Peptide biomarker candidates for the respiratory tract pathogens Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis and Staphylococcus aureus are presented. First, bacterial cultures representing the genetic variability in each of the four species, were analyzed. The peptide biomarker candidates were then experimentally verified to be present in a clinical situation by analyzing true positive clinical samples. The most promising peptide biomarkers were used in a targeted MS mode, demonstrating their use for future clinical implementation.
Keywords: Biomarker: diagnostic, bacteria, early diagnosis, tandem mass spectrometry, infectious disease, microbiology, peptides
Graphical Abstract
Highlights
Discovery of peptide biomarker candidates of respiratory tract pathogens S. pneumoniae, H. influenzae, M. catarrhalis and S. aureus as target pathogens.
Peptide biomarker candidates were experimentally verified in clinical samples.
Targeted MS using promising peptide biomarker candidates shown as proof-of-concept.
Abstract
Mass spectrometry (MS) and proteomics offer comprehensive characterization and identification of microorganisms and discovery of protein biomarkers that are applicable for diagnostics of infectious diseases. The use of biomarkers for diagnostics is widely applied in the clinic and the use of peptide biomarkers is increasingly being investigated for applications in the clinical laboratory. Respiratory-tract infections are a predominant cause for medical treatment, although, clinical assessments and standard clinical laboratory protocols are time-consuming and often inadequate for reliable diagnoses. Novel methods, preferably applied directly to clinical samples, excluding cultivation steps, are needed to improve diagnostics of infectious diseases, provide adequate treatment and reduce the use of antibiotics and associated development of antibiotic resistance. This study applied nano-liquid chromatography (LC) coupled with tandem MS, with a bioinformatics pipeline and an in-house database of curated high-quality reference genome sequences to identify species-unique peptides as potential biomarkers for four bacterial pathogens commonly found in respiratory tract infections (RTIs): Staphylococcus aureus; Moraxella catarrhalis; Haemophilus influenzae and Streptococcus pneumoniae. The species-unique peptides were initially identified in pure cultures of bacterial reference strains, reflecting the genomic variation in the four species and, furthermore, in clinical respiratory tract samples, without prior cultivation, elucidating proteins expressed in clinical conditions of infection. For each of the four bacterial pathogens, the peptide biomarker candidates most predominantly found in clinical samples, are presented. Data are available via ProteomeXchange with identifier PXD014522. As proof-of-principle, the most promising species-unique peptides were applied in targeted tandem MS-analyses of clinical samples and their relevance for identifications of the pathogens, i.e. proteotyping, was validated, thus demonstrating their potential as peptide biomarker candidates for diagnostics of infectious diseases.
Respiratory tract infections (RTIs)1 are a major reason for hospital admissions and are often treated with antibiotics (1). Today, a clinical assessment performed by the physician, is mainly based on symptoms, together with supporting clinical laboratory microbiological confirmation (2). Microbiological characterization of a clinical sample traditionally relies on cultivation of bacteria, which not only takes precious time, but in many cases is inconclusive because of the difficulty to recover viable bacteria. For example, in only ∼50% of the cases, are Streptococcus pneumoniae, a responsible agent for pneumococcal infections, recovered by culturing (3). Because bacterial infection can lead rapidly to invasive life-threatening situations, physicians may prescribe broad-spectrum antibiotics before knowing whether the infection is caused by bacteria or virus. Overuse of broad-spectrum antibiotics is a significant contributor to the emergence of anti-microbial resistance (AMR). One of the key counter-measures in the battle against AMR will be the development of improved, rapid, accurate and comprehensive diagnostic methods.
DNA-based diagnostic approaches, such as real-time polymerase chain reaction (RT-PCR) is currently implemented in the routine protocols of the clinical microbiology laboratory and whole-genome sequencing is increasingly applied. However, PCR is a targeted approach and, thus, detects and identifies only the known and selected targets, which can lead to biased results and insufficient species resolution and characterization. One example is in the differentiation of closely related species within the Mitis Group of the genus Streptococcus, using PCR-based analyses of house-keeping genes or virulence factors (4–6).
Matrix-Assisted Laser Desorption/Ionization-Time-Of-Flight (MALDI-TOF) MS-based microbial species identification has emerged as an alternative to traditional phenotypic- or genotypic-based methods (7–10). Demonstrating benefits, such as reliable species-level resolution, in most cases, ease-of-use and speed of processing samples, as well as low cost per analysis, MALDI-TOF MS identification is now used in clinics world-wide. However, a significant drawback of MALDI-TOF MS analyses is that it, in most cases, requires time-consuming cultivation and isolation of the relevant microorganisms. Further drawbacks include limitations in discriminating closely related species, including some species of the Mitis Group of the genus Streptococcus, and, except in some limited cases (11–13), it has proven ineffective for obtaining information on characteristic features, such as AMR and virulence (14).
To increase the discriminative power and resolution for differentiating closely related species, even to strain-level typing, tandem MS approaches at the peptide level have been employed (14–21). Peptide biomarker discovery has been facilitated by development of MS-instruments performing bottom-up “high-resolution accurate-mass (HRAM)” tandem MS proteomics, enabling identification of thousands of peptides simultaneously, in a single analysis (16). At the peptide level, tandem MS has the power to elucidate expressed point mutations (22), enabling high levels of resolution. Biomarkers for resistance and virulence factors can be detected simultaneously in the same analysis, providing crucial information for diagnoses and proper treatments (23, 24).
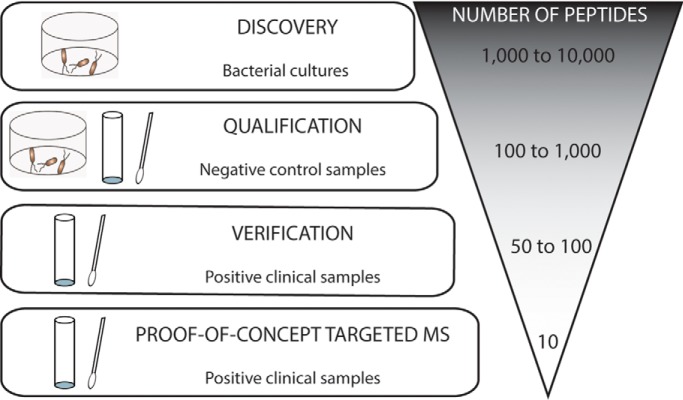
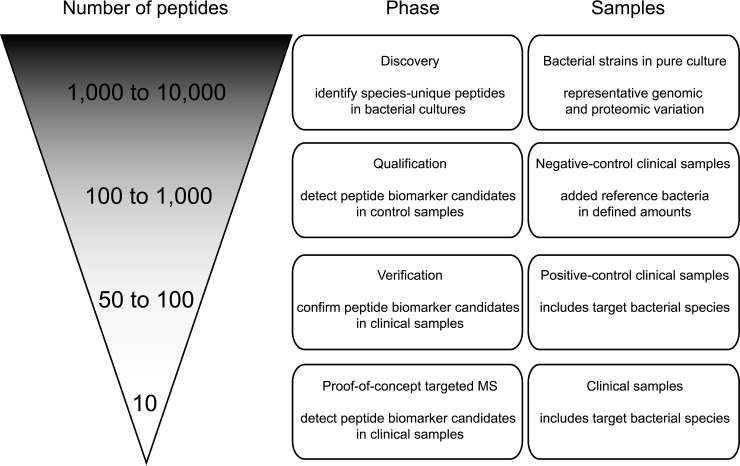
Previously, we have shown that peptide biomarkers have the power to differentiate bacterial species (14), as well as strains within the same species (18). This “proteotyping” approach (14, 25, 26) can also be used for differentiating taxonomically-close species, such as the pathogen S. pneumoniae from commensal species, S. pseudopneumoniae and S. mitis of the Mitis Group of the genus Streptococcus (14). In the present study, the workflow combines HRAM tandem MS and the TCUP (Typing and Characterization of bacteria Using bottom-up tandem mass spectrometry Proteomics) bioinformatics pipeline (27) in the search for novel species-unique peptides as potential biomarkers for the respiratory tract pathogens, Staphylococcus aureus, Moraxella catarrhalis, Haemophilus influenzae and Streptococcus pneumoniae. In contrast to traditional cultivation-based methodologies, proteotyping is not relying on recovery of cultivable cells, but can be applied directly to clinical samples. The purpose of this study was to initially identify species-unique peptides as potential peptide biomarker candidates from bacterial cultures of reference strains of the target bacterial species and then to confirm these biomarker candidates in clinical respiratory-tract samples without any cultivation step (Fig. 1).
