Abstract
Post‐transcriptional regulation of cytokine production is crucial to ensure appropriate immune responses. We previously demonstrated that poly‐rC‐binding protein‐1 (PCBP1) can act as a trans‐acting factor to stabilize transcripts encoding sortilin, which mediates cytokine trafficking. Here, we report that PCBP2, which strongly resembles PCBP1, can stabilize sortilin transcripts in macrophages using the same mechanism employed by PCBP1. PCBP2 recognized the C‐rich element in the 3′ UTR of sortilin mRNA, and PCBP2 knockdown decreased sortilin transcripts, indicating that PCBP2 stabilizes sortilin mRNA by binding to its 3′ UTR. Zn2+ reversibly inhibited the nucleotide binding ability of PCBP2 in vitro. These findings suggest that both PCBP2 and PCBP1 may control the stability of sortilin transcripts by sensing intracellular Zn2+ levels in immune cells.
Keywords: cytokines, innate immunity, post‐transcriptional regulation, RNA‐binding proteins, Sort1, zinc
We analyzed the function of poly‐rC‐binding protein‐2 (PCBP2), a paralog of PCBP1, in stabilizing the mRNA of sortilin, a cytokine trafficking mediator. PCBP2 binds to C‐rich elements in the 3′ UTR of sortilin mRNA; Zn2+ interferes with this interaction, suggesting that both PCBP1 and PCBP2 may regulate the stability of sortilin mRNA by sensing the intracellular Zn2+ levels.
Abbreviations
- cDC
conventional dendritic cell
- CRE
C‐rich element
- PCBP
Poly‐rC‐binding protein
- pDC
plasmacytoid dendritic cell
- PRR
pattern‐recognition receptor
- RBP
RNA‐binding protein
- TLR
Toll‐like receptor
Recognition of pathogen‐associated molecular patterns by pattern‐recognition receptors (PRRs) and the subsequent production of proinflammatory cytokines play an important role in the innate immune system 1. Excessive immune responses are characterized by the overproduction of cytokines, which can cause tissue damage by chronic inflammation and lead to autoimmune disease. Therefore, tight control of the signaling pathways regulating cytokine production is important for appropriate immune responses.
RNA‐binding proteins (RBPs) contribute to the post‐transcriptional regulation of immunity‐related mRNA 2; this regulation is important for the repression of excessive immune responses 3. Specific cis‐elements for post‐transcriptional regulation identified in the 3′UTRs of many immunity‐related mRNA contribute to their degradation or stabilization; moreover, many different RBPs, as trans‐acting factors involved in this regulation, have been identified 2, 4. The ARE‐binding proteins tristetraprolin, AUF1, and HuR recognize and bind to mRNA containing AU‐rich elements, which results in the control of target mRNA stability. Roquin and Regnase‐1 bind to stem‐loop structures followed by destabilization of target mRNA 2, 3, 4. The functions of these RBPs are regulated by a number of kinase pathways, resulting in the coordinated expression of their target mRNA 2, 3.
Poly‐rC‐binding proteins (PCBPs) are a group of multifunctional RBPs. They recognize C‐rich elements (CREs) of single‐stranded RNA and have diverse functions affecting RNA processing, translation, and stability through their binding to CREs 5, 6. A recent study demonstrated that Pcbp1‐deficient mouse embryos die at the peri‐implantation stage 7, indicating an essential role of PCBP1 in mouse embryonic development. PCBPs are also important for iron metabolism, acting as iron chaperones 8, 9, 10, 11, 12, 13, 14. Moreover, PCBP1 mediates proinflammatory cytokine production via stabilization of mRNA in iron‐promoted CD4+ T‐cell pathogenicity 15. In microRNA processing, PCBP2 functions by sensing cytosolic iron status 16. These studies suggest that PCBP1 and PCBP2 link RNA stability and processing to iron metabolism.
We previously demonstrated that PCBP1 is involved in the regulation of sortilin, which mediates cytokine trafficking via stabilization of its mRNA 17. Interestingly, the binding of Zn2+ to PCBP1 reversibly interfered with the nucleotide binding activity of PCBP1 in vitro 17. Two major isoforms, PCBP1 and PCBP2, are ubiquitously expressed 18. They are highly homologous; however, little is known about the involvement of PCBP2 in the post‐transcriptional regulation of sortilin or whether Zn2+ affects the nucleotide binding activity of PCBP1 paralogs. In this study, we measured the protein expression levels of PCBP1 and PCBP2 in multiple types of immune cells and examined the involvement of PCBP2 in the stabilization of sortilin mRNA. We describe the interaction of PCBP2 with the CRE of the 3′ UTR of sortilin mRNA, which leads to the stabilization of the sortilin mRNA. Zn2+ reversibly interferes with PCBP2 nucleotide binding to the CRE. We previously observed that loading primary macrophages with Zn2+ leads to a reduction of sortilin mRNA 17, suggesting that PCBP2, like PCBP1, may control sortilin mRNA stability by sensing intracellular Zn2+ levels in immune cells. Our observations provide insights into the roles of PCBP1 and PCBP2 in mRNA regulation, metal homeostasis, and innate immunity via the post‐transcriptional control of sortilin expression.
