Abstract
Cell fate specification defines the earliest steps towards a distinct cell lineage. Neural crest, a multipotent stem cell population, is thought to be specified from the ectoderm, but its varied contributions defy canons of segregation potential and challenges its embryonic origin. Aiming to resolve this conflict, we have assayed the earliest specification of neural crest using blastula stage chick embryos. Specification assays on isolated chick epiblast explants identify an intermediate region specified towards the neural crest cell fate. Furthermore, low density culture suggests that the specification of intermediate cells towards the neural crest lineage is independent of contact mediated induction and Wnt-ligand induced signaling, but is, however, dependent on transcriptional activity of β-catenin. Finally, we have validated the regional identity of the intermediate region towards the neural crest cell fate using fate map studies. Our results suggest a model of neural crest specification within a restricted epiblast region in blastula stage chick embryos.
1. Introduction
Through the evolution of trilaminar, bilaterian embryos, a unique cell population arose in vertebrates termed the neural crest (NC). NC is a transient, early embryonic multipotent stem cell population that contributes, among other derivatives, to the craniofacial skeleton and peripheral neurons and glia. NC are considered to be at the center of vertebrate evolution and diversity by defining vertebrates through its contribution to key features for the predatory lifestyle including a larger brain enclosure, jaws, and paired sense organs (Gans and Northcutt, 1983; Northcutt, 2005; Le Douarin, 1980; Le Douarin and Kalcheim, 1999). Improper NC development leads to a host of pathologies known as neurocristopathies (Bolande, 1996, 1974; Etchevers et al., 2006; Farlie et al., 2004). NC cells are thought to be derived from the ectoderm, which is consistent with their contribution to skin melanocytes and peripheral neurons and glia. However, their ectomesenchymal contributions in the head region – including bone, cartilage, and fat cells, which in other parts of the body are derived from the mesoderm – have been a topic of scientific focus and discourse. If NC cells are truly ectodermally derived, then their contribution to mesodermal-like derivatives suggests that they uniquely defy current assumptions of sequential segregation and restriction of potential. A break from the conceptual cannons of NC induction came in 2006, with the suggestion that NC specification is ongoing during gastrulation, and that it occurs independent from mesodermal or neural contributions (Basch et al., 2006). Accordingly, a pre-gastrula origin of NC would not be subjected to the expected fate restrictions imposed on the three germ layers. Additional support for pre-gastrula specification of the NC emerged from other researchers (Patthey et al., 2008a, 2008b). Furthermore, mammalian work using rabbit embryos and a human model of NC formation based on embryonic stem cells (Betters et al., 2018; Leung et al., 2016), also suggest that early anterior NC is specified prior to gastrulation and is independent from definitive neural and mesodermal tissues. Finally, recent work in Xenopus proposed that prospective NC retain stemness markers and pluripotency from epiblast cells, alluding to a pre-gastrula origin of NC (Buitrago-Delgado et al., 2015). These findings highlight the need to understand the precise origins of NC cells.
Here, we report for the first time the earliest known specification of NC in chick blastula embryos. We have identified a restricted intermediate territory of the blastula epiblast as capable of generating NC when cultured in isolation under non-inductive conditions. This territory contains prospective NC (pNC) cells which develop independently from apparent mesoderm or neural contributions. Importantly, low density cell cultures of dissociated epiblast explants suggest that early specification has been established prior to the culture of the cells, and that it can proceed in a cell autonomous and contact independent fashion. We find that cells within this region require continued transcriptional activity of β-catenin for the expression of the NC marker Pax7. Finally, the contribution of the intermediate epiblast territory to the NC lineage in vivo is supported by fate mapping analysis. Our results suggest that precursors to the most anterior NC in amniotes arise from the pluripotent epiblast, and prior to definitive germ layer formation, which is in alignment with the multipotent character of NC.
2. Results
2.1. Restricted region of the chick blastula epiblast is specified towards the neural crest cell fate
To understand the ontogeny of the NC, we sought to investigate the earliest cell fate decisions that segregates the pNC cells from other cell fates. Previously, we reported on a specified population of NC in the chick gastrula (Basch et al., 2006). In the current study, we asked whether NC cells are specified in blastula stage avian embryos, the stage preceding gastrulation and germ layer commitment. Here, specification refers to an initiated path of differentiation towards a cell fate. While the specified cell does not initially express known markers of the tested fate (pre-migratory and migratory NC), they are able to do so after continuing with the specified program. The continuation of the specified program relies on permissive conditions, which if compromised, could prevent the originally specified fate.
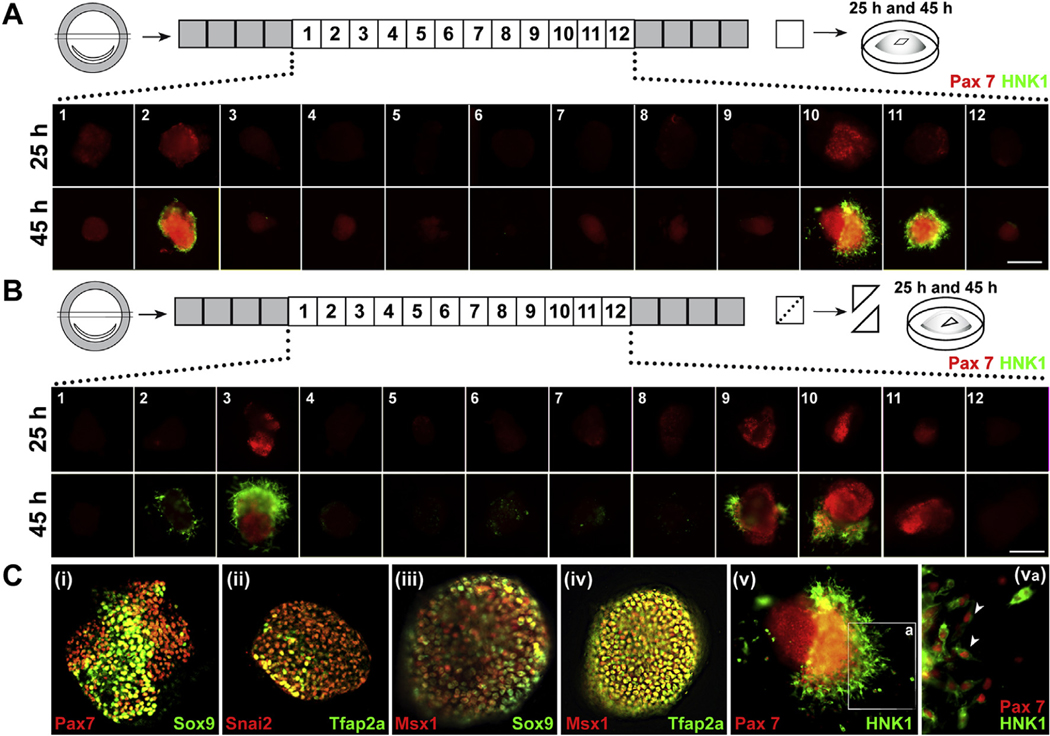
Specification can be assessed by culturing isolated regions of the epiblast under a neutral, serum free, and defined non-inductive environment. Using this assay, we analyzed whether chick epiblast explants from stage XII (Eyal-Giladi Staging) (Eyal-Giladi and Kochav, 1976) embryos are specified towards the NC cell fate. Accordingly, blastula embryos were collected, and the underlying hypoblast layer removed. A horizontal strip from the equatorial plane ~250μm above the Koller sickle was then dissected and further cut into 12 explants of ~80–100 μm2 each (Fig. 1A). The explants were cultured in isolation in a collagen gel under non-inductive conditions (Basch et al., 2006) for 25h or 45h (corresponding to approximately stage HH4+ and HH8, respectively) and assessed for markers of the NC cell fate. In the chick, the expression of the transcription factor Pax7 has been reported to begin at gastrula stage HH4+, where it is used as a key marker of early NC development (Basch et al., 2006). We observed that intermediate explants, and not those taken from the most lateral or medial regions, display clear features of NC specification. Unlike explants from other regions, intermediate explants expressed nuclear Pax7 after 25h of culture, and acquire migratory characteristics with positive HNK1 epitope staining and maintained Pax7 expression after 45h of culture (pattern observed in 5/6 embryos, Fig. 1A). To verify that migratory NC at 45h arise from the same Pax7-positive intermediate explants at 25h, we generated a series of explants in which each explant was bisected diagonally, and the resulting half fragments were cultured for either 25 or 45h (Fig. 1B, n = 10). Immunostaining on these explants indicate that the intermediate explants positive for Pax7 expression after 25h of culture are the same explants that express the migrating NC cell marker HNK1 at 45h (pattern of expression observed in 10/11 embryos). To further confirm the nature of the proposed pNC specified at blastula stages, we assayed for the expression of additional NC markers in intermediate explants. We observed the expression of Sox9, Snai2, Msx1 and Tfap2a in the same intermediate explants (Fig. 1C). Taken together, these results suggest that a specific population of intermediate epiblast cells are specified in the blastula epiblast, prior to gastrulation, towards a NC cell fate.