Fig. 1.
Illustration showing the process employed for identifying species-unique peptides as potential peptide biomarker candidates. During the process, bacterial cultures, representing genomic and proteomic variation within the species, as well as clinical samples, were analyzed. The purpose of this workflow was to initially identify as many species-unique peptides as possible and in later phases narrow down the number of peptides to the most promising peptide biomarker candidates to be used for diagnostic analyses.
EXPERIMENTAL PROCEDURES
Cultivation and Classification of Bacteria
Bacterial strains were selected of each of four common respiratory-tract infectious bacterial species: S. aureus (12 strains), M. catarrhalis (11 strains), H. influenzae (9 strains), and S. pneumoniae (7 strains); obtained from the Culture Collection, University of Gothenburg, Gothenburg, Sweden (CCUG; www.ccug.se) (supplemental Table S1). Cultures were grown overnight in the following way: S. aureus was grown on Blood Agar, at 37 °C, aerobically; M. catarrhalis and S. pneumoniae were grown on Blood Agar, at 37 °C, with 5% CO2; H. influenzae strains were grown on Chocolate Agar medium, at 36 °C, with 5% CO2. The classifications of the selected strains of H. influenzae, M. catarrhalis were confirmed by 16S rRNA gene sequence determinations and comparative sequence analyses (28). Classifications of the selected strains of S. aureus were confirmed by 16S rRNA gene and sodA sequence analyses (29). Classifications of the selected strains of S. pneumoniae were confirmed by whole genome sequence Average Nucleotide Identity based on BLAST (ANIb) analyses (30), using JSpeciesWS (31), against the genome sequence of S. pneumoniae NCTC 7465T (GenBank accession number: LN831051).
Peptide Generation from Bacterial Cultures
Bacterial biomass was collected from fresh cultures and suspended in phosphate-buffered saline (PBS). The bacteria were washed with PBS and lysed, by bead beating (14). The bacterial lysates were frozen until further analysis. The Lipid-based Protein Immobilization (LPI®) methodology was employed for generating peptides from the cultured bacteria, as described previously (14, 18, 27). Each strain of the four bacteria, S. aureus, M. catarrhalis, H. influenzae, and S. pneumoniae, were digested in triplicates (supplemental Fig. S1).
To digest bacterial proteins into peptides, the cell lysate was injected into a LPI Hexalane FlowCell (Nanoxis Consulting AB, Gothenburg, Sweden, www.nanoxisconsulting.com; Patent Application No. WO2006068619), using a pipette to fill the FlowCell channel (channel volume of ∼30 μl). Proteins were immobilized to the FlowCell surface, after incubation for 1 h, at room temperature. The FlowCell channels were washed with 400 μl of ammonium bicarbonate, using a syringe pump, with a flow rate of 100 μl/min. Enzymatic digestion of the proteins was performed by injecting trypsin (V5111, Promega, Madison, WI) (2 μg/ml in 20 mm ammonium bicarbonate, pH 8.0) into the FlowCell channels and incubating for 1 h at room temperature. The generated peptides were eluted by injecting 200 μl ammonium bicarbonate buffer (20 mm, pH 8.0) into the channels. The eluted peptides were collected at the outlet ports, using a pipette, and transferred into tubes (2.0 ml, Axygen, Corning Life Sciences, MA). The peptide solutions were incubated at room temperature overnight and subsequently frozen at −20 °C until analysis by MS. The peptide samples were not reduced or alkylated prior to MS analysis.
Clinical Samples
Clinical respiratory tract samples (nasopharyngeal and nasal swabs, n = 218), analyzed and reported as positive by the Clinical Microbiology Laboratory (Sahlgrenska University Hospital, Gothenburg, Sweden), were collected in Amies media (eSwab, Copan Diagnostics, Inc, CA). The clinical samples were reported to contain at least one of the four pathogens included in the study (S. aureus, M. catarrhalis, H. influenzae and/or S. pneumoniae). In many cases, the samples displayed co-infection with two or more of these pathogens. The pathogens in clinical samples were confirmed by the standard, accredited clinical microbiology laboratory protocols for selective and differential isolation of bacteria, including subsequent identification by MALDI-TOF MS analysis. Samples were supplemented with STGG (Skim milk, Tryptone, Glucose, Glycerol) to bolster the viability of bacteria as well as recovery of bacterial proteins during storage of respiratory tract samples and frozen until processing (32). Only samples that were collected as part of the standard diagnostic protocols were included in this study; no additional or extra sampling from patients was carried out and no patient identifiable information was collected; hence, informed consent was not required.
In the qualification phase, clinical respiratory tract samples, reported to be negative for bacteria by cultivation-based protocols and MALDI-TOF-MS, were spiked with cells of the type strains of the four species H. influenzae (CCUG 23945T), M. catarrhalis (CCUG 353T), S. aureus (CCUG 41582T) and S. pneumoniae (CCUG 28588T), to select the most promising peptide biomarker candidates for the validation phase. The number of added cells to the negative clinical samples ranged from 100 cells/ml to 1 million cells/ml (supplemental Fig. S2).
Peptide Generation from Clinical Samples
The MolYsis kit (MolYsis Basic5 kit, Molzym GmbH & Co. Bremen, Germany) was used for removal of human biomass, according to the supplier's protocol, with minor modifications. After sample treatment, the resulting bacterial pellets were re-suspended in 120 μl ammonium bicarbonate (20 mm pH 8) and bacteria were lysed, using bead beating (14). For digestion of proteins, to generate peptides, sodium deoxycholate (SDC, 5% in 20 mm ammonium bicarbonate, pH 8) was added to 1% (w/v) final concentration. Trypsin (2 μg/ml, 100 μl ammonium bicarbonate, 20 mm pH 8) was added and samples were digested for ∼8 h at 37 °C. SDC was removed by precipitation by addition of formic acid (FA) followed by centrifugation at 13,000 × g for 10 min. Supernatants containing the peptides were stored at −20 °C until analysis. The peptide samples were not reduced or alkylated prior to MS analysis (supplemental Fig. S3).
