Abstract
Successful colonization of the intestine requires that bacteria interact with the innate immune system and, in particular, neutrophils. Progression of inflammatory bowel diseases (IBD) is associated with alterations in gut microbiota, and dysbiosis in Crohn’s disease (CD) patients is often associated with an expansion of Escherichia coli. Here, we investigated the ability of such E. coli isolates to avoid neutrophil activation and to utilize reactive oxygen species. Neutrophil activation was detected in vitro in normal human blood via luminol chemiluminescence (CL) induced by reactive oxygen and halogen species generated by neutrophils. No significant difference in neutrophil activation in vitro was detected between isolates from inflamed (23 isolates) vs healthy intestines (5 isolates), with 10‐fold variation within both groups (2.9–61.2 mV). CL activity of isolates from the same patient differed by 1.5–5 times. Twenty‐four isolates from ileal aspirate, biopsy, and feces of seven patients with CD and one patient with no intestine inflammation were tested for extracellular peroxidase and catalase activity and cell surface hydrophobicity. Average values between patients varied from 26 ± 3 to 73 ± 18 µmol·g−1 of air dry weight for peroxidase activity, from 15 ± 2 to 189 ± 56 mmol·g−1 of air dry weight for catalase activity, and from 5 ± 3 to 105 ± 9 a.u. for the hydrophobic probe fluorescence. Extracellular peroxidase activity and hydrophobicity of bacterial cell surface correlated negatively with stimulated neutrophil CL. The ability of some isolates to avoid neutrophil activation and to utilize reactive oxygen species may provide a strategy to survive assault by the innate immune system.
Keywords: chemiluminescence, Crohn’s disease, E. coli, fluorescent probes, neutrophil activation, peroxidase
The ability of Escherichia coli isolates from the intestines of Crohn’s disease (CD) patients to avoid neutrophil activation and to utilize reactive oxygen species was analyzed. Neutrophil activation was detected in vitro in normal human blood via luminol chemiluminescence (CL). Extracellular peroxidase activity and hydrophobicity of bacterial cell surface correlated negatively with stimulated neutrophil CL.
Abbreviations
- ANS
1‐anilino‐8‐naphthalenesulfonate
- CD
Crohn’s disease
- CFU
colony‐forming units
- CL
chemiluminescence
- DMC
4‐dimethylaminochalcone
- DMEM
Dulbecco’s modified Eagle’s medium
- DW
dry weight
- FBS
fetal bovine serum
- IBD
inflammatory bowel disease
- KRB
Krebs–Ringer bicarbonate
- OD
optical density
- PBS
phosphate‐buffered saline
- PMA
phorbol 12‐myristate 13‐acetate
- RHS
reactive halogen species
- ROS
reactive oxygen species
Progression of inflammatory bowel diseases (IBD) is associated with alteration in gut microbiota. Dysbiosis often observed in CD patients is associated with an expansion of Escherichia coli capable of various adaptive reactions 1, 2, 3. One of these reactions is surviving the attack of human immune system. Thus, antibodies to E. coli were found in biopsy samples from ulcer, erosion, and granuloma of patients with Crohn’s disease, suggesting the presence of bacteria at the very spot of acute inflammation 4.
It is hard to estimate whether it would be more profitable for bacteria to avoid neutrophil stimulation, or to provoke it. On the one hand, negative correlation between neutrophil‐stimulating capacity and virulence was shown for uropathogenic E. coli 5. At the same time, ROS/RHS produced by activated neutrophils at the site of inflammation can enhance damage of gut wall 6, 7, 8 facilitating further bacterial expansion 3.
Upon activation, neutrophils produce reactive oxygen (•O2ˉ, H2O2, •OH, etc.) and halogen (HOCl, HOBr, etc.) species (ROS/RHS), which can be detected by chemiluminescence (CL) 9. One of the key products of activated neutrophil, hydrogen peroxide, can be decomposed by bacterial hydrogenases (HPI and HPII) 10, 11, 12, 13. Unlike HPII, HPI shows not only catalase but also peroxidase activity and oxidizes o‐dianisidine in the presence of H2O2 14, 15. The expression of hydrogenases is regulated by E. coli interaction with phagocytes and reorganization of inner and outer cell membrane 16.
Activation of neutrophils could depend on bacterial properties such as hydrophobicity and surface charge of bacterial cell membranes 17, 18, and expression of mannose‐sensitive adhesins 19, 20, 21 and of ligands for Toll‐4 receptor of neutrophils 22. The hydrophobicity of bacterial cell envelope can be detected by the affinity of E. coli for the fluorescent membrane probes. One of them, amphiphilic 1‐anilino‐8‐naphthalenesulfonate (ANS) 23, does not penetrate the bacterial cell 24 because of its negatively charged sulfo group. Another fluorescent probe used in this study, 4‐dimethylaminochalcone (DMC), is neutral and also sensitive to the hydrophobic sites on bacterial cell surface 25.
The aim of our work was to study the interplay of adaptive mechanisms based on peroxidase/catalase activity, surface hydrophobicity, and neutrophil‐stimulating capacity of E. coli from the inflamed gut. Whole blood of healthy donors was used in CL assays of ROS/RHS production in order to exclude effects of patients’ immune status. We previously demonstrated that E. coli isolates from the healthy or inflamed human intestine were able to induce CL response in neutrophils in these conditions 26.
Materials and methods
Chemicals
Lysogeny broth (LB) medium, Krebs–Ringer bicarbonate (KRB) buffer with CaCl2, pH 7.4, phorbol 12‐myristate 13‐acetate (PMA), o‐dianisidine, luminol, lucigenin, horseradish peroxidase (HRP), hydrogen peroxide (H2O2) solution (30 wt, %), potassium iodide (KI), 1‐anilino‐8‐naphthalenesulfonate (ANS), and phosphate‐buffered saline, pH 7.4 (PBS), were purchased from Sigma‐Aldrich (St. Louis, MO, USA). A working solution of H2O2 was prepared immediately before use by diluting the stock with PBS. Its concentration was controlled by measuring the absorbance at 240 nm (molar extinction coefficient of 43.6 m−1·cm−1 27), HCl, and H3PO4. Tris and NaCl were from Chimmed, Russia. 4‐(Dimethylamino)chalcone (DMC) was kindly provided by Krasowitsky B.M. (Institute of Monocrystals, Kharkov, Ukraine).
Patients
Crohn’s patient donors of ileum content, biopsy, or feces (n = 7, 5 male, two female, age 23–47, median 33), healthy feces donors (n = 4, 3 male, one female, age 19–28, median 19.5), and healthy blood donors (n = 5, female, age 21–64, median 38) were enrolled in the study and gave written informed consent (Table 1). For one person (male, age 14), the informed content was given by legal representatives.
Table 1.
