Abstract
Objective
Charcot-Marie-Tooth disease (CMT) is most commonly caused by duplication of a chromosomal segment surrounding Peripheral Myelin Protein 22, or PMP22 gene, which is classified as CMT1A. Several candidate therapies reduce Pmp22 mRNA levels in CMT1A rodent models, but development of biomarkers for clinical trials in CMT1A is a challenge given its slow progression and the difficulty in obtaining nerve samples. Quantitative PCR measurements of PMP22 mRNA in dermal nerves were performed using skin biopsies in human clinical trials for CMT1A, but this approach did not show increased PMP22 mRNA in CMT1A patients compared to controls. One complicating factor is the variable amounts of Schwann cells (SC) in skin. The objective of the study was to develop a novel method for precise evaluation of PMP22 levels in skin biopsies that can discriminate CMT1A patients from controls.
Methods
We have developed methods to normalize PMP22 transcript levels to SC-specific genes that are not altered by CMT1A status. Several CMT1A-associated genes were assembled into a custom Nanostring panel to enable precise transcript measurements that can be normalized to variable Schwann cell content.
Results
The digital expression data from Nanostring analysis showed reproducible elevation of PMP22 levels in CMT1A vs. control skin biopsies, particularly after normalization to SC-specific genes.
Interpretation
This platform should be useful in clinical trials for CMT1A as a biomarker of target engagement that can be used to optimize dosing, and the same normalization framework is applicable to other types of CMT.
Keywords: myelin, Schwann neuropathy, biomarker, Nanostring, biopsy
Introduction
Charcot Marie Tooth disease type 1A (CMT1A) is the most common inherited peripheral neuropathy affecting approximately 1:5,000 individuals1. CMT1A is an autosomal dominant demyelinating neuropathy and is caused by a duplication of a 1.4 Mb segment on chromosome 17, containing the Peripheral Myelin Protein 22, PMP22, gene. Rodent models have shown that neuropathy is reversible upon reduction of PMP22 mRNA levels, as was shown in transgenic mice that over-expressed PMP22 via an inducible promoter2. A recent study employing antisense oligonucleotides showed dramatic improvement in a severe mouse model of CMT1A as well as a more mild rat model of the disease3. While it remains to be seen if reversibility can be expected in CMT1A patients, therapeutic approaches are currently being developed to decrease PMP22 expression4.
Clinical outcome assessments such as the Rasch modified CMT Neuropathy or Exam Scores (CMTNS-R/CMTES-R) measure severity of the neuropathy in adults and children with CMT1A5, 6. However, since neuropathy is slowly progressive, there is a clear need for sensitive biomarkers that measure disease progression and target engagement in patients. Recent studies have tested for biomarkers of disease impairment and/or progression in CMT1A. For example, MRI measurements of intramuscular fat accumulation (IMFA) fraction of calf muscles are a sensitive biomarker that increases over 12 months in patients with CMT1A7, 8. In addition, neurofilament L protein (NEFL) levels are elevated in CMT1A plasma samples and correlate with severity as measured by the CMTNS-R and CMTES-R9.
Since several candidate therapeutic strategies are aimed to reduce PMP22 mRNA levels3, 10, developing a robust method for establishing PMP22 mRNA levels from skin biopsies would be a valuable marker of target engagement for clinical trials in CMT1A. Dermal skin biopsies allow for the relatively non-invasive evaluation of myelinated nerve fibers innervating structures such as Meissner’s corpuscles11. Immuno-EM showed elevated PMP22 protein in CMT1A patient skin samples compared to control, although the levels were more variable than in non-CMT1A samples 11, 12. Quantitative PCR measurements of PMP22 RNA were employed in clinical trials for CMT1A 13, 14, but these assays were unable to discriminate PMP22 levels in CMT1A patients from controls 15. However, in the antisense oligonucleotide study, the improvement of neuropathy was accompanied by a measurable reduction of Pmp22 by quantitative PCR of foot pad skin3. The loss of dermal nerve fibers in CMT1A patients16–18 likely hampers the ability to establish a reliable assay, and Schwann cell numbers in skin can be affected by age and sex19. Therefore, it is important to develop optimal normalization criteria to address the inherent variability in skin biopsy analysis. In addition, several technologies are now available to digitally assess gene expression level, such as RNA-seq, as well as other methods with increased precision, including droplet digital PCR and Nanostring20–23. For the relatively subtle differences in PMP22 levels between CMT1A and control Schwann cells (1.5 fold), the use of alternate technologies may present considerable advantages over more traditional methods.
Methods
Patient recruitment and consent
Ethics approval was obtained from University of Iowa and University of Wisconsin, and written informed assent/consent were provided by participants under a protocol approved by the ethics board of the NIH Rare Diseases Clinical Research Network (Protocol INC6611).
RNA-seq
Two 2 mm punch skin biopsies were performed 9 cm proximal to the ulnar crease and placed in RNAlater (ThermoFisher) prior to freezing as previously described 12, 13, 24–26. Total RNA isolated from the samples were prepared for RNA-seq using NuGEN ovation library prep kits and the libraries were loaded on the Illumina HiSeq 2500 at the UW Biotechnology Center. Analysis of RNA-seq data was performed by the Bioinformatics Resource Center at Univ. of Wisconsin using methods that adhere to ENCODE guidelines and best practices for RNA-Seq (Encode Consortium, 2016). Illumina HiSeq reads were trimmed 27 to remove sequencing adaptors and low quality bases. Reads shorter than 25 nucleotides after trimming were removed from downstream analysis. The trimmed sequences were aligned to the annotated (Ensembl 38.86) human reference genome using splice-junction aware alignment software, STAR28.
Quantitative RT-PCR
RNA samples from skin biopsies (300ng) were converted to cDNA using an M-MLV reverse transcriptase kit (Invitrogen #28025–013), RNase inhibitor (Promega #N251A), and a 50 μM mix containing random hexamers and oligo dT primer. Subsequent to cDNA synthesis, samples were diluted 1/10 for use in quantitative PCR (qPCR) reactions. qPCR reaction mixes were prepared using 2X SYBR green master mix (Thermo Fisher #4367659) and 300 nM primers (final concentration), then aliquoted (13μl) onto a 384-well reaction plate. Each reaction received 2μl of diluted cDNA for a final volume of 15μl. qPCR was performed using the ViiA 7 platform (Thermo Fisher) under the following cycling conditions: Hold1 at 50°C for 2min; Hold 2 at 95°C for 10min; 40 cycles at 95°C for 15s and 60°C for 1min. Data were analyzed using the relative ΔΔCT method 29.