Fig. 1. Neural crest cells are specified at blastula stage in a restricted epiblast region.
(A) Schematic showing hypoblast-free epiblast explants (≈80 μm) from EGK Stage XII chick embryos generated from an equatorial stripe, immersed in collagen gels and cultured under defined non-inducing conditions in isolation. After 25h of culture, lateral explants displayed a robust Pax7+ expression not seen in other explants (n = 5). After 45h, a similar intermediate location displays Pax7+/HNK-1+ and clear double positive migratory cells likely to be NCC (n = 5; scale bar 100 μm). (B) Diagram of a bisection approach to monitor different conditions in original explants. Bisected explants from the same original region generate Pax7+ expressing cells at 25h and migrating Pax7+/HNK-1+ cells at 45h (n = 10; scale bar 100 μm). (C) Images of intermediate explants with colocalization of neural crest markers at 45h (i) Pax7/Sox9, (ii) Snail2/AP2, (iii) Msx1/2/Sox9, (iv) Msx1/2/AP2, (v) Pax7/HNK-1 explant and (va) magnification of the migrating Pax7+/HNK-1+ cells at 45h.
2.2. Intermediate explants are devoid of neuroectodermal and mesodermal markers
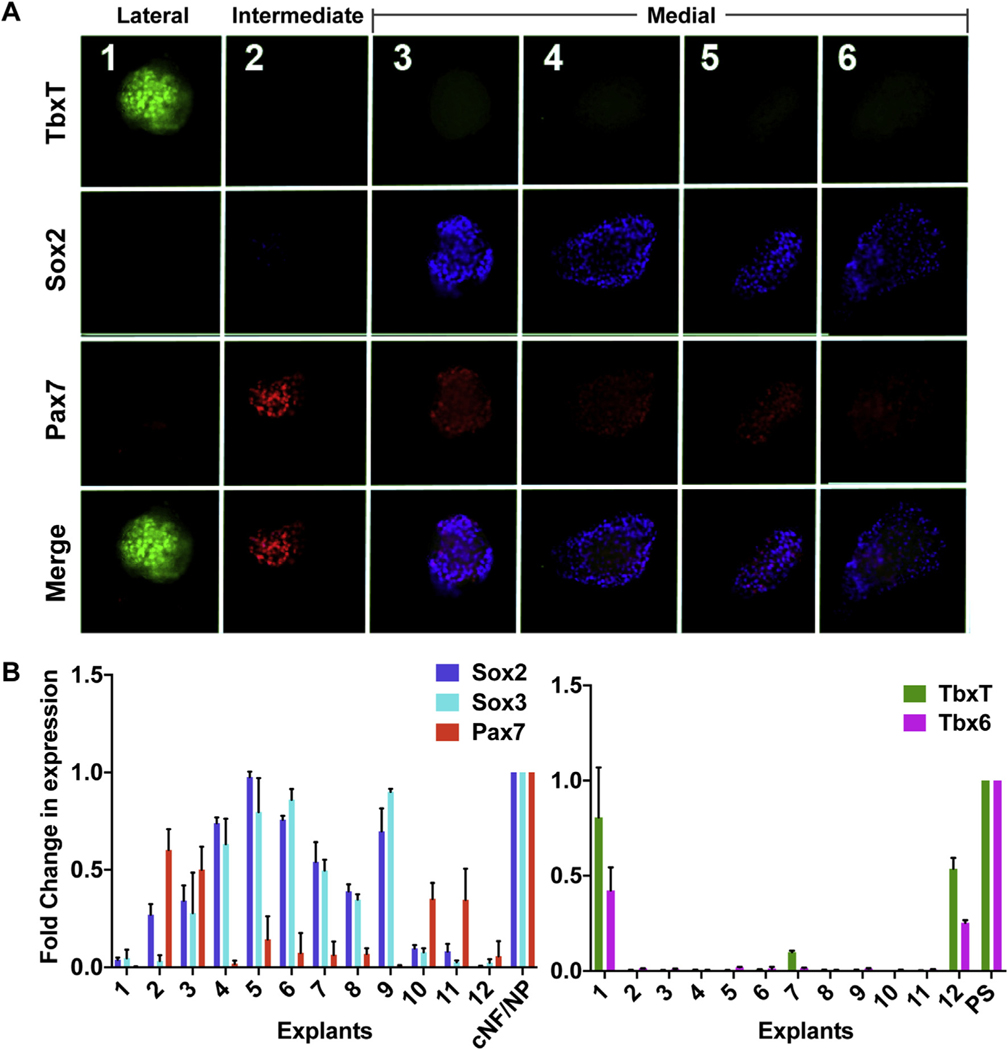
Previous work has reported on cell fate specification prior to gastrulation, including neural ectoderm, non-neural ectodermal and mesodermal cell fates (Hatada and Stern, 1994; Onjiko et al., 2015; Patthey et al., 2008b, 2008a; Pegoraro et al., 2015; Shin et al., 2011; Trevers et al., 2018; Wilson et al., 2001). We therefore interrogated the relationship between pNC, neural, and mesodermal tissue in the blastula stage chick embryo. Specification of neuroectodermal (Sox2), mesodermal (TBXT), and NC (Pax7) fates were simultaneously assessed in restricted blastula stage epiblast regions using specification assays described in Fig. 1A. We observed Sox2 expression predominately in medial explants devoid of robust Pax7 signal (Fig. 2A). In the intermediate explants (#2), clear Pax7+/Sox2-expression was identified. In a small number of embryos (n = 2/10), we noted medial explants with very low levels of Pax7 expression along with robust Sox2 expression. Furthermore, only the lateral-most explants (#1) displayed TBXT expression, and these explants did not display Pax7 signal (Fig. 2A). This pattern of expression was observed in 90% of the embryos (n = 4/5) analyzed for NC, neural and mesodermal cell fate markers. The expression of these genes was further assessed quantitatively using RT-qPCR in the twelve explants after 25hrs of culture. Consistent with the immunostaining, intermediate explants expressed strong Pax7, minimal Sox2/-Sox3, and no TbxT/Tbx6 (lateral mesodermal markers) (Fig. 2B). Lower levels of Sox2 expression was also observed in intermediate explants (explants 2/3 or 10/11). Together this evidence strongly supports a distinct regional specification towards NC within the intermediate epiblast explant, with medial and lateral epiblast explants displaying neural and mesodermal specification, respectively.
Fig. 2. Intermediate region of epiblast is specified towards neural crest cell fate independent of neuroectodermal and mesodermal cell fates.
(A) Explants from EGK Stage XII chick embryos, collected after 25h and analyzed for expression of different cell fates; mesodermal (TbxT, (Brachyury) lateral-most), neural (Sox2, medial), and NC (Pax7, intermediate explant) (n = 4). (B) Graphs showing RT-qPCR analysis of twelve explants after 25h of culture, for the expression of Pax7 (NC marker), Sox2/Sox3 (neural marker), and TbxT/Tbx6 (a lateral mesodermal markers). Fold change in expression is represented by normalizing to GAPDH (reference gene) and relative to the positive control region for each gene: cranial neural fold (NF)/neural plate (NP) for Pax7 and Sox2/Sox3 expression and primitive streak (PS) for TbxT/Tbx6. Data from four individual sets of embryos, each with 12 explants and controls, are represented in the RT-qPCR graphs are shown, with error bars representing standard error of mean between four biological replicates (4 embryos).