Parameters of Escherichia coli isolates. CD patients and non‐IBD donors of feces and blood used in the study.
| Patient | Isolate | CL max, Lum, LB | Px, μmol·g−1 dry weight | Cat, nmol H2O2·g−1 dry weight | Groups of the isolates according to their CL‐stimulating capacity |
Escherichia coli genome |
Escherichia coli phylogroup b 29 | F (ANS). a.u. | F (DMC). a.u. | Localization 30 | Clinical activity 30 | Endoscopic activity 31 | Type of material for preparation of isolates | E. coli DNA copies per ml aspirate/fecesa | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli donors | CD | G | G | 24 ± 7 | 36 ± 1.8 | 77.0 ± 4.1 | 2 | RCE05‐01 | D | 32.8 | 26.2 | Ileitis–jejunitis | Severe disease | 9 | Feces |
2.1 x 109 stool |
| A | A1 | 2.5 ± 1 | 51.89 ± 3.2 | 108.4 ± 5 | 1 | RCE01‐05 | A | 41 ± 6 | 105 ± 9 | Ileitis | Mild disease | 10 | Lumen |
1.25 x 107 stool |
||
| A2 | 3 ± 1.2 | 59.9 ± 2.5 | 60.8 ± 4.1 | 1 | RCE01‐02 | A | Ileum biopsy | |||||||||
| A3 | 2.8 ± 1.5 | 86.4 ± 3.3 | 86.9 ± 5 | 1 | RCE01‐06 | A | Feces | |||||||||
| A4 | 3.5 ± 1.3 | 51.0 ± 2.1 | 24.4 ± 3.7 | 1 | RCE01‐04 | A | Lumen | |||||||||
| B | B1 | 42.3 ± 12 | 28.7 ± 3 | 16.7 ± 2 | 3 | RCE03‐01 | D | 24 ± 3 | 5 ± 3 | Ileocolitis | Moderate disease | 14 | Ileum biopsy |
2.8 x 105 aspirate |
||
| B2 | 61.2 ± 10 | 22.1 ± 2 | 14.4 ± 3.2 | 3 | RCE03‐02 | D | ||||||||||
| M | M1 | 14 ± 6 | 33.7 ± 3.4 | 14.4 ± 1.6 | 1 | RCE06‐01 | B2 | 30 ± 6 | 85 ± 12 | Ileocolitis | Mild disease | 15 | Lumen |
2 x 105 aspirate |
||
| M2 | 18 ± 6 | 30.0 ± 1.8 | 8.3 ± 2.1 | 1 | RCE06‐02 | B2 | ||||||||||
| M3 | 11.8 ± 5 | 73.8 ± 2.2 | 33.8 ± 3.7 | 2 | RCE06‐03 | B2 | ||||||||||
| M4 | 8 ± 3 | 43.7 ± 2.5 | 13.1 ± 2.4 | 2 | RCE06‐04 | B2 | ||||||||||
| M5 | 3.2 ± 2 | 39.7 ± 3.3 | 11.5 ± 3.5 | 2 | RCE06‐05 | B2 | ||||||||||
| Z | Z | 31 ± 6 | 20.8 ± 2.8 | 78.6 ± 5 | 3 | RCE07‐01 | A | 20.2 | 25.5 | Ileocolitis | Remission | 3 | Lumen |
1.3 x 108 aspirate |
||
| V | V1 | 9 ± 2.7 | 20.1 ± 2.4 | 273.1 ± 3 | 1 | RCE02‐02 | B1 | 21 ± 5 | 36 ± 8 | Ileocolitis | Mild disease | 13 | Lumen |
5 x 104 aspirate |
||
| V2 | 41.8 ± 14 | 26.7 ± 1.8 | 175.8 ± 12 | 2 | RCE02‐01 | B1 | ||||||||||
| V3 | 15.1 ± 4 | 25.8 ± 3.5 | 150.3 ± 8.1 | 2 | ‐ | ‐ | ||||||||||
| V4 | 17.6 ± 4 | 38.0 ± 3.2 | 158.1 ± 6.3 | 3 | RCE02‐03 | B1 | ||||||||||
| P | P1 | 18.7 ± 5 | 18.8 ± 2.6 | 105.5 ± 15 | 1 | RCE04‐01 | A | 35 ± 15 | 33 ± 9 | Ileocolitis–perianal | Severe disease | 0 | Lumen |
5.2 x 104 aspirate |
||
| P2 | 12.2 ± 5 | 40.9 ± 3.6 | 130.0 ± 3.5 | 1 | RCE04‐02 | A | ||||||||||
| P3 | 7.2 ± 3 | 45.5 ± 2.8 | 170.4 ± 8 | 2 | RCE04‐03 | A | ||||||||||
| P4 | 9.7 ± 3 | 45.5 ± 3.6 | 180.0 ± 10 | 2 | RCE04‐04 | A | ||||||||||
| P5 | 20.4 ± 8 | 24.7 ± 3 | 125.1 ± 8 | 2 | RCE04‐05 | A | Ileum biopsy | |||||||||
| P6 | 10.5 ± 5 | 32.9 ± 2.7 | 130.1 ± 7 | 2 | RCE04‐06 | A | ||||||||||
| Healthy | C | C | 59 ± 13 | 3.2 ± 0.7 | 29.1 ± 0.6 | 3 | 15 | 16 | No IBD | 0 | Lumen |
4 x 102 aspirate |
||||
| h‐1 | H1 | 41.6 ± 5 | No IBD | Feces | ||||||||||||
| h‐2 | H2 | 10 ± 3.2 | No IBD | Feces | ||||||||||||
| h‐3 | H3 | 58.5 ± 5.1 | No IBD | Feces | ||||||||||||
| h‐4 | H4 | 5 ± 2.1 | No IBD | Feces | ||||||||||||
| Blood donors | Blood donor 1 | No IBD | ||||||||||||||
| Blood donor 2 | No IBD | |||||||||||||||
| Blood donor 3 | No IBD | |||||||||||||||
| Blood donor 4 | No IBD | |||||||||||||||
| Blood donor 5 | No IBD | |||||||||||||||
Values are given as M ± SD, where M is mean value between the isolates from one patient. SD—standard deviation.
References to the genomes of E. coli isolates are given according to Ref. 29
Localization, clinical activity, and endoscopical activity of inflammation were determined according to Ref. 30, 31
The amount of E. coli DNA was measured in feces or aspirate even if some E. coli strains for study were sampled from biopsy. From CD samples, only those with E. coli DNA content 1 × 104 copies per ml and above were selected for the study
Phylogroups of E. coli isolates were determined from their genomes (sequencing performed in our previous work 29).
The inclusion criteria for CD patients were endoscopically and radiologically diagnosed, and histologically confirmed Crohn’s disease and the content of E. coli DNA in the lumen liquid or feces above 1 × 104 copies per mL. The exclusion criteria were signs of indeterminate colitis, infectious diseases, anamnesis of total colectomy, presence of stoma, and recent (less than 2 months) antibiotic treatment. The inclusion criteria for healthy individuals donating blood or feces were no intestinal or autoimmune diseases in anamnesis, no infection diseases, and no recent antibiotic treatment.
Small intestine aspirates and bowel biopsies were obtained during diagnostic endoscopy procedures, which aim was to confirm the IBD diagnosis or to evaluate the state of relapse or remission. For one person (patient C), the diagnostic endoscopy revealed no signs of IBD/inflammation; therefore, this patient was referred as ‘healthy’ in regard to IBD. Stool samples were collected prior to endoscopy for those subjected to it and as a morning stool sampling procedure for healthy volunteers—donors of feces. Samples were stored in sterile containers at +4 °C for up to several hours prior to E. coli isolation.
Blood samples from healthy volunteers were obtained by venipuncture using heparin as an anticoagulant. Blood was used for experiments immediately after the draw.
The study methodologies were conformed to the standards set by the Declaration of Helsinki and approved by Ethics committees of Federal Research and Clinical Center of Physical‐Chemical Medicine of FMBA and Moscow Clinical Scientific Center, Central Scientific Institute of Gastroenterology.
Escherichia coli concentration measurement
Escherichia coli concentration in ileal and fecal microbiota samples from patients was estimated using SEPTOSKRIN Kit OneStep Strip (Lytech, Moscow, Russia) according to the manufacturer’s protocol. Total DNA from samples was isolated according to CTAB DNA extraction protocol described by Ref. 28 with some modifications: 0.5 mL of material was suspended in 0.5 mL CTAB Buffer, incubated at 60 °C for 30 min, and mixed with 0.5 mL of chloroform. After centrifugation (9000 g, 10 min at 4 °C), the aqueous phase was mixed with an equal volume isopropanol and 0.1 volume 3M NaOAc, pH 5.2, and placed on −20 °C for 1 h. DNA was collected by centrifugation (10 000 g for 10 min), washed with cold 80% ethanol, and resuspended in 0.2 mL of H2O. Total DNA concentration was measured using NanoDrop ND‐100 (Thermo Fisher Scientific, Waltham, MA, USA) and equalized to 200 ng·µL−1. Escherichia coli DNA amount in the samples was detected by quantity RT‐PCR with specific primers provided by SEPTOSKRIN Kit (Lytech).
