs from known interactions (FKI) using six E. coli MS2-MS3 XL-MS fractions from Liu et al. (12). E, Overlap between interprotein CSMs from MaXLinker and XlinkX at 1% FDR showing the respective FKI values in the parentheses. F, Overlap between interprotein CSMs at 1%FDR from MaXLinker and additional filtering on 1% FDR with “ΔXlinkX score” ≥ 50 for XlinkX, showing the respective FKI values in the parentheses. (Significance was determined by a two-sided Z-test; The error bars represent the estimated standard error of mean).
Fig. 4.
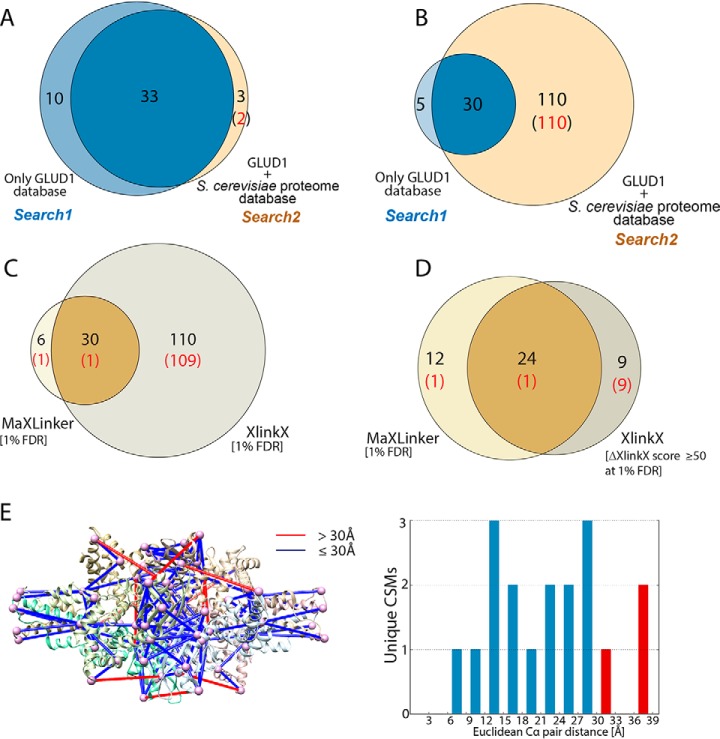
Validation and Comparison of MaXLinker's performance with that of XlinkX using bovine GLUD1 XL-MS. Search1 was performed using sequence of only GLUD1 protein as the search database and Search2 was performed using a concatenated database consisting sequence for GLUD1 along with the entire S. cerevisiae proteome. A, Overlap between MaXLinker's identifications from Search1 and Search2 at 1% FDR. B, Overlap between identifications from XlinkX from Search1 and Search2 at 1% FDR. C, Overlap of Search2 identifications at 1% FDR from MaXLinker and XlinkX. D, Overlap of Search2 identifications from MaXLinker at 1% FDR and with additional filtering (“ΔXlinkX Score” ≥ 50) at 1% FDR from XlinkX. E, Validation of cross-links from GLUD1 identified using MaXLinker, by mapping them onto its three-dimensional structure (PDB: 5K12). Cross-links exceeding theoretical distance constraint for DSSO (30Å) is shown in red. The cross-links are shown in default mode, where all possible mappings are visualized for a homo-oligomer (GLUD1 is a homo-hexamer). The histogram shows distance distribution for all the cross-links mapped on to the structure (cross-links with distance >30Å shown in red). The structure mappings were performed using Xlink Analyzer(55) implemented in UCSF Chimera (56).
Description of MaXLinker
MaXLinker runs in two steps: (1) pre-processing generates a “.MS2_rescue.mgf” file, which is needed for the PSM search in PD 2.1 to be eventually used in the main search. (2) cross-link search accepts .mgf files with different levels of MS spectra(MS1, MS2, and MS3), and two files containing the list of PSMs from PD2.1 SEQUEST search on MS3 spectra and MS2_rescue spectra. The key steps of the search process are described in Fig. 2 and manual available at https://www.yulab.org/resources/MaxLinker/ (along with MaXLinker download). After the search, a final MaXLinker score is assigned to each cross-link and it is derived using the following equation:
| (4) |
where WXL were systematically optimized to minimize mis-identifications through a rigorous training procedure. Moreover, MaXLinker utilizes the target-decoy strategy to establish the FDR. A concatenated database consisting target and decoy (random) sequences is used for the PSM search and the FDR is calculated using the following equation (11):
| (5) |
where, FP denote false positive hits and TP denote true positive hits. For cross-link identification, TP represent the number of cross-links with both linked peptides from the target database and FP represent the number of cross-links with at least one of the linked peptides from decoy database. After the search is complete, the identified cross-links are annotated as “interprotein” if neither of the linked peptides were derived from a common protein (with the exception where, both the linked peptides from a common protein, were identical or one of them was a complete subset of the other and the peptide occurs only once in the protein sequence). Cross-links that did not satisfy the aforementioned criteria were annotated as “intraprotein.”
Fig. 2.
Overview of MaXLinker's strategy for identification of cross-links from XL-MS.
For a potential protein-protein interaction analysis using the identified set of cross-links, we suggest filtering at the desired FDR cutoff and utilize the unambiguous interprotein cross-links to infer and obtain a non-redundant list of protein-protein interactions. In the current study, for cross-link search on E. coli proteome-wide XL-MS fractions (12) using MaXLinker, we also performed an FDR estimation using equation from Fischer and Rappsilber (22) (an adapted version for FMI calculation is shown as Eq. 2) separately, and found that the list of cross-links was identical to the one obtained using Eq. 5.
Statistics
Statistical analyses were performed using a two-sided Z test or a one-sided Welch Two Sample t test, as indicated in the figure captions. Exact p values are provided for all compared groups.
Code Availability
MaXLinker software is freely available for download as Supplementary Software at “https://www.yulab.org/resources/MaxLinker/”.