NanoLC-MS/MS Analysis
Peptide samples were desalted, using PepClean C18 spin columns (Thermo Fisher Scientific, MA), according to the manufacturer's guidelines. MS analyses were carried out, using Q Exactive or a QExactive HF MS (Thermo Fisher Scientific) interfaced with an Easy nLC 1200 liquid chromatography system (Thermo Fisher Scientific). Peptides were trapped on an Acclaim Pepmap 100 C18 trap column (100 μm × 2 cm, particle size 5 μm, Thermo Fischer Scientific) and separated on an in-house packed analytical column (75 μm × 300 mm, particle size 3 μm, Reprosil-Pur C18, Dr. Maisch, Germany), using a gradient from 7% to 35% B over 35, 50, or 75 min followed by an increase to 100% B for 5 min at a flow of 300 nL/min. Solvent A was 0.2% formic acid and solvent B was 80% acetonitrile in 0.2% formic acid. The instrument operated in data-dependent mode where the precursor ion mass spectra were acquired at a resolution of 70,000 (QE) or 60,000 (QEHF), the 10 most intense multiply charged ions were isolated in a 2.0 Da isolation window and fragmented using collision energy HCD settings at 27. MS2 spectra were recorded at a resolution of 35,000 (QE) or 30,000 (QEHF). Dynamic exclusion was set to 20–30 s with 10 ppm tolerance. Inclusion lists, containing the candidate peptide biomarkers for each species, were used in the qualification and verification phases together with pick others to improve the sensitivity of the MS-method. The m/z ratios (a maximum of 50 for each MS-analysis) corresponding to specific peptide biomarkers were prioritized for fragmentation even if they were not among the Top10 most abundant peptides.
TCUP Bioinformatics Pipeline
Raw data were evaluated using the TCUP bioinformatics pipeline (27) to identify species-unique peptides. The LC-MS/MS output was converted from the proprietary Thermo Xcalibur RAW format to the open-source mzXML format (33), using ReAdW (34) (version 201411.xcalibur), with command-line arguments: “–nocompress -gzip.” The X! Tandem spectrum search engine (version VENGEANCE Dec. 15, 2015) (35, 36) was used to identify peptides from the mass spectra with the following settings: fragment monoisotopic mass error = 20; parent monoisotopic mass error plus = 5; parent monoisotopic mass error minus = 5; fragment mass type monoisotopic, dynamic range = 100.0; total peaks = 50; maximum parent charge = 4; minimum parent m+h = 800.0; minimum fragment m/z = 100.0, minimum peaks = 15, potential modification mass = 16.0@M, maximum valid expectation value = 1.0. In addition, X! Tandem peptides were also filtered to only allow peptides with a hyperscore of >30 in downstream analyses (37). Values for all X!Tandem settings are available in supplemental file 1. The reference database used in this step was a customized database consisting of 56,967,781 non-redundant proteins from the NCBI GenBankTM NR (38) and 6,320,906 peptide sequences from the reference genomes archived within the Human Microbiome Project (39). All sequences containing unidentified peptides (“X”), as well as duplicates of sequences shared between the two databases, were removed. The resulting database used with X! Tandem contained a total of 59,349,300 distinct protein sequences. The taxonomic hierarchy used in TCUP was based on the complete NCBI Taxonomy (40) (taxdump downloaded Nov. 17, 2015) and each reference genome in the reference database was associated with a unique node in the taxonomic tree. The search parameters were set, according to Boulund et al. (27). All peptides presented in Tables I–IV were mapped against RefSeq sequences (Oct 2018) using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table I. The peptide biomarker candidates of S. aureus and the proteins from which they originate.
| Peptide sequence | Number of times detected in 36 MS analyses of S. aureus cultures | Number of times detected in unique clinical samples | Protein (GenBank accession number and description) | |
|---|---|---|---|---|
| TVQPIDVDTIVASVEK | 36 | 22 | AKJ16950.1 | 2-oxoisovalerate dehydrogenase |
| QAGVGAAVVAELSER | 36 | 18 | ||
| ELINNIQSGQR | 36 | 15 | AKJ17520.1 | Preprotein translocase subunit YajC |
| LGISDGDVEETEDAPK | 36 | 16 | AKJ17148.1 | Recombinase RecA |
| ALLNNMVQGVSQGYVK | 36 | 14 | AKJ18065.1 | 50S ribosomal protein L6 |
| SNVNDATDYSSETPEGK | 36 | 12 | AKJ17216.1 | Transketolase |
| ANNVATDANHSYTSR | 36 | 13 | AKJ17623.1 | Hypothetical protein |
| ILAESPNLAISSSSR | 35 | 10 | AKJ16422.1 | HAD family hydrolase |
| NVVEIPLNDEEQSK | 31 | 9 | AKJ16109.1 | Lactate dehydrogenase |
| ATEATNATNNQSTQVSQATSQPINFQVQK | 24 | 7 | AKJ16987.1 | Heme transporter IsdA |
| IHLVGDEIANGQGIGR | 35 | 8 | AKJ17576.1 | Pyruvate kinase |
| NISNNVLVTIDAAQGK | 13 | 6 | ||
| TAKPVAEVESQTEVTE | 26 | 10 | AKJ16406.1 | DNA-directed RNA polymerase subunit beta' |
| SQGVSEEELNESIDR | 29 | 1 | AKJ16022.1 | Acetaldehyde dehydrogenase |
| AEENGLTVVDAFNFEAPK | 16 | 7 | AKJ18079.1 | 50S ribosomal protein L4 |
| LLGINATIVMPETAPQAK | 1 | 1 | AKJ17317.1 | Threonine dehydratase |
Table II. The peptide biomarker candidates of M. catarrhalis and the proteins from which they originate.
| Peptide sequence | Number of times detected in number of 33 MS analyses of M. catarrhalis cultures | Number of times detected in unique clinical samples | Protein (GenBank accession number and description) | |
|---|---|---|---|---|
| VVLAGDTVVSDR | 33 | 14 | WP_003666427.1 | TonB-dependent receptor |
| QIVSNAGDEASVIVNEVK* | 33 | 18 | WP_063454121.1 | Chaperonin GroEL |
| AIAQVGSISANSDATIGELISK | 29 | 16 | ||
| ELSNTAAETQPK | 33 | 18 | WP_003659702.1 | 30S ribosomal protein S1 |
| VDATVDAQNPTK | 24 | 16 | WP_003660336.1 | Hypothetical protein |
| QSDVGQLTGK | 5 | 9 | ||
| FNATAALGGYGSK | 31 | 12 | WP_063454085.1 | Cell surface protein |
| THTSALAEENQQASIPR | 33 | 12 | WP_063454087.1 | Cell division protein FtsZ |
| YVVEGANMPLDAQAIDIVR | 17 | 11 | WP_049156084.1 | NADP-specific glutamate dehydrogenase |
| SQIYQTTASVSGAR | 33 | 9 | WP_003657351.1 | Ohr family peroxiredoxin |
| LLNETTGQVVPK | 33 | 8 | WP_003657987.1 | DUF4377 domain-containing protein |
| SSENVVVVSVR | 33 | 10 | WP_063454071.1 | Electron transfer flavoprotein subunit beta |
| AISYGNSADAQPYVGAK | 33 | 10 | WP_003658939.1 | Porin family protein |
| GLPVSNSGAPISVPVGQATLGR | 31 | 8 | WP_003658974.1 | F0F1 ATP synthase subunit beta |
| VNYNGDTDTVTLSGVAK | 33 | 13 | WP_003656943.1 | Peptidoglycan-binding protein LysM |
| AVATQQATVSAEYLQK | 5 | 10 | WP_003657125.1 | ABC transporter substrate-binding protein |
| ADSGLSESEIEEMIR | 32 | 12 | WP_003669031.1 | Molecular chaperone DnaK |
| LGAQEAELVSNSK | 33 | 7 | WP_003660298.1 | CTP synthase |
*This peptide was also found in samples spiked with the fewest number of cells (supplemental Table S2).