Only CD samples with E. coli DNA content of 1 x 104 copies per ml of initial sample and above were selected for downstream experiments.
Bacterial isolates
Escherichia coli isolates were achieved as described in Ref. 29. Briefly, aspirates and feces were diluted by 106 in sterile PBS; biopsy fragments were shaken in 0.2 mL of sterile PBS. Aliquots (100 µL) of the resulting liquid were plated on Petri dishes coated with LB agar and incubated overnight at 37 °C. The species affiliation was determined with matrix‐assisted laser desorption/ionization (MALDI) using a mass spectrometer Bruker Microflex combined with the software Biotyper from Bruker Daltonics, Germany.
Preparation of bacterial cell suspension
Escherichia coli cultures were grown on an orbital shaker (150 rpm) at 37 °C by inoculating isolated colony of bacteria into LB medium. Overnight culture of bacteria was collected by centrifugation (3500 g, 10 min) and washed twice with PBS. The washed cells were suspended in PBS, and their optical density (OD) at 540 nm was measured. To estimate the dry weight (DW, mg·mL−1), bacterial suspensions were washed with distilled water and dried. The OD of 1.0 corresponded to DW of 0.16 ± 0.03 mg·mL−1. Bacterial pellets were stored at −80 °C.
Adhesion and invasion assay
The assay was performed according to Ref. 32 with slight alterations. Isolates were cultivated on LB (37 °C, 200 RPM) till mid‐log phase. Bacteria were harvested by centrifugation (3500 g, 15 min), and the pellet was washed with PBS and resuspended in PBS. Bacterial suspension (150 μL, OD at 540 = 0.2) was mixed with 5 mL DMEM + 20% fetal bovine serum (FBS). Bacteria in DMEM + FBS (500 μL) were added to CaCo cells grown in 24‐well plate till the monolayer formation (about 500 000 CaCo cells per well). Control bacteria suspension was collected at this point and plated onto LB agar plates at 10−3–10−4 dilution (control value). The 24‐well plates with CaCo and bacteria were incubated for 3 h at 37 °C. For invasion test, cell suspension was additionally treated with 1 mL of DMEM (20% FBS) with 300 mg·L−1 of gentamicin for 1 h at 37 °C. After incubation, the monolayer was gently washed twice with PBS to remove unbound bacteria and cells were removed from the plate by trypsin (200 μL). Cells were lysed by mixing with 0.5% Triton X‐100 in DMEM (20% FBS). The lysates were plated on LB agar at 10−2–10−3 dilution (5 μL per agar plate). After 24 h of incubation at 37 °C, colony counts were performed to determine colony‐forming units (CFU).
Adhesion + invasion values were determined as:
the number of bacteria adhered–invaded per CaCo cell = CFU per agar plate after adhesion*dilution/500 000 (the number of CaCo cells per well);
% of adhered–invaded bacteria in bacterial suspension = CFU per agar plate after adhesion/CFU per agar plate in control * 100 (considering corresponding dilutions);
Invasion value was determined as % of adhered–invaded bacteria surviving gentamicin treatment.
Chemiluminescence measurements of neutrophil activation
Chemiluminescence measurements were performed using a luminometer LKB Wallac 1251 according to the method described by Ref. 33 with modifications 26. Chemiluminescence was tracked at 37 °C under continuous agitation of a sample, 20 μL of healthy volunteer’s blood and 1 mL of 0.2 mm luminol or lucigenin in KRB solution (pH 7.4) were placed into a luminometer cuvette, and spontaneous chemiluminescence was continuously recorded for one minute. Then, a bacterial suspension in PBS was added to a final concentration of 70 ± 10 μg DW·mL−1 (bacteria:neutrophil cell ratio was 4000 CFU/cell), and a chemiluminescence response was recorded. Results were expressed in voltage values (mV) corresponding to maximum CL levels. The responses of donors’ blood to standard activators PMA (100 nm) and opsonized zymosan (0.2 mg·mL−1) were 35 ± 19 mV and 80 ± 15 mV, respectively.
To rule out the possible toxic effects of E. coli on neutrophils, in a number of experiments, 156 nm PMA was added to the luminometer cuvette at the end of CL analysis, cuvette after the level of E. coli‐induced CL had reached its maximum value.
Extracellular bacterial peroxidase activity
Extracellular bacterial peroxidase activity was estimated by the oxidation of o‐dianisidine by bacterial cells in the presence of H2O2, by method described in Ref. 14 with modifications according to Ref. 34. Two hundred microlitre of 100 mm citrate buffer (pH 5.5) containing 0.32 mm o‐dianisidine and 2.3 mm H2O2 was added to 200 μL of bacterial suspension (3 mg of DW per ml in PBS), and the reaction mixture was incubated for 5 min at ambient temperature. The reaction was stopped by addition of 1 mL of 35% phosphorous acid. The cells were precipitated by centrifugation at 900 g for 20 min, and the OD of supernatants was read at 560 nm. The control probes contained PBS instead of bacterial suspension. The concentration of oxidized o‐dianisidine was calculated by using its molar extinction coefficient of 20.040 m −1·cm−1 34. The amount of oxidized o‐dianisidine was normalized to a dry‐weight basis. To make sure that the bacterial peroxidase activity is related to the cell wall, bacterial suspensions were incubated with 1% Triton X‐100 for 1 h, and then, the cells were precipitated at 900 g (30 min), and peroxidase activity was measured in supernatant.
Bacterial catalase activity
Extracellular bacterial catalase activity was measured by iodometric determination of H2O2 decomposed by bacterial cells as described in Ref. 35, with modifications. The assay was carried out as follows: 400 μL of cell suspension (or 400 μL of PBS in control probe) was mixed with 100 μL of 51 mm H2O2 solution in PBS and allowed to stand for 10 min at ambient temperature. For each E. coli sample, three dilutions in PBS (up to OD of 0.15, 0.30, and 0.60 at 540 nm) were used; the consumption of H2O2 by bacteria and the suspension optical density were directly proportional. Then, 20 μL of 6 N HCl and 500 μL of 245 mm solution of KI in H2O were added. Low pH provided the reaction of KI with H2O2 and inhibition of catalase activity. After 15 min of centrifugation at 600 g, the OD of supernatants was read at 492 nm 35. The difference between the OD at 492 nm in the control (5 mm H2O2) and in the test probes was expressed in μmoles of H2O2 and normalized to the dry‐weight value.
Fluorescence study of the hydrophobicity of cell envelope
The measurements of fluorescence (F) were performed according to Ref. 24 using a Hitachi F‐4000 fluorescence spectrophotometer and a 0.5 × 0.5 cm quartz cuvette. Fluorescence spectra of ANS (60 µm) and DMC (30 µm) were recorded at excitation wavelength of 360 nm and 420 nm, respectively. PBS or bacterial suspension was mixed with ANS or DMC before analysis; the final concentration of bacterial suspension was 0.8 ± 0.1 mg of DW·mL−1. The maximum fluorescence intensity was normalized to a dry‐weight basis.