2.3. Neural crest specification at blastula stage does not require cell-cell contact mediated signaling and is contingent on continued β-catenin activity
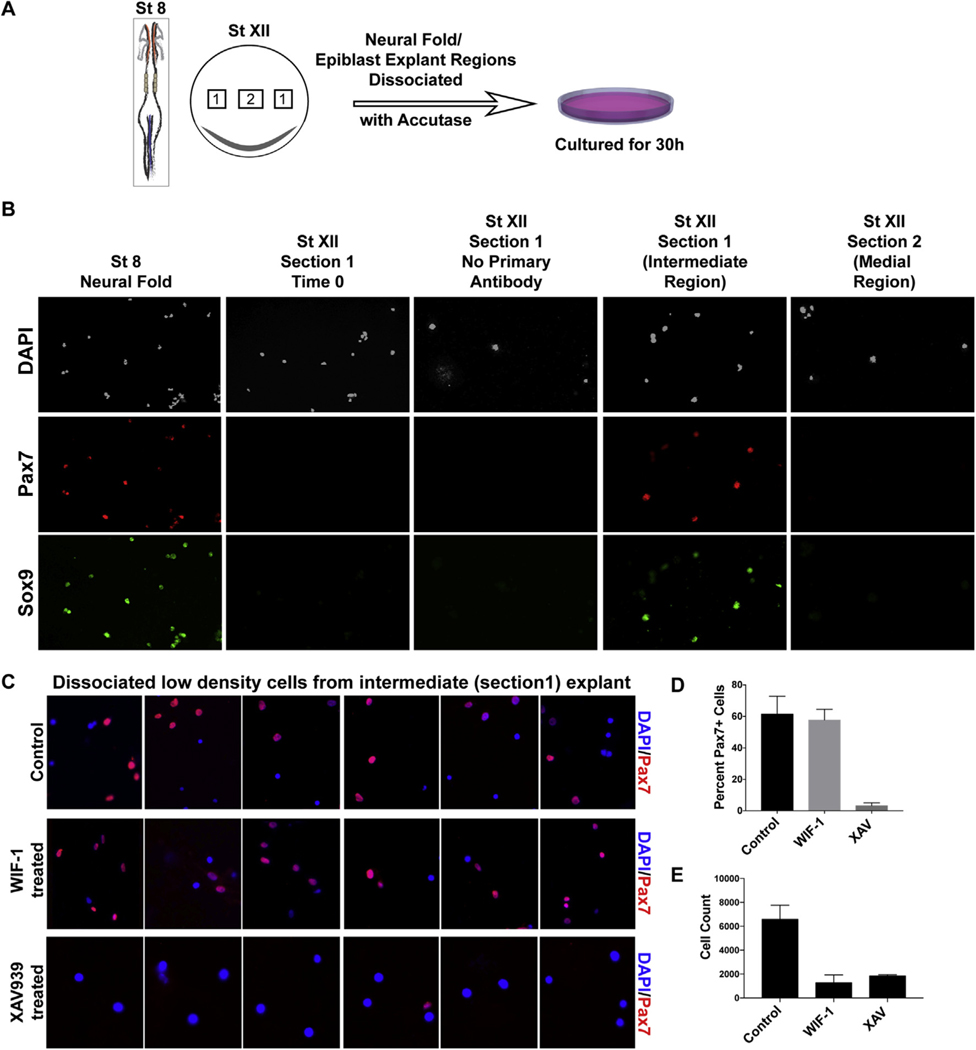
The regional specification of prospective NC in intermediate epiblast explants reveal clear heterogeneity of epiblast cells. This is further exposed by the non-homogeneous expression of NC markers in the intermediate explants and suggests the identified specification could be the result of contact mediated interactions between these cells. To address this possibility, we assessed cell specification in low density cultures from dissociated epiblast explants. This approach minimizes or removes contact mediated induction events and limits signaling from immediately adjacent cells. The intermediate and medial regions of epiblast from EKG Stage XII blastula embryos were dissociated into single cells using accutase, plated on a layer of collagen at very low density (10–20 cells/ cm2) and either fixed immediately (time 0), or cultured for 30h in non-inductive media (Fig. 3A). These cultures were then evaluated for Pax7 and Sox9 expression via immunofluorescence. As a control, neural fold cells from an HH stage 8 embryo were dissociated and processed for immunofluorescence without culture. As expected, cells from the low-density cultures of HH Stage 8 NPB region display robust Pax7/Sox9 expression. No Pax7/Sox9 expression was observed in intermediate or medial low-density cells at time 0 (right after plating). Instead, 38% of the cells from the intermediate epiblast region expressed Pax7, and 24% expressed Sox9 (n = 4), while cells from the medial region contained only 8% Pax7 and 6% Sox9 positive cells (Fig. 3B). These results reveal that a significant number of intermediate epiblast cells express NC markers in the absence of cell-cell contact, suggesting a potential cell autonomous specification of neural crest when cultured in vitro in the absence of in vivo repressive cues. This experiment further suggests that a NC specification program has already been initiated within single cells of the heterogenous blastula epiblast.
Fig. 3. Low density isolated cell analysis of intermediate epiblast region identifies cell autonomous specification of neural crest in culture.
(A) Schematic showing HH stage 8 and EGK stage XII embryos used for low density isolated cell analysis and the workflow. Cells were dissociated from Stage 8 embryo marked by Pax7 expressing neural fold region (boxes sections within the red region) (positive control), plated, fixed and immunostained for Pax7/Sox9. Stage XII embryo explants marked as 1 (intermediate) and 2 (medial) regions were dissociated and cultured at very low density (10–20 cells/cm2) on a thin layer of collagen gel under non-inducing conditions for 30h. (B) Isolated cells in the low density culture were immunostained for Pax7/Sox9 expression from neural fold region, intermediate region (section 1) at time 0 (immediately after plating), intermediate region (section 1) used as no primary control, and from sections 1 and 2. Intermediate region (section 1) had the highest Pax7 positive cells (38%) with 8% Pax+ cells in medial region (n = 4). (C) Explants from EGK St. XII epiblast were dissected, dissociated and cultured for 25h as described in (A) in presence or absence of Wnt-ligand inhibitor (WIF-1) or β-catenin inhibitor (XAV939) and processed for Pax7 immunofluorescence. Six panels from 4 individual embryo explants show distant cells cultured in isolation express Pax7 under control and WIF-1 treated conditions, while no Pax7 expression was seen under XAV939 treatment. (D) Average cell counts for Pax7 positive cells from the three different conditions, untreated control, WIF-1-treated and XAV939-treated (n = 3), and (E) Total DAPI positive cells under these conditions (n = 3). Error bars represent SEM from three separate embryo. Cell counts are provided in supplemental Table 1.
If the specification of NC has already been initiated in the intermediate explant, then we would expect that the inhibition of extrinsic signaling should not affect the specified cells. Wnt/β-catenin signaling has been shown to play a crucial role in the specification of NC (Garcia-Castro et al., 2002; Ikeya et al., 1997; LaBonne and Bronner-Fraser, 1998; Saint-Jeannet et al., 1997; Sasai et al., 2001). To assess the contribution of Wnt/β-catenin signaling in early epiblast NC specification, we used small molecules WIF-1 (Wnt ligand inhibitor) or XAV939 (a WNT inhibitor that stabilizes Axin, a key component of the β-catenin destruction complex) in our low-density cultures. Immunofluorescence after 30h of culture with WIF-1 (Wnt-ligand inhibition) did not affect the total number Pax7 expressing distant cells compared to control samples (n = 3/3) (Fig. 3C and D). However, XAV treatment appeared to block Pax7 expression (n = 3/3) (Fig. 3C and D). To address possible effects on survival or proliferation triggered by Wnt-inhibition (WIF-1 and XAV condition) we counted the total number of cells using DAPI staining. In comparison to control cultures, WIF-1 and XAV treated cultures display a considerable reduction in cell numbers (≥75%) (Fig. 3E). Given that both treatments reduced cell counts, but only XAV prevented Pax7 expression in isolated cells, these results suggest that specified cells do not require Wnt-ligand but still depend on transcriptional activity of β-catenin to advance the NC program.