PMP22 (total) F: GCTACAGTTCTGCCAGAGA R: CTCCTCCTGTTGCTGAGTATC
PMP22 (P1) F: GCTGCAAAGAAATCTGCTTGG R: GATGATACTCAGCAACAGGAGG
Gliomedin F: AACTGCCATTCAGAAGTGAGG R: GAGAGTCTGCTAACAAGAGTGA
Nanostring Analysis
The custom 54 gene Codeset was designed by Nanostring to maximize detection of most transcript variants, with the exception of the probes targeted to exons 1A and 1B of the PMP22 gene. SC-specific expression was also assessed using profiling of sorted cell types in mouse embryonic skin30. Total RNA was extracted from skin biopsies by homogenization/purification with the Trizol reagent, and then further purified using the Qiagen RNeasy MinElute Cleanup Kit (Cat# 74204). The samples were assessed for concentration, purity, and quality by NanoDrop and Bioanalyzer (Agilent RNA6000 PicoChip). The nCounter gene expression assay was performed by Nanostring (Seattle, WA). Each RNA sample (100 ng) was added to the CodeSet in hybridization buffer and incubated at 65°C for 16 hours. The custom CodeSet consists of Reporter and Capture probes that hybridize to the target sequences of interest, forming a tripartite complex. After hybridization/washing, transcript counts were obtained using the MAX System Digital Analyzer. For most samples, the two punches were combined prior to purification of RNA, but for selected samples, the two punches were purified separately. The same reagent batches were used in 3 runs as follows: Round 1: 3 control, 21 CMT1A; Round 2: 3 control in duplicate (+1 carryover), 4 CMT1A in duplicate; Round 3: 5 control (+1 carryover), 7 CMT1A in duplicate.
Statistical analysis
Transcriptome alignments of RNA-seq data from STAR were used as RSEM (RNASeq by Expectation Maximization) input for expression estimation 31. Genes with low or undetectable expression were independently filtered by requiring at least 3 samples to have a minimum of 1 count per million reads sequenced (cpm>1). The un-normalized read counts from RSEM were provided to EdgeR 32–34 for normalization and differential expression using the generalized linear model (GLM) method. Significance of the negative-binomial test was adjusted with a Benjamini-Hochberg FDR correction using a 5% threshold35.
Skin biopsy Nanostring data were analyzed by nSolver Analysis Software v4.0. Raw counts were normalized to the geometric means of the specified combinations of normalization genes before assessing changes between CMT1A vs. control. P values were adjusted using the Benjamini-Yekutieli FDR correction method. Displayed normalizations used either two genes (PLLP, VGLL3), 5 genes (CADM4, CNP, GLDN, GPM6B, PLLP) or 6 genes (CUEDC2, ERBB3, GLDN, PLEKHA1, PLLP, VGLL3). Pearson correlation coefficients were calculated for each gene relative to neuropathy score (CMTES-R) to produce the correlogram using HMISC version 4.0 36 at a 95% (or 90%) confidence interval.
Results
RNA-seq analysis of Human Skin Biopsies
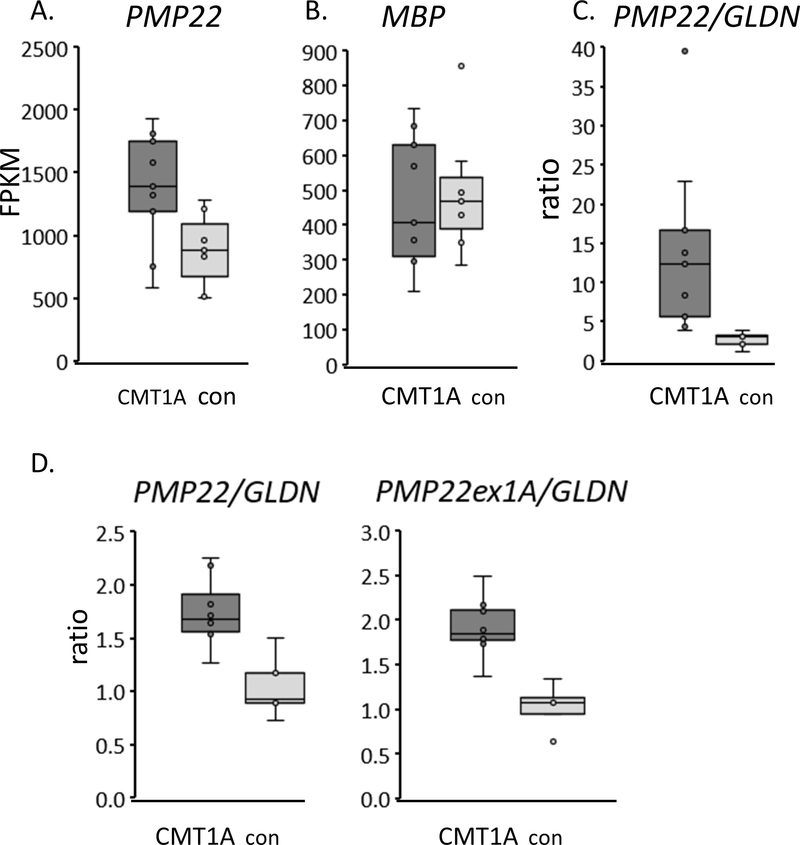
To improve mRNA analysis in skin biopsies, we attempted to identify changes in PMP22 and other Schwann cell genes by performing RNA-seq analysis of skin biopsies from nine CMT1A patients and seven controls. The punch biopsies were collected 9 cm from the ulnar crease, and Illumina sequencing and mapping to the hg38 genome yielded ~9–12 M mapped reads/sample. This analysis revealed relatively few genes that were found to be differentially expressed between groups with a FDR-adjusted p-value of <0.05 (Supplementary Table 1). Analysis of the most significantly induced genes in CMT1A samples only revealed one gene, SOX5, that is SC-specific by comparison to a gene expression database of sorted skin cell types 30. This was somewhat expected given that Schwann cells are a minority component of skin. However, we analyzed these data sets to determine if changes in PMP22 and other SC-specific genes could be detected in CMT1A skin biopsy samples relative to controls. After normalizing the RNA-seq data to each sample’s read depth, the PMP22 levels (measured in FPKM) are ~1.55 fold higher in CMT1A samples than in control samples (Figure 1, p=0.016, t-test, not FDR corrected). The same difference was not observed for MBP (myelin basic protein). It is striking that using a more digital method of RNA detection—compared to quantitative PCR used in previous studies 15—revealed an elevated level of PMP22. However, levels were more variable in the CMT1A samples, which may reflect variable amounts of Schwann cells in CMT1A skin biopsies as described above.
Figure 1. RNA-seq analysis of CMT1A skin biopsies.
A) RNA was purified from skin biopsies from 9 CMT1A patients and 7 controls for RNA-seq analysis. After normalizing for read depth and identifying differentially expressed genes (See Supplementary Table 1), the FPKM’s (fragments per kilobase per million mapped fragments) are shown for PMP22 gene in a box and whisker plot (box shows 25%/75% values). PMP22 levels were ~1.5 fold higher in CMT1A patient samples compared to control skin biopsies.
B) FPKM levels of another major myelin protein, Myelin Basic Protein, are shown.
C) To normalize RNA-seq data for Schwann cell content, the FPKM ratio of PMP22 to gliomedin (GLDN, which encodes a nodal component) is shown for CMT1A and control skin biopsies. GLDN levels were more variable at this read depth since this gene is expressed at a lower level than PMP22.
D) Quantitative RT-PCR was used on a subset of the same samples (8 CMT1A, 5 control) to show the ratio of total PMP22 to GLDN in CMT1A vs. control skin biopsies. A second primer set for the Schwann cell-specific transcript of PMP22 (containing exon1A initiating from the P1 promoter) gave similar results when normalized to GLDN.