Table III. The peptide biomarker candidates of H. influenzae and the proteins from which they originate.
| Peptide sequence | Number of times detected 26 MS analyses of H. influenzae cultures | Number of times detected in unique clinical samples | Protein (GenBank accession number and description) | |
|---|---|---|---|---|
| GVAADAISATGYGK* | 22 | 22 | WP_038441355.1 | Porin OmpA |
| ANLKPQAQATLDSIYGEMSQVK | 5 | 6 | ||
| ADSVANYFVAK | − | 5 | ||
| GSYEVLDGLDVYGK | 12 | 3 | ||
| LSQERADSVANYFVAK** | − | 2 | ||
| AVVYNNEGTNVELGGR* | 22 | 14 | WP_058222193.1 | Porin |
| YDANNIIAGIAYGR* | 13 | 6 | ||
| ATHNFGDGFYAQGYLETR | 15 | 5 | ||
| AVVYNNEGTKVELGGR | − | 5 | ||
| QQVNGALSTLGYR | 18 | 1 | ||
| YVPTNGNTVGYTFK | − | 4 | ||
| LSVIAEQSNSTR* | 4 | 1 | ||
| SADLTNEVAVGDVVEAK | 4 | 6 | WP_011272719.1 | 30S ribosomal protein S1 |
| SADLTSEVAVGDVVEAK | 11 | 2 | ||
| TSPTQNLSLDAFVAR | 9 | 5 |
WP_058222202.1 WP_050846043.1 |
ShlB/FhaC/HecB family hemolysin secretion/activation protein |
| AQYIVEQVIGQAR | 26/29 | 2 | WP_011272712.1 | Pyruvate dehydrogenase (acetyl-transferring), homodimeric type |
*This peptide was also found in samples spiked with the fewest number of cells (supplemental Table S2).
**Peptide with missed cleavage includes ADSVANYFVAK.
Table IV. The peptide biomarker candidates of S. pneumoniae and the proteins from which they originate.
| Peptide sequence | Number of times detected in 21 MS analyses of S. pneumoniae cultures | Number of times detected in unique clinical samples | Protein (GenBank accession number and description) | |
|---|---|---|---|---|
| VSDVAESTGEFTSEQFEK* | 21 | 22 | WP_000064115.1 | Asp23/Gls24 family envelope stress response protein |
| GAANGVVSHENTR* | − | 9 | ||
| EEAPVASQSK | − | 9 | WP_001035310.1 | Hypothetical protein |
| SADQQAEEDYAR | − | 8 | ||
| APLQSELDTK | − | 3 | ||
| LKEIDESDSEDYVK | − | 3 | ||
| NVEIIEDDKQGVIR | 1 | 10 | WP_000245505.1 | 30S ribosomal protein S8 |
| NLPVGSDGTFTPEDYVGR | 20 | 8 | WP_001291372.1 | Methionine–tRNA ligase |
| TLELEIAESDVK | − | 5 | WP_000458177.1 | Hypothetical protein |
| DIGLANDGSIVGINYAK | 12 | 5 | WP_000927809.1 | Sugar ABC transporter substrate-binding protein |
| IAELEYEVQR | − | 6 | WP_001008677.1 | Asp-tRNA(Asn)/Glu-tRNA(Gln) amidotransferase subunit GatB |
| AVAAADAADAGAAK | 3 | 3 | WP_001196960.1 | 50S ribosomal protein L7/L12 |
| GQDWVIAAEVVTKPEVK | 16 | 5 | WP_000116461.1 | Trigger factor |
| TLSPEEYAVTQENQTER | − | 6 | WP_000998307.1 | Peptide-methionine (R)-S-oxide reductase |
| KDEAEAAFATIR | − | 3 | WP_001284361.1 | Thiol-activated toxin pneumolysin |
| SQPSSETELSGNKQEQER | 16 | 2 | WP_078148305.1 | Sialidase |
| IGVISVVEDGDEALAK | − | 2 | WP_000808063.1 | Elongation factor Ts |
| VAYFNEIDTYSEVK | − | 2 | WP_000685088.1 | Nucleotide sugar dehydrogenase |
*This peptide was also found in samples spiked with the fewest number of cells (supplemental Table S2).
Generation of Targeted Database and MS-inclusion Lists
A targeted database was compiled, including 15,417 species-unique peptides identified by TCUP in at least one of the MS analyses of the representative strains of the four bacterial species (Table V, supplemental Table S2). The smaller targeted database was used for increasing the probability of positive identification of the relevant peptide biomarkers. Additionally, MS-inclusion lists used in the later qualifications phase were generated. Peptides that were detected in all strains and all MS analyses were ranked the highest in the lists (supplemental Fig. S1). The lists contain the 100 highest-ranked peptides for each species.
Table V. Number of strains analyzed, corresponding number of MS analyses and the resulting number of species-unique peptides found for each of the species.
| Species | Number of strains | Number of MS analyses | Number of species-unique peptides |
|---|---|---|---|
| S. aureus | 12 | 36 | 5,847 |
| M. catarrhalis | 11 | 33 | 5,810 |
| H. influenzae | 9 | 26 | 2,978 |
| S. pneumoniae | 7 | 21 | 782 |
These peptide lists were revised after the qualification phase and the peptides detected in the samples with a lower number of spiked pathogenic bacteria were ranked the highest. In verification phase, the clinical samples were analyzed in batches and the peptide lists were again revised after each batch, according to the following criteria: (1) Identified peptides from the list were verified as a peptide biomarker candidate by its presence in clinical samples; (2) Peptides identified in the clinical samples by TCUP, but not present in the inclusion lists, were added to updated versions of the inclusion lists; (3) Peptide biomarker candidates present in the initial inclusion lists, but not detected in the clinical samples, were removed from updated inclusion lists or were given a lower ranking (supplemental Data S1–S4). After ranking, the final lists of peptides for each of the bacterial species were reduced to the top 16–18 peptide biomarker candidates (supplemental Fig. S3, Tables I–IV).
Database Matching
In parallel with the TCUP bioinformatics pipeline, the data was matched, using Proteome Discoverer (Thermo Fisher Scientific, version 1.4), against the targeted database. Mascot 2.5 (Matrix Science, MA) was used as a search engine with precursor mass tolerance of 5 ppm and fragment mass tolerance of 200 mmu and variable methionine oxidation. Fixed Value with a maximum delta Cn of 0.05 was employed in the database matching and the peptides used for protein identification were filtered at 1% FDR. The fragmentation spectra and ion series for all detected peptides in the clinical samples were inspected manually to verify correct identifications.
Targeted MS (PRM) Analyses
For each of the four bacterial species, the top 16–18 peptide biomarker candidates (Tables I–IV, supplemental Data S5–S8), most prominently found in clinical samples were analyzed, using parallel reaction monitoring (PRM) on a Q Exactive HF (Thermo Fisher Scientific). Separation was performed, using a 50 min gradient, as stated above and the precursor ions of the peptides were targeted without scheduling. The QEHF orbitrap resolution was 30 000, a quadrupole isolation window of 1.2 Da and collision energy HCD settings at 27 were used. PRM data were analyzed using Skyline (version 4.2.0) (39). Peak picking was manually checked and corrected in accordance with the retention time, transitions and mass accuracy to confirm the identities of peptides. For this proof-of-concept, the PRM method used here was employed to bias toward detection of the peptide biomarkers of interest (Tier 3, as defined in the MCP guidelines).