Results and Discussion
PCBP1 and PCBP2 are expressed in multiple types of immune cells
To investigate the expression levels of PCBP1 and PCBP2 proteins in murine immune cells, we carried out immunoblot analysis, indicating that PCBP1 and PCBP2 were expressed at high levels in CD4+ and CD8+ splenocytes (Fig. 1A). These proteins were moderately expressed in B220+ splenocytes; peritoneal macrophages; and bone marrow‐derived conventional dendritic cells (cDCs), plasmacytoid DCs (pDCs), and eosinophils (Fig. 1A). PCBP1 was modestly expressed in bone marrow‐derived mast cells and basophils; in contrast, there was no visible PCBP2 expression in mast cells or basophils. We detected endogenous PCBP2 as a doublet band. A doublet band has been reported using other antibodies, and immunoblot analysis with mouse PCBP2 knockout embryos has shown complete loss of the doublet band 7. This doublet band was depleted in PCBP2 knockdown cells with siRNA (Fig. 3A), indicating that the two detected bands were derived from endogenous PCBP2 rather than being nonspecific bands. Various Pcbp2 transcript variants registered in the database supports this view. Interestingly, we previously found lowered expression levels of sortilin in mast cells 17, correlating with these PCBP1 and PCBP2 expression levels, suggesting that these proteins are not important for cytokine secretion from mast cells. Quantitative real‐time PCR (qRT‐PCR) with tissue samples showed that Pcbp1 mRNA was expressed at high levels in the brain, lungs, and spleen, with lower levels in the heart and liver (Fig. 1B). Pcbp2 mRNA was predominantly expressed in the lung and spleen, while very similar Pcbp2 mRNA expression levels were observed in the brain, heart, and liver (Fig. 1C). Overall, the expression patterns of Pcbp1 and Pcbp2 are similar to that of sortilin as we previously reported 17.
Figure 1.
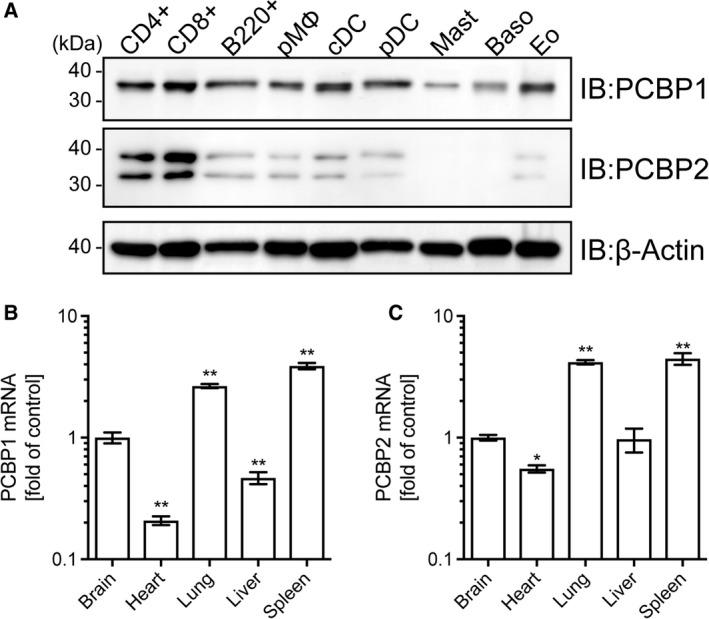
Expression profiles of PCBP1 and PCBP2 in immune cells and tissues. (A) Expression of PCBP1 and PCBP2 protein. β‐Actin was used as an internal control. CD4+, splenic CD4+ cells; CD8+, splenic CD8+ cells; B220+, splenic B220+ cells; Baso, bone marrow‐derived basophils; Eo, bone marrow‐derived eosinophils; Mast, bone marrow‐derived mast cells; pMØ, peritoneal macrophages. IB denotes the antibody used for immunoblotting. (B, C) Expression of Pcbp1 (B) and Pcbp2 (C) mRNA in mouse organs quantified by qRT‐PCR, normalized to Gapdh expression. Data are the means ± SD (n = 3). *P < 0.05 **P < 0.01 (two‐way ANOVA).
Figure 3.