Because of the potential variations in Schwann cell content, we sought to identify ways to normalize the data using SC-specific genes. Several resources are available to identify SC-specific genes, including a) expression profiles of peripheral nerve development and response to injury 37–44, b) profiling of sorted cell types in mouse embryonic skin 30 and c) RNA-seq profiling of >400 human tibial nerve samples 45 along with many other tissues (Broad Institute, gtexportal.org). These resources were used to filter the skin RNA-seq data to identify a series of SC-specific genes that are relatively unchanged in nerve injury and CMT1A models. In a trial analysis to normalize PMP22 levels to SC content, we used Gliomedin (GLDN), which is a SC-specific gene (Supplementary Figure 2) involved in the formation of nodes of Ranvier 46, 47. Many Schwann cell genes associated with myelination change dramatically after peripheral nerve injury in rodent models 37, 42, but Gldn expression does not change after injury. Interestingly, normalization to GLDN appeared to greatly decrease the variability in PMP22 levels in the control group. In addition, the normalized PMP22 level in the CMT1A samples is still more variable, yet statistically different from the control samples (p=0.02, t-test). The increased variability of the normalized PMP22 in CMT1A skin biopsies is similar to the relative variabilities of PMP22 protein observed in the immuno EM studies for CMT1A vs. control 12. Nonetheless, our data suggest that employing more digital methods of analysis coupled with normalization to appropriate genes may create a more reliable and precise method for discriminating PMP22 levels in skin biopsies of CMT1A patients for clinical trials.
Validation of PMP22 Levels
The same samples were analyzed by qRT-PCR (Figure 1D), which showed a similarly increased level of PMP22 in the CMT1A samples after normalization to GLDN. While PMP22 is expressed at lower levels in most other cell types, we also measured levels of the Schwann cell-specific isoform containing exon 1A from the P1 promoter. While both exon 1A- and exon 1B-containing transcripts are developmentally induced in Schwann cells during the myelination period 48, the exon 1A-containing transcript is exclusively expressed in Schwann cells, and accounts for ~50% of PMP22 in human peripheral nerve 48, and this was corroborated by analysis of human tibial nerve data sets (gtexportal.org) provided by the Broad Institute 45. Exon 1B-containing transcripts from the P2 promoter are the major form in fibroblasts 49 and most other tissues 45. Increased levels of the exon 1A-driven transcript of PMP22 was similarly elevated in CMT1A samples compared to controls after normalization to GLDN.
Nanostring Detection of SC-specific Transcripts in Skin Biopsies
With the ultimate goal of developing a clinically applicable platform for analysis of skin biopsies, we developed a custom gene expression panel using the Nanostring platform, which uses a barcode-based probe design to count RNA transcripts 23. Since no amplification or cDNA synthesis is involved, the precision of the assay is substantially better than qPCR techniques, and it can detect transcripts that are present at <1 copy per cell. This would suggest that it would also be able to detect transcripts in the minor fraction of Schwann cells that are found in skin biopsies. Nanostring technology has been used in clinical trial biomarker assays 20, 21, 50.
Our initial RNA-seq studies were used to assemble a pilot 52 gene panel of SC genes that are a) dysregulated in CMT1A rodent models, or b) SC-specific genes that are relatively stable and therefore will serve as normalization controls (Table 1). SC-specific expression was assessed using the aforementioned profiling of sorted cell types in mouse embryonic skin 30 and RNA-seq datasets of >400 human tibial nerve samples 45. The panel includes probes to detect total PMP22 and also the two major transcripts with alternate first exons (exons 1A and 1B, driven by P1 and P2 promoters, respectively). The set includes a number of known myelin genes (Mpz, Mag, Mbp, Pmp2, S100B), many of which are regulated by the SOX10/EGR2 transcriptional network that is required for Schwann cell differentiation 51, 52. Most of the selected Schwann cell genes are not expressed in other skin cell types, although several are expressed in oligodendrocytes of the CNS, partly due to the common regulation by the SOX10 transcription factor 53, 54. Several of the genes are mutated in other types of CMT (e.g. MPZ, PMP2, NDRG1, PRX, EGR2). We also included transcripts of Schwann cell injury genes, many of which are normally induced through the action of JUN in Schwann cells after demyelinating nerve injury 40: NGFR, GDNF, OLIG1, SHH, BDNF, RUNX2, HMGA2, FGF5, CCL2, GFAP, and UCN2.
Table 1.
Selection of Nanostring Genes for Skin Biopsy Analysis
| Myelin Genes | |
| PMP22 | MPZ |
| -exonla | MAG |
| -exonlb | MBP |
| CLDN19 | PMP2 |
| MAL | PLP1 |
| CD9 | CNTF |
| IL16 | PRX |
| AATK | NDRG1 |
| L1CAM | SOX10 |
| UGT8 | EGR2 |
| GPM6A | S100B |
| GPR37L1 | DHH |
| Injury Genes | |
| GDNF | OLIG1 |
| SHH | BDNF |
| RUNX2 | HMGA2 |
| FGF5 | CCL2 |
| GFAP | UCN2 |
| TMPRSS5 | |
| CMT1A Genes | |
| POU3F1 | NGFR |
| CXCL14 | ID2 |
| EDNRB | SOX4 |
| SOX5 | CUEDC2 |
| TTYH1 | BZW2 |
| PLEKHA1 | |
| Stable Genes | |
| GPM6B | CNP |
| CADM4 | PLLP |
| PLEKHB1 | LGI4 |
| NR4A2 | VGLL3 |
| ERBB2 | ERBB3 |
| GLDN |
Table 1. Custom Nanostring panel design.
The indicated list of genes were submitted to Nanostring for capture/detection probe design. Indicated categories are major myelin genes, genes induced in Schwann cells after nerve injury, genes shown to be induced rodent CMT1A models, and also Schwann cell genes that are relatively unchanged in nerve injury and/or neuropathy model profiles. Most genes were chosen based on evidence that their expression was Schwann cell-specific using published profiles of sorted cells and/or specific to human tibial nerve. Many peripheral nerve genes are also expressed in oligodendrocytes in the CNS. The complete list of genes and Refseq accession number used for probe design is in Supplemental Table 1.
As shown in Table 1, we incorporated additional candidate biomarkers that robustly reflect PMP22 expression levels and their effect on Schwann cell function. In the recent study employing antisense oligonucleotides (ASO), RNA-seq analysis of the mouse model revealed several hundred differentially expressed genes (DEG) compared to nerves from wild type animals3. Interestingly, ASO treatment to reduce PMP22 mRNA in the C22 mice resulted in a significant normalization of only a subset of DEG, suggesting that there are “rapid response” genes that are sensitive to restoration of normal levels of PMP22. Many of these genes correlated with disease severity as defined by electrophysiology in the mice 3. We prioritized those that are Schwann cell-specific as described above, including BZW2, POU3F1, NGFR, CXCL14, ID2, EDNRB, SOX4, CUEDC2, TTYH1 and PLEKHA1.
Finally, the panel was designed to normalize PMP22 levels to those of other Schwann cell specific genes (e.g. Gliomedin) that are not affected by CMT1A status, and we identified several candidate genes that are not apparently modulated in rodent models of CMT 3, 55, but nonetheless are SC-specific. In addition to gliomedin, we selected several such genes that are preferentially expressed in Schwann cells relative to other skin cell types: DHH, GPM6B, CNP, CADM4, PLLP, PLEKHB1, LGI4, NR4A2, VGLL3, and ERBB3.