To study the influence of the ionic strength of solution on the interaction between the fluorescent probes and bacteria, fluorescence intensity of the bound probe was measured twice in each sample: at the ionic strength of 0.157 m and 0.407 m. The ionic strength of the solution was increased by addition of 0.25 m NaCl to the mixture of the probes and bacterial cells in 0.01 m PBS. The ratio F(0.407)/F(0.157) was calculated, where F(0.157) and F(0.407) are the fluorescence intensity of the probe at the solution′s ionic strength of 0.157 m and 0.407 m, respectively 25. For negatively charged probe, ANS F(0.407)/F(0.157) >1 indicated negatively charged surface and F(0.407)/F(0.157) <1 suggested positively charged surface. DMC fluorescence was not expected to depend on the ionic strength of buffer solution since its molecule is uncharged.
Microscopic study
Morphological changes in neutrophils in blood incubated with bacteria at a 1:6 ratio of neutrophils to bacteria were examined by light microscopy according to Ref. 36. About 3 μL of E. coli suspension in PBS (OD at 540 = 2.5) was mixed with 17 μL of a healthy volunteer’s blood. Incubation continued for 1 h at 37 °C with periodical shaking, and then, the smears were prepared. The smears were stained by the Romanowsky–Giemsa technique, and microscopic examination of stained cells was carried out using a Motic B3 light microscope.
Statistics
The data are shown as a mean value ± standard deviation if not indicated otherwise. Correlations were calculated by the Pearson method; for nonlinear relations, Pearson’s approximation method was applied. The software statistica 6.0 was used for statistical analysis of data (Mann–Whitney U‐test or Student’s t‐test).
Results
CL analysis of neutrophil‐stimulating capacity and hydroperoxidase activity of Escherichia coli
Neutrophil CL was activated by bacterial suspensions in diluted blood of healthy volunteers, without leukocyte isolation 37 in order to imitate in vivo conditions. CL responses varied for different isolates from 2 to 70 mV (Fig. 1, Table 1). Isolates collected from the same patient differed in CL‐stimulating capacity by 1.5–5 times, and for some patients (M, V, and P), the difference was significant. The difference between isolates did not depend on blood sample used for analysis and was reproducible for at least three batches of each isolate. No significant difference in CL was detected between isolates from CD patients (23 isolates) vs healthy individuals (five isolates) (Fig. 1, Table 1).
Figure 1.
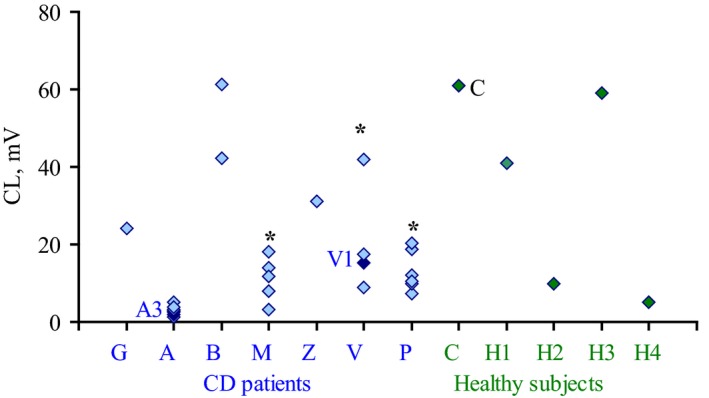
Chemiluminescence responses of neutrophils stimulated by E. coli isolates from CD (in blue) and healthy patients (in green) in whole blood of healthy volunteers. Each point represents CL maximum value for one E. coli isolate averaged for 3–5 independent experiments with 3–5 different blood samples. *Indicates patients with a significant difference between CL values of isolates within patient, P < 0.05 (Student’s t‐test).
Since bacterial toxins can influence neutrophil activity 38, isolates were tested for their ability to inhibit CL response induced in diluted blood. Phorbol ester (PMA) was added to the luminometer cuvette at the end of CL analysis, when the maximum value of CL induced by E. coli was achieved, and the second CL maximum was registered (Fig. 2). No significant effect of E. coli on subsequent PMA‐stimulated CL response was found, so the difference between the isolates could not result from any toxic effect of bacteria toward neutrophils.
Figure 2.
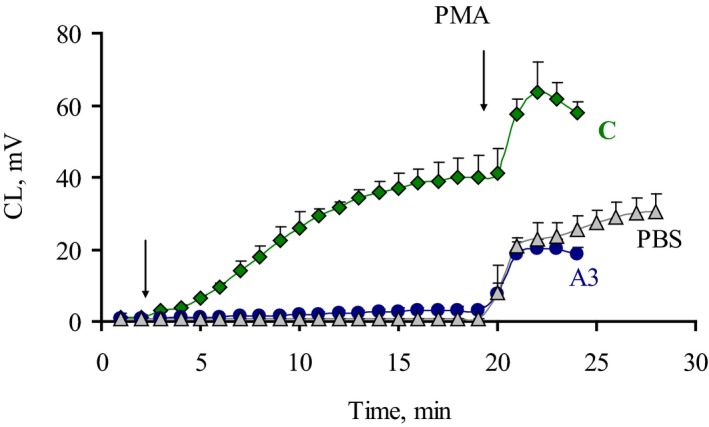
Time development and phorbol ester (PMA) stimulation effect on CL responses of blood neutrophils stimulated by E. coli. Typical time curves for PBS and isolates with low (A3) and high (C) CL values are shown, moments of stimulant addition indicated by arrows. Error bars represent SD.
Luminol‐enhanced CL stimulated in blood by E. coli isolates correlated with lucigenin‐enhanced CL, which is sensitive only to superoxide anion production (Fig. 3). Therefore, E. coli isolates that did not induce CL response in neutrophils probably managed it by disruption of superoxide produced by neutrophils.
Figure 3.
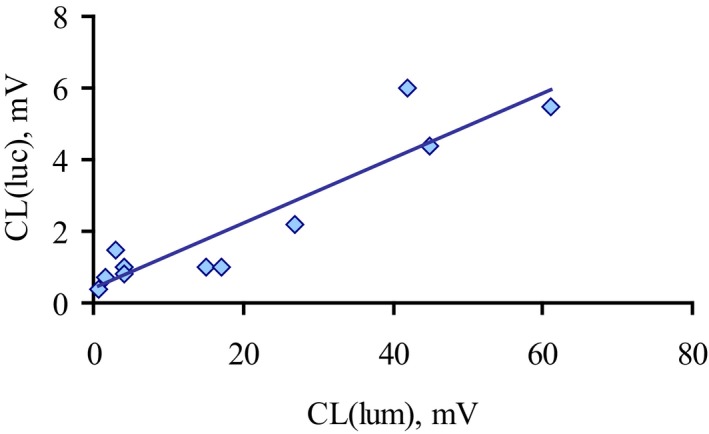
Correlation between luminol and lucigenin‐enhanced chemiluminescence of neutrophils stimulated by E. coli isolates from CD patients (n = 12). Luminol‐enhanced CL response of neutrophils in whole blood (CL (lum)) is plotted versus superoxide‐sensitive lucigenin‐enhanced CL (luc). Correlations were calculated by the Pearson method (r = 0.8, P < 0.05, n = 12).
In order to evaluate the ability of bacteria to protect themselves from ROS produced by neutrophils, catalase and peroxidase activities of E. coli isolates were assessed without cell destruction. Less than 10% of extracellular peroxidase activity was solubilized by Triton X‐100, indicating that the major part of enzymes was cell wall‐bound (tested for 12 E. coli isolates, data not shown).
We found that E. coli isolates with the highest peroxidase activity induced low CL response, while the isolates with significant CL‐stimulating capacity showed the lowest peroxidase activity, and this negative correlation was significant (P < 0.05; Fig. 4). There was no isolate combining high peroxidase‐ and CL‐stimulating activities. Bacterial catalase activity showed the same tendency, but correlation between catalase activity and CL values was not significant.
Figure 4.