The MS proteomics data (MS/MS-spectra for all species-unique peptides presented in Tables I–IV, as well as raw-files and PD1.4 search files of representative clinical samples) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifiers PXD014522.
Experimental Design and Statistical Rationale
The workflow of how to discover, qualify and verify the species-unique peptides is shown in Fig. 1 and supplemental Fig. S4. The number of bacterial strains included in the discovery phase of finding species-unique peptides were S. aureus (12 strains), M. catarrhalis (11 strains), H. influenzae (9 strains) and S. pneumoniae (7 strains), all analyzed in triplicate, resulting in, at least, 21 MS analyses per species (Table V). The number of identified species-unique peptides increased with the number of analyzed strains. However, at a certain stage, analyses of additional strains did not contribute to further increase in the number of species-unique peptides and the number of strains selected per species was concluded to be satisfactory (supplemental Figs. S5–S8).
In the verification phase, to verify the presence of the species-unique peptides in patient samples, without prior culturing, the number of clinical samples included was 218. As this study was focused on the discovery of species-unique peptides no replicate analyses were performed at this stage, as it was deemed more important to analyze many individual clinical samples.
RESULTS
In summary, the workflow of discovering, qualifying and verifying the species-unique peptides as promising peptide biomarker candidates was divided into four phases (Fig. 1 and supplemental Fig. S4). First, in the discovery phase, species-unique peptides were identified from pure bacterial cultures. Subsequently, in the qualification phase, negative clinical samples were spiked with bacterial cells in order to ensure that the peptides could be detected in the context of a realistic clinical sample. In the verification phase, positive clinical samples were analyzed to verify the species-unique peptides most frequently found in clinical samples. Last, as a proof-of-concept, positive clinical samples were analyzed using a targeted MS approach with the selected candidate peptide biomarkers as targets.
In the discovery phase, several representative strains from each of the four target species, S. aureus (13 strains), M. catarrhalis (11 strains), H. influenzae (9 strains) and S. pneumoniae (7 strains) were selected to reflect the genetic variation within the species (supplemental Table S1). Each species was analyzed with a minimum of 21 MS runs resulting in identified species-unique peptides (Table V). The largest number of species-unique peptides were found in S. aureus and M. catarrhalis (5847 and 5810, respectively), H. influenzae strains comprised 2978 species-unique peptides, whereas the fewest number of species-unique peptides (782) was detected in strains of S. pneumoniae. The peptides were ranked, based on the number of strains in which they were detected. These results from the MS-analyses were compiled to a database containing the 15,417 species-unique peptides (supplemental Table S2). The most promising peptide biomarker candidates, based on the number of strains they were found in, were reduced to lists of 100 peptides for each species (supplemental Fig. 1).
In the qualification phase, the suitability of the species-unique peptides as potential peptide biomarker candidates was evaluated. Negative clinical samples were spiked with varying concentrations of bacterial cells (supplemental Fig. S2) and the MS analyses were performed using inclusion lists with the hundred highest ranking species-unique peptides identified in the discovery phase. The number of bacterial cells per ml of sample ranged from 100 to 1 million cells/ml, reflecting the variation in the number of bacteria cells typical for nasopharyngeal/nasal swab samples; bacterial loads vary during different phases of infection and are also dependent on the pathogen (41, 42). The selected range was considered to realistically reflect both weakly- and strongly-infected samples. The species-unique peptides detected in samples containing the lowest number of bacterial cells, ranging from 1000 to 10,000, were deemed to be promising peptide biomarker candidates (supplemental Table S3). The ranking of the peptides in the respective inclusion lists were revised in accordance with the results from the qualification phase (supplemental Fig. S2).
In the verification phase, 218 clinical respiratory tract samples (312 MS injections) were included, from which isolations of S. aureus, M. catarrhalis, H. influenzae and/or S. pneumoniae, were reported. These samples were thus used as “positive control” samples for MS analyses. The clinical samples were reported by the Clinical Microbiology Laboratory to contain at least one of the pathogens included in the study, but in many cases, samples displaying co-infection with two or more of these four pathogens, were included. By analyzing these samples, peptide biomarker candidates were detected and identified, confirming that the proteins from which the peptide biomarker candidates originate, are present in vivo. The peptides most prominently detected in clinical samples, and their corresponding proteins, as well as the number of times they were detected in the cultures of bacterial reference strains, are presented in Tables I, II, III, and IV. In order to verify the identities of the peptides in the clinical samples, all fragmentation spectra were inspected manually. The fragmentation spectra and ion series for a top ranked peptide for each of the four species are shown in supplemental Figs. S9–S12. During the analysis of the clinical samples in the verification phase, the lists containing the peptide biomarker candidates were continuously revised according to the ranking of the peptide. The final lists were reduced to contain only the 15–20 most promising peptide biomarker candidates for the proof-of-concept targeted MS analyses (supplemental Fig. 3, supplemental Data S1–S4).
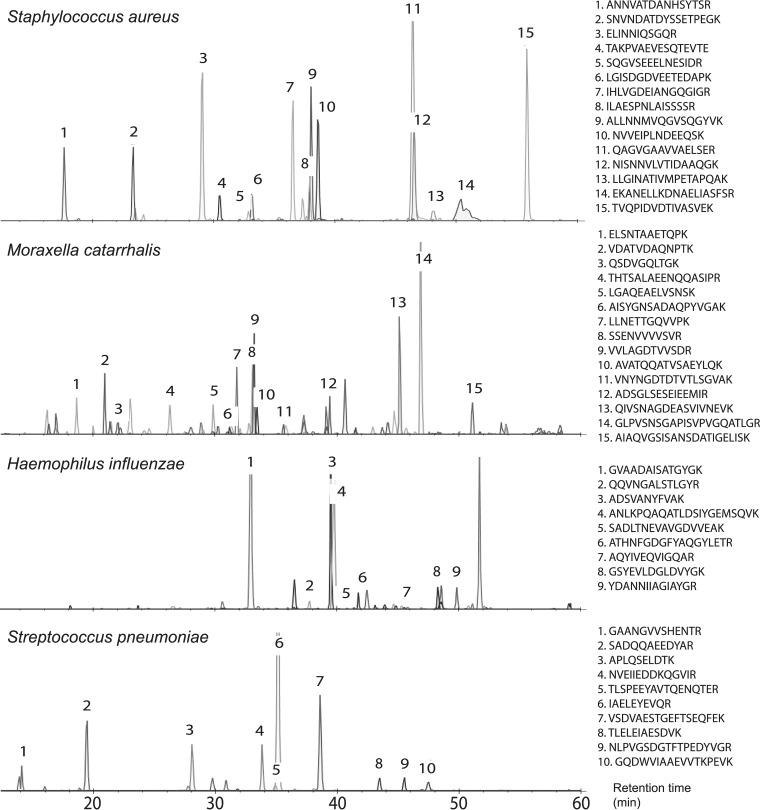
Finally, as a proof-of-concept, a PRM method was developed, offering increased sensitivity and high selectivity (22), by targeting the most suitable peptide biomarker candidates identified in the verification phase (Tables I–IV, supplemental Data S5–S8, supplemental Table S4, Fig. 2). The peptide identities were verified by aligning the retention time together with correct transitions and mass accuracy.
Fig. 2.