Optimizing Detection of Differential PMP22 Levels in CMT1A Skin Samples vs. Controls
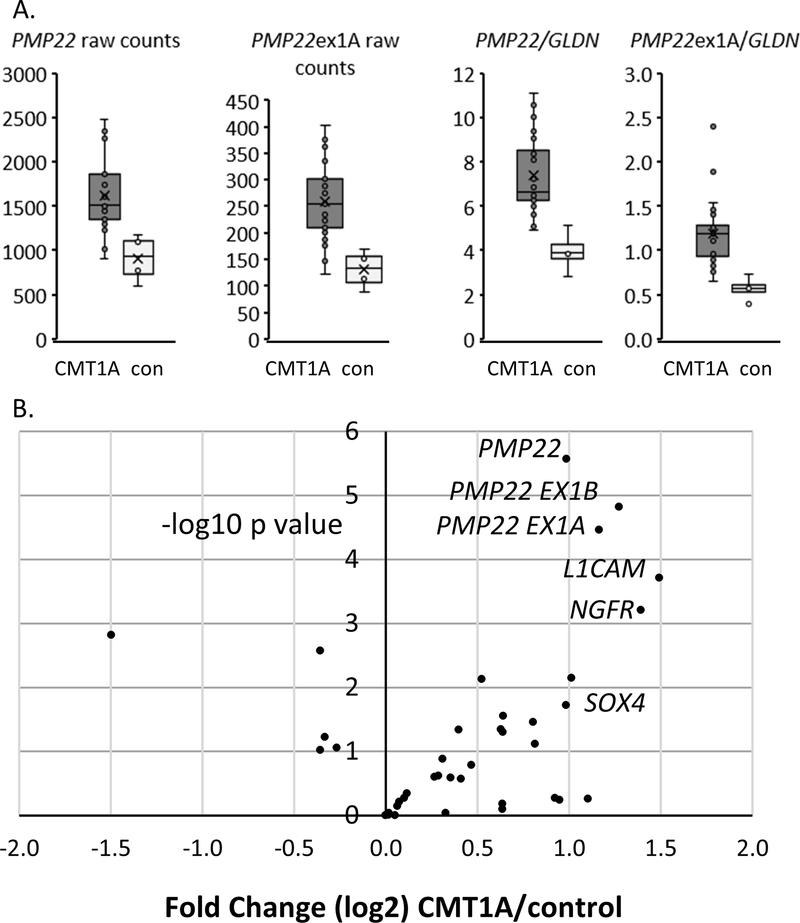
For the initial Nanostring run, we obtained a set of 21 CMT1A skin biopsies (ages 23–68, median 50) and 4 controls. Although the number of controls was relatively low, our previous experience using RNA-seq suggested that PMP22 levels are relatively consistent among controls. The resulting data (Figure 2A) showed that the CMT1A skin biopsies had elevated levels of total PMP22 and its SC-specific exon 1A containing transcript relative to controls. The elevation was observed with both raw count data as well as after normalization to GLDN. Consistent with previous analysis of RNA-seq data, the normalization to GLDN appeared to reduce the variability among control samples, although there was still enhanced variability of the CMT1A samples relative to controls.
Figure 2. Nanostring analysis of CMT1A skin biopsies.
A) Equal amounts of skin biopsy RNA’s were applied to the custom Nanostring panel, and the plots show raw counts of total PMP22 as well as the Schwann cell-specific Exon 1A form of PMP22. In addition, the ratio of both forms to the Schwann cell-specific GLDN is shown. (21 CMT1A and 4 control, avg. age 48.2 and 51.2 yr, respectively)
B) The volcano plot shows each of the genes (dots) ranked with the most statistically significant changes between control and CMT1A skin biopsies in the upper right quadrant. The results are normalized to the geometric mean of six SC-specific normalizing genes including GLDN using Nanostring nSolver software, and the plot shows levels (log 2, x-axis) plotted against the significance of the changes (negative log10 p value) between control and CMT1A. The plot shows elevation of total PMP22 in CMT1A samples along with specific probes for its two major transcripts (Ex 1A and Ex 1B), and also increased levels of NGFR and L1CAM. (21 CMT1A, 4 controls).
After normalizing to six SC-specific genes including GLDN, the Nanostring panel results are shown as a volcano plot in Figure 2B, in which the most statistically significant increases in CMT1A biopsies relative to controls are shown in the upper right quadrant. Consistent with our transcript data, we saw an ~2-fold increase in PMP22 levels (and its two major transcripts in Schwann cells) in CMT1A vs. control, which were statistically significant even after FDR correction of the p-values (adj. p<0.01). In addition, we also saw elevated levels of L1CAM, which is a potentially novel biomarker of CMT1A, as well as increased levels of two treatment responsive SC-specific genes from the recent ASO study 3: SOX4 and NGFR/p75.
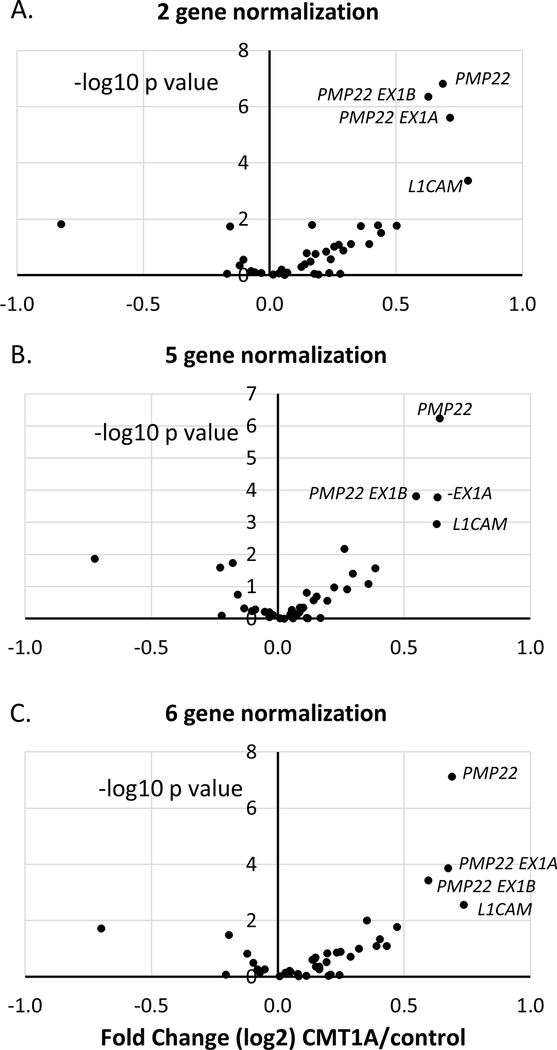
To provide a more robust set for testing of the panel, we obtained additional CMT1A and control skin samples for a total of 32 cases and 11 controls. The distribution of age/sex, and neuropathy score across the sample groups is shown in Table 2. The overall data set was normalized to different combinations of SC-specific genes, and we found that several independent normalization methods highlighted total PMP22 (and the exon1A/1B transcripts) as being the most significant changes in the CMT1A samples relative to controls (see volcano plots in Figure 3). The other most consistently elevated gene across different normalizations was L1CAM.
Table 2.