RESULTS
Current MS2-centric Approach for DSSO Cross-link Identification Is Limited in Its Sensitivity and Specificity
When compared with traditional PSM searches, the identification of CSMs from a proteome-wide study is markedly more complex. This fact motivated us to thoroughly examine the MS2-centric approach (where the search is initiated at MS2 level even when data from MS3 level is available) for XL identification from proteome-wide XL-MS studies. XlinkX (12) is currently the most popular MS2-centric search engine for DSSO (39–43). It is important to note that other purely MS2-based software packages are available that presumably perform better than XlinkX on MS2-only acquisitions (such as MeroX for MS-cleavable linkers (44)). However, we define MS2-centric approaches as the ones that have the capability to process and utilize fragment ions from multiple MS levels (MS2 and MS3) for cross-link identification, and start their search from the MS2 spectra. Hence, to the best of our knowledge XlinkX is the only MS2-centric search engine available for the identification of DSSO cross-linked peptides on a proteome-scale. Thus, we introduce a new quality metric called “fraction of mis-identifications” (FMI) to perform a systematic quality comparison of cross-links identified by XlinkX from data across multiple XL-MS acquisition strategies described in Liu et al. (12) (Experimental Procedures). First, we obtained corresponding raw files for the three fragmentation schemes CID-MS2-ETD-MS2, CID-MS2-MS3 and CID-MS2-MS3-ETD-MS2 (through E-mail request to Dr. Fan Liu). Then we performed cross-link search using XlinkX software (Proteome Discoverer 2.2) at 1% FDR with a concatenated database containing sequences from E. coli proteome (true search space) and S. cerevisiae (false search space). It is important to note that XlinkX by default, generates a reversed version of the input database and uses it as a decoy database to estimate FDR. In other words, the target database would consist of protein sequences from E. coli proteome and S. cerevisiae proteome. On the other hand, the decoy database would consist of reversed version of sequences from E. coli proteome and S. cerevisiae proteome. As a next step, we compared the three fragmentation strategies in terms of the number of incorrect unique CSMs (CSMs with at least one peptide from the S. cerevisiae search space, i.e. mis-identifications). However, CSMs with shared peptides between homologous proteins from E. coli and S. cerevisiae were considered as true identifications to avoid over-estimation of mis-identifications. The aim of this search is to re-assess the quality of cross-links at 1% FDR, with expected fraction of incorrect CSMs involving unambiguous peptides from S. cerevisiae to be less than 1%. Surprisingly, the fraction of incorrect CSMs range from 14.8% to as high as 26.9% across the three acquisition strategies (Fig. 1A). Upon closer examination, we observed that among the three strategies, CID-MS2-MS3 showed significantly lower proportion of incorrect CSMs (14.8%) followed by CID-MS2-ETD-MS2 (25.1%), and CID-MS2-MS3-ETD-MS2 (26.9%) strategies. This analysis clearly indicates that the methodology implemented in XlinkX does not adequately evaluate the quality of the identified CSMs. Therefore, utilizing only the number of identifications for comparative evaluations (12) might not yield accurate conclusions about the capability of different acquisition strategies. We further repeated the analysis at redundant CSM level and observed results consistent with what was found at the unique CSM level (supplemental Fig. S3). The high false positive rate for XlinkX identifications observed in our analysis was corroborated by a recent independent study that employed a spike-in strategy using cross-linked bovine serum albumin samples (45).
The Most Reliable Sequence Information for Cross-linked Peptides Come from the MS3-Level
We also evaluated the quality of identifications from CID-MS2-MS3 strategy, with sequence information obtained exclusively from MS3 spectra (MS3-only) (Fig. 1A). Strikingly, we observed a drastically lower fraction of incorrect CSMs for MS3-Only (3.3%), which is a subset of CID-MS2-MS3 (with 14.8% FMI). This result clearly demonstrates that MS3 which is the most advanced MS level, provides higher quality sequence information compared with MS2-level. Furthermore, we explored different parameters available in the XlinkX output based on their descriptions and selected “Δ XlinkX score” (a measure of confidence for each CSM) to further filter the identifications, aiming to obtain a set with higher quality compared with that of the original 1% FDR set. As a next step, we filtered the CSMs using five different Δ XlinkX score cutoffs in the increasing order of stringency (namely ≥10, ≥20, ≥30, ≥40, and ≥50) and re-assessed their quality across different acquisition strategies. We observed that, overall, increasing the stringency based on Δ XlinkX score significantly reduced the number of incorrect CSMs for all three acquisition strategies (supplemental Fig. S4). However, even after filtering by Δ XlinkX score, the trend across the different strategies was similar to what was observed before the filtering (Fig. 1A and supplemental Fig. S4), with data from the MS3-level yielding the highest fraction of reliable CSMs.
Fraction of Interprotein Cross-links from Known Interactions Is a Reliable Metric for Comparative Quality Assessment for Proteome-wide XL-MS Data Sets
To perform a more comprehensive and rigorous quality evaluation, we next utilized the “fraction of interprotein cross-links from known interactions” (FKI) to compare the quality across the three acquisition strategies (EXPERIMENTAL PROCEDURES). FKI is analogous to a widely used metric in machine learning known as “precision” which has been previously utilized for evaluating the quality of large-scale interaction screens, where it is derived using known interactions (as training set) (1). Furthermore, some of the previous studies utilized the known protein-protein interaction networks to visualize and infer biological insights from XL-MS datasets (46, 47). However, none of the reported XL-MS studies have adapted it as a quality estimate for their data sets to the best of our knowledge. Here we present FKI as a measure to assess quality of cross-link data sets from proteome-wide XL-MS studies. FKI for XL-MS essentially represents the fraction of identified interprotein cross-links that correspond to known protein-protein interactions. Remarkably, FKI complements the result obtained in the above FMI analysis using additional S. cerevisiae search space (Fig. 1B, supplemental Fig. S4).