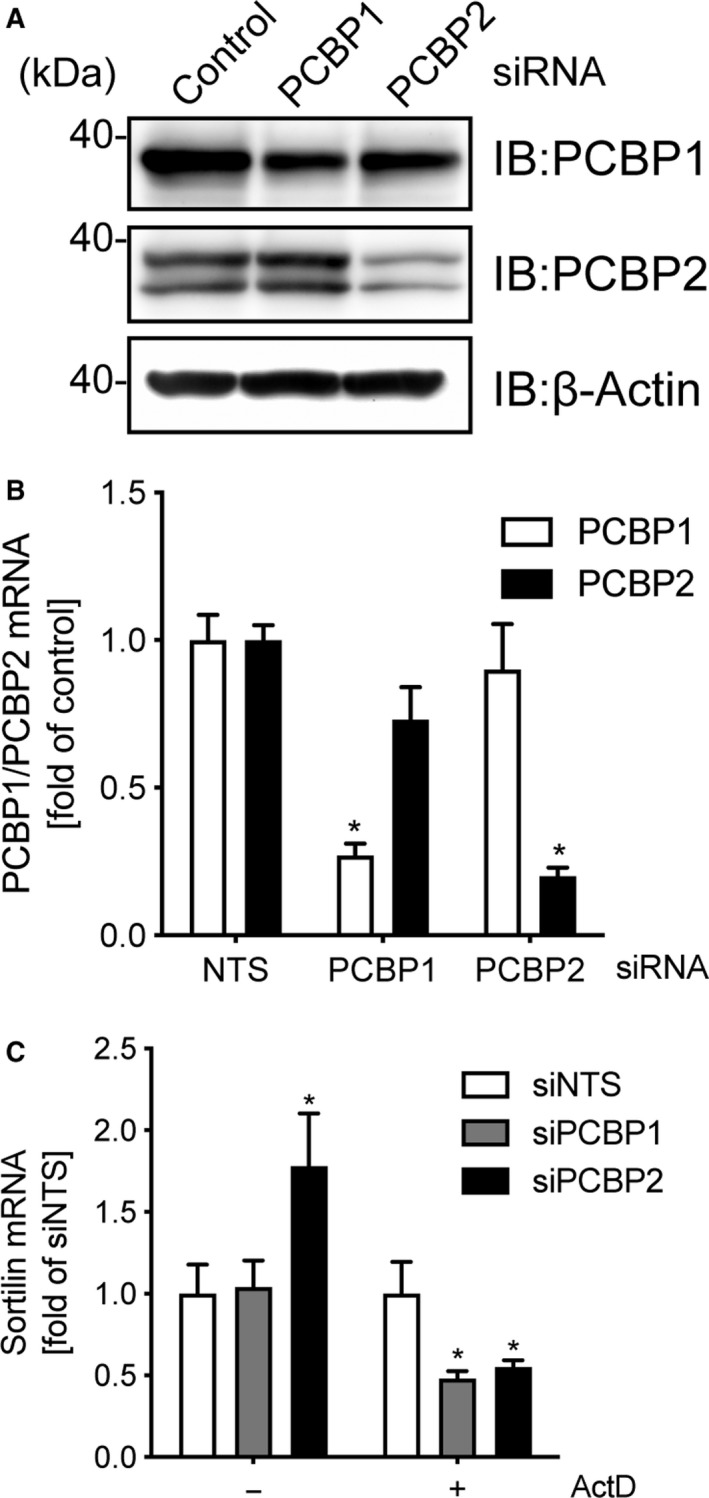
PCBP2 depletion affects sortilin mRNA levels. (A, B) Depletion of PCBP1 and PCBP2 in RAW264.7 cells. Cells were transfected with siRNA targeting Pcbp1, Pcbp2, or a control nontargeting siRNA, then harvested 48 h after transfection. Cell lysates were subjected to immunoblotting (A) with the indicated antibodies. Total RNA were subjected to qRT‐PCR (B) to quantify Pcbp1 and Pcbp2 mRNA. Data are the means ± SD (n = 3). *P < 0.01 (two‐way ANOVA). (C) Destabilization of sortilin mRNA by PCBP1 depletion in cells. RAW264.7 cells were transfected with control (siNTS), PCBP1 (siP1), or PCBP2 (siP2) siRNA for 48 h and, then, treated with 1 µg·mL−1 ActD for 9 h. Total RNA were subjected to qRT‐PCR to quantify sortilin mRNA. Data are the means ± SD (n = 3). *P < 0.01 (two‐way ANOVA).