Distribution of Age/Sex and Neuropathy Score Across Sample Groups
| Age (range) | Sex | CMTES-R (range) | |
|---|---|---|---|
| control | 42.8 (25–58) | 6 F, 5M | NA |
| CMT1A | 46.4 (17–70) | 18F, 14 M | 11 (0–19) |
CMTES-R = Rasch modified CMT Exam Score; NA = not applicable.
Figure 3. Nanostring analysis with alternate normalization methods.
Nanostring results from skin biopsies are shown for 11 controls and 32 cases, from three separate runs. The volcano plot shows levels (log 2, x-axis) plotted against the significance of the changes (negative log10 p value) between control and CMT1A. The results are normalized to the geometric mean of two Schwann cell specific genes (A), 5 SC-specific genes (B) or six SC-specific normalizing genes (C).
Analyzing Variability of PMP22 levels in CMT1A Skin Samples
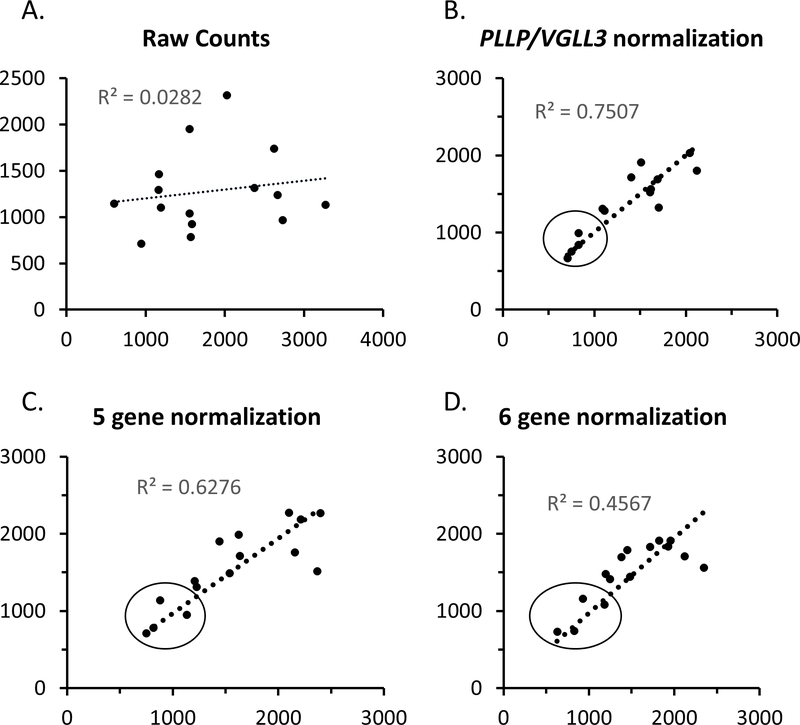
As noted above, the increased variability among CMT1A samples12 could arise from inherent variability of sample collection sites within a given subject. Since the samples were originally pooled from two separate skin punches, we also purified each skin punch independently for some selected cases and controls. For these samples, we could use them to assess intrasubject variability of skin biopsy analysis as well as test if our normalization methods are able to account for differential Schwann cell content of the two skin punches. As shown in Figure 4, there was fairly poor correlation between raw counts of duplicate samples. However, after normalization to SC-specific genes, the correlation was much improved. This demonstrates the importance of normalization to SC content and shows that the apparent variability of normalized PMP22 levels among CMT1A patients is not due to sampling variability in skin biopsies.
Figure 4. Nanostring Analysis of PMP22 levels in duplicate skin biopsies.
For selected control and CMT1A cases, the two skin punches were processed independently and submitted for Nanostring analysis in order to test the intra-subject variability of the results. The correlation plots show the PMP22 values for the two samples from each subject using A) raw counts or B-D) using several alternate normalization methods employing different combinations of Schwann cell-specific genes. The line shows the best fit of the data, and the r2 value is indicated. The circled points show those that were obtained from control subjects, which have consistently lower levels of PMP22 than the remaining CMT1A cases.
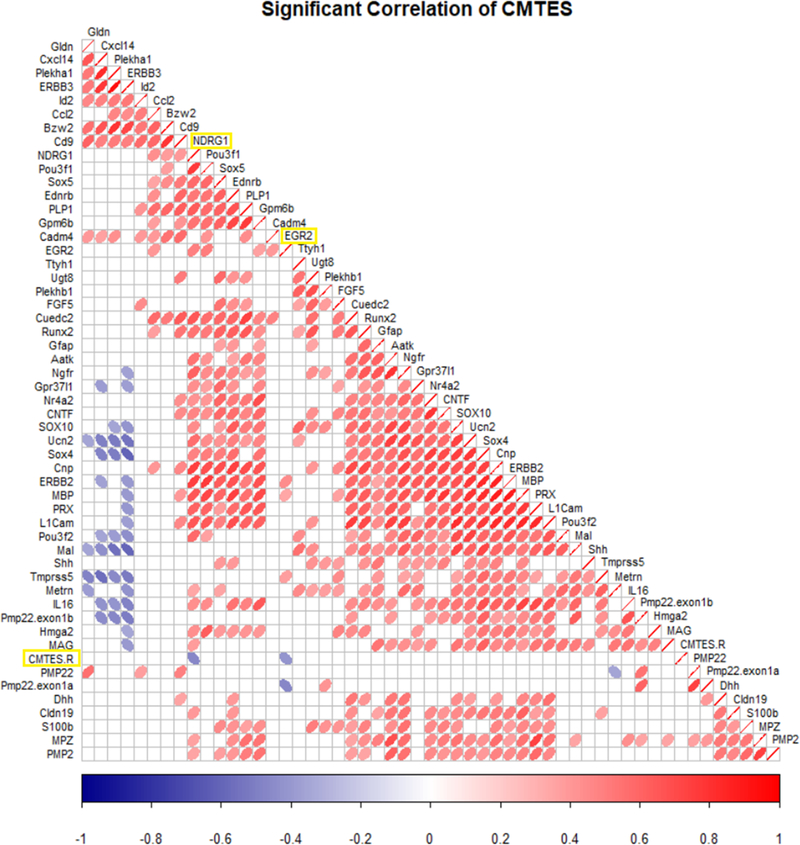
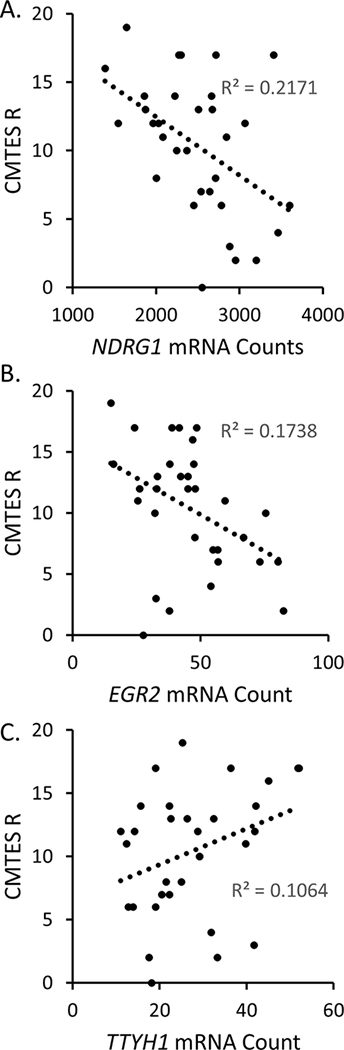
The normalized data were tested for correlations at a 95% confidence interval between individual genes and the CMTES-R neuropathy score and age (Figure 5). There was no correlation between PMP22 level and neuropathy score, consistent with previous immuno-EM data 11, 12. However, we did observe an inverse correlation between the neuropathy score and the levels of EGR2 and NDRG1. Both genes are mutated in other forms of CMT, and in fact EGR2 is a transcription factor that controls the level of NDRG1 56. The NDRG1 and EGR2 correlations with CMTES-R were also observed with the alternately normalized data sets that were used in Figure 3. The correlation analysis performed at a 90% confidence interval (not shown) revealed an additional positive correlation between neuropathy score and the TTYH1 gene. In the C22 CMT1A model, TTYH1 responded to treatment with antisense oligonucleotides for PMP22 3. Finally, there is an inverse correlation between age and the EGR2, NDRG1, CCL2, and CADM4 genes at the 95% confidence interval, consistent with the known age-dependent progression of CMT1A.