Most of the Reliable Cross-link Identifications Are Contributed by CID-MS2-MS3 Methodology
It is important note that CID-MS2-MS3-ETD-MS2 (combination CID-MS2-MS3 and CID-MS2-ETD-MS2 methodologies) resulted in higher FMI when compared with CID-MS2-MS3 strategy (Fig. 1A). Upon closer examination of the quality of CSMs identified by the inherent CID-MS2-MS3 and CID-MS2-ETD-MS2 methodologies, we observed that at 1% FDR, CSMs identified exclusively by CID-MS2-ETD-MS2 contains almost 2-fold higher FMI compared with exclusive identifications by CID-MS2-MS3 (supplemental Fig. S5). We repeated the analysis after filtering the CSMs at different Δ XlinkX score cut-offs. It is interesting to note that, as the cut-off score increases, the number of identifications contributed exclusively by CID-MS2-ETD-MS2 reduces consistently, to as low as 6% when compared with the exclusive identifications by CID-MS2-MS3 (at Δ XlinkX score ≥ 50) (supplemental Fig. S5). These results reveal that, for CID-MS2-MS3-ETD-MS2, at higher quality cut-offs, CID-MS2-ETD-MS2 fails to yield additional cross-links than what were already captured by CID-MS2-MS3. Nonetheless, given that our analysis is based on the results obtained from XlinkX, which is the only available software for processing complex and ensemble acquisition strategies such as CID-MS2-MS3-ETD-MS2 utilizing DSSO, we cannot completely rule out the utility of ETD fragmentation for cross-linked peptides. Future studies may explore the potential of ETD in conjunction with other cross-linkers, modified acquisition strategies and improved cross-link identification pipelines. Moreover, the performance of such pipelines can be evaluated using our proposed quality metrics and validation approaches.
Our observations provide captivating evidence that, among the three widely used strategies, CID-MS2-MS3 results in cross-links with significantly better quality, most of which rely on MS3 spectra for sequence information. However, the high number of incorrect identifications for CID-MS2-MS3 strategy at 1% FDR strongly demonstrates the need for an improved search algorithm that can efficiently eliminate false positives while maintaining a minimum number of false negatives.
MaXLinker: A Novel MS3-centric Approach for Cross-link Identification
To address the limitations of current MS2-centric approach for DSSO MS2-MS3 fragmentation strategy, we designed a novel MS3-centric approach (Fig. 2). XlinkX starts the search at MS2-level by calculating the potential precursor mass for the linked peptides and attempts to identify CSMs exclusively from the MS2 spectrum, for cases with no available sequence information from MS3-level. However, our analyses revealed that such MS2-centric approach could lead up to 14.8% false identifications (Fig. 1A) with ∼26% of the identified cross-links not relying on MS3 information at all (Experimental Procedures). On the contrary, our approach starts the search from MS3-level (which is confirmed through our analyses to be most informative level for the sequences of cross-linked peptides; Fig. 1) and requires at least one of the two peptides to be identified from the MS3 level. Additionally, our approach utilizes MS2-level to rescue candidate CSMs (MS2 Rescue node) only if one of the two cross-linked peptides could be reliably identified from the MS3 spectra (Fig. 2 Node “C”). Finally, we require all cross-link candidates to pass through an additional validation filter that performs theoretical reconstruction of cross-link using the identified peptide sequences (Fig. 2 Node “D”) and perform correction for mis-assigned monoisotopic MS1 precursor masses (Fig. 2 Node “B”). It is important to note that the novel aspect of MaXLinker does not claim utilizing only MS3 fragment ions for cross-link identification (it has been previously implemented by Huang lab's in-house algorithm for DSSO (7) and Bruce lab's ReACT algorithm for PIR (48)). The most important novel feature of MaXLinker is its MS3-centric workflow that involves efficient and prioritized utilization of fragment ions from MS3 spectra over that of the MS2 spectra. Other crucial components of our workflow include its MS2 rescue module, and thorough validation filters to eliminate potential false positives. Furthermore, a final MaXLinker score is assigned to each cross-link, which is designed using a machine learning approach to integrate various measurements of the confidences of the PSM for each peptide, and the occurrence of the peptide pair. Key feature of the MaXLinker score is its systemically optimized weights for the utilized parameters through a rigorous training procedure. One of the major advantages of this scoring scheme is its versatility and adaptability that allows incorporation of new features for further optimization and improvement in better ranking the identified cross-links based on their quality. Overall, although the MS2 rescue module contributes to MaXLinker's high sensitivity (rescues cross-link candidates that otherwise would not be identified), its high specificity is accorded by its stringent MS3-centric design (described in the following text) and optimized scoring scheme which labels candidate pairs that lack reliable information as potential false positives and discards them. Most importantly, it is the combination of all the individual modules/components that make the MaXLinker's work-flow unique, and its reliability has been thoroughly demonstrated using our new quality metrics and validation approaches.
The well-established general experimental methodology for MS2-MS3 acquisition strategy for DSSO (7) in a typical Tribrid mass spectrometer involves precursor selection at multiple stages of mass spectrometry. First, ions above certain threshold charge state (typically ≥ +3 or +4) will be selected for fragmentation at MS2 stage to yield signature ions with predefined mass difference (Δm = 31.97 for DSSO). Further, an iterative search known as “targeted inclusion” is performed by mass spectrometer on-the-fly to select ion pairs with signature Δm, following certain prioritization criteria to perform fragmentation at MS3-level to yield two MS3 spectra per peptide in an ideal scenario.
To perform the XL search, MaXLinker accepts “.mgf” files consisting different levels of MS spectra exported using Proteome discover (PD), along with PSM annotations from PD as input (Experimental Procedures). MaXLinker initiates the search from MS3-level by performing the mandatory precursor-based mass validation (Fig. 2 Node “A”). Initiating the search from MS3, the most informative level in terms of the peptide sequence information, provides a key advantage to MaXLinker in eliminating potential false positives. If a set of MS3 spectra representing a potential cross-link pass the precursor-based mass validation step (Fig. 2 Node “A”) (Case 1 in Fig. 2), it is verified through multiple validation filters (Fig. 2 Node “D”). It is important to note that typically larger size of crosslinked peptides can often result in the mis-assignment of the monoisotopic MS1 precursor mass (49), thus for cases that fail to pass through the precursor mass-based filter (Fig. 2 Node “A”), MaXLinker inspects the corresponding MS1 spectrum to verify mis-assignment of the monoisotopic MS1 precursor mass (Fig. 2 Node “B”). Such cases are systematically examined and passed on to the next filter if they satisfy the mass validation step with the adjusted precursor mass. The remaining failed candidates are sent to the MS2 Rescue Module (Fig. 2 Node “C”).