PCBP2 stabilizes sortilin mRNA
We next investigated the involvement of PCBP2 in the post‐transcriptional regulation of sortilin expression. We carried out an RNA electromobility shift assaywith macrophage lysates and an RNA probe containing the sortilin CREs as previously described 17. Major doublet band was found in the negative control sample (lane N in Fig. 2A). Interestingly, the observed upper or lower band was mostly supershifted following incubation with anti‐PCBP1 or anti‐PCBP2, respectively, and co‐incubation with both antibodies supershifted these doublet bands. Meanwhile, the control incubated with normal IgG did not show supershifted bands (Fig. 2A). These observations indicate that the upper or lower protein–RNA complex mainly contains PCBP1 or PCBP2, respectively, suggesting both PCBP1 and PCBP1 can independently bind CREs. We observed, by RNA immunoprecipitation (RIP), that CRE‐containing fragments in the 3′ UTR of sortilin mRNA were amplified in the samples immunoprecipitated with anti‐PCBP2, as were those immunoprecipitated with anti‐PCBP1 (Fig. 2B), indicating that endogenous PCBP2 directly interacts with the sortilin CRE. Moreover, immunoprecipitation with macrophage cell lysate coprecipitated endogenous PCBP2 in addition to endogenous PCBP1 with BrUTP‐labeled RNA derived from the 3′ UTR of the sortilin mRNA (Fig. 2C), indicating the capability of both PCBP1 and PCBP2 to bind. These data strongly suggest that PCBP2 directly interacts with the CRE in the 3′ UTR of sortilin mRNA.
Figure 2.
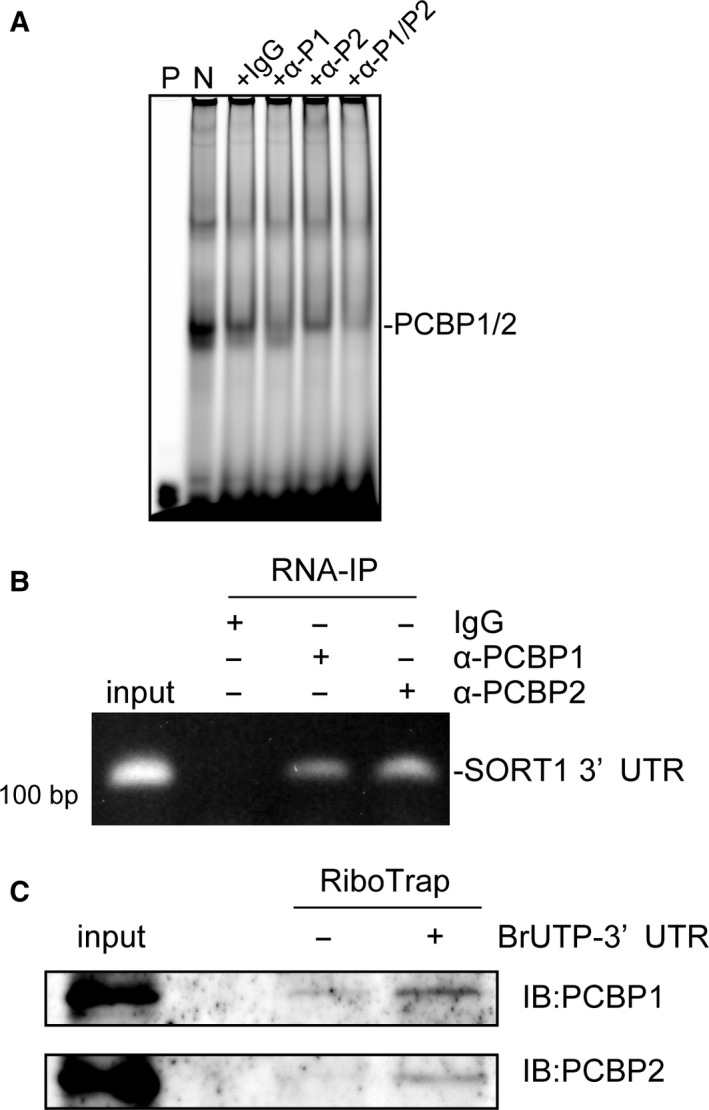
PCBP2 binds to CREs in the 3′ UTR of sortilin mRNA. (A) RNA EMSA analysis. P, CRE probe mixed with binding buffer alone; N, whole‐cell macrophage lysates mixed with labeled WT probe and 50‐fold molar excess of nonspecific competitor; +IgG, negative control; +α‐P1, anti‐PCBP1 antibody supershift; +α‐P2, anti‐PCBP2 supershift; +α‐P1/P2, anti‐PCBP1 and anti‐PCBP2 supershift. (B) Sortilin mRNA coimmunoprecipitates with both PCBP1 and PCBP2. (C) RiboTrap analysis. Macrophage cytosol was mixed with the BrUTP‐labeled 3′ UTR synthesized in vitro. Protein/RNA complexes were precipitated with protein A/G magnetic beads bound to anti‐BrdU or rabbit IgG at 4 °C for 3 h. The BrUTP‐labeled RNA–protein complex was eluted with BrdU in PBS, followed by immunoblotting.