Figure 5. Correlation analysis of CMTES-R with gene expression levels.
Nanostring gene expression levels normalized to PLLP/VGLL3 were correlated with CMTES-R to determine significant correlations. Correlation values using Pearson’s method are represented in the correlogram. The strength of the correlation is indicated by the heat map scale for ellipses, showing positive (red, right leaning), and negative correlations (blue, left leaning). The row labeled as CMTES-R shows two inverse correlations with EGR2 and NDRG1, which are significant at a 95% confidence interval. There was no significant correlation of CMTES-R with PMP22 levels.
Discussion
Development of biomarkers for clinical trials in CMT1A is a major challenge given the slowly progressive nature of the disease. Biomarkers can be used to not only measure disease process (NEFL levels) and disease burden (MRI) but also target engagement for a given therapy 57. NEFL assays in CMT has provided an important marker of axonal degeneration in CMT1A 9, but there is currently a lack of biomarkers to assess disease processes in the Schwann cells that are most directly affected by the PMP22 gene duplication. Several candidate therapies reduce PMP22 levels in CMT1A rodent models and thereby ameliorate the symptoms of PMP22 overexpression 3, 10, 58. However, assessing PMP22 mRNA reduction in human trials is problematic given that such analysis has not demonstrated clear elevation of PMP22 mRNA in CMT1A patients compared to controls 15.
The analysis of skin biopsies is hampered by variable amounts of Schwann cells (SC) in skin, as well as the variable amount of PMP22 in SC as previously established by immuno-EM in CMT1A skin biopsies 11, 12, 16, 17. Therefore, it is important to develop optimal normalization criteria to address the variability inherent in skin biopsy analysis. Previous RT-PCR studies of human skin biopsies have used SC-specific genes (S100B or MBP) or ubiquitous genes (GAPDH, SDHA, TBP) for normalization 13, 15, 59. The ideal normalization gene would be Schwann cell-specific and be unaffected by injury or CMT1A status. Therefore, we employed data sets from CMT1A rodent models 3, 55, 60 that have been filtered to identify SC-specific genes 30 so that gene expression analysis can focus on the Schwann cell compartment of skin.
Another important consideration in assay development is the use of more digital gene expression platforms. The theoretical increase of PMP22 levels is 1.5 for CMT1A, and the precision of quantitative PCR is limited in this range 61. This is a particularly important point as the precision must be adequate for measurement of both PMP22 and normalization genes. Compared to hybridization-based microarrays, RNA-seq has much improved dynamic range as well as better precision particularly for less abundant transcripts. The pilot RNA-seq analysis of skin biopsies showed that PMP22 levels were ~1.5 fold higher in CMT1A patient samples compared to control skin biopsies after normalization to read depth, indicating that a precise method of gene expression analysis combined with robust normalization to Schwann cell-specific genes could provide a much improved technology. To construct such an assay, we turned to another digital gene expression platform based on Nanostring technology, since the performance/analysis costs of RNA-seq analysis are substantial. This platform directly detects RNA transcripts without cDNA synthesis and amplification, and its sensitivity has obvious advantages for development of a robust analysis platform for skin biopsies20, 21, 50.
Using various combinations of SC-specific genes for normalization, the custom Nanostring platform was able to reduce the apparent variability and optimize the differential levels of PMP22 between CMT1A and control skin biopsy samples. We obtained similar results with probes designed for total PMP22 as well as the two major Schwann cell transcripts with alternate first exons (1A and 1B). Nanostring data were analyzed with several independent normalization methods, all of which have been able to distinguish PMP22 levels in CMT1A vs. control skin biopsies. Based on our data, we suggest that this platform could be a valuable tool in clinical trials for CMT1A as a measure of target engagement that could be used within several weeks. In addition, this platform could also be used to establish optimal dosing, since over-suppression of PMP22 would have the risk of inducing another neuropathy phenotype similar to HNPP. In addition, while it would seem optimal to target a ~30% reduction of PMP22 levels in CMT1A to bring it to the normal level, the significance of the results suggests that it would be possible to detect more subtle changes in PMP22 levels (e.g. 10–20%), which could lead to improvement in the neuropathy, although perhaps a somewhat larger patient sample could be required to establish the significance of the effect. The normalization framework may also be applicable to other types of CMT, although it will need to be validated in a larger sample set.
The increased variability of PMP22 measurements found previously in CMT1A skin samples 12 was also evident in our study, and it has been proposed that other mechanisms besides elevated PMP22 mRNA per se are triggers for the neuropathy 15, 62. To test if increased variability could be due to sampling variability, we processed duplicate skin biopsies from the same patient and found very good concordance between them after normalization to SC-specific genes. Therefore, the range of PMP22 levels in CMT1A patients appears to reflect true variability between cases rather than variability between sites of biopsy.
In addition to measuring PMP22, we incorporated analysis of other SC-specific genes that may reflect the dysregulation of CMT1A Schwann cells. Although there are hundreds of altered genes in rodent models of CMT1A 3, the most effective biomarkers will be those that respond quickly to normalization of PMP22 levels. The recent identification of an effective PMP22 ASO provided an opportunity to rank dysregulated genes that are most responsive to therapeutic reduction of PMP22 expression levels. RNA-seq analysis of sciatic nerve from the C22 mouse model of CMT1A identified a small subset of 76 differentially expressed genes between wild type and mutant nerves that returned towards wild type levels following treatment with ASO to reduce PMP22 mRNA levels3. These genes were mined to identify those that are SC-specific and several of these were included in the Nanostring panel. Of the candidate markers included in the panel, the most promising were L1CAM and NGFR, which were found to be increased in CMT1A skin. In rodent studies, Ngfr and L1cam are elevated in CMT1A models3, 63, 64. In normal nerve, both genes are normally expressed in nonmyelinating Schwann cells, but both L1CAM and NGFR are expressed in myelinating Schwann cells of human skin19. It is anticipated that reduction of PMP22 in a clinical trial could be assessed relatively early through this method, and normalization of PMP22 levels and restoration of Schwann cell homeostasis would also hopefully reduce the levels of L1CAM and/or NGFR. It is notable that most of the Schwann cell injury genes were not elevated in CMT1A. The JUN transcription factor is a driver of demyelination after injury 40, and while it is detected in CMT1A skin biopsies 65, its levels may not be sufficient to drive an injury program. JUN was not included in this panel since it is not specific to Schwann cells in skin, but these and other data 3 suggest that CMT1A Schwann cells are distinct from injury-associated Schwann cells. Indeed deletion of Jun from a CMT1A mouse model does not mitigate neuropathy but rather exacerbates it 66.