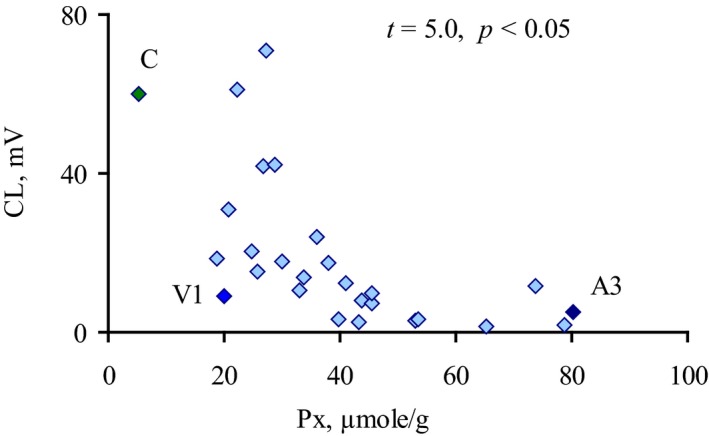
Correlation between CL response of whole blood neutrophils to E. coli isolates and their peroxidase activity. Peroxidase activity (Px) is given in µmol of oxidized o‐dianisidine per g of bacterial pellet dry weight. Significance (P) was quantified by the Pearson method for nonlinear correlation (t 0.5 = 2.05). Values for isolates C, A3, and V1 (analyzed in microscopic assay with neutrophils below) are indicated by black diamonds.
Since luminol‐enhanced CL depends on H2O2 and H2O2‐derived HOCl 9, the direct effect of peroxidase activity on CL should be evaluated. Exogenous peroxidase (HRP) was added to the CL probes with isolate C (with the lowest peroxidase activity of its own; Table 1) so that its activity was equal to the highest demonstrated by E. coli isolates in the current study (86.4 ± 3.3 µmol·g−1 dry weight; Table 1). This did not affect CL response of blood neutrophils, suggesting that the low CL response is not due to chemiluminescence inhibition by ROS eliminating.
Morphological analysis of neutrophil activation
Microscopic study of morphological alterations of stimulated neutrophils was performed to detect whether there are other signs of activation induced in neutrophils by E. coli isolates with low CL‐inducing activity. As such, two CD isolates with low CL activity were selected—A3 and V1. Isolate C (from healthy patient) was selected as a control isolate with the one of the highest neutrophil‐stimulating activities (regarding CL response induction).
Figure 5 demonstrates that all isolates induced signs of neutrophil activation: swelling of neutrophils nuclei, formation of vacuoles, and cell aggregation (Fig. 5A).
Figure 5.
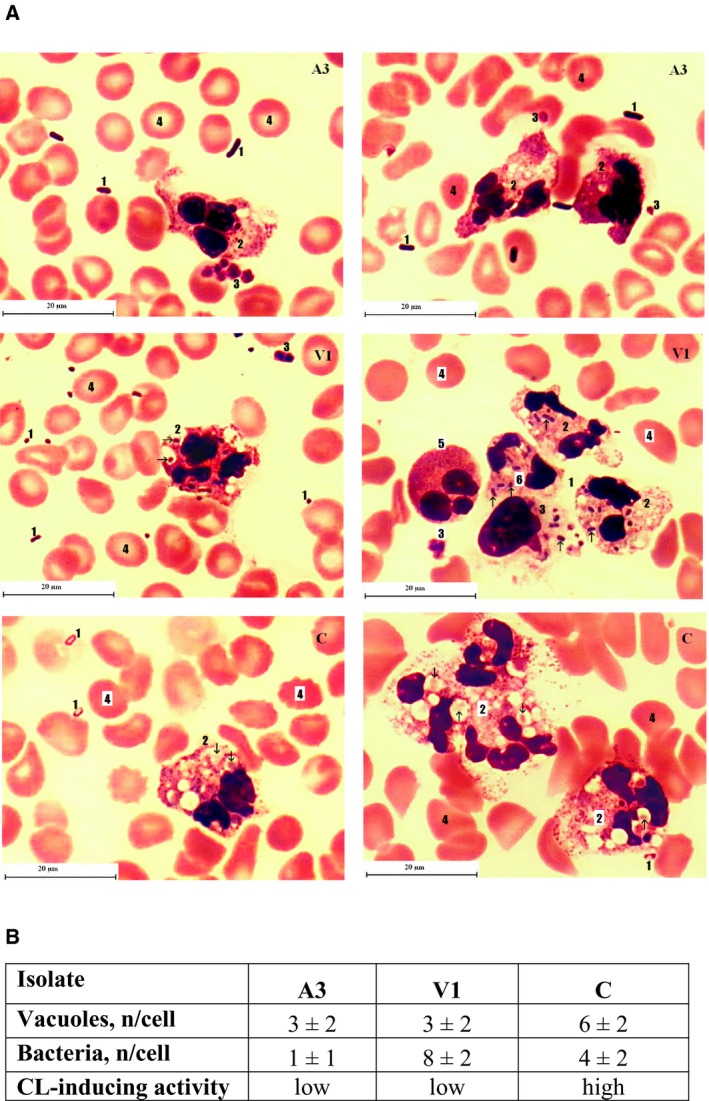
Effect of E. coli isolates with diverse CL‐stimulating activity on the morphological features of neutrophils. Whole blood samples were incubated with E. coli isolates that have induced low (A3 and V1) and high (C) CL response. The Romanowsky–Giemsa staining was used to visualize the details. At least 50 microscope fields of view per isolate were evaluated for picture and calculations. (A) Typical picture of activated neutrophil for each isolate: 1—E. coli cells; 2—neutrophils/neutrophil aggregates containing vacuoles (arrows indicate the captured bacterial cells); 3—platelets, 4—erythrocytes; 5—eosinophil; and 6—monocyte with captured bacteria. The length of the scale bars is 20 μm. (B) Parameters of phagocytic activity of neutrophils stimulated by E. coli isolates. Mean values are given ± SD.
Isolate with the lowest CL‐stimulating activity (A3) was the least susceptible to the phagocytosis (Fig. 5B), even though the neutrophils incubated with it showed signs of activation. Only the cells of isolate with the highest CL‐stimulating activity (C) showed signs of degradation in phagolysosome (the engulfed bacterial cells differ in form and staining from the free ones). This isolate also induced higher number of vacuoles formed within neutrophil cells. As for the isolate V, the vacuoles with bacteria were rather phagosomes, smaller than for isolate C, and with no signs of bacterial digestion.
Fluorescent analysis of hydrophobicity of Escherichia coli cell surface
As shown by the experiments above, the difference in neutrophil capturing capacity toward E. coli could not be entirely attributed to the activity of bacterial hydrogenases or to the direct inhibiting effect of bacteria on neutrophils. Another factor contributing in that is the possible difference in the cell surface properties of various isolates.
Escherichia coli cell surface properties were studied by fluorescent probes ANS (negatively charged amphiphilic) and DMC (neutral hydrophobic). Addition of bacterial suspensions significantly increased probes’ fluorescence intensity: from 2.4 ± 0.3 to 16–64 a.u. for ANS and from 1.4 ± 0.1 to 2 – 110 a.u. for DMC. The averaged values for isolates within patients are given in Table 1.
Binding of the probes to the bacterial surface and transition from an aquatic to a hydrophobic environment were proved by the blue shift of the fluorescence maxima 39, 40, 41 registered for ANS (from 512 ± 1 to 490 ± 2nm) and for DMC (from 522 ± 1 nm to 499 ± 1 nm) in bacterial suspensions.
Fluorescence intensity of both probes negatively correlated with CL values: r = −0.46 in the experiments with ANS (data not shown) and r = −0.63 in the experiments with DMC, where the correlation was significant at P < 0.05 (Fig. 6).
Figure 6.
