Abstract
The enzyme co-substrate S-adenosylmethionine exhibits two general modes of reactivity at its sulphur center: methyl group donation and radical generation. The discovery of a novel metabolite reveals a third way in which this amazingly versatile molecule can react.
In a process known as post-transcriptional modification, biology streamlines protein synthesis to modify the structure of the four naturally occurring nucleotides after they have been chemically stitched together to form ribonucleic acid (RNA). These alterations to the nucleotides impart unique structural features that make them more recognizable to the molecules that mediate the translation of genetic information encoded in RNA into proteins. On page xxx of this issue, Kim et al. report their findings on a post-transcriptional modification that occurs in bacteria of an already modified nucleotide, 5-hydroxyuridine (ho5U) to generate a novel molecule called 5-oxyacetyluridine (cmo5U). The authors find that this modification is carried out by two enzymes, named CmoA and CmoB, in a series of steps involving the synthesis of cmo5U from two precursor molecules found in the cell that each exhibit never before seen modes of reactivity.
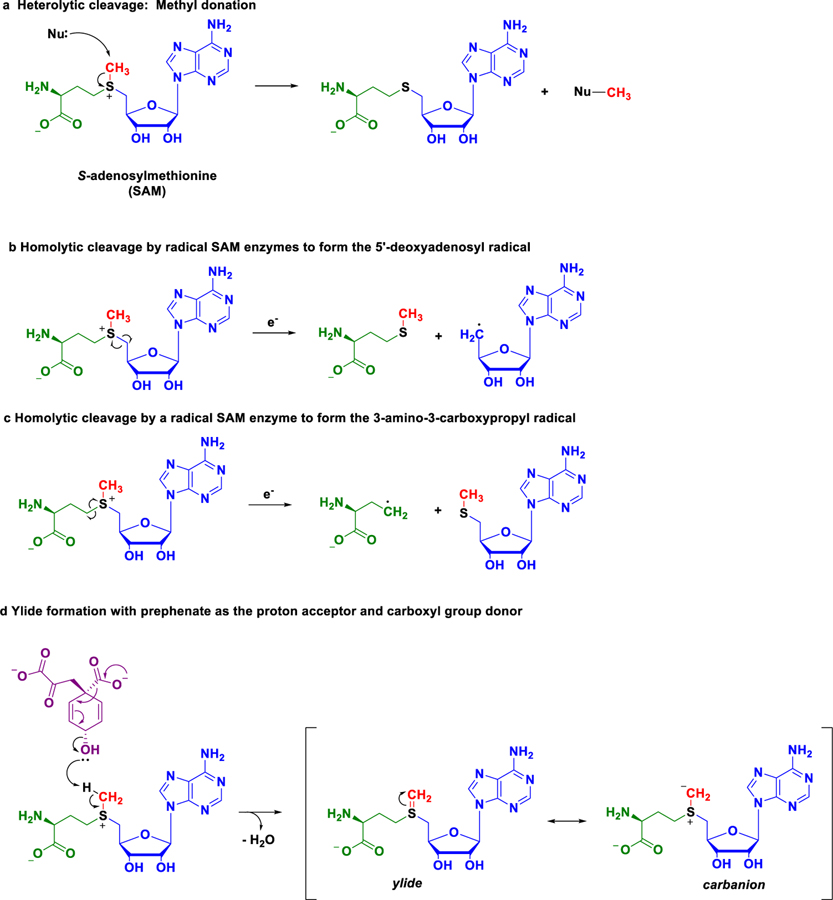
The formation of cmo5U by CmoA and CmoB is dependent on a co-substrate called S-adenosylmethionine (SAM), which exhibits two modes of reactivity that originate at its positively charged sulphur atom. This sulphur atom is bonded to three different chemical groups: a methyl group, a 5’-deoxyadenosyl group, and a 3-amino-3-carboxypropyl group (Fig. 1a). The positive charge on the sulphur atom attracts the electrons from the adjacent carbon atoms of these three groups, making them electron-deficient, or electrophilic. Other molecules with electron-donating properties, known as nucleophiles, can donate their electrons to these electrophilic carbon atoms, resulting in the simultaneous breaking of SAM’s sulphur-carbon (S-C) bond and formation of a new carbon-nucleophile bond. This type of bond breaking, in which both of the electrons in the S-C bond end up on the sulphur atom, is known as heterolytic cleavage (Fig. 1a). In the cell, it is the bond to the methyl group that is most frequently cleaved, which is why SAM has historically been known as a methyl group donor. However, an array of chemical transformations occur through enzyme-mediated cleavage of SAM’s other two S-C bonds to generate highly potent free radicals.
Figure 1. Modes of reactivity for an enzyme co-substrate.
Enzymes commonly use S-adenosylmethionine (SAM) as a co-substrate during catalysis. SAM contains a positively charged sulphur atom bonded to a methyl group (red), a 5’-deoxyadenosyl group (blue), and a 3-amino-3-carboxypropyl group (green). a, The most commonly observed enzyme-catalyzed reaction of SAM in which the sulphur-carbon (S-C) bond to the methyl group is broken. The heterolytic cleavage of the bond results in both of the electrons residing on the sulphur atom. Curled arrows indicate the movement of pairs of electrons, and Nu represents a nucleophile (an electron pair donor). b and c, Radical SAM enzymes use SAM and an electron (e-) to form (b) the 5’-deoxyadenosyl radical or (c) the 3-amino-3-carboxypropyl radical by breaking their respective S-C bonds. The homolytic cleavage of these S-C bonds results in the sulphur and carbon atoms each receiving one electron. Fish hook arrows represent the movement of one electron. d, The findings of Kim et al. show the enzyme CmoA catalyzes a novel reaction mode for SAM with the molecule prephenate (violet). Prephenate acts as a base (proton acceptor) to remove a proton from the methyl group of SAM to form a sulphur-carbon double bond (S=C), resulting in a structure called an ylide (pronounced ill-id). The ylide is represented as two structures, called resonance structures (denoted by brackets), which differ only by the location of electrons on the molecule. The resonance structure of the ylide is a carbanion, whereby the electron pair from the S=C is localized on the carbon atom. The formation of the ylide completely transforms the reactivity of the methyl group carbon atom from electrophilic (electron-pair acceptor) to nucleophilic (electron-pair donor), which the enzyme CmoA facilitates and exploits, in conjunction with CmoB, in the biosynthesis of 5-oxyacetyluridine.
The group of enzymes that activate SAM for radical formation, the so-called radical SAM enzymes, use a reduced cluster of iron and sulphur atoms to inject an electron into the S-C bond to the 5’-deoxyadenosyl or 3-amino-3-carboxypropyl groups of SAM. Injection of the electron causes a scission of the bond such that both the sulphur and carbon atoms receive one electron, a type of bond-breaking known as homolytic cleavage. This reaction results in a net gain of two electrons by the sulphur atom and one electron, or a radical, on the carbon atom to form the corresponding 5’-deoxyadenosyl (Fig. 1b) or 3-amino-3-carboxypropyl radical (Fig. 1c) Such carbon-centered radicals are considered to be ‘hot’ because they are highly reactive and can break incredibly strong bonds to initiate complex and difficult chemical reactions.
Kim et al. now show that SAM exhibits yet another mode of reactivity in the biosynthesis of cmo5U in which a second bond is formed between the sulphur atom and the carbon atom of the methyl group (S=C) to form what is known as an ylide (pronounced ill-id). Ylides have two forms, or resonance structures: the first can be depicted as a pair of electrons that is shared between the sulphur and carbon atoms to form the S=C, and the second localizes the pair of electrons on the methyl group carbon atom to form what is known as a carbanion (Fig. 1d). The formation of the ylide completely changes the reactivity of the carbon atom from electrophilic to nucleophilic, a transformation that is key to the formation of cmo5U. The additional electrons needed to form the SAM-based ylide come from prephenate (Fig. 1d), a molecule found in the cell. Prephenate plays unprecedented roles in the biosynthesis of cmo5U. Not only does it acts as a base, or proton acceptor, in the formation of the ylide, it also donates its carboxyl group to synthesize the novel metabolite, carboxy-S-adenosylmethionine (Cx-SAM), which contains a synthesized oxyacetyl group. To complete the post-transcriptional modification, the enzyme CmoB mediates the transfer of the oxyacetyl group from Cx-SAM to the hydroxyl group of 5-hydroxyuridine to form 5-oxyacetyluridine.