Using the CMTES-R score, we did not see correlation of PMP22 mRNA levels (or NGFR/L1CAM) with the severity of the neuropathy. This was expected given that impairment in patients correlates more with axonal degeneration than demyelination 67. In addition, the distribution of PMP22 protein levels as measured by immuno-EM in CMT1A skin biopsies did not correlate with the severity of the neuropathy12. Nonetheless, we did observe inverse correlations of neuropathy score with EGR2 and NDRG1. EGR2 is a transcription factor that activates many myelin genes during Schwann cell development and itself is mutated in CMT1D and some more severe peripheral myelinopathies51, 56, 68, 69. NDRG1 is also mutated in a recessive form of CMT (CMT4E). While not described before, it could be that progression of CMT is due in part to decreased activation of the EGR2-driven transcriptional network, which includes NDRG1, as continued expression of EGR2 is required for myelin maintenance70. Alternatively, the association may reflect the loss of myelinated fibers that occurs during progression, although one would have expected a commensurate loss of MPZ expression if that was the explanation. The correlation analysis performed at a 90% confidence interval also revealed a correlation of CMTES-R with TTYH1, which was one of the ASO-responsive genes in the recent treatment study of the C22 mouse model of CMT1A3.
Recent studies have employed a quantitative PCR analysis of human skin biomarkers based on gene expression studies of skin in rat model of CMT1A59, 71. While this approach has been successful in measuring progression of CMT1A patients over a 2–3 year trial period, this analysis was not designed to detect SC-specific genes and PMP22 levels were also not incorporated. Therefore, some of the changes may occur in other cell types in response to CMT1A-associated denervation, and therefore may not be as immediately responsive to successful treatment. Our studies provide a complementary framework for gene expression analysis targeted to the Schwann cell compartment of skin biopsies, and provide an assay platform with enhanced precision to measure PMP22 levels that is designed to detect early changes in PMP22 and other CMT1A-associated genes. Further optimization of the Nanostring panel may enhance the ability to detect changes in CMT1A gene signatures.
Supplementary Material
Figure 6. CMTES-R neuropathy scores correlate with EGR2, NDRG1, and TTYH1.
Regression models (scatterplots) of significantly correlated genes to CMTES-R are shown, including the linear regression function. The correlation with TTYH1 was found only at the 90% confidence interval A) NDRG1, B) EGR2, and C) TTYH1
Acknowledgments
We thank the University of Wisconsin Biotechnology Center Gene Expression Center for providing RNA quality analysis and library preparation and the DNA Sequencing Facility for their sequencing services. We thank Chris DeHeer for help with analysis of Nanostring data and Vijay Modur for helpful conversations.
This work was supported by U54NS065712 provided by NINDS/NCATS-ORD, a core grant to the Waisman Center from NICHD (U54 HD090256), a grant from the Charcot-Marie-Tooth Association and funds provided by the Stahl family.
Footnotes
Potential Conflicts of Interest
There are no conflicts of interest.
REFERENCES
- 1.Fridman V, Bundy B, Reilly MM, et al. CMT subtypes and disease burden in patients enrolled in the Inherited Neuropathies Consortium natural history study: a cross-sectional analysis. J Neurol Neurosurg Psychiatry. 2015. August;86(8):873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huxley C, Passage E, Robertson A, et al. Correlation between varying levels of PMP22 expression and the degree of demyelination and reduction in nerve conduction velocity in transgenic mice. Hum Mol Genet. 1998. March;7(3):449–58. [DOI] [PubMed] [Google Scholar]
- 3.Zhao HT, Damle S, Ikeda-Lee K, et al. PMP22 antisense oligonucleotides reverse Charcot-Marie-Tooth disease type 1A features in rodent models. The Journal of clinical investigation. 2018. January 2;128(1):359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attarian S, Vallat JM, Magy L, et al. An exploratory randomised double-blind and placebo-controlled phase 2 study of a combination of baclofen, naltrexone and sorbitol (PXT3003) in patients with Charcot-Marie-Tooth disease type 1A. Orphanet J Rare Dis. 2014 Dec;9:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadjadi R, Reilly MM, Shy ME, et al. Psychometrics evaluation of Charcot-Marie-Tooth Neuropathy Score (CMTNSv2) second version, using Rasch analysis. J Peripher Nerv Syst. 2014. September;19(3):192–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandarakas MR, Menezes MP, Rose KJ, et al. Development and validation of the Charcot-Marie-Tooth Disease Infant Scale. Brain. 2018. December;141(12):3319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrow JM, Evans MRB, Grider T, et al. Validation of MRC Centre MRI calf muscle fat fraction protocol as an outcome measure in CMT1A . Neurology. 2018. September;91(12):e1125–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrow JM, Sinclair CD, Fischmann A, et al. MRI biomarker assessment of neuromuscular disease progression: a prospective observational cohort study. Lancet Neurol. 2016. November;15:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandelius Å, Zetterberg H, Blennow K, et al. Plasma neurofilament light chain concentration in the inherited peripheral neuropathies. Neurology. 2018. January;90:e518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chumakov I, Milet A, Cholet N, et al. Polytherapy with a combination of three repurposed drugs (PXT3003) down-regulates Pmp22 over-expression and improves myelination, axonal and functional parameters in models of CMT1A neuropathy. Orphanet J Rare Dis. 2014. December;9:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Bai Y, Ghandour K, et al. Skin biopsies in myelin-related neuropathies: bringing molecular pathology to the bedside. Brain. 2005;128(Pt 5):1168–77. Epub 2005 Mar 17. [DOI] [PubMed] [Google Scholar]
- 12.Katona I, Wu X, Feely SM, et al. PMP22 expression in dermal nerve myelin from patients with CMT1A . Brain. 2009. July;132(Pt 7):1734–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis RA, McDermott MP, Herrmann DN, et al. High-dosage ascorbic acid treatment in Charcot-Marie-Tooth disease type 1A: results of a randomized, double-masked, controlled trial. JAMA Neurol. 2013. August;70(8):981–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pareyson D, Reilly MM, Schenone A, et al. Ascorbic acid in Charcot-Marie-Tooth disease type 1A (CMT-TRIAAL and CMT-TRAUK): a double-blind randomised trial. Lancet Neurol. 2011. April;10(4):320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobbio L, Visigalli D, Radice D, et al. PMP22 messenger RNA levels in skin biopsies: testing the effectiveness of a Charcot-Marie-Tooth 1A biomarker. Brain. 2014. June;137(Pt 6):1614–20. [DOI] [PubMed] [Google Scholar]
- 16.Manganelli F, Nolano M, Pisciotta C, et al. Charcot-Marie-Tooth disease: New insights from skin biopsy. Neurology. 2015. October;85(14):1202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolano M, Manganelli F, Provitera V, et al. Small nerve fiber involvement in CMT1 A. Neurology. 2015. January 27;84(4):407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebenezer GJ, McArthur JC, Thomas D, et al. Denervation of skin in neuropathies: the sequence of axonal and Schwann cell changes in skin biopsies. Brain. 2007. October;130(Pt 10):2703–14. [DOI] [PubMed] [Google Scholar]