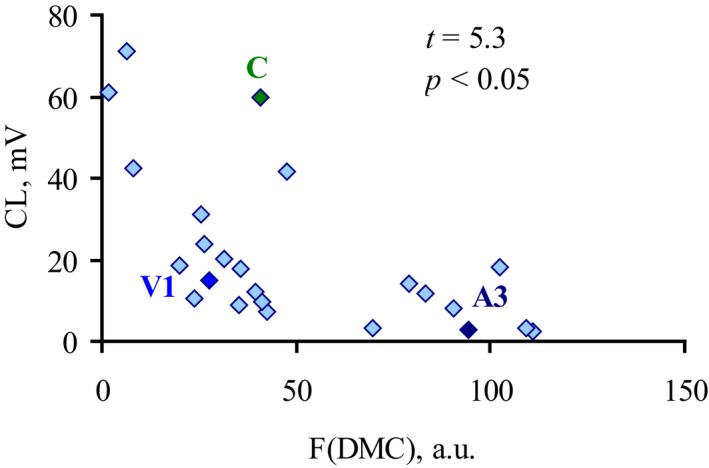
Correlation between CL response of whole blood neutrophils induced by E. coli isolates and their DMC fluorescence intensity reflecting bacterial surface hydrophobicity. DMC hydrophobic fluorescent probe was added to 24 isolate bacterial suspensions (Table 1), and F(DMC) was recorded at 499 nm. Significance (P) was quantified by the Pearson method for nonlinear correlation (t 0.5 = 2.05).
To establish the role of electrostatic forces in the interaction of the probes with E. coli, the effect of the increase in ionic strength was studied. We detected approximately 10% elevation of ANS fluorescence intensity, F(0.407)/F(0.157) = 1.07 ± 0.08. Thus, the contribution of electrostatic forces to the binding of the probe to E. coli was not significant. Fluorescence values in experiments with DMC were not influenced by ionic forces and therefore resulted from hydrophobic binding of the probe to the bacterial cell surface.
Analysis of the adhesive and invasive ability of Escherichia coli isolates
Analysis of the ability of E. coli isolates to adhere and invade CaCo cells was performed for eight isolates from seven CD patients that demonstrated high, low, and intermediate values of neutrophil‐stimulating peroxidase and catalase activities (Table 1), two isolates from healthy individuals, and one laboratory strain MG1655 (Table 2). Higher adhesion ability (5–30 times more than the laboratory strain) was demonstrated by four isolates (V2, V4, M4, and G). Invasion was demonstrated by three isolates (V2, M4, and P2). Therefore, adhesive–invasive activity was observed for two isolates (V2 and M4).
Table 2.
Adhesion and invasion assay
| V2 | V4 | M4 | G | P2 | A1 | B1 | H1 | C | MG1655 | |
|---|---|---|---|---|---|---|---|---|---|---|
| CFU per agar plate in control ×104 | 19.5 ± 1.92 | 22.87 ± 2.23 | 18.13 ± 2.03 | 13.25 ± 1.91 | 55.67 ± 4.59 | 74.67 ± 4.03 | 76.17 ± 4.07 | 46.83 ± 4.07 | 50 ± 4 | 18.875 ± 2.53 |
| CFU per agar plate after adhesion. ×103 | 9.75 ± 1.83 | 18.37 ± 6.57 | 59.33 ± 9.09 | 11 ± 0.93 | 3.67 ± 0.82 | 3.5 ± 0.55 | 8.83 ± 1.47 | 1.5 ± 0.55 | 4.33 ± 0.82 | 1.9875 ± 0.23 |
| CFU per agar plate after gentamicin treatment | 12.5 ± 4.63 | 0 | 47.5 ± 11.65 | 0 | 13.33 ± 5.16 | 0 | 41.67 ± 9.83 | 0 | 0 | 0 |
| Number of adhered–invaded bacteria per CaCo cell | 0.02 | 0.04 | 0.12 | 0.02 | 0.007 | 0.007 | 0.018 | 0.003 | 0.009 | 0.004 |
| % of adhered–invaded bacteria in bacterial suspension | 5 | 8.03 | 32.74 | 8.3 | 0.66 | 0.47 | 1.16 | 0.32 | 0.87 | 1.05 |
| % of invaded bacteria | 0.13 | 0 | 0.08 | 0 | 0.36 | 0 | 0.47 | 0 | 0 | 0 |
Values are given ± SD (standard deviation).
No correlation was observed between adhesive and invasive ability of E. coli isolates and the intensity of neutrophil stimulation (Tables 1 and 2). Isolates with the lowest CL values (A1) and with the highest (C) demonstrated no adhesion and invasion activity. Isolates with maximum observed adhesive–invasive isolates demonstrated both high (V2) and low CL values (M4).
Escherichia coli isolates fall into three groups
The isolates could be divided into three groups on the basis of their CL‐stimulating capacity: group 1 with CL values of 1−10 mV, group 2 with CL values of 11–30 mV, and group 3 with CL of more than 31 mV (Table 1).
As Fig. 7 demonstrates, cell surface hydrophobicity (F(DMC)) and peroxidase activity (Px) significantly differed between these groups.
Figure 7.
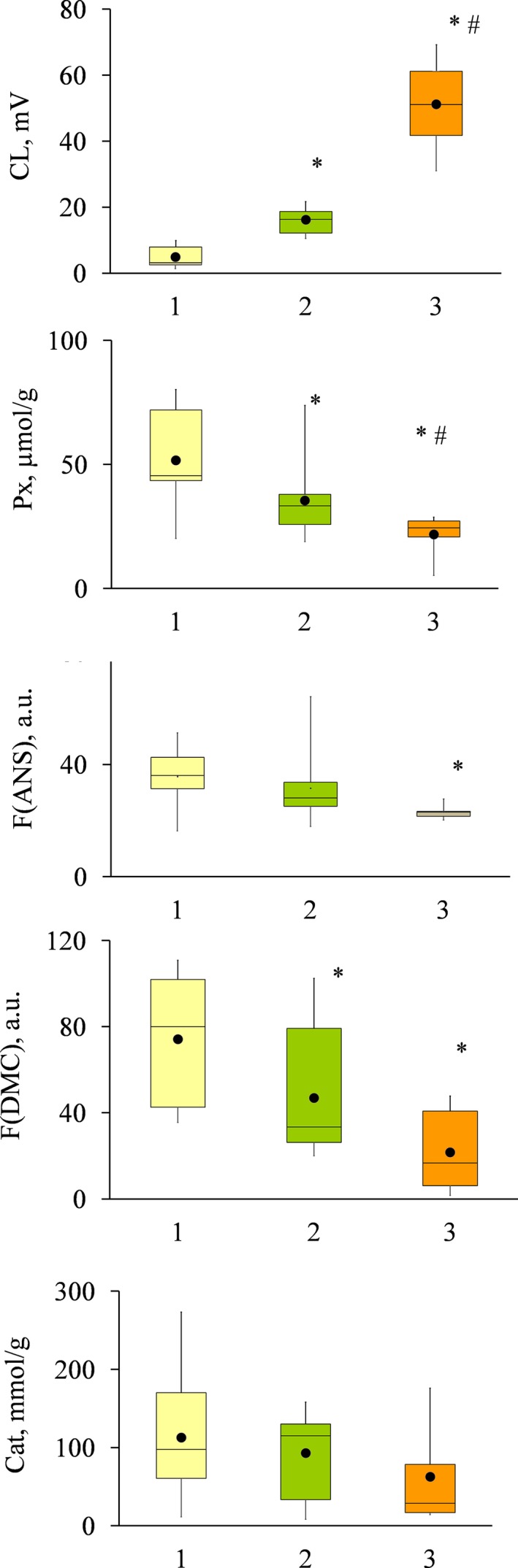
Three groups of E. coli isolates. E. coli isolates were arranged in three groups according to the CL‐inducing activity (from the lowest to the highest, 1, 2, and 3). The following parameters were compared within/between the groups: extracellular peroxidase activity (Px, μmol·g−1 DW), cell surface hydrophobicity defined by probe fluorescence (F(ANS), a. u., and F(DMC), a.u.), and extracellular catalase activity (catalase, nmol·g−1). Significant difference is indicated: *—in comparison with group 1; #—in comparison with group 2 (P < 0.05, Mann–Whitney test).