Direct analyses of clinical respiratory tract samples, using PRM, targeting the most promising peptide biomarker candidates, presented in Tables I–IV. The peptide intensities are summed up fragment ion intensities of the peptides′ most abundant charge state. Whenever peptides contain a methionine the more abundant oxidized form is shown in the spectra. The peptide peaks are labeled with numbers corresponding to their sequences.
DISCUSSION
The selection of species-unique peptides was performed throughout the phases of discovery, qualification and verification (Fig. 1, supplemental Fig. S4). The purpose of these phases was to narrow down the number of species-unique peptides for determining the most suitable peptide biomarker candidates, from the starting point of analyses of bacterial reference cultures in the discovery phase, representing the genomic and proteomic variation of the species included in the study. In the qualification phase, the species-unique peptides detected in the lowest number of spiked cells displayed suitable properties for ionization, fragmentation and detection in the MS-analyses. Also, they were not suppressed by contaminating peptides of human origin from the clinical samples. Furthermore, the results show that sufficient amounts of bacterial cells were recovered during the removal of the human biomass, also suggesting that the limited amount of bacterial pathogen material in clinical respiratory tract samples can be recovered for detection in the MS-analysis.
The results from the subsequent verification phase demonstrate the importance of confirming data stemming from cultures of bacterial reference strains, by analyses of clinical samples. As expected, not all the species-unique peptides identified in the bacterial cultures were detected in the clinical samples. During traditional protocols including cultivation, the conditions are selected to best promote growth for recovery of enough biomass for downstream analyses. However, during invasion of the host, pathogens are known to experience stress, such as nutrient limitation, low pH, etc. Exposure to host environments also triggers virulence responses by pathogens and, thus, virulence factors may be expressed and present in high levels in clinical samples, whereas they may be present at limited levels in culture. Therefore, pathogens display different protein profiles in vivo, compared with what is observed in defined cultivation conditions (43, 44).
Differences in protein profiles for cultured bacteria and clinical samples can be seen clearly in the analyses of S. pneumoniae and H. influenzae (Table III and IV). For these two species, 4 of the most promising species-unique peptides of H. influenzae, and 13 for S. pneumoniae, identified in the clinical samples, were not found in the analyses of any of the cultured bacterial reference strains. For M. catarrhalis and S. aureus, many of the peptide biomarker candidates originated from highly abundant cytosolic proteins, including ribosomal proteins. Because cytosolic house-keeping proteins, in general, are present in relatively high levels, regardless of growth conditions, the most prominent peptide biomarker candidates would most likely originate from the house-keeping proteins when analyzing clinical samples. These results are consistent with traditional gene-based approaches and MALDI-TOF MS, which both commonly use house-keeping genes and proteins as targets for identification. In contrast, many of the proteins identified from the peptide biomarker candidates for S. pneumoniae and H. influenzae include those associated with the surface of the cells. This might be because of the differences in taxonomic structure of the different species. M. catarrhalis and S. aureus are phylogenetically more distant from their closest related species and as a result their house-keeping proteins, including ribosomal proteins, do not display substantial sequence homology of the species closest to them. However, for S. pneumoniae and H. influenzae, the taxonomic structures around these species are more complex and the house-keeping proteins, including ribosomal proteins, display a higher degree of sequence homology to closely related species. Therefore, it may be more difficult to find peptide biomarker candidates originating from their house-keeping proteins. Surface-associated proteins have different functions, helping the bacteria survive in diverse and dynamic ecological niches and, particularly, these proteins are often involved in host-pathogen interactions, effectively functioning, as virulence factors (18). Many of the proteins identified from S. pneumoniae and H. influenzae by their respective peptide biomarker candidates belong to the group of surface-associated virulence factors. This can be explained by the fact that these proteins are the ones differentiating them from their closest relatives, as well as being expressed significantly in clinical samples.
In conclusion, the aim of this study was to initially identify species-unique peptides in cultures of bacterial reference strains from respiratory tract infectious bacteria (S. aureus, M. catarrhalis, H. influenzae and S. pneumoniae) and subsequently determine the most promising and applicable peptide biomarker candidates in clinical samples. Previous proteomic studies, focused on discovery of peptide biomarker candidates for infectious disease diagnostics, have mostly been performed using in vitro model system samples, mainly because of analytical challenges such as recovery of sufficient amount of bacterial proteins from human clinical samples and the high background of human contaminating proteins obstructing the detection of peptide biomarkers from bacteria. In this study, a simple workflow was developed, including removal of human material from clinical respiratory tract samples, whereas still being able to recover sufficient amounts of bacteria for detection of peptide biomarker candidates. Importantly, several hundreds of clinical respiratory tract samples were analyzed directly, without any culturing, thus confirming the presence of peptide biomarker candidates in the clinical samples and, at the same time, their relevance for identifications of the pathogens and as diagnostic biomarkers.
In further studies, the peptide biomarker candidates, will be employed in the development of a targeted MS-approach, as demonstrated here (Fig. 2). Targeted approaches, such as PRM and SRM/MRM (Selected Reaction Monitoring/Multiple reaction monitoring) have several advantages, compared with discovery phase studies, such as higher sensitivity and specificity, simplified MS-analysis and data evaluation (22). In this study, MS proteomics analyses of clinical samples that were confirmed to be positive for a respiratory tract pathogen, determined by standard clinical microbiology methodologies, was employed as a cost-effective approach for identifying peptides from the relevant pathogens included in this study. Notably, samples were frozen until correct identifications could be confirmed by standard means, although, that freezing step may have had a negative effect on some species, sensitive to freezing, thus reducing the number of intact cells prior to the sample preparation for the MS analysis workflow. For comparison of the peptide biomarker approach versus traditional culture-based methods for clinical microbiology diagnostics, the experimental design would be different, i.e. samples would not be frozen prior to processing and more of the sample volume would be dedicated and processed for proteotyping. Furthermore, targeted MS approaches, such as parallel reaction monitoring (PRM) would be employed, as demonstrated here by the proof-of-concept experiment, shown in Fig. 2, wherein a small sub-set of positive clinical samples were analyzed, targeting only the peptide biomarker candidates presented in Tables I–IV. In the continued development of the targeted approach, a larger ensemble of peptide biomarkers (selected from the species-unique peptides in supplemental Table S2) could be employed. Further studies are necessary to compare the use of MS-based peptide biomarkers for identifying respiratory tract pathogens to traditional methodologies - including cultivation-dependent techniques such as MALDI-TOF MS - in terms of sensitivity, specificity, speed, and cost. However, as demonstrated here with the proof-of concept PRM analysis, we show the value of a targeted approach for future high throughput and specific detection of bacteria within complex samples, such as clinical respiratory tract samples, i.e. without prior cultivation steps.
DATA AVAILABILITY
The mass spectrometry proteomics data (MSMS-spectra for all species-unique peptides presented in Tables I–IV, as well as raw-files and PD1.4 search files of representative clinical samples) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifiers PXD014522.