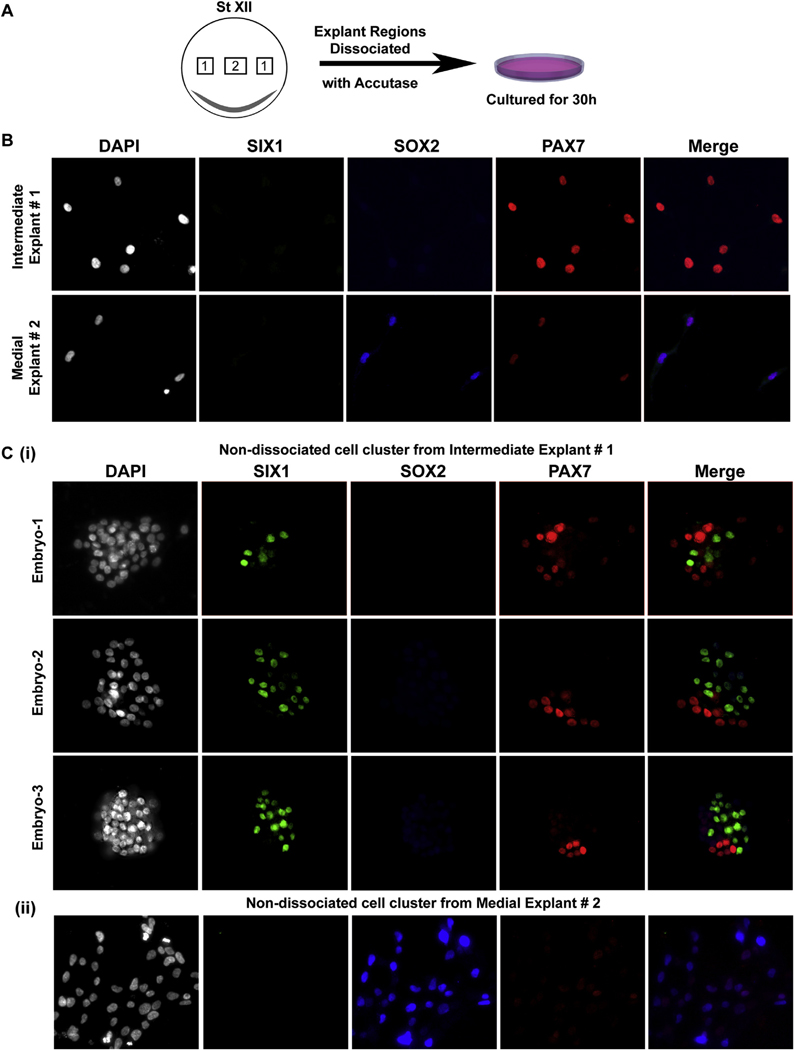
2.4. Intermediate epiblast consists of precursors of NC and placodal cell fates
In addition to NC, precursors for neural and cranial placodal cells are thought to emerge from the NPB territory. Hence, we assessed the possible presence of placodal ectodermal precursors along with neuroectodermal fates within the intermediate explants. We assessed co-expression of Pax7, Sox2 and Six1 (markers of NC, neural, and placodal fates, respectively) in low-density cultures of dissociated St. XII epiblast explants from intermediate and medial regions cultured for 30h. Here, we chose to use dissociated explants instead of whole explants to better expose the cellular heterogeneity within the intermediate region. Dissociated cells from intermediate and medial explants from St. XII embryos were cultured for 30h under non-inductive conditions. Our immunostaining results confirm the expression of Pax7 in distant low-density cells from dissociated intermediate explants (n = 6/6) (Fig. 4B). On the contrary, Sox2 appears in distant low-density cells from dissociated medial explants (n = 6/6) (Fig. 4B). However, we did not observe expression of Six1 in distant low-density cells from medial or intermediate explants (n = 6/6) (Fig. 4B). Interestingly, a few remaining undissociated clusters from the low-density culture of the intermediate explants do reveal Six1 expression in 8% of the cells (average from 6 embryos) (Fig. 4C(i)). It is noteworthy that in these clusters, while we do see cells expressing Pax7, we are unable to identify co-expression of Six1 with Pax7, suggesting that while these fates may coexist adjacently, they appear as distinct cell populations at least after 30h of culture (n = 6/6) (Fig. 4C(i)). This could suggest that a community effect, or contact mediated interaction is necessary to sustain the progression towards the acquisition of this placodal marker, or that the dissociation triggers an inhibiting signal preventing its progression. However, as shown in Figs. 3 and 4, neither of these possible mechanisms are at play for the acquisition of the earliest restricted anterior NC marker, Pax7. Resolving this intriguing result for the ectodermal fate requires further investigation. The undissociated cluster from medial explant had similar expression as dissociated cells, with robust Sox2 expression and much weaker Pax7 expression with no detectable Six1 expression at this time point (Fig. 4C(ii)). These results support our model suggesting a distinct NC specification of epiblast cells within a restricted intermediate epiblast region, which additionally also contain cells fated towards the placodal cell fate, but not shared with neuroectoderm.
Fig. 4. Intermediate epiblast consists of precursors of NC and placodal cell fates.
(A) Schematic showing intermediate and medial explant regions from EGK stage XII used for the analysis. (B) Explants from EGK St. XII epiblast were dissociated and cultured for 36hrs. Distant cells in this low-density culture from intermediate explant (explant #1) express Pax7, no ectodermal marker was expressed in these cells. Distant cells in low density culture from medial explants (explant #2) expressed Sox2 and lower levels of Pax7. Expression of placodal gene, Six1, in distant isolated cells was not seen. (C) (i) Images from 3 individual embryos, from few remaining undissociated clusters of cells in the culture corresponding to intermediate epiblast region, analyzed for Six1, Sox2 and Pax7 expression. Six1 is expressed in cells present in clusters. Six1 and Pax7 expression is mutually exclusive. (ii) Undissociated clusters of cells in the culture corresponding to medial epiblast region, display robust Sox2 expression and weak Pax7 expression. This expression pattern was observed in dissociated intermediate explants from 6 individual embryos. Cell counts for Six1+ cells are provided in supplemental Table 1.
2.5. Intermediate epiblast cells contribute to the neural crest lineage in vivo
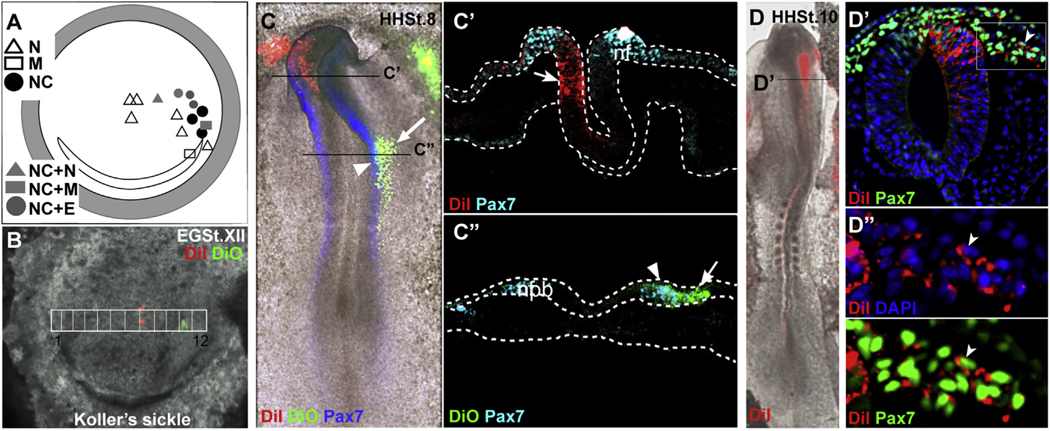
The explant studies provide a clear and precise approach towards temporal and spatial identification of NC specification. However, to assess the contribution of the specified cells within the explants towards the NC lineage, in vivo analysis of the epiblast cells is required. We therefore explored the in vivo contribution of the St. XII intermediate epiblast cells towards NC using lineage tracing analysis with DiI and DiO. Following injection, embryos were cultured from 16 to 36h (EC culture) (Chapman et al., 2001) and subsequently fixed and immunostained for Pax7. Labeled regions corresponding to the intermediate explants (Fig. 5B) contributed to the Pax7-expressing neural plate border (NPB) (Fig. 5C’) and neural folds (Fig. 5C”), matching the expected location of NC cells (n = 8/11) (Fig. 5D”). In a few cases, targeted regions labeled simultaneously neural fold/NC along with mesoderm (n = 1/11), neural (n = 1/11), and non-neural ectoderm (n = 3) (summarized in Fig. 5A). In contrast, labeled medial cells contributed to the neural tube (n = 3/3) (Fig. 5C’). Additional neural contribution was also seen from regions close, but medial (two open triangles), to the intermediate region (solid black dots) (n = 2/11), and in one embryo from a region posterior-lateral (open triangle) to the intermediate region (n = 1/11) (Fig. 5A). While mixed contributions were observed from few regions, most of the intermediate epiblast cells populated the NPB and neural folds expressing Pax7, lending in vivo support to the notion that at blastula stages the intermediate epiblast harbors pNC cells. A Chi-squared test of observed NC contribution from the intermediate region compared to medial region revealed a statistically significant contribution of intermediate epiblast cells towards the NC lineage (p-value = 0.024). These results are in agreement with previous lineage tracing studies in the blastula embryo (Hatada and Stern, 1994). Taken together, our explant specification experiments, and lineage tracing studies strongly suggest that cells in a restricted “intermediate” domain of the avian blastula epiblast are already poised to initiate the NC developmental program.