These data suggest the existence of three different E. coli phenotypes differing in their ability to evade ROS production by neutrophils, extracellular peroxidase activity, and cell surface hydrophobicity. Escherichia coli isolates of the same origin can belong to different phenotypes (patients ‘M’, ‘V’, and ‘P’), or to one (patient ‘A’ and ‘B’).
Parameters such as the source of isolate, the severity of disease, inflammation localization, and sex/age of patients and isolates phylogroup showed no correlation with the intensity of neutrophil stimulation or bacterial hydroperoxidase activity (Table 1, File S1).
Discussion
The role of E. coli in the acute inflammation during Crohn’s disease is still disputable 42, 43, 44. However, multiple observations of E. coli intensive growth in the afflicted intestine suggest that bacteria proliferating under such conditions have to develop some mechanisms of resistance to ROS/RHS or avoiding their formation. The aim of the current study was to evaluate various isolates of E. coli in regard to their interaction with inflammation agents, such as neutrophils and ROS.
We have observed high variability in these parameters in E. coli from CD patients. In particular, a phenotype was detected combining low neutrophil activation with high ROS utilization activity. About 39% of the total number of E. coli isolates from CD patients (9 isolates of 23) induced CL response below 10 mV, and four of seven CD patients involved into the study had in their intestine at least one isolate with low CL‐stimulating activity. At the same time, 17.4% of CD isolates (4 out of 23) induced high CL responses and showed much lower peroxidase activity.
The low CL‐inducing isolates were not completely ‘invisible’ to neutrophils, as was shown by microscopic study in the experiments with the isolates A3 and V1.
It is necessary to mention that low CL values were indeed correct indications of decreased neutrophil activation, and not a result of bacterial utilization of ROS produced by activated neutrophils. This was confirmed by the fact that luminol CL values correlated with lucigenin CL independent of H2O2 production by neutrophils, and was insensitive to exogenous peroxidase (HRP 45). Presumably, isolates with low CL‐stimulating capacity were able to avoid the induction of ROS/RHS production at all during neutrophil activation.
Previously published researches suggested that bacterial cell surface hydrophobicity could influence the scenario of neutrophil activation, and bacteria with high hydrophobicity should have the most stimulating capacity 17, 18. Unexpectedly, in the studied E. coli isolates, the higher hydrophobicity corresponded to the lower CL values and the higher peroxidase activity.
Alignment of E. coli genome sequences from different intestine parts of one patient performed earlier 29 revealed only minor heterogeneity and some deletions (usually a deletion of one of the smaller contigs, probably a plasmid). At the same time, sequences of genes identified as encoding catalases and peroxidases of isolates from one patient were identical, since none of the found polymorphisms was located in those genes. Therefore, one may suggest that the difference in phenotype of genetically close isolates from one patient observed in the current study (patients V, M, and P) could be due to the mobile elements of genome that could be achieved via horizontal transfer.
Hence, we report that E. coli isolates from CD patients are rather variable regarding the interaction with inflammation agents. One of the phenotypes observed combines the ability for low neutrophil activation with comparatively high ability to utilize extracellular ROS and increased cell surface hydrophobicity.
Author contributions
MAM, TVV, JPB, LJB, SAG, AAG, NVS, and GED performed the laboratory section of the study. EVM analyzed the data. PLS, AIP, and NAF performed the clinic part of the study. EVM, DVR, OMP, OVP, and VMG were involved in study designing, writing, and editing of the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
File S1. Parameters of E. coli isolates are presented on 3D plot: CL (Chemiluminescent responce intensity, lum), Cat (extracellular catalase activity, nmol H2O2/ g dry weight) and Px (peroxidase activity, µmol of oxidized o‐dianisidine per g of bacterial pellet dry weight).
Acknowledgements
The authors thank Professor Bettye Chambers for her assistance in the proofreading of the manuscript. This study was supported by the Russian Science Foundation (Grant No 16‐15‐00258). The funding body had no role in the design of the study and collection, in analysis and interpretation of data, and in writing the manuscript.
References
- 1. Conte MP, Schippa S, Zamboni I, Penta M, Chiarini F, Seganti L, Osborn J, Falconieri P, Borrelli O and Cucchiara S (2006) Gut‐Associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 55, 1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gevers D, Kugathasan S, Denson LA, Vazquez‐Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M et al (2014) The treatment‐naive microbiome in new‐onset Crohn’s disease. Cell Host Microbe 15, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang Y and Jobin C (2014) Microbial imbalance and intestinal pathologies: connections and contributions. Dis Mod Mech 7, 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carrière J, Darfeuille‐Michaud A and Nguyen HT (2014) Infectious etiopathogenesis of Crohn’s disease. World J Gastroenterol 20, 12102–12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Demirel I, Kinnunen A, Önnberg A, Söderquist B and Persson K (2013) Comparison of host response mechanisms evoked by extended spectrum beta lactamase (ESBL)‐ and non‐ESBL‐producing uropathogenic E. coli . BMC Microbiol 31, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mouzaoui S, Djerdjouri B, Makhezer N, Kroviarski Y, El‐Benna J and Dang PM (2014) Tumor necrosis factor‐α‐induced colitis increases NADH oxidase 1 expression, oxidative stress, and neutrophil recruitment in the colon: preventive effect of apocynin. Mediat Inflamm 2014, 312484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alzoghaibi M (2013) Concepts of oxidative stress and antioxidant defense in Crohn’s disease. World J Gastroenterol 19, 6540–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kruidenier L, Kuiper I, Lamers CB and Verspaget HW (2003) Intestinal oxidative damage in inflammatory bowel disease: semi‐quantification, localization, and association with mucosal antioxidants. J Pathol 201, 28–36. [DOI] [PubMed] [Google Scholar]
- 9. Vladimirov YA and Proskurnina EV (2009) Free radicals and cell chemiluminescence. Biochemistry (Moscow) 74, 1545–1566. [DOI] [PubMed] [Google Scholar]
- 10. Visick JE and Clarke S (1997) RpoS‐ and OxyR‐independent induction of HPI catalase at stationary phase in Escherichia coli and identification of rpoS mutations in common laboratory strains. J Bacteriol 179, 4158–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shiratsuchi A, Shimamoto N, Nitta M, Tuan TQ, Firdausi A, Gawasawa M, Yamamoto K, Ishihama A and Nakanishi Y (2014) Role for s38 in prolonged survival of Escherichia coli in Drosophila melanogaster . J Immunol 192, 666–675. [DOI] [PubMed] [Google Scholar]
- 12. Farr SB and Kogoma T (1991) Oxidative stress responses in Escherichia coli and Salmonella typhimurium . Microbiol Rev 55, 561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loewen PC, Switala J and Triggs‐Raine BL (1985) Catalases HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys 243, 144–149. [DOI] [PubMed] [Google Scholar]
- 14. Claiborne A and Fridovich I (1979) Purification of the o‐dianisidine peroxidase from Escherichia coli B. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J Biol Chem 254, 4245–4252. [PubMed] [Google Scholar]