Supplementary Material
Acknowledgments
The CCUG and the staff are acknowledged for providing reference strains and expert characterization analyses. The CCUG is supported by the Department of Clinical Microbiology, Sahlgrenska University Hospital. The staff of the Bacteriology laboratory of the Department of Clinical Microbiology of Sahlgrenska University Hospital are acknowledged for providing clinical samples and for expert identification analyses. We acknowledge the expertise and effort of the Proteomics Core Facility, Sahlgrenska Academy, University of Gothenburg. We thank Dr. Vincent Collins, BioKonsult Göteborg, for critical discussions and proof-reading the manuscript. Beatriz Piñeiro Iglesias and Shora Yazdanshenas are acknowledged for technical assistance. Chantal van Houten and Louis Bont at the Division of Pediatric Immunology and Infectious Diseases, University Medical Centre Utrecht, The Netherlands and Dan Engelhardt at the Division of Pediatric Infectious Disease Unit, Hadassah-Hebrew University Medical Centre, Jerusalem, Israel, are acknowledged for fruitful discussion regarding collection of clinical samples during the Tailored Treatment project. Authors A.K. and R.K. are affiliated to a company, Nanoxis Consulting AB. The Company did not have influence on the collection, analysis, or interpretation of data, the writing of the paper, or the decision to submit for publication.
Footnotes
* R.K., F.S.-S., H.E.J., L.G.-S., D.J.-L., F.B., A.J., E.K., and E.R.B.M. acknowledge support from the European Commission 7th Framework Programme: “Tailored-Treatment”, EU Grant Agreement No.: HEALTH-F3-602860-2013. Swedish Västra Götaland regional funding, project nos. ALFGBG-437221 supported R.K., F.S.-S., E.R.B.M. and ALFGBG-720761 supported R.K., F.S.-S., L.G.-S., and E.R.B.M. The Swedish Västra Götaland Region, FoU grant number VGFOUREG-665141 and Lab Medicine Project number 51060-6258 supported (R.K., S.S., E.K., and E.R.B.M.). F.S.-S., H.E.J., D.J.-L., S.S., and E.R.B.M. acknowledge support from the Swedish Västra Götaland Region, Lab Medicine Project number 51060-6268. R.K., F.S.-S., D.J.-L., A.J., E.K., and E.R.B.M. acknowledge support from the Center for Antibiotic Resistance Research (CARe) at the University of Gothenburg. R.K. and D.J.-L. were supported by stipends for Basic and Advanced Research from the Culture Collection of the University of Gothenburg (CCUG), through the Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg. The authors have declared a conflict of interest.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- RTI
- respiratory tract infection
- AMR
- antimicrobial resistance
- TCUP
- typing and characterization of bacteria using bottom-up tandem MS proteomics
- HRAM
- high-resolution accurate-mass
- LPI
- lipid-based protein immobilization
- ANI
- average nucleotide identity
- BLAST
- Basic Local Alignment Search Tool
- SDC
- sodium deoxycholate
- NCBI
- National Center for Biotechnology Information
- PRM
- parallel reaction monitoring
- SRM
- selected reaction monitoring
- MRM
- multiple reaction monitoring.
REFERENCES
- 1. Kronman M. P., Zhou C., and Mangione-Smith R. (2014) Bacterial prevalence and antimicrobial prescribing trends for acute respiratory tract infections. Pediatrics 134, e956–e965 [DOI] [PubMed] [Google Scholar]
- 2. van Houten C. B., de Groot J. A. H., Klein A., Srugo I., Chistyakov I., de Waal W., Meijssen C. B., Avis W., Wolfs T. F. W., Shachor-Meyouhas Y., Stein M., Sanders E. A. M., and Bont L. J. (2017) A host-protein based assay to differentiate between bacterial and viral infections in preschool children (OPPORTUNITY): a double-blind, multicentre, validation study. Lancet Infect. Dis. 17, 431–440 [DOI] [PubMed] [Google Scholar]
- 3. Song J. Y., Eun B. W., and Nahm M. H. (2013) Diagnosis of pneumococcal pneumonia: current pitfalls and the way forward. Infect. Chemother. 45, 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnston C., Hinds J., Smith A., van der Linden M., Van Eldere J., and Mitchell T. J. (2010) Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. J. Clin. Microbiol. 48, 2762–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rolo D. A. S. S, Domenech A., Fenoll A., Linares J., de Lencastre H., Ardanuy C., and Sa-Leao R. (2013) Disease isolates of Streptococcus pseudopneumoniae and non-typeable S. pneumoniae presumptively identified as atypical S. pneumoniae in Spain. PloS One 8, e57047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simoes A. S., Sa-Leao R., Eleveld M. J., Tavares D. A., Carrico J. A., Bootsma H. J., and Hermans P. W. (2010) Highly penicillin-resistant multidrug-resistant pneumococcus-like strains colonizing children in Oeiras, Portugal: genomic characteristics and implications for surveillance. J. Clin. Microbiol. 48, 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Erhard M., von Dohren H., and Jungblut P. (1997) Rapid typing and elucidation of new secondary metabolites of intact cyanobacteria using MALDI-TOF mass spectrometry. Nat. Biotechnol. 15, 906–909 [DOI] [PubMed] [Google Scholar]
- 8. Welker M., and Moore E. R. (2011) Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Systematic Appl. Microbiol. 34, 2–11 [DOI] [PubMed] [Google Scholar]
- 9. Singhal N., Kumar M., Kanaujia P. K., and Virdi J. S. (2015) MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 6, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Florio W., Tavanti A., Barnini S., Ghelardi E., and Lupetti A. (2018) Recent advances and ongoing challenges in the diagnosis of microbial infections by MALDI-TOF mass spectrometry. Front. Microbiol. 9, 1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hrabak J., Walkova R., Studentova V., Chudackova E., and Bergerova T. (2011) Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49, 3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jung J. S., Eberl T., Sparbier K., Lange C., Kostrzewa M., Schubert S., and Wieser A. (2014) Rapid detection of antibiotic resistance based on mass spectrometry and stable isotopes. Eur. J. Clin. Microbiol. Infect. Dis. 33, 949–955 [DOI] [PubMed] [Google Scholar]
- 13. Sparbier K., Schubert S., Weller U., Boogen C., and Kostrzewa M. (2012) Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J. Clin. Microbiol. 50, 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karlsson R., Gonzales-Siles L., Gomila M., Busquets A., Salva-Serra F., Jaen-Luchoro D., Jakobsson H. E., Karlsson A., Boulund F., Kristiansson E., and Moore E. R. B. (2018) Proteotyping bacteria: Characterization, differentiation and identification of pneumococcus and other species within the Mitis Group of the genus Streptococcus by tandem mass spectrometry proteomics. PloS One 13, e0208804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen S. H., Parker C. H., Croley T. R., and McFarland M. A. (2019) Identification of Salmonella taxon-specific peptide markers to the serovar level by mass spectrometry. Anal. Chem. 91, 4388–4395 [DOI] [PubMed] [Google Scholar]
- 16. Chenau J., Fenaille F., Caro V., Haustant M., Diancourt L., Klee S. R., Junot C., Ezan E., Goossens P. L., and Becher F. (2014) Identification and validation of specific markers of Bacillus anthracis spores by proteomics and genomics approaches. Mol. Cell. Proteomics 13, 716–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dworzanski J. P., Deshpande S. V., Chen R., Jabbour R. E., Snyder A. P., Wick C. H., and Li L. (2006) Mass spectrometry-based proteomics combined with bioinformatic tools for bacterial classification. J. Proteome Res. 5, 76–87 [DOI] [PubMed] [Google Scholar]