MS2 Rescue Module is another important and unique feature of MaXLinker. As mentioned earlier, this module is triggered if the candidate spectra failed to pass the precursor-based mass validation step (Fig. 2 Node “A”) and could not be validated through precursor mass re-assignment. We found that failure to pass these filters often coincided with poor or “uninformative” MS3 spectral data for one of the cross-linked peptides (case 2 in Fig. 2). In this case, considering a scenario where the mass spectrometer picked an incorrect Δm pair from the MS2-level having the signature just by chance, MaXLinker attempts to “rescue” sequence information for the peptide by utilizing fragment ions from the corresponding MS2 spectrum (Fig. 2 Node “C”). First, precursor masses for the peptide with poor MS3 spectra are derived using MS2 precursor mass and MS3 precursor masses of the “informative” MS3 spectra (with account for the linker long and short arm modifications) (supplemental Fig. S6). An additional validation search is performed on the ions of the corresponding MS2 spectrum to confirm presence of the derived MS3 precursor masses. Subsequently, a PSM search is performed on the deconvoluted MS2 spectrum with the derived masses (both long and short) as the precursor mass. If the search returns at least one reliable PSM, the cross-link candidate (along with sequence information for the “rescued” peptide) is directed to the general validation pipeline for further evaluation (Fig. 2 Node “D”). Additionally, the MS2 Rescue module also accounts for cases where the mass spectrometer selects two pairs with signature Δm for MS3, however both pairs represent different charge states of one of the two cross-linked peptides (supplemental Fig. S7). Upon completion of the search, a unique list of cross-links is obtained by merging the redundant CSM entries, and a confidence score is assigned to each identification (equation 2 in Experimental Procedures). Finally, a target-decoy strategy is employed to establish the FDR at the level of unique cross-linked peptide pairs.
MaXLinker Outperforms XlinkX in Both Specificity and Sensitivity
We evaluated the performance of MaXLinker utilizing MS2-MS3 XL-MS raw files for six E. coli fractions from Liu et al. (12). First, we utilized the strategy employed in Fig. 1A and performed the search using MaXLinker at 1% FDR. We noted that the FMI was less than 1% (supplemental Table S1), and for majority of the identifications (∼80%), the peptide sequence information was obtained from MS3 spectra, which agrees with MaXLinker's fundamental algorithmic design. Next, we compared the results with unique CSMs identified using XlinkX at 1% FDR on the same set of raw files (Fig. 3A). Our analysis showed that MaXLinker evidently outperforms XlinkX, indicated by the extremely significant difference (18-fold lower) in the fraction of mis-identifications (Fig. 3B). We then examined the overlap between identifications from the two search engines. It clearly reveals that the overlapping fraction from XlinkX has only 0.6% mis-identifications, whereas the non-overlapping CSMs which were identified exclusively by XlinkX contained a large fraction (33.1%) of mis-identifications. Further, using FKI as a complementary quality metric, we observed similar results (Fig. 3D, 3E). When we repeated these quality analyses by filtering the identifications from XlinkX at different Δ XlinkX score cutoffs, we observed that MaXLinker consistently finds 13–31% more cross-links than XlinkX at comparable quality (supplemental Fig. S8). Importantly, the CSMs identified exclusively by MaXLinker are of 3-fold higher quality than the exclusive identifications by XlinkX, even at the highly stringent cutoff Δ XlinkX score ≥ 50 (Fig. 3C, 3F). All these results demonstrate that MaXLinker outperforms XlinkX for CSM identifications in both specificity and sensitivity.
Next, we cross-linked commercially available Bovine Glutamate Dehydrogenase 1 (GLUD1) using DSSO and performed a CID-MS2-HCD-MS3 experiment in our own lab (Experimental Procedures). We employed MaXLinker to perform two individual CSM searches at 1% FDR, search1: using Bovine GLUD1 sequence as the search database yielding 43 unique CSMs, and search2: with a concatenated database with Bovine GLUD1 and a full proteome of Saccharomyces cerevisiae, yielding 36 unique CSMs. We then examined the overlap between CSMs from search1 and search2 to inspect MaXLinker's ability to find true CSMs from single protein in a plethora of false search space. Strikingly, we observed that 33 of 36 (92%) CSMs from search2 were overlapping with the ones from search1 (Fig. 4A). Out of the remaining 3 CSMs, 2 were known mis-identifications having one of the peptides in the pair from S. cerevisiae proteome (false search space). Of note, 10 CSMs were identified exclusively in search1. Upon close examination, we noted that MaXLinker rejected those 10 CSM candidates because of either (1) its stringent validation filters or (2) lower confidence in their PSM assignments, attributable to the drastic increase in the number of competing candidate peptides for individual spectra.
On the other hand, when we performed similar analysis using XlinkX at 1% FDR, search1 and search2 yielded 35 and 140 unique CSMs, respectively. Out of the 140 CSMs from search2, 30 were overlapping with search1 and the remaining 110 had at least one of the peptides unambiguously from S. cerevisiae proteome (known mis-identifications) (Fig. 4B). We examined the overlap between search2 identifications from MaXLinker and XlinkX, and observed that most of the mis-identifications from XlinkX (109 of 110) were not found by MaXLinker (Fig. 4C). Further, we filtered CSMs from XlinkX using Δ XlinkX score ≥ 50 and re-inspected the overlap with MaXLinker's identifications. This filtering step resulted in drastic elimination of false positives (Fig. 4D). However, all the non-overlapping CSMs from XlinkX were observed to be mis-identifications. On the other hand, MaXLinker identified 12 CSMs (containing 11 true CSMs) that were missed by XlinkX. For further validation of MaXLinker's identifications, we mapped CSMs from search1 on to a three-dimensional structure (Fig. 4E) of Bovine GLUD1. We observed that 15 of the 18 mapped CSMs were within the theoretical distance constraint (30Å), and the remaining three CSMs were within 38Å, validating reliable quality of our identifications. This analysis serves as a revealing case study for MaXLinker's unique ability to identify cross-links with high sensitivity and specificity. Furthermore, estimation of FDR at redundant CSM-level might be a potential reason for high fraction of false positives from XLinkX (∼2-fold increase in mis-identifications at the peptide pair level compared with the redundant-CSM level; supplemental Fig. S3 and Fig. 1). Moreover, it is unclear how XlinkX handles FDR at different levels, which would greatly influence the number of residue pairs passing a given FDR threshold (22, 50).