- 15. Loewen P (1996) Probing the structure of catalase HPII of Escherichia coli – a review. Gene 179, 39–44. [DOI] [PubMed] [Google Scholar]
- 16. Alteri CJ, Lindner JR, Reiss DJ, Smith SN and Mobley HL (2011) The broadly conserved regulator PhoP links pathogen virulence and membrane potential in Escherichia coli . Mol Microbiol 82, 145–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Magnusson KE, Davies J, Grundström T, Kihlström E and Normark S (1980) Surface charge and hydrophobicity of Salmonella, E. coli, Gonococci in relation to their tendency to associate with animal cells. Scand J Infect Dis Suppl 24, 135–140. [PubMed] [Google Scholar]
- 18. Williams P, Lambert PA, Haigh CG and Brown MR (1986) The influence of the O and K antigens of Klebsiella aerogenes on surface hydrophobicity and susceptibility to phagocytosis and antimicrobial agents. J Med Microbiol 21, 125–132. [DOI] [PubMed] [Google Scholar]
- 19. Lock R, Dahlgren C, Linden M, Stendahl O, Svensbergh A and Ohman L (1990) Neutrophil killing of two type 1 fimbria‐bearing Escherichia coli strains: dependence on respiratory burst activation. Infect Immun 58, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blomgran R, Zheng L and Stendahl O (2004) Uropathogenic Escherichia coli triggers oxygen‐dependent apoptosis in human neutrophils through the cooperative effect of type 1 fimbriae and lipopolysaccharide. Infect Immun 72, 4570–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dreux N, Denizot J, Martinez‐Medina M, Mellmann A, Billig M, Kisiela D, Chattopadhyay S, Sokurenko E, Neut C, Gower‐Rousseau C et al (2013) Point mutations in FimH adhesin of Crohn’s disease‐associated adherent‐invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog 9, e1003141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park BS and Lee JO (2013) Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 45, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nivea‐Gomez D and Gennis RB (1997) Affinity of intact Escherichia coli for hydrophobic membrane probes is a function of the physiological state of the cells. Proc Natl Acad Sci USA 74, 1811–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phillips SK and Cramer WA (1973) Properties of the fluorescence probe response associated with the transmission mechanism of colicin E1. Biochemistry 12, 1170–1176. [DOI] [PubMed] [Google Scholar]
- 25. Dobretsov GE, Gularyan SK, Panasenko OM, Melnichenko AA and Orekhov AN (2006) The charge of the surface layer of multiply modified low‐density lipoproteins. Biochem (Moscow), Series A Memb Cell Biol 23, 420–425. [Google Scholar]
- 26. Mikhalchik E, Balabushevich N, Vakhrusheva T, Sokolov A, Baykova J, Rakitina D, Shcherbakov P, Gusev S, Gusev A, Kharaeva Z et al, (2019) Mucin adsorbed by E. coli can affect neutrophil activation in vitro. FEBS Open Bio 10, 180–196.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beers RF and Sizer IW (1952) A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem 195, 133–140. [PubMed] [Google Scholar]
- 28. Porebski S, Bailey GL and Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15, 8–15. [Google Scholar]
- 29. Rakitina DV, Manolov AI, Kanygina AV, Garushyants SK, Baikova JP, Alexeev DG, Ladygina VG, Kostryukova ES, Larin AK, Semashko TA et al (2017) Genome analysis of E. coli isolated from Crohn's disease patients. BMC Genom 18, 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Silverberg MS, Satsangi J, Ahmad T, Arnott ID, Bernstein CN, Brant SR, Caprilli R, Colombel J‐F, Gasche C, Geboes K et al (2005) Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a working party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol 19 (supplA), 5–36. [DOI] [PubMed] [Google Scholar]
- 31. Best WR, Becktel JM, Singleton JW and Kern F Jr (1976) Development of a Crohn's disease activity index. Nat Cooperat Crohn's Dis Study Gastroenterol 70, 439–444. [PubMed] [Google Scholar]
- 32. Kondratyeva K, Wollman A, Gerlitz G and Navon‐Venezia S (2017) Adhesion and invasion to epithelial cells and motility of extended‐spectrum b‐lactamase‐producing Escherichia coli reveal ST131 superiority: a comparative in vitro study of extraintestinal pathogenic E. coli lineages. J Med Microbiol 66, 1350–1357. [DOI] [PubMed] [Google Scholar]
- 33. Easmon CSF, Cole PJ, Williams AJ and Hastings M (1980) The measurement of opsonic and phagocytic function by Luminol‐dependent chemiluminescence. Immunology 41, 67–74. [PMC free article] [PubMed] [Google Scholar]
- 34. Andrews PC, Parens C and Krinsky NI (1984) Comparison of myeloperoxidase and hemi‐myeloperoxidase with respect to catalysis, regulation and bactericidal activity. Arch Biochem Biophys 228, 439–442. [DOI] [PubMed] [Google Scholar]
- 35. Bukharin OV, Cherkasov SV, Sgibnev AV, Zabirova TM and Ivanov YB (2000) Effects of microbial metabolites on catalase activity and growth of Staphylococcus aureus 6538P. Bull Exp Biol Med 130, 679–681. [DOI] [PubMed] [Google Scholar]
- 36. Brest P, Bétis F, Çuburu N, Selva E, Herrant M, Servin A, Auberger P and Hofman P (2004) Increased rate of apoptosis and diminished phagocytic ability of human neutrophils infected with Afa/Dr diffusely adhering Escherichia coli strains. Infect Immun 72, 5741–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lindena J, Burkhardt H and Dwenger A (1987) Mechanisms of non‐opsonized zymosan‐induced and luminol‐enhanced chemiluminescence in whole blood and isolated phagocytes. J Clin Chem Clin Biochem 11, 765–778. [DOI] [PubMed] [Google Scholar]
- 38. Russo TA, Davidson BA, Genagon SA, Warholic NM, Macdonald U, Pawlicki PD, Beanan JM, Olson R, Holm BA and Knight PR 3rd (2005) E. coli virulence factor hemolysin induces neutrophil apoptosis and necrosis/lysis in vitro and necrosis/lysis and lung injury in a rat pneumonia model. Am J Physiol Lung Cell Mol Physiol 289, L207–L216. [DOI] [PubMed] [Google Scholar]
- 39. Stryer L (1965) The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non‐polar binding sites. J Mol Biol 13, 482–495. [DOI] [PubMed] [Google Scholar]
- 40. Dobretsov GE, Petrov VA, Mishijev VE, Klebanov GI and Vladimirov YuA (1977) 4‐Dimethylaminochalcone and 3‐methoxybenzanthrone as fluorescent probes to study biomembranes. I. Spectral characteristics. Stud Biophys 65, 91–98. [Google Scholar]
- 41. Bakhshiev NG, Gularyan SK, Dobretsov GE, Kirillova AYu and Svetlichnyi VYu (2006) Solvatochromism and solvatofluorochromism of the intramolecular charge transfer band of 4‐dimethylaminochalcone in the electronic spectra of its solutions. Opt Spectrosc 100, 700–708. [Google Scholar]
- 42. Strober W (2011) Adherent‐invasive E. coli in Crohn disease: bacterial “agent provocateur”. J Clin Invest 121, 841–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Agus A, Massier S, Darfeuille‐Michaud A, Billard E and Barnich N (2014) Understanding host‐adherent‐invasive Escherichia coli interaction in Crohn's disease: opening up new therapeutic strategies. Biomed Res Int 2014, 567929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martinez‐Medina M and Garcia‐Gil LJ (2014) Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity. World J Gastrointest Pathophysiol 5, 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kopprasch S, Kohl M and Graessler J (1998) Different effects of exogenous horseradish peroxidase on luminol‐amplified chemiluminescence induced by soluble and particulate stimuli in human neutrophils. J Biolumin Chemilumin 13, 267–271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1. Parameters of E. coli isolates are presented on 3D plot: CL (Chemiluminescent responce intensity, lum), Cat (extracellular catalase activity, nmol H2O2/ g dry weight) and Px (peroxidase activity, µmol of oxidized o‐dianisidine per g of bacterial pellet dry weight).