- 18. Karlsson R., Davidson M., Svensson-Stadler L., Karlsson A., Olesen K., Carlsohn E., and Moore E. R. (2012) Strain-level typing and identification of bacteria using mass spectrometry-based proteomics. J. Proteome Res. 11, 2710–2720 [DOI] [PubMed] [Google Scholar]
- 19. Misra R. V., Ahmod N. Z., Parker R., Fang M., Shah H., and Gharbia S. (2012) Developing an integrated proteo-genomic approach for the characterisation of biomarkers for the identification of Bacillus anthracis. J. Microbiol Methods 88, 237–247 [DOI] [PubMed] [Google Scholar]
- 20. Wang H., Drake S. K., Yong C., Gucek M., Lyes M. A., Rosenberg A. Z., Soderblom E., Arthur Moseley M., Dekker J. P., and Suffredini A. F. (2017) A Genoproteomic approach to detect peptide markers of bacterial respiratory pathogens. Clin. Chem. 63, 1398–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Semanjski M., and Macek B. (2016) Shotgun proteomics of bacterial pathogens: advances, challenges and clinical implications. Exp. Rev. Proteomics 13, 139–156 [DOI] [PubMed] [Google Scholar]
- 22. Ronsein G. E., Pamir N., von Haller P. D., Kim D. S., Oda M. N., Jarvik G. P., Vaisar T., and Heinecke J. W. (2015) Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. J. Proteomics 113, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cecchini T., Yoon E. J., Charretier Y., Bardet C., Beaulieu C., Lacoux X., Docquier J. D., Lemoine J., Courvalin P., Grillot-Courvalin C., and Charrier J. P. (2018) Deciphering multifactorial resistance phenotypes in Acinetobacter baumannii by genomics and targeted label-free proteomics. Mol. Cell. Proteomics 17, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Charretier Y., Dauwalder O., Franceschi C., Degout-Charmette E., Zambardi G., Cecchini T., Bardet C., Lacoux X., Dufour P., Veron L., Rostaing H., Lanet V., Fortin T., Beaulieu C., Perrot N., Dechaume D., Pons S., Girard V., Salvador A., Durand G., Mallard F., Theretz A., Broyer P., Chatellier S., Gervasi G., Van Nuenen M., Ann Roitsch C., Van Belkum A., Lemoine J., Vandenesch F., and Charrier J. P. (2015) Rapid bacterial identification, resistance, virulence and type profiling using selected reaction monitoring mass spectrometry. Sci. Rep. 5, 13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grenga L. P. O, and Armengaud J. (2019) Pathogen proteotyping: A rapidly developing application of mass spectrometry to address clinical concerns. Clin. Mass Spectrom. 14, 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karlsson R., Gonzales-Siles L., Boulund F., Svensson-Stadler L., Skovbjerg S., Karlsson A., Davidson M., Hulth S., Kristiansson E., and Moore E. R. (2015) Proteotyping: Proteomic characterization, classification and identification of microorganisms–A prospectus. Syst. Appl. Microbiol. 38, 246–257 [DOI] [PubMed] [Google Scholar]
- 27. Boulund F., Karlsson R., Gonzales-Siles L., Johnning A., Karami N., Al-Bayati O., Ahren C., Moore E. R. B., and Kristiansson E. (2017) Typing and characterization of bacteria using bottom-up tandem mass spectrometry proteomics. Mol. Cell. Proteomics 16, 1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lane D. J. (1991) 16S/23S sequencing. In Nucleic acid Techniques in Bacterial Systematics, pp. 115–175, John Wiley, Chichester, UK [Google Scholar]
- 29. Ghebremedhin B., Layer F., Konig W., and Konig B. (2008) Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J. Clin. Microbiol. 46, 1019–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., and Tiedje J. M. (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. System. Evolutionary Microbiol. 57, 81–91 [DOI] [PubMed] [Google Scholar]
- 31. Richter M., Rossello-Mora R., Oliver Glockner F., and Peplies J. (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaijalainen T., Ruokokoski E., Ukkonen P., and Herva E. (2004) Survival of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis frozen in skim milk- tryptone-glucose-glycerol medium. J. Clin. Microbiol. 42, 412–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., and Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 34. Seattle Proteome Center. (2009) ReAdW (Internet) (cited July 10, 2015) http://tools.proteomecenter.org/wiki/index.php?titleSoftware:ReAdW
- 35. Bjornson R. D., Carriero N. J., Colangelo C., Shifman M., Cheung K. H., Miller P. L., and Williams K. (2008) X!!Tandem, an improved method for running X!tandem in parallel on collections of commodity computers. J. Proteome Res. 7, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Craig R., and Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 37. Gupta N., Bandeira N., Keich U., and Pevzner P. A. (2011) Target-decoy approach and false discovery rate: when things may go wrong. J. Am. Soc. Mass Spectrom. 22, 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benson D. A., Boguski M. S., Lipman D. J., Ostell J., Ouellette B. F., Rapp B. A., and Wheeler D. L. (1999) GenBank. Nucleic Acids Res. 27, 12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Human Microbiome Project, C. (2012) Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Federhen S. (2012) The NCBI Taxonomy database. Nucleic Acids Res. 40, D136–D143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baggett H. C., Watson N. L., Deloria Knoll M., Brooks W. A., Feikin D. R., Hammitt L. L., Howie S. R. C., Kotloff K. L., Levine O. S., Madhi S. A., Murdoch D. R., Scott J. A. G., Thea D. M., Antonio M., Awori J. O., Baillie V. L., DeLuca A. N., Driscoll A. J., Duncan J., Ebruke B. E., Goswami D., Higdon M. M., Karron R. A., Moore D. P., Morpeth S. C., Mulindwa J. M., Park D. E., Paveenkittiporn W., Piralam B., Prosperi C., Sow S. O., Tapia M. D., Zaman K., Zeger S. L., O'Brien K. L., and Group P. S. (2017) Density of upper respiratory colonization with Streptococcus pneumoniae and its role in the diagnosis of pneumococcal pneumonia among children aged <5 Years in the PERCH Study. Clin. Inf. Dis. 64, S317–S327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park D. E., Baggett H. C., Howie S. R. C., Shi Q., Watson N. L., Brooks W. A., Deloria Knoll M., Hammitt L. L., Kotloff K. L., Levine O. S., Madhi S. A., Murdoch D. R., O'Brien K. L., Scott J. A. G., Thea D. M., Ahmed D., Antonio M., Baillie V. L., DeLuca A. N., Driscoll A. J., Fu W., Gitahi C. W., Olutunde E., Higdon M. M., Hossain L., Karron R. A., Maiga A. A., Maloney S. A., Moore D. P., Morpeth S. C., Mwaba J., Mwenechanya M., Prosperi C., Sylla M., Thamthitiwat S., Zeger S. L., Feikin D. R., and Group P. S. (2017) Colonization density of the upper respiratory tract as a predictor of pneumonia- Haemophilus influenzae, Moraxella catarrhalis, Staphylococcus aureus, and Pneumocystis jirovecii. Clin. Inf. Dis. 64, S328–S336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Diaz-Pascual F., Ortiz-Severin J., Varas M. A., Allende M. L., and Chavez F. P. (2017) In vivo Host-Pathogen interaction as revealed by global proteomic profiling of zebrafish larvae. Front. Cell Infect. Microbiol. 7, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rossetti C. A., Drake K. L., Lawhon S. D., Nunes J. S., Gull T., Khare S., and Adams L. G. (2017) Systems biology analysis of temporal In vivo Brucella melitensis and bovine transcriptomes predicts host:pathogen protein-protein interactions. Front. Microbiol. 8, 1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data (MSMS-spectra for all species-unique peptides presented in Tables I–IV, as well as raw-files and PD1.4 search files of representative clinical samples) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifiers PXD014522.