Our Proteome-wide K562 XL-MS Study
Having established the MaXLinker software and optimized the experimental pipeline in our lab, we carried out a comprehensive proteome-wide XL-MS study on human K562 cell lysates, using the CID-MS2-HCD-MS3 acquisition strategy. Previous proteome-wide XL-MS studies implemented the strong cation exchange chromatography (SCX) for pre-fractionation of crosslinked proteome samples. Here, to capture a more comprehensive set of cross-links, we employed both SCX and hydrophilic interaction chromatography (HILIC) for our proteome-wide XL-MS study (Experimental Procedures). We then employed MaXLinker for cross-link identification. Our study yielded 9319 unique cross-links at 1% FDR (8051 intraprotein and 1268 interprotein, representing 585 unambiguous interactions with 74.2% FKI; among the 585 unambiguous interactions, 410 were inferred through single cross-links and the remaining were inferred using cross-links in the range of 2–21 per interaction; supplemental Table S3). In comparison, some of the previous proteome-wide studies on human reported cross-links in the range of few hundreds to few thousands. Specifically, Liu et al. (12) reported ∼3300 cross-links from HeLa cell lysates.
Systematic Experimental Validation of Novel Protein-Protein Interactions Identified in Our Proteome-wide XL-MS Study
Currently, a common way of confirming the quality of cross-links is to map them onto available three-dimensional structures of representative complexes and identify the fraction of cross-links that satisfy the theoretical restraint of the cross-linker. Although we believe that such a structure-based validation approach often under-estimates the underlying error rate of proteome-wide cross-link data sets (51), we still mapped our identified cross-links from 26S proteasome onto its available three-dimensional structure (Fig. 5A, 5B). Out of the 100 cross-links mapped to the structure, 90 were within the theoretical constraint i.e. 30Å. Additionally, we observed that one cross-link that was exceeding 30Å, was within the distance constraint in a different structure (Fig. 5C), suggesting potential conformational changes in the corresponding subunits. Six out of the remaining nine cross-links were within 35Å, and all the others were within 50Å.
Fig. 5.
Validation of cross-links and novel interactions identified in the proteome-wide human K562 XL-MS study at 1% FDR. A, Mapping cross-links from 26S proteasome complex on to a recently published structure (PDB: 5GJQ; cross-links exceeding maximum theoretical constraint 30Å are shown in red). B, A circular plot showing interprotein cross-links between various subunits from 26S proteasomal complex.(cross-links exceeding maximum theoretical constraint 30Å are shown in red; the plot was generated using Circos (57).) C, Validation of a cross-link from 26S proteasome that violate distance constraints (>30Å) in one structure (PDB: 5GJQ) and obey in a different structure (PDB: 5T0J), suggesting potential conformational changes. D, Network map showing protein-protein interactions identified in the current study. (known interactions are shown in green and novel interactions are shown in orange). E, Experimental validation of a representative set of 49 novel interactions identified in the current study using Protein-fragment complementation assay (PCA) (mean fraction positive: 0.130) (PRS: Positive Reference Set (45 interactions; mean fraction positive: 0.133); RRS: Random Reference Set (45 interactions; mean fraction positive: 0.029); The error bars represent the standard deviation; Significance was determined by a one-sided Welch Two Sample t test; 95% confidence interval; t-statistic 0.53 for “PRS - This Study,” and 164.75 for “RRS - This Study”; 2 degrees of freedom).
Furthermore, it should be noted that FDR would be significantly higher in more convoluted levels (e.g. unique sites and protein-protein interactions) compared with that of lower levels such as CSM and unique peptide pair (22, 50). Hence in order to evaluate the quality of our dataset at the level of protein-protein interactions and to address the limitations of the current structure-based validation approach in validating the interprotein cross-links (51), we performed an orthogonal experimental validation of the novel protein-protein interactions identified in our study, thereby confirming the quality of the interprotein cross-links from which they were inferred. Such an orthogonal experimental validation is indispensable since majority fraction of false positive cross-links tends to be interprotein (50, 52). Moreover, the true positive and false positive interprotein cross-links do not have equal probability of successfully mapping to an existing 3D structure, leading to massive underestimation of false positives (51). In other words, the distance-based validation effectively pre-filters the data set, ignoring most of the potential false positive cross-links during the validation procedure.
We tested a representative subset of 49 interactions (randomly-chosen out of the 160 novel interactions identified in our study) individually using a Protein Complementation Assay (PCA). PCA facilitates high-throughput validation of novel protein-protein interactions in a mammalian cellular environment (Experimental Procedures). The fraction of PCA-positive interactions among the novel interactions identified in our XL-MS study is statistically indistinguishable (p = 0.325) from that of the positive reference set containing well-established interactions in the literature, but significantly higher (p = 1.8 × 10−5) than that of a negative reference set containing random protein pairs (Fig. 5E) (53). This large set of experimental results demonstrate the high quality of the novel cross-links and the corresponding interactions identified in our proteome-wide XL-MS study, and further confirm the reliability and accuracy of MaXLinker.
DISCUSSION
Machine learning approaches have been an integral part of conventional mass spectrometry-based methods (54). Here, we extended their applications for comparative quality assessment among multiple proteome-wide XL-MS data sets. In addition to using search space from an un-related organism (FMI), we demonstrated fraction of interprotein cross-links from known interactions (FKI) as an effective additional metric for comparative quality assessments. It should be noted that, because a large fraction of true protein interactions is yet to be discovered, FKI should not be used as an absolute measure for data quality. Nevertheless, it can be an orthogonal and reliable quality metric for comparative assessments of proteome-wide XL-MS studies. Moreover, even though we performed FKI-based comparative analysis at the level of unique peptide pair in this study, it is noteworthy that FKI can also be adapted to be estimated at the level of unique sites and interactions.