- 19.Reinisch CM, Tschachler E. The dimensions and characteristics of the subepidermal nerve plexus in human skin--terminal Schwann cells constitute a substantial cell population within the superficial dermis. J Dermatol Sci. 2012. March;65(3):162–9. [DOI] [PubMed] [Google Scholar]
- 20.Veldman-Jones MH, Brant R, Rooney C, et al. Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res. 2015. July;75(13):2587–93. [DOI] [PubMed] [Google Scholar]
- 21.Wallden B, Storhoff J, Nielsen T, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC medical genomics. 2015. August 22;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day E, Dear PH, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods. 2013. January;59(1):101–7. [DOI] [PubMed] [Google Scholar]
- 23.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008. March;26(3):317–25. [DOI] [PubMed] [Google Scholar]
- 24.Saporta MA, Katona I, Lewis RA, Masse S, Shy ME, Li J. Shortened internodal length of dermal myelinated nerve fibres in Charcot-Marie-Tooth disease type 1A . Brain. 2009. December;132(Pt 12):3263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan KM, Bai Y, Pisciotta C, et al. Absence of Dystrophin Related Protein-2 disrupts Cajal bands in a patient with Charcot-Marie-Tooth disease. Neuromuscular disorders : NMD. 2015. October;25(10):786–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pisciotta C, Bai Y, Brennan KM, et al. Reduced neurofilament expression in cutaneous nerve fibers of patients with CMT2E . Neurology. 2015. July 21;85(3):228–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads . BMC Bioinformatics. 2014. June;15:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013. January;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 30.Sennett R, Wang Z, Rezza A, et al. An Integrated Transcriptome Atlas of Embryonic Hair Follicle Progenitors, Their Niche, and the Developing Skin. Dev Cell. 2015. September;34(5):577–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome . BMC Bioinformatics. 2011. August;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010. January;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, McCarthy DJ, Chen Y, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013. September;8(9):1765–86. [DOI] [PubMed] [Google Scholar]
- 34.Lun AT, Chen Y, Smyth GK. It’s DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol Biol. 2016;1418:391–416. [DOI] [PubMed] [Google Scholar]
- 35.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003. February;19(3):368–75. [DOI] [PubMed] [Google Scholar]
- 36.Harrell FE. R Software. Regression Modeling Strategies: with Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis, 2nd Edition. Cham: Springer International Publishing Ag; 2015. p. 127–42. [Google Scholar]
- 37.Nagarajan R, Le N, Mahoney H, Araki T, Milbrandt J. Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proc Natl Acad Sci U S A. 2002. June 25;99(13):8998–9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrette B, Calvo E, Vallières N, Lacroix S. Transcriptional profiling of the injured sciatic nerve of mice carrying the Wld(S) mutant gene: identification of genes involved in neuroprotection, neuroinflammation, and nerve regeneration. Brain Behav Immun. 2010. November;24(8):1254–67. [DOI] [PubMed] [Google Scholar]
- 39.Bosse F, Hasenpusch-Theil K, Küry P, Müller HW. Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J Neurochem. 2006. March;96(5):1441–57. [DOI] [PubMed] [Google Scholar]
- 40.Arthur-Farraj PJ, Latouche M, Wilton DK, et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012. August;75(4):633–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verdier V, Csárdi G, de Preux-Charles AS, et al. Aging of myelinating glial cells predominantly affects lipid metabolism and immune response pathways. Glia. 2012. May;60(5):751–60. [DOI] [PubMed] [Google Scholar]
- 42.Arthur-Farraj PJ, Morgan CC, Adamowicz M, et al. Changes in the Coding and Non-coding Transcriptome and DNA Methylome that Define the Schwann Cell Repair Phenotype after Nerve Injury. Cell Rep. 2017. September;20(11):2719–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma KH, Duong P, Moran JJ, Junaidi N, Svaren J. Polycomb Repression regulates Schwann Cell Proliferation and Axon Regeneration after Nerve Injury. Glia. 2018;66:2487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clements MP, Byrne E, Camarillo Guerrero LF, et al. The Wound Microenvironment Reprograms Schwann Cells to Invasive Mesenchymal-like Cells to Drive Peripheral Nerve Regeneration. Neuron. 2017. September;96(1):98–114.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium G The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013. June;45(6):580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amor V, Feinberg K, Eshed-Eisenbach Y, et al. Long-term maintenance of Na+ channels at nodes of Ranvier depends on glial contact mediated by gliomedin and NrCAM. J Neurosci. 2014. April;34(15):5089–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feinberg K, Eshed-Eisenbach Y, Frechter S, et al. A glial signal consisting of gliomedin and NrCAM clusters axonal Na+ channels during the formation of nodes of Ranvier. Neuron. 2010. February;65(4):490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suter U, Snipes GJ, Schoener-Scott R, et al. Regulation of tissue-specific expression of alternative peripheral myelin protein-22 (PMP22) gene transcripts by two promoters. J Biol Chem. 1994. October 14;269(41):25795–808. [PubMed] [Google Scholar]
- 49.Visigalli D, Castagnola P, Capodivento G, et al. Alternative Splicing in the Human PMP22 Gene: Implications in CMT1A Neuropathy. Hum Mutat. 2016. January;37(1):98–109. [DOI] [PubMed] [Google Scholar]
- 50.Payton JE, Grieselhuber NR, Chang LW, et al. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. The Journal of clinical investigation. 2009. June;119(6):1714–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srinivasan R, Sun G, Keles S, et al. Genome-wide analysis of EGR2/SOX10 binding in myelinating peripheral nerve. Nucleic Acids Res. 2012. April;40:6449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones EA, Jang S-W, Mager GM, et al. Interactions of Sox10 and Egr2 in myelin gene regulation. Neuron Glia Biology. 2007 2007;3:377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopez-Anido C, Sun G, Koenning M, et al. Differential Sox10 genomic occupancy in myelinating glia. Glia. 2015. May;63:1897–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weider M, Reiprich S, Wegner M. Sox appeal - Sox10 attracts epigenetic and transcriptional regulators in myelinating glia. Biol Chem. 2013. December;394(12):1583–93. [DOI] [PubMed] [Google Scholar]
- 55.Fledrich R, Abdelaal T, Rasch L, et al. Targeting myelin lipid metabolism as a potential therapeutic strategy in a model of CMT1A neuropathy. Nat Commun. 2018. 08;9(1):3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le N, Nagarajan R, Wang JY, Araki T, Schmidt RE, Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc Natl Acad Sci U S A. 2005. February 15;102(7):2596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao X, Modur V, Carayannopoulos LN, Laterza OF. Biomarkers in Pharmaceutical Research . Clin Chem. 2015. November;61(11):1343–53. [DOI] [PubMed] [Google Scholar]
- 58.Sereda MW, Meyer zu Horste G, Suter U, Uzma N, Nave KA. Therapeutic administration of progesterone antagonist in a model of Charcot-Marie-Tooth disease (CMT1A). Nat Med. 2003. December;9(12):1533–7. [DOI] [PubMed] [Google Scholar]
- 59.Fledrich R, Mannil M, Leha A, et al. Biomarkers predict outcome in Charcot-Marie-Tooth disease