To address the question whether PCBP2 stabilizes sortilin mRNA, we depleted PCBP2 in the murine macrophage cell line RAW264.7 using siRNA, followed by quantitation of sortilin mRNA by qRT‐PCR. We confirmed that siRNA‐transfected cells exhibited reduced PCBP1 and PCBP2 expression at both the mRNA and protein levels (Fig. 3A,B). We observed that both PCBP1 and PCBP2 knockdown with actinomycin D (ActD) treatment efficiently decreased sortilin mRNA levels compared with non‐targeting siNTS; however, this difference was attenuated without ActD treatment (Fig. 3C), consistent with the destabilization of sortilin mRNA by PCBP2 depletion. Collectively, these data suggest that PCBP2 promotes the stabilization of sortilin transcripts by binding to the CRE in the 3′ UTR of sortilin mRNA.
Zn2+ affects the nucleotide binding activity of PCBP2
We previously reported reversible inhibition of nucleotide binding ability of PCBP1 by Zn2+ 17. To investigate whether the poly(C)‐binding activity of PCBP2 is also affected by Zn2+, we performed electrophoretic mobility shift assay (EMSA) on yeast cell lysates expressing FLAG‐PCBP2 in the presence of Zn2+. As expected, the shifted band derived from the complex of FLAG‐PCBP2 with radiolabeled probe was decreased when the lysate was incubated with zinc, whereas the signals were unchanged when the lysate was incubated with iron, indicating the Zn2+ specificity of PCBP2 binding (Fig. 4). Incubation with an excess amount of EDTA attenuated this binding (Fig. 4), indicating a reversible inhibition of PCBP2 binding. This reversible inhibition is consistent with the requirement of Zn2+ for PCBP2 poly(C) binding.
Figure 4.
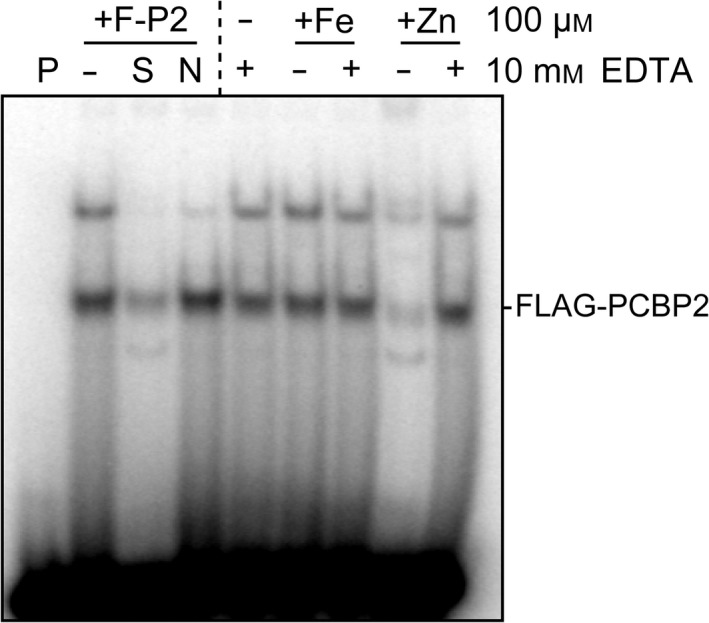
Zinc ions affect the poly(C)‐binding ability of PCBP2. EMSA analysis of PCBP2. 32P‐labeled poly(C) oligonucleotide probe (100 nm) was mixed with buffer alone (P) or yeast lysates expressing FLAG‐PCBP2. Samples were separated by PAGE. A 10‐fold molar excess of unlabeled poly(C) oligonucleotide or control mutated oligonucleotide was added as specific competitor (S) and nonspecific competitor (N), respectively. Zinc sulfate and ferrous ammonium sulfate (both 100 μm) with or without 10 mm EDTA were incubated with samples prior to incubation with the probes.
Pathogen infection and cellular stimuli induce PRR signaling, resulting in cytokine and chemokine production by immune cells. These inflammatory responses protect the body against harmful stimuli; however, chronic inflammation and/or autoimmune diseases are primarily attributed to excessive immune responses caused by cytokine overproduction. Therefore, the appropriate regulation of cytokine production is crucial for accurate immune responses. Here, we demonstrate that PCBP2, a paralog of PCBP1, can control the stability of sortilin mRNA through the binding to the CRE in 3′ UTR, along with PCBP1. So far, it has been reported that sortilin plays a key role in exocytic trafficking of cytokines in multiple types of immune cells 17, 19, 20, suggesting the involvement of both PCBP2 and PCBP1 in the post‐translational regulation of cytokine production through sortilin.