Our systematic analyses revealed for the first time, the limitations of current quality assessment strategies and the drawbacks of the current MS2-centric cross-link identification approach for DSSO MS2-MS3 strategy with high mis-identification rates (Fig. 1A). Our analyses also revealed that for MS2-MS3 strategy, the MS3-level provides sequence information with significantly higher quality when compared with that of the MS2-level, and identification of cross-links exclusively from MS2-level could result in alarmingly high mis-identification rate. To address these issues, we designed and implemented a novel MS3-centric approach (MaXLinker) (Fig. 2). The current MS2-centric methods for DSSO MS2-MS3 strategy such as XlinkX start the search from the MS2-level and attempts cross-link identifications without any information from MS3-level, resulting in high fraction of false positives. On the contrary, MaXLinker starts the search from MS3-level and discards any cross-link candidate without reliable sequence information from MS3-level for at least one of the two cross-linked peptides. Furthermore, the MS2-Rescue module utilizes MS2-level information to rescue XLs that have partial information because of selection of incorrect signature pairs by the mass spectrometer (supplemental Fig. S6, supplemental Fig. S7), which provides a key advantage to MaXLinker in terms of sensitivity. MS2-Rescue module adds more than 20% to the number of XLs identified by just considering the MS3 information alone, with comparable quality (p = 0.09; supplemental Table S2). The MS2 Rescue module along with other novel features including the strict validation filters and thoroughly optimized score (Fig. 2), play a crucial role in MaXLinker's superior sensitivity over the previously established approach, without compromising on the specificity. Overall, MaXLinker significantly outperforms XlinkX with 18-fold lower mis-identification rate and up to 31% higher number of identifications.
One can argue that our observations might suggest XlinkX's drastically different efficiency in identifying cross-links from MS2 spectra compared with that from MS3 spectra with the MS2-MS3 acquisition strategy. We hypothesize that such drastic difference in cross-link quality could be because of the inherently distinct information available in MS3 spectra for individual peptides constituting the cross-link (two individual spectra for each cross-linked peptide; with “long” and “short” modification of DSSO). However, we do not completely disregard the scope for an improved methodology in the future that better utilizes the MS2 information from strategies such as CID-MS2-MS3-ETD-MS2 DSSO acquisitions to significantly minimize the error rates while achieving comparable sensitivity. Moreover, XlinkX is the only publicly available search engine capable of processing DSSO cross-linked peptides with composite acquisition strategies such as CID-MS2-MS3 and CID-MS2-MS3-ETD-MS2, enabling us to perform such analyses. Furthermore, all observations from our analyses were from DSSO cross-linked samples, and other cleavable linkers such as DSBU, PIR, CDI etc. might behave differently given their drastically different chemical properties.
Having MaXLinker in hand, we report a comprehensive set of 9319 cross-links at 1% FDR (supplemental Table S3), representing 160 unambiguous novel interactions (supplemental Table S4). Furthermore, considering the limitations of current structure-based validation approach, we used an orthogonal experimental approach to validate the identified novel interactions and thereby affirming the quality of the interprotein cross-links reported by our study. Moreover, to our knowledge, this is the first study that performed a large-scale orthogonal experimental validation of novel interactions identified from a proteome-wide XL-MS study. Overall, we believe that our robust cross-link search engine along with the new quality assessment metrics and validation approaches constitute a significant contribution of this study.
With the constant technical advancements in XL-MS methodologies, reliable search algorithms such as MaXLinker will play a highly significant role in the success of future cross-linking studies. Moreover, the expanding size of cross-link datasets would allow researchers to investigate interaction networks in many disease phenotypes more thoroughly, thereby enabling us to better understand the underlying molecular mechanisms.
DATA AVAILABILITY
All cross-links are reported in the Supplementary information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE(38) partner repository with the dataset identifier PXD013928.
Supplementary Material
Acknowledgments
We thank Rosa Viner for support in data processing with XlinkX workflow in Proteome Discoverer and Shayne Wierbowski for helpful suggestions regarding data representation. We thank Robert Fragoza for the assistance with PCA experiments. We thank Elizabeth Anderson and Robert Sherwood for their technical support in sample preparation.
Footnotes
* The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- XL-MS
- cross-linking mass spectrometry
- DSSO
- disuccinimidyl sulfoxide
- DSBU
- disuccinimidyl dibutyric urea
- PIRs
- protein interaction reporters
- FDR
- false discovery rate
- PSM
- peptide spectrum matches
- CSM
- cross-link spectrum match
- SCX
- strong cation exchange.