Fig. 5. In vivo lineage tracing validates the contribution of intermediate epiblast region to neural crest lineage.
(A) Schematic summary identifying regions in the epiblast that contributed to NC (n = 8) alone (4) or in combination with other fates (N-neural, NC-neural crest, E-epidermal, M-mesodermal). Chi-Squared analysis identified statistically significant (p<0.05) NC contributions of the cells within the intermediate epiblast region. Embryo labeled with DiI/DiO at time 0 (B) and after 28h of culture at St. 8 (C); sections demonstrating contribution to the CNS by cells in the middle of the embryo labeled with DiI (C′, C”) and contribution to the lateral neural fold (nf) and neural plate border (nbp) by cells in the intermediate epiblast labeled with DiO, colocalized with Pax7 expression or lateral to it (arrowhead and arrow respectively, C′, C”). (D) Second embryo labeled with DiI on intermediate region of epiblast, demonstrating anterior neural fold localization of DiI (St. 10). (D′) Neural tube cross-section at denoted anterior axial level with colocalization of DiI/Pax7 expression domains in migratory and premigratory neural crest cells and some cells in neural epithelium. (D″) An inset of migratory NC region from D′, labeled with DiI in the cell membrane and co-labeled with nuclear DAPI or Pax7.
3. Discussion
According to the model of sequential segregation of plasticity, pluripotent epiblast cells differentiate and give rise to the three germ layers, endoderm, mesoderm, and ectoderm, each with a distinct potential restricted in comparison to their progenitor. In turn, each of the germ layers differentiate into progenitors with progressively more restricted potential, ultimately generating the specific cell types that constitute the building blocks of the vertebrate body. A recognized exception is the primordial germ cell lineage, which arises independently from gastrulation (Magnusdottir and Surani, 2013; Saitou and Yamaji, 2012). Classic models of NC formation suggest that NC arises from the ectoderm, and therefore, one would expect them to be devoid of mesoderm and endoderm associated potential. However, NC generated ectomesenchymal derivatives encompass ectoderm and mesoderm capacities, and thus represent a difficult paradigm. Efforts to resolve this issue include the suggestion that the NC constitute a fourth germ layer (Hall, 2018, 2000), and a recent model proposing that NC retains stemness markers and the same potential as pluripotent stem cells (Buitrago-Delgado et al., 2015). Our work presented in this report points to a model of blastula stage specification of NC as a segregated population of cells distinct from other cell fates.
We previously showed that anterior cranial neural crest specification is on-going during gastrulation, well before the overt expression of Pax7, the earliest restricted marker associated with NC in chick and rabbit embryos (Basch et al., 2006; Betters et al., 2018). This specification appears to be independent from either mesoderm or neural ectoderm (Basch et al., 2006). In this report, we expose the earliest known specification of anterior cranial NC in the chick blastula embryo. Using epiblast explants at high spatial resolution, we demonstrate NC specification in a restricted intermediate region of epiblast consisting of pNC cells. We also observed heterogeneity within the intermediate epiblast region, as our low-density explant dissociation experiments revealed that 38% of the cells within the intermediate explants go on to express Pax7. Amongst the low density dissociated cells we were unable to identify the expression of early placodal maker Six1. However, in the remaining undissociated cell clusters from the intermediate region, we did identify ~8% of cells expressing Six1. These placodal precursors display an independent identity from the prospective NC (no coexpression of Pax7) and seem to require cell-contact mediated signals for their specification. Our results are in agreement with a recent study using NPB sections from chick embryos, where in general Pax7 and Six1 expression profiles appear to be independent in most NPB cells (Roellig et al., 2017). During later developmental stages, the neural plate border is known to contain both NC and placodal cells. Given that we observed precursors of NC as well as placodal cell fates within this intermediate epiblast region, we refer to this region as the pre-Border (pB) region, the precursor to the NPB region that gives rise to NC and placodes. The heterogeneity of the early epiblast marked by a progressive loss of pluripotency has previously been documented in multiple species, including chick and human (Chen et al., 2018; De Paepe et al., 2014; Shi et al., 2015). Furthermore, low density culture of cells within this pB region suggest NC specification is independent of cell-cell contact mediated inductive interactions in the absence of repressive cues experienced by these cells in vivo from surrounding tissues. Inhibition of Wnt-ligand based signaling in low density cultures also suggest a specified state of NC within the pB region. However, the specification is dependent on continued transcriptional activity of β-catenin. We provide further in vivo support for NC specification using fate map studies and validate that the intermediate epiblast region contains pNC cells that contribute to NC in vivo. Our study found contributions from several of our injections to mesoderm, neural ectoderm, and endoderm (with or without co-contribution to NC) which is in agreement with the broad allocation for those fates provided by the seminal work of Hatada and Stern (1994). Together, our results from explants, low density single cells, and fate mapping very clearly suggest an early specification of prospective NC at blastula stage in chick embryos. Importantly, we observe a distinct regional predisposition of epiblast cells to adopt neural, mesodermal, placodal, or NC fates. However, our assay clearly demonstrates the propensity of the blastomeres in the intermediate epiblast region to be uniquely specified towards the NC cell fate.
Early cell fate specification studies offer additional support for our model. Cell fate analysis has provided early evidence on neuroectoderm and mesendoderm cell fate restriction in blastomeres of Xenopus embryos from 16, 32 and 64-cell stages (Hemmati-Brivanlou and Melton, 1997; Jacobson and Hirose, 1981; Moody, 1987a, 1987b), and fate map analyses in Xenopus and zebrafish has attributed this early segregation to positional identity (Kimmel et al., 1990; Moody and Kline, 1990). The pre-gastrula stage specification of neuroectoderm (Streit et al., 2000; Wilson et al., 2001, 2000) and non-neural ectoderm (Wilson et al., 2001) has been documented in chick embryos. These studies have suggested that medial epiblast territories are specified towards the neural cell fate, while the intermediate epiblast explants are fated towards neural plate border (Wilson et al., 2001). Recently, it was also suggested that cranial placodes, fated towards lens, as well as NC are specified in the pre-gastrula epiblast; however, regional variations in cell fate specification were not observed (Trevers et al., 2018). Our work points towards the blastula stage specification of NC, neuroectoderm, and non-neural ectodermal cell fates in distinct epiblast regions of the chick embryo (Fig. 6). Our model is in agreement with previous studies that have observed regional predisposition of epiblast cells towards NPB and neural cell fates (Patthey et al., 2008a; Wilson et al., 2001, 2000).
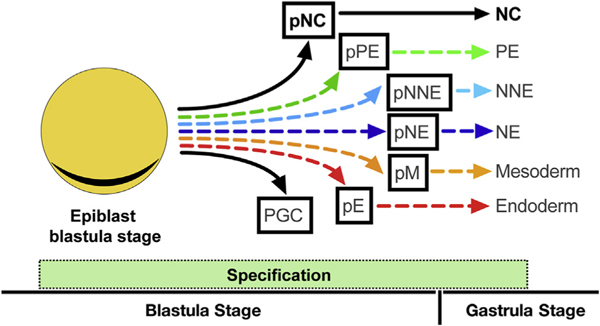
Fig. 6. Model of NC cell fate specification from the epiblast.
Beginning with early blastula, we propose a model of early NC cell fate specification from epiblast cells (depicted in orange with underlying hypoblast layer in grey). Based on our study, the model suggests specification of prospective NC (pNC) from the epiblast during blastula stages of embryonic development. The earlier specification of ectodermal and mesodermal fates suggested by various studies are also reflected in the model. pNE, prospective neural ectoderm; pNNE, prospective non-neural ectoderm; pPE, pre-placodal ectoderm; pM, prospective mesoderm; pE, prospective endoderm; PGC, primordial germ cells.