Toll‐like receptor (TLR) stimulation leads to a decrease in the amount of sortilin mRNA that is stabilized by PCBP1 17 and PCBP2 (Fig. 3), suggesting that both PCBPs have important roles in the regulation of sortilin mRNA by TLR signals. We observed that protein levels of both PCBP1 and PCBP2 were unchanged by TLR stimulation (data not shown), suggesting that mRNA destabilization may be caused by the dissociation of PCBP1 and PCBP2 with CRE. Although previous studies have demonstrated that phosphorylation of PCBP1 results in the loss of polyribonucleotide binding ability 21, 22, we did not observe any phosphorylated PCBP1 or PCBP2 after TLR9 stimulation in macrophages (data not shown), suggesting that phosphorylation of PCBP1 and/or PCBP2 did not trigger destabilization of sortilin mRNA in response to TLR signals.
Both PCBPs are iron chaperones that can bind and deliver iron, in addition to their functions as RBPs 23. Interestingly, we observed no relationship between the poly(C)‐ and iron‐binding abilities of either PCBP1 17 or PCBP2 (Fig 4). We have previously shown that Zn2+ affected the poly(C)‐binding ability of PCBP1 17; in this study, we obtained similar results with PCBP2 (Fig. 4), suggesting that both PCBP1 and PCBP2 can bind Zn2+. We have also demonstrated that cells, which were loaded with Zn2+, have decreased sortilin levels 17. Given that TLR signals induce an increase in the cytosolic free Zn2+ content 24, 25, we propose a model in which a TLR‐zinc signaling axis triggers dissociation of the PCBP1/2‐mRNA complex by the binding of Zn2+ to PCBP1/2, destabilizing sortilin mRNA to attenuate the immune response by preventing cytokine overproduction. We infer that both PCBP1 and PCBP2 may function as sensors of intracellular Zn2+ levels to control multiple genes, in addition to Sort1, by the binding to their CREs; however, further studies are required to better understand the underlying mechanisms. In summary, we show that the post‐transcriptional regulation of sortilin mRNA by PCBP2 and PCBP1 is important for the post‐translational control of cytokine production.
Methods
Mice
C57BL/six mice (female, 6–7 weeks old) were purchased from Sankyo Labo Service Corporation (Tokyo, Japan). All animal experimental protocols were approved by the Animal Research Committee of the Medical Research Institute, Kanazawa Medical University. All animal experiments were carried out in accordance with institutional guidelines.
Cell culture and reagents, preparation of primary cells from mice
All cells were cultured at 37 °C in a humidified, 5% CO2 atmosphere. Media for primary culture were described previously 17. The preparation of primary cells from mice was done as described previously 17. RAW 264.7 cells were grown in Dulbecco's modified Eagle's medium (Wako Chem., Osaka, Japan) supplemented with 10% FCS (Sigma‐Aldrich, St Louis, MO, USA), 100 U·mL−1 penicillin, and 100 U·mL−1 streptomycin (Wako Chem.).
Protein depletion by siRNA and immunoblotting
PCBP1 depletion and PCBP2 depletion in RAW264.7 cells were performed by transfection of siRNA (siGENOME SMARTpool siRNA; Dharmacon, Lafayette, CO, USA) using ScreenFect siRNA (Wako Chem.) according to the manufacturer's instructions. A nontargeting sequence siRNA pool (siGENOME Non‐Targeting siRNA Pool #2; Dharmacon Inc) was used as a control. The preparation of whole‐cell lysate and immunoblotting was carried out as previously described 17. The blots were incubated with anti‐PCBP1 (1 : 10 000) 9, anti‐PCBP2 (MBL, Nagoya, Japan; 1 : 1000), and anti‐actin (Sigma‐Aldrich; 1 : 10 000) antibodies. Enhanced chemiluminescent detection was carried out as previously described 17.
Quantitative real‐time PCR
Total RNA from 5 × 105 cells was isolated with ReliaPrep RNA Cell Miniprep System (Promega, Madison, WI, USA), and cDNA was synthesized using 500 ng of total RNA with ReverTra Ace qPCR RT Master Mix (Toyobo, Ohtsu, Japan), according to the manufacturers' instructions. qRT‐PCR was performed as previously described 17. Cycle threshold values were normalized to the housekeeping gene Gapdh. The primer set for the detection of Pcbp1, Sort1, and Gapdh was described previously 17. The primer set for detection of PCBP2 was designed using Primer3Plus (https://primer3plus.com/cgi-bin/dev/primer3plus.cgi): forward, 5′‐TATGCCATTCCACAGCCAGA‐3′; reverse, 5′‐CTGCCCAATAGCCTTTCACC‐3′.