REFERENCES
- 1. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J.-F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A.-S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A.-L., Tavernier J., Hill D. E., and Vidal M. (2008) High quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X., Wei X., Thijssen B., Das J., Lipkin S. M., and Yu H. (2012) Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat. Biotechnol. 30, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rappsilber J. (2011) The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Structural Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leitner A., Faini M., Stengel F., and Aebersold R. (2016) Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 [DOI] [PubMed] [Google Scholar]
- 5. Yugandhar K., Gupta S., and Yu H. (2019) Inferring protein-protein interaction networks from mass spectrometry-based proteomic approaches: a mini-review. Computational Structural Biotechnol. J. 17, 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sinz A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrometry Rev. 25, 663–682 [DOI] [PubMed] [Google Scholar]
- 7. Kao A., Chiu C. L., Vellucci D., Yang Y., Patel V. R., Guan S., Randall A., Baldi P., Rychnovsky S. D., and Huang L. (2011) Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10, M110 002212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Müller M. Q., Dreiocker F., Ihling C. H., Schäfer M., and Sinz A. (2010) Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal. Chem. 82, 6958–6968 [DOI] [PubMed] [Google Scholar]
- 9. Tang X., and Bruce J. E. (2010) A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol. bioSystems 6, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sinz A. (2017) Divide and conquer: cleavable cross-linkers to study protein conformation and protein–protein interactions. Anal. Bioanal. Chem. 409, 33–44 [DOI] [PubMed] [Google Scholar]
- 11. Liu F., Rijkers D. T. S., Post H., and Heck A. J. R. (2015) Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179. [DOI] [PubMed] [Google Scholar]
- 12. Liu F., Lössl P., Scheltema R., Viner R., and Heck A. J. R. (2017) Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 8, 15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meyer M. J., Beltrán J. F., Liang S., Fragoza R., Rumack A., Liang J., Wei X., and Yu H. (2018) Interactome INSIDER: a structural interactome browser for genomic studies. Nat. Methods 15, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bastos de Oliveira Francisco M., Kim D., Cussiol José R., Das J., Jeong Min C., Doerfler L., Schmidt Kristina H., Yu H., and Smolka Marcus B. (2015) Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol. Cell 57, 1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bastos de Oliveira F. M., Kim D., Lanz M., and Smolka M. B. (2018) Quantitative analysis of DNA damage signaling responses to chemical and genetic perturbations. In Genome Instability: Methods and Protocols (Muzi-Falconi M., and Brown G. W., eds.), pp. 645–660, Springer New York, New York, NY: [DOI] [PubMed] [Google Scholar]
- 16. Yang X., Boehm J. S., Yang X., Salehi-Ashtiani K., Hao T., Shen Y., Lubonja R., Thomas S. R., Alkan O., Bhimdi T., Green T. M., Johannessen C. M., Silver S. J., Nguyen C., Murray R. R., Hieronymus H., Balcha D., Fan C., Lin C., Ghamsari L., Vidal M., Hahn W. C., Hill D. E., and Root D. E. (2011) A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H., Sahalie J. M., Murray R. R., Roncari L., de Smet A.-S., Venkatesan K., Rual J.-F., Vandenhaute J., Cusick M. E., Pawson T., Hill D. E., Tavernier J., Wrana J. L., Roth F. P., and Vidal M. (2008) An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods 6, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venkatesan K., Rual J.-F., Vazquez A., Stelzl U., Lemmens I., Hirozane-Kishikawa T., Hao T., Zenkner M., Xin X., Goh K.-I., Yildirim M. A., Simonis N., Heinzmann K., Gebreab F., Sahalie J. M., Cevik S., Simon C., de Smet A.-S., Dann E., Smolyar A., Vinayagam A., Yu H., Szeto D., Borick H., Dricot A., Klitgord N., Murray R. R., Lin C., Lalowski M., Timm J., Rau K., Boone C., Braun P., Cusick M. E., Roth F. P., Hill D. E., Tavernier J., Wanker E. E., Barabási A.-L., and Vidal M. (2008) An empirical framework for binary interactome mapping. Nat. Methods 6, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabasi A. L., Tavernier J., Hill D. E., and Vidal M. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simonis N., Rual J.-F., Carvunis A.-R., Tasan M., Lemmens I., Hirozane-Kishikawa T., Hao T., Sahalie J. M., Venkatesan K., Gebreab F., Cevik S., Klitgord N., Fan C., Braun P., Li N., Ayivi-Guedehoussou N., Dann E., Bertin N., Szeto D., Dricot A., Yildirim M. A., Lin C., de Smet A.-S., Kao H.-L., Simon C., Smolyar A., Ahn J. S., Tewari M., Boxem M., Milstein S., Yu H., Dreze M., Vandenhaute J., Gunsalus K. C., Cusick M. E., Hill D. E., Tavernier J., Roth F. P., and Vidal M. (2009) Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat. Methods 6, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fischer L., and Rappsilber J. (2017) Quirks of error estimation in cross-linking/mass spectrometry. Anal. Chem. 89, 3829–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., Bridge A., Briganti L., Brinkman F. S. L., Cesareni G., Chatr-aryamontri, Chautard A. E., Chen C., Dumousseau M., Goll J., Hancock R. E. W., Hannick L. I., Jurisica I., Khadake J., Lynn D. J., Mahadevan U., Perfetto L., Raghunath A., Ricard-Blum S., Roechert B., Salwinski L., Stümpflen V., Tyers M., Uetz P., Xenarios I., and Hermjakob H. (2012) Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods 9, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., Duesbury M., Dumousseau M., Feuermann M., Hinz U., Jandrasits C., Jimenez R. C., Khadake J., Mahadevan U., Masson P., Pedruzzi I., Pfeiffenberger E., Porras P., Raghunath A., Roechert B., Orchard S., and Hermjakob H. (2012) The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40, D841–D846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E., Sacco F., Palma A., Nardozza A. P., Santonico E., Castagnoli L., and Cesareni G. (2012) MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 40, D857–D861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salwinski L., Miller C. S., Smith A. J., Pettit F. K., Bowie J. U., and Eisenberg D. (2004) The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 32, D449–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chatr-aryamontri A., Breitkreutz B.-J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L., Reguly T., Nixon J., Ramage L., Winter A., Sellam A., Chang C., Hirschman J., Theesfeld C., Rust J., Livstone M. S., Dolinski K., and Tyers M. (2015) The BioGRID interaction database: 2015 update. Nucleic Acids Res. 43, D470–D478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keshava Prasad T. S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., Balakrishnan L., Marimuthu A., Banerjee S., Somanathan D. S., Sebastian A., Rani S., Ray S., Harrys Kishore C. J., Kanth S., Ahmed M., Kashyap M. K., Mohmood R., Ramachandra Y. L., Krishna V., Rahiman B. A., Mohan S., Ranganathan P., Ramabadran S., Chaerkady R., and Pandey A. (2009) Human Protein Reference Database—2009 update. Nucleic Acids Res. 37, D767–D772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pagel P., Kovac S., Oesterheld M., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Mark P., Stümpflen V., Mewes H.-W., Ruepp A., and Frishman D. (2005) The MIPS mammalian protein–protein interaction database. Bioinformatics 21, 832–834 [DOI] [PubMed] [Google Scholar]