Lateral/intermediate regions of the chick epiblast have been reported to display Wnt/β-catenin activity, which is required for the expression of the NPB/NC marker Msx1/2 (Wilson et al., 2001). This suggests a potential early role of Wnt/β-catenin during NC specification. We have further validated the role of Wnt/β-catenin signaling in the blastula stage specification of NC using low-density epiblast cultures. We observed that inhibition of Wnt-ligand has no effect on NC specification (Pax7 expression). Our results suggest that cells within the intermediate epiblast region have already been exposed to Wnt-signaling, and have initiated the specification program prior to dissection of the explants. However, downstream inhibition of the transcriptional effector of Wnt, β-catenin, blocked NC specification, suggesting a continued requirement of β-catenin transcriptional activity during NC specification. The mechanism by which specified epiblast cells are able to proceed towards the NC cell fate in the presence of a Wnt-ligand inhibitor, but not through β-catenin inhibition, is intriguing, and suggests that enough active β-catenin is already accumulated in these cells. Alternatively, a recent article suggests a GSK3β independent specification of NC mediated through Dkk2 and β-catenin during neural plate border stages (Devotta et al., 2018). The role of other signaling pathways during these blastula stage of NC specification also requires further evaluation. Furthermore, blastula stage specification of NC does not disregard the possibility of signaling contributions that might be required for continued NC development from surrounding tissues in vivo.
An important aspect of cell fate specification are the transcriptional changes that govern different cell fates. For the past four decades studies have revealed the regional segregation of cell fate specifiers during pre-gastrula stages of development, in particular using the Xenopus model. The regional localization of maternal factors such as Vg1 and VegT have been well studied in mesendodermal specification (Weeks and Melton, 1987; Zhang and King, 1996; reviewed in Heasman, 2006). The positional identity of the cells has also been attributed to localization of maternal factors, such as the dorsalization of the embryo due to nuclear β-catenin during mid-blastula stage (Wylie et al., 1996). Similarly, experiments using dissociated blastula cells to address the cell fate decisions based on maternally inherited factors and positional identity of the blastomeres revealed a heterogeneity within the blastula stage Xenopus embryo shared between ectodermal and endodermal lineages (Sargent et al., 1986). Chick embryo studies also suggested specification of skeletal muscle cells prior to gastrulation using expression markers and culture of epiblast explants (George-Weinstein et al., 1996; Gerhart et al., 2011). Recent studies have started to decipher the transcriptional network involved during early cell fate specification. A study in Xenopus eludes to the role of maternally derived factors, FoxD4l1 and Zic2 in specifying neural fate in blastomeres by biasing them towards the neuroectodermal cell fate (Gaur et al., 2016). Thus, further analysis of transcriptional changes leading to NC specification from epiblast cells will help in identifying the regulatory network involved during these early steps of NC induction. It is essential to assess this early NC regulatory network, especially under the light of new studies exposing a predisposition of blastomeres towards specific cell fate, based on the relative expression ratios of different lineage specifiers (Shi et al., 2015). To better understand the cell fate specification of different lineages, positional single cell transcriptional analysis at blastula stage is needed to identify the network structure of individual cells to ascertain their predisposed cell fates.
Based on our experiments in chick, we propose prospective NC cells as the earliest NC cell state specified during blastula stages from epiblast cells, prior to the formation and concomitant fate segregation associated with the three germ layers. It is believed that chick epiblast and human ES cells are primed, or more advanced than the naïve state (Mak et al., 2015), and display differentiation bias towards certain cell fates (Mak et al., 2015; Shin et al., 2011), suggesting an early specification of cell fates prior to gastrulation in multiple species (Sheng, 2015). Our data from chick suggests a similar model for NC specification from epiblast cells prior to gastrulation. This model establishes a direct lineage between epiblast and NC, which are endowed with multipotent potential to form mesectodermal derivatives. Our model provides a parsimonious perspective underscoring the formation of this important vertebrate cell type in amniotes.
4. Materials & methods
4.1. Chicken embryos
Fertile hen eggs were obtained from Hardy’s Hatchery (Massachusetts, USA) and Sunstate Ranch (Sylmar, CA). Embryos were staged according to Hamburger and Hamilton (Hamburger and Hamilton, 1951) for stages 1 and over or Eyal-Giladi and Kochav (EGK) for preprimitive streak (prestreak) stages, Stage IX to XII. Embryos were cultured at 38 °C in humidified incubator.
4.2. Chick embryo explant culture
The hypoblast of pre-streak embryos at stage XII was mechanically removed using glass needles, and a horizontal strip of epiblast tissue was cut from the center of the embryo. This strip was trimmed to include only the area pellucida and dissected into 12 equivalent-sized squares (each approximately 100 μm2) and kept in PB1 buffer (5.97 g/L NaCl, 0.2 g/L KCl, 1.142 g/L NaH2PO4, 0.19 g/L KH2PO4, 0.04 g/L Sodium Pyruvate, 1 g/L Glucose, 0.1 g/L MgCl2–6 H20, 0.14 g/L CaCl2–2H20, 0.06 g/L Penicillin, 0.05 g/L Streptomycin, 0.01 g/L Phenol Red, 4 mg/L BSA (added just prior to use)) till next step. These squares were immobilized in separate collagen gels in four-well plates. Collagen gels were prepared by combining 90 μl of 3.68 mg/ml collagen(BD Biosciences), 10 μl of 10X DMEM (Gibco) and 3.7 μl of 7.5% sodium hydrogen carbonate. Collagen gels were immersed in DMEM/F12 containing N2 supplement (Gibco) for 35–48 h at 37 °C. Explants were fixed in 4% paraformaldehyde for 15 min before immunostaining.
4.3. Single cell dissociation experiment
The hypoblast of pre-streak embryos at stage XII was mechanically removed using glass needles, and a horizontal strip of epiblast tissue was cut from the center of the embryo. Horizontal epiblast strip was further dissected into 3 sections (2 intermediate and one medial) of ~150 cells each (Fig. 2A). The sections were collected in PB1 buffer and treated with accutase for 5min, rinsed in PB1 buffer and dissociated via pipetting. The dissociated cells were washed 2x in PB1 and then transferred in minimal PB1 onto collagen sheets in a chamber slide and was placed in incubator for 15min. Cell density after plating was around 10–20 cells per cm2. The chambers were then covered with neutral media (DMEM/F12 + N2 + Pen/Strep + 0.1%BSA) and placed back in the incubator for 30h. The chambers were washed with PBS and fixed in 4% paraformaldehyde for 30min before immunostaining. The number of cells obtained from each section after the complete dissociation and culture procedure were ~50 due to loss during dissociation and plating.
For assessing the role of Wnt/β-catenin signaling in NC specification using low density cultures, we used the same assay as described above. Dissociated intermediate epiblast cells were plated at low density in media containing WIF-1 (Wnt ligand inhibitor, 1.5 μg/ml; R&D Systems #1341-WF-050) or XAV939 (5 μM based on (Huang et al., 2009), Cayman Chemicals # 13596) or under control untreated conditions. Cells were harvested for immunostaining 30h after culture.
4.4. In vivo lineage tracing
At the pre-streak stage, Stage XII, embryos were injected with DiI and DiO (Molecular Probes) into cells of the lateral and medial region of the epiblast layer. Lateral region corresponds to the explant #2–3 described above, while medial regions correspond to explant # 6–7. Embryos were cultured at 37° C for 25–48 h in EC culture (Chapman et al., 2001), then fixed in 4% paraformaldehyde for 15 min before immunostaining. Embryos were mounted in gelatin and sectioned at 12 μm using a Leica CM1900 Cryostat. Sections were mounted with Permafluor (Thermo Scientific). Images were acquired on Nikon Eclipse 80i microscope and processed in Adobe Photoshop.