Electrophoretic mobility shift assay
RNA EMSA with macrophage cell lysate and EMSA with a yeast pep4Δ strain transformed with pYES2 FLAG‐PCBP2 were performed as described previously 17. Antibody supershift experiments were performed by incubation with 1 μg of anti‐PCBP1 or anti‐PCBP2 antibody (MBL) on ice for 5 min after the RNA/protein binding reaction. Normal rabbit IgG (Merck Millipore, Billerica, MA, USA) was used as a negative control. Ferrous ammonium sulfate or zinc sulfate (both 100 μm) was incubated with cell lysate prior to incubation with probes. Samples were separated on 6% polyacrylamide DNA gels (Invitrogen, Waltham, MA, USA) and analyzed by the Odyssey system or phosphorimaging.
RNA immunoprecipitation and RiboTrap analysis
RNA immunoprecipitation was carried out as described previously 17. Briefly, 60 µL of Magnetic Beads Protein A/G (Merck Millipore) bound to 15 µg of anti‐PCBP2 (MBL) or rabbit IgG (Merck Millipore) was incubated with lysate at 4 °C for 18 h. RNA precipitated by antibodies was purified using an miRNeasy Mini Kit (QIAGEN, Hilden, Germany), followed by cDNA synthesis with iScript Advanced cDNA Synthesis Kit (Bio‐Rad Laboratories, Richmond, CA, USA). Real‐time PCR was carried out using a GoTaq Master Mix (Promega).
The cytosolic fraction of macrophages for RiboTrap analysis was prepared from 5 × 107 macrophages using a RiboCluster Profiler RiboTrap kit (MBL), according to the manufacturer's instructions. For in vitro transcription with T7 RNA polymerase, the 3′ UTR of sortilin mRNA was amplified from a plasmid containing full‐length sortilin cDNA (DNAFORM, Yokohama, Japan), and the resulting amplicon was cloned into a pGEM‐4Z vector (Promega) using the In‐Fusion HD Cloning Kit (Clontech, Palo Alto, CA, USA). The BrUTP‐labeled 3′ UTR of sortilin mRNA was prepared using an RNA Riboprobe System‐T7 kit (Promega). Magnetic Beads Protein A/G (Merck Millipore) bound to anti‐BrdU (MBL) or rabbit IgG (Merck Millipore) were incubated with the mixture of cytosol and BrUTP‐labeled 3′ UTR at 4 °C for 3 h. The BrUTP‐labeled RNA–protein complex bound to magnetic beads was eluted with PBS containing BrdU, followed by immunoblotting.
Statistical analysis
Statistical analysis was performed by ANOVA using graphpad prism software (GraphPad Software, La Jolla, CA, USA). One‐way and two‐way ANOVA were used for experiments with one and two variables, respectively. A P value of < 0.05 was considered statistically significant.
Conflict of interests
The authors declare no conflict of interest.
Author contributions
TY‐W conceived the project. TY‐W, CCP, and NO supervised the project. TY‐W performed the experiments and analyzed the results. TY‐W, CCP, and NO wrote and edited the manuscript.
Acknowledgements
We thank H. Nakamura and C. Ogasawara for technical support. We thank S. Matsuba and F. Saito for stimulating discussion. This work was supported, in part, by Grants‐in‐aid for Scientific Research 16K18518 and 19K06550 to TY‐W and 15H04717 and 18H02644 to NO from the Japan Society for the Promotion of Science, and by the Takeda Science Foundation (to TY‐W and NO). These studies were also supported, in part, by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, USA (TY‐W and CCP).
Contributor Information
Toshiki Yabe‐Wada, Email: tyw5510@kanazawa-med.ac.jp.
Nobuyuki Onai, Email: onai@kanazawa-med.ac.jp.