- 30. Turner B., Razick S., Turinsky A. L., Vlasblom J., Crowdy E. K., Cho E., Morrison K., Donaldson I. M., and Wodak S. J. (2010) iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database 2010, baq023-baq023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruepp A., Waegele B., Lechner M., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., and Mewes H. W. (2010) CORUM: the comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res. 38, D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alfarano C., Andrade C. E., Anthony K., Bahroos N., Bajec M., Bantoft K., Betel D., Bobechko B., Boutilier K., Burgess E., Buzadzija K., Cavero R., D'Abreo C., Donaldson I., Dorairajoo D., Dumontier M. J., Dumontier M. R., Earles V., Farrall R., Feldman H., Garderman E., Gong Y., Gonzaga R., Grytsan V., Gryz E., Gu V., Haldorsen E., Halupa A., Haw R., Hrvojic A., Hurrell L., Isserlin R., Jack F., Juma F., Khan A., Kon T., Konopinsky S., Le V., Lee E., Ling S., Magidin M., Moniakis J., Montojo J., Moore S., Muskat B., Ng I., Paraiso J. P., Parker B., Pintilie G., Pirone R., Salama J. J., Sgro S., Shan T., Shu Y., Siew J., Skinner D., Snyder K., Stasiuk R., Strumpf D., Tuekam B., Tao S., Wang Z., White M., Willis R., Wolting C., Wong S., Wrong A., Xin C., Yao R., Yates B., Zhang S., Zheng K., Pawson T., Ouellette B. F. F., and Hogue C. W. V. (2005) The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 33, D418–D424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown K. R., and Jurisica I. (2005) Online Predicted Human Interaction Database. Bioinformatics 21, 2076–2082 [DOI] [PubMed] [Google Scholar]
- 34. Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., and Mering Christian v. (2018) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 36. The UniProt Consortium. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Käll L., Canterbury J. D., Weston J., Noble W. S., and MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923. [DOI] [PubMed] [Google Scholar]
- 38. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yılmaz Ş Tiwary. S, Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., and Vizcaíno J. A. (2018) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu F., Lössl P., Rabbitts B. M., Balaban R. S., and Heck A. J. R. (2018) The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol. Cell. Proteomics 17, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fasci D., van Ingen H., Scheltema R. A., and Heck A. J. R. (2018) Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol. Cell. Proteomics, mcp.RA118.000924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogawa T., Saijo S., Shimizu N., Jiang X., and Hirokawa N. (2017) Mechanism of catalytic microtubule depolymerization via KIF2-tubulin transitional conformation. Cell Reports 20, 2626–2638 [DOI] [PubMed] [Google Scholar]
- 42. Fux A., Korotkov V. S., Schneider M., Antes I., and Sieber S. A. (2019) Chemical cross-linking enables drafting ClpXP proximity maps and taking snapshots of in situ interaction networks. Cell Chem. Biol. 26, 48–59.e47 [DOI] [PubMed] [Google Scholar]
- 43. Stieger C. E., Doppler P., and Mechtler K. (2019) Optimized fragmentation improves the identification of peptides cross-linked by MS-cleavable reagents. J. Proteome Res. 18, 1363–1370 [DOI] [PubMed] [Google Scholar]
- 44. Iacobucci C., Götze M., Ihling C. H., Piotrowski C., Arlt C., Schäfer M., Hage C., Schmidt R., and Sinz A. (2018) A cross-linking/mass spectrometry workflow based on MS-cleavable cross-linkers and the MeroX software for studying protein structures and protein–protein interactions. Nat. Protocols 13, 2864–2889 [DOI] [PubMed] [Google Scholar]
- 45. Ser Z., Cifani P., and Kentsis A. (2019) Optimized cross-linking mass spectrometry for in situ interaction proteomics. J. Proteome Res. 18, 2545–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chavez J. D., Schweppe D. K., Eng J. K., Zheng C., Taipale A., Zhang Y., Takara K., and Bruce J. E. (2015) Quantitative interactome analysis reveals a chemoresistant edgotype. Nat. Commun. 6, 7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keller A., Chavez J. D., Eng J. K., Thornton Z., and Bruce J. E. (2019) Tools for 3D interactome visualization. J. Proteome Res. 18, 753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weisbrod C. R., Chavez J. D., Eng J. K., Yang L., Zheng C., and Bruce J. E. (2013) In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res. 12, 1569–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lenz S., Giese S. H., Fischer L., and Rappsilber J. (2018) In-search assignment of monoisotopic peaks improves the identification of cross-linked peptides. J. Proteome Res. 17, 3923–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Götze M., Iacobucci C., Ihling C. H., and Sinz A. (2019) A simple cross-linking/mass spectrometry workflow for studying system-wide protein interactions. Anal. Chem. 91, 10236–10244 [DOI] [PubMed] [Google Scholar]
- 51. Yugandhar K., Wang T.-Y., and Yu H. (2019) Structure-based validation can drastically under-estimate error rate in proteome-wide cross-linking mass spectrometry studies. bioRxiv, 617654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keller A., Chavez J. D., Felt K. C., and Bruce J. E. (2019) Prediction of an upper limit for the fraction of interprotein cross-links in large-scale in vivo cross-linking studies. J. Proteome Res. 18, 3077–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu H., Tardivo L., Tam S., Weiner E., Gebreab F., Fan C., Svrzikapa N., Hirozane-Kishikawa T., Rietman E., Yang X., Sahalie J., Salehi-Ashtiani K., Hao T., Cusick M. E., Hill D. E., Roth F. P., Braun P., and Vidal M. (2011) Next-generation sequencing to generate interactome datasets. Nat. Methods 8, 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207. [DOI] [PubMed] [Google Scholar]
- 55. Kosinski J., von Appen A., Ori A., Karius K., Müller C. W., and Beck M. (2015) Xlink Analyzer: Software for analysis and visualization of cross-linking data in the context of three-dimensional structures. J. Structural Biol. 189, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 57. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S. J., and Marra M. A. (2009) Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All cross-links are reported in the Supplementary information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE(38) partner repository with the dataset identifier PXD013928.