4.5. Immunostaining for chick embryos and explants
Immunostaining for chick embryos and explants were performed as previously described (Basch et al., 2006; Stuhlmiller and Garcia-Castro, 2011). Collagen gels containing explants were fixed with 4% paraformaldehyde for 10 min and then washed three times with PBS. Gels were blocked with PBS containing 1% BSA and 0.1%Tween-20 (PBST) for 1 h at room temperature. Double or triple staining was performed. Primary antibodies for mouse IgG1anti-Pax7 (1:50; Developmental Studies Hybridoma Bank (DSHB)); mouse IgG1 anti-Msx1,2 (1:50; 4G1, DSHB), mouse IgG1 anti-Snail2 (1:100; 62.1E6, DSHB), mouse IgG2b anti-AP2 (1:50; 3B5, DSHB), mouse IgM anti-HNK-1 (1:100; 1C10, DSHB), goat IgG anti-Sox2 (1:100; R&D AF2018), rabbit IgG anti-Six1 (1:200; Sigma HPA001893), GP IgG anti-Sox9 (1:100; gift from Dr. Vivian Lee) and rabbit IgG anti-Brachyury (1:10; gift from Dr. Susan Mackem) were diluted in PBST and incubated at 4 °C overnight. Primary antibody was washed 3 × 10 min with PBST. Gels were then incubated with secondary antibodies (goat anti-mouse IgG1Alexa 568, 1:2500; goat anti-mouse IgG2b Alexa 488, 1:2500; donkey anti-IgM Cy5, 1:500; donkey anti-goat IgG 488 or 633, 1:2500 and goat rabbit anti-IgG Alexa 488, 1:2500) diluted in PBST and incubated at 4 °C overnight. Secondary antibody was washed 3 × 10 min with PT and then stained with DAPI (5 μg/mL) for 5 min and washed again 3 × 10 min with PBS before imaging. Primary antibodies used for chick immunostaining are listed in key resource table.
4.8.
Key resources table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Msx1/2 | DSHB | 4G1 |
| Snai2 | DSHB | 62.1E6 |
| Tfap2a | DSHB | 3B5 |
| Sox2 | R&D | AF2018 |
| Sox9 | Gift from Dr. Vivian Lee |
NA |
| HNK-1 | DSHB | 1C10 |
| TbxT (Brachyury) | Gift from Dr Susan Mackem |
NA |
| Six1 | Sigma | HPA001893 |
| Pax7 | DSHB | Pax7 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| WIF-1 | R&D Systems | 1341-WF-050 |
| XAV939 | Cayman Chemicals |
13596 |
| Oligonucleotides Pax7 |
This paper | F: CCAGAGACAATAGGTTGGGTACATTTA R: GGTCTTTAGCATGGGCAGACA |
| Sox2 | This paper | F: CAGGCTAAAGTAGTTTGAATG R: CTGTTCTTCTGGTTGTTCG |
| Sox3 | This paper | F: TTCGCTTCCGAGTCTTAAAGATG R: GGTCGGTTTTTCGTTTTGTCA |
| TbxT | This paper | F: CGCTGAGGAATCACCGTTCT R: GGAAGGCAGGCAGAGGAGTTA |
| Tbx6 | This paper | F: CTCCCTTGTCGCACCTATGTC R: GTGAGCTTCAGTTTCTGGAAGGA |
| GAPDH | This paper | F: CTGTTGTTGACCTGACCTGC R: AGACAACCTGGTCCTCTGTGT |
| Software and Algorithms | ||
| Nikon Elements | Nikon | NA |
| General Analysis | ||
4.6. Image processing for chick explants
Images were taken using a Nikon Eclipse 80i or Nikon Eclipse Ti inverted microscope and processed in Adobe Photoshop CC version 14.2.1. Images of each experiment were taken in a fluorescent scope (Nikon) using the same settings for each fluorophore channel. Images were compiled in a Photoshop grid image and intensity levels were adjusted at the same time. The threshold levels were set using a positive reference when a clear nuclear staining was detected in an explant of the series. Nikon Elements software (with General Analysis) (Nikon) was used for cell counts and cell intensity analysis. For cell count analysis, under general analysis, bright spot detection method was used, with clustered object parameter, with variable object (cell) sizes depending on the sample, determined manually for each sample to detect every cell in the image under each channel. No primary control was used to subtract the background fluorescence for all the samples in an experiment equally. Unmodified images were used to obtain total cell counts and plotted as bar graphs from each individual experiment. Each experiment was done in 3 biological replicates.
4.7. Gene expression analysis
After the 25h of culture, explants were collected in a solution for RNA extraction using the provider specifications (Zymoresearch Direct-zol RNA Microprep kit). Total RNA was collected in 14 μl and reverse-transcribed using PrimeScript reverse transcriptase kit (Takara Bio, cat. RR014B). Quantitative polymerase chain reaction (QPCR) was carried out using the SYBR Premix ExTaqII SYBR Green premix (Takara Bio, cat. RR82WR) on Applied Biosystems StepOne Plus System (Applied Biosystems). Expression of Pax7, Sox2, Sox3, TbxT, Tbx6 along with GAPDH as the reference gene, were analyzed in each of the 12 explants. Positive controls of neural plate/neural folds from HH stage 8 embryo were used to compare the expression of Sox2 and Pax7, respectively, while primitive streak region was used as positive control for mesodermal genes, TbxT and Tbx6. Fold change in expression was determined using the ΔΔCT formula. 12 explants from 4 separate embryos were used for RT-qPCR and data was represented as average of the 4 embryos.
Statistical analysis
Chi-Squared test for observed statistical significance for lineage tracing experiments in chick embryo were done using two-way Contingency table, with one-degree of freedom and represented as p-value significance for observed NC contribution of cells labeled within intermediate and medial epiblast regions.
Supplementary Material
Acknowledgements
Imaging was performed at the UCR Stem Cell Core Facility (California Institute for Regenerative Medicine (CIRM) funded shared facility). We are grateful to Dr. Changjun Yu (Sunstate Ranch, Sylmar, CA) for providing fertilized chicken eggs for this study. We thank Dr. Ken Cho (UC Irvine) and Dr. Ira Blitz (UC Irvine) for helpful conversations, suggestions and critical reading of the manuscript.
Funding
This work was funded by NIH grant R01DE017914 to M.I.G-C. and F32DE027862 to R.M.C.
Footnotes
Data and materials availability
All data is available in the main text or the supplementary materials.
Declaration of competing interest
Authors declare no competing financial interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2019.10.007.