References
- 1. Takeuchi O and Akira S (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820. [DOI] [PubMed] [Google Scholar]
- 2. Kafasla P, Skliris A and Kontoyiannis DL (2014) Post‐transcriptional coordination of immunological responses by RNA‐binding proteins. Nat Immunol 15, 492–502. [DOI] [PubMed] [Google Scholar]
- 3. Akira S (2013) Regnase‐1, a ribonuclease involved in the regulation of immune responses. Cold Spring Harb Symp Quant Biol 78, 51–60. [DOI] [PubMed] [Google Scholar]
- 4. Anderson P (2010) Post‐transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol 10, 24–35. [DOI] [PubMed] [Google Scholar]
- 5. Chaudhury A, Chander P and Howe PH (2010) Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA 16, 1449–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makeyev AV and Liebhaber SA (2002) The poly(C)‐binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghanem LR, Kromer A, Silverman IM, Chatterji P, Traxler E, Penzo‐Mendez A, Weiss MJ, Stanger BZ and Liebhaber SA (2016) The poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol Cell Biol 36, 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frey AG, Nandal A, Park JH, Smith PM, Yabe T, Ryu MS, Ghosh MC, Lee J, Rouault TA, Park MH et al (2014) Iron chaperones PCBP1 and PCBP2 mediate the metallation of the dinuclear iron enzyme deoxyhypusine hydroxylase. Proc Natl Acad Sci USA 111, 8031–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leidgens S, Bullough KZ, Shi H, Li F, Shakoury‐Elizeh M, Yabe T, Subramanian P, Hsu E, Natarajan N, Nandal A et al (2013) Each member of the poly‐r(C)‐binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J Biol Chem 288, 17791–17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nandal A, Ruiz JC, Subramanian P, Ghimire‐Rijal S, Sinnamon RA, Stemmler TL, Bruick RK and Philpott CC (2011) Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab 14, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi H, Bencze KZ, Stemmler TL and Philpott CC (2008) A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryu MS, Zhang D, Protchenko O, Shakoury‐Elizeh M and Philpott CC (2017) PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J Clin Invest 127, 1786–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ryu MS, Duck KA and Philpott CC (2018) Ferritin iron regulators, PCBP1 and NCOA4, respond to cellular iron status in developing red cells. Blood Cells Mol Dis 69, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel SJ, Frey AG, Palenchar DJ, Achar S, Bullough KZ, Vashisht A, Wohlschlegel JA and Philpott CC (2019) A PCBP1‐BolA2 chaperone complex delivers iron for cytosolic [2Fe‐2S] cluster assembly. Nat Chem Biol 15, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Z, Yin W, Zhu L, Li J, Yao Y, Chen F, Sun M, Zhang J, Shen N, Song Y et al (2018) Iron drives T helper cell pathogenicity by promoting RNA‐binding protein PCBP1‐mediated proinflammatory cytokine production. Immunity 49, 80–92.e7. [DOI] [PubMed] [Google Scholar]
- 16. Li Y, Lin L, Li Z, Ye X, Xiong K, Aryal B, Xu Z, Paroo Z, Liu Q, He C et al (2012) Iron homeostasis regulates the activity of the microRNA pathway through poly(C)‐binding protein 2. Cell Metab 15, 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yabe‐Wada T, Matsuba S, Takeda K, Sato T, Suyama M, Ohkawa Y, Takai T, Shi H, Philpott CC and Nakamura A (2016) TLR signals posttranscriptionally regulate the cytokine trafficking mediator sortilin. Sci Rep 6, 26566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW 3rd et al (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mortensen MB, Kjolby M, Gunnersen S, Larsen JV, Palmfeldt J, Falk E, Nykjaer A and Bentzon JF (2014) Targeting sortilin in immune cells reduces proinflammatory cytokines and atherosclerosis. J Clin Invest 124, 5317–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herda S, Raczkowski F, Mittrücker H‐W, Willimsky G, Gerlach K, Kühl AA, Breiderhoff T, Willnow TE, Dörken B, Höpken UE et al (2012) The sorting receptor Sortilin exhibits a dual function in exocytic trafficking of interferon‐gamma and granzyme A in T cells. Immunity 37, 854–866. [DOI] [PubMed] [Google Scholar]
- 21. Meng Q, Rayala SK, Gururaj AE, Talukder AH, O'Malley BW and Kumar R (2007) Signaling‐dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci USA 104, 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL and Howe PH (2010) TGF‐β‐mediated phosphorylation of hnRNP E1 induces EMT via transcript‐selective translational induction of Dab2 and ILEI. Nat Cell Biol 12, 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Philpott CC and Jadhav S (2019) The ins and outs of iron: escorting iron through the mammalian cytosol. Free Radic Biol Med 133, 112–117. [DOI] [PubMed] [Google Scholar]
- 24. Brieger A, Rink L and Haase H (2013) Differential regulation of TLR‐dependent MyD88 and TRIF signaling pathways by free zinc ions. J Immunol 191, 1808–1817. [DOI] [PubMed] [Google Scholar]
- 25. Haase H, Ober‐Blöbaum JL, Engelhardt G, Hebel S, Heit A, Heine H and Rink L (2008) Zinc signals are essential for lipopolysaccharide‐induced signal transduction in monocytes. J Immunol 181, 6491–6502. [DOI] [PubMed] [Google Scholar]