References
- Basch ML, Bronner-Fraser M, Garcia-Castro MI, 2006. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature 441, 218–222. 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Betters E, Charney RM, García-Castro MI, 2018. Early specification and development of rabbit neural crest cells. Dev. Biol 10.1016/j.ydbio.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolande RP, 1996. Neurocristopathy: its growth and development in 20 years. Pediatr. Pathol. Lab. Med 17, 1–25. [PubMed] [Google Scholar]
- Bolande RP, 1974. The neurocristopathies: a unifying concept of disease arising in neural crest maldevelopment. Hum. Pathol 5, 409–429. [DOI] [PubMed] [Google Scholar]
- Buitrago-Delgado E, Nordin K, Rao A, Geary L, LaBonne C, 2015. Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science 248, 1332–1335. 10.1126/science.aaa3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman SC, Collignon J, Schoenwolf GC, Lumsden A, 2001. Improved method for chick whole embryo culture using a filter paper carrier. Dev. Dynam 220, 284–289.. [DOI] [PubMed] [Google Scholar]
- Chen Q, Shi J, Tao Y, Zernicka-Goetz M, 2018. Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo. Nat. Commun 1–11. 10.1038/s41467-018-04155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe C, Krivega M, Cauffman G, Geens M, Van de Velde H, 2014. Totipotency and lineage segregation in the human embryo. MHR: Basic Sci. Reproductive Med. 20, 599–618. 10.1093/molehr/gau027. [DOI] [PubMed] [Google Scholar]
- Devotta A, Hong CS, Saint-Jeannet JP, 2018. Dkk2 promotes neural crest specification by activating wnt/β-catenin signaling in a GSK3β independent manner. eLife 7, e34404 10.7554/eLife.34404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchevers HC, Amiel J, Lyonnet S, 2006. Molecular bases of human neurocristopathies. Adv. Exp. Med. Biol 589, 213–234. 10.1007/978-0-387-46954-6_14. [DOI] [PubMed] [Google Scholar]
- Eyal-Giladi H, Kochav S, 1976. From cleavage to primitive streak formation: a complementary normal table and a new look at the first stages of the development of the chick. Dev. Biol 49, 329–337. [DOI] [PubMed] [Google Scholar]
- Farlie PG, McKeown SJ, Newgreen DF, 2004. The neural crest: basic biology and clinical relationships in the craniofacial and enteric nervous systems. Birth Defect Res C 72, 173–189. 10.1002/bdrc.20013. [DOI] [PubMed] [Google Scholar]
- Gans C, Northcutt RG, 1983. Neural crest and the origin of vertebrates: a new head. Science 220, 268–273. 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M, 2002. Ectodermal Wnt function as a neural crest inducer. Science 297, 848–851. [DOI] [PubMed] [Google Scholar]
- Gaur S, Mandelbaum M, Herold M, Majumdar HD, Neilson KM, Maynard TM, Mood K, Daar IO, Moody SA, 2016. Neural transcription factors bias cleavage stage blastomeres to give rise to neural ectoderm. Genesis 54, 334–349. 10.1002/dvg.22943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George-Weinstein M, Gerhart J, Reed R, Flynn J, Callihan B, Mattiacci M, Miehle C, Foti G, Lash JW, Weintraub H, 1996. Skeletal myogenesis: the preferred pathway of chick embryo epiblast cells in vitro. Dev. Biol 173, 279–291. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Scheinfeld VL, Milito T, Pfautz J, Neely C, Fisher-Vance D, Sutter K, Crawford M, Knudsen K, George-Weinstein M, 2011. Myo/Nog cell regulation of bone morphogenetic protein signaling in the blastocyst is essential for normal morphogenesis and striated muscle lineage specification. Dev. Biol 359, 12–25. 10.1016/j.ydbio.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BK, 2018. Germ layers, the neural crest and emergent organization in development and evolution. Genesis 56 10.1002/dvg.23103e23103–9. [DOI] [PubMed] [Google Scholar]
- Hall BK, 2000. The neural crest as a fourth germ layer and vertebrates as quadroblastic not triploblastic. Evol. Dev 2, 3–5. 10.1046/j.1525-142x.2000.00032.x. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton H, 1951. A series of normal stages in the development of the chick embryo. J. Morphol 88, 49–92. 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hatada Y, Stern CD, 1994. A fate map of the epiblast of the early chick embryo. Development 120, 2879–2889. [DOI] [PubMed] [Google Scholar]
- Heasman J, 2006. Patterning the early Xenopus embryo. Development 133, 1205–1217. 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA, 1997. Vertebrate neural induction. Annu. Rev. Neurosci 20, 43–60. [DOI] [PubMed] [Google Scholar]
- Huang S-MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F, 2009. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620. 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SMK, Johnson JE, McMahon AP, Takada S, 1997. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 389, 966–970. 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Jacobson M, Hirose G, 1981. Clonal organisation of the central nervious system of the frog. J. Neurosci 1, 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF, 1990. Origin and organization of the zebrafish fate map. Development 108, 581–594. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M, 1998. Neural crest induction in Xenopus: evidence for a two-signal model. Development 125, 2403–2414. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, 1980. The ontogeny of the neural crest in avian embryo chimaeras. Nature 286, 663–669. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Kalcheim C, 1999. The Neural Crest. Cambridge Univ. Press, Cambridge. [Google Scholar]
- Leung AW, Murdoch B, Salem AF, Prasad MS, Gomez GA, García-Castro MI, 2016. WNT/β-catenin signaling mediates human neural crest induction via a pre-neural border intermediate. Development 143, 398–410. 10.1242/dev.130849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusdottir E, Surani MA, 2013. How to make a primordial germ cell. Development 141, 245–252. 10.1242/dev.098269. [DOI] [PubMed] [Google Scholar]
- Mak SS, Alev C, Nagai H, Wrabel A, Matsuoka Y, Honda A, Sheng G, Ladher RK, 2015. Characterization of the finch embryo supports evolutionary conservation of the naive stage of development in amniotes. eLife 11 (4), e07178 10.7554/eLife.07178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SA, 1987a. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol 119, 560–578. [DOI] [PubMed] [Google Scholar]
- Moody SA, 1987b. Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev. Biol 122, 300–319. 10.1016/0012-1606(87)90296-X. [DOI] [PubMed] [Google Scholar]
- Moody SA, Kline MJ, 1990. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat. Embryol 182, 347–362. [DOI] [PubMed] [Google Scholar]
- Northcutt GR, 2005. The new head hypothesis revisited. J. Exp. Zool 304B, 274–297. 10.1002/jez.b.21063. [DOI] [PubMed] [Google Scholar]
- Onjiko RM, Moody SA, Nemes P, 2015. Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc. Natl. Acad. Sci 112, 6545–6550. 10.1073/pnas.1423682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthey C, Edlund T, Gunhaga L, 2008a. Wnt-regulated temporal control of BMP exposure directs the choice between neural plate border and epidermal fate. Development 136, 73–83. 10.1242/dev.025890. [DOI] [PubMed] [Google Scholar]
- Patthey C, Gunhaga L, Edlund T, 2008b. Early development of the central and peripheral nervous systems is coordinated by Wnt and BMP signals. PLoS One 3 10.1371/journal.pone.0001625e1625–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro C, Figueiredo AL, Maczkowiak F, Pouponnot C, Eychène A, Monsoro-Burq AH, 2015. PFKFB4 controls embryonic patterning via Akt signalling independently of glycolysis. Nat. Commun 6, 5953 10.1038/ncomms6953. [DOI] [PubMed] [Google Scholar]
- Roellig D, Tan-Cabugao J, Esaian S, Bronner ME, 2017. Dynamic transcriptional signature and cell fate analysis reveals plasticity of individual Neural Plate border cells. eLife. 10.7554/eLife.21620.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jeannet J-P, He X, Varmus HE, Dawid IB, 1997. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc. Natl. Acad. Sci 94, 13713–13718. 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Yamaji M, 2012. Primordial germ cells in mice. Cold Spring Harb Perspect Biol 4 10.1101/cshperspect.a008375a008375-a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent TD, Jamrich M, Dawid IB, 1986. Cell interactions and the control of gene activity during early development of Xenopus laevis. Dev. Biol 114, 238–246. [DOI] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K, Sasai Y, 2001. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development 128, 2525–2536. [DOI] [PubMed] [Google Scholar]
- Sheng G, 2015. Epiblast morphogenesis before gastrulation. Dev. Biol 401, 17–24. 10.1016/j.ydbio.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Shi J, Chen Q, Li X, Zheng X, Zhang Y, Qiao J, Tang F, Tao Y, Zhou Q, Duan E, 2015. Dynamic transcriptional symmetry-breaking in pre-implantation mammalian embryo development revealed by single-cell RNA-seq. Development 142, 3468–3477. 10.1242/dev.123950. [DOI] [PubMed] [Google Scholar]
- Shin M, Alev C, Wu Y, Nagai H, Sheng G, 2011. Activin/TGF-beta signaling regulates Nanog expression in the epiblast during gastrulation. Mech. Dev 128, 268–278. 10.1016/j.mod.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD, 2000. Initiation of neural induction by FGF signalling before gastrulation. Nature 406, 74–78. 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI, 2011. FGF/MAPK signaling is required in the gastrula epiblast for avian neural crest induction. Development 139, 289–300. 10.1242/dev.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevers KE, Prajapati RS, Hintze M, Stower MJ, Strobl AC, Tambalo M, Ranganathan R, Moncaut N, Khan MAF, Stern CD, Streit A, 2018. Neural induction by the node and placode induction by head mesoderm share an initial state resembling neural plate border and ES cells. Proc. Natl. Acad. Sci 115, 355–360. 10.1073/pnas.1719674115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks DL, Melton DA, 1987. A maternal mRNA localized to the vegetal hemisphere in xenopus eggs codes for a growth factor related to TGF-β. Cell 51, 861–867. 10.1016/0092-8674(87)90109-7. [DOI] [PubMed] [Google Scholar]
- Wilson S, Rydstrom A, Trimborn T, Willert K, Nusse R, Jessell TM, Edlund T,€ 2001. The status of Wnt signalling regulates neural and epidermal fates in the chick embryo. Nature 411, 325–330. 10.1038/35077115. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Graziano E, Harland R, Jessell TM, Edlund T, 2000. An early requirement for FGF signalling in the acquisition of neural cell fate in the chick embryo. Curr. Biol 10, 421–429. 10.1016/S0960-9822(00)00431-0. [DOI] [PubMed] [Google Scholar]
- Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, Heasman J, 1996. Maternal β-catenin establishes a “dorsal signal” in early Xenopus embryos. Development 122, 2987–2996. [DOI] [PubMed] [Google Scholar]
- Zhang J, King ML, 1996. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development 122, 4119–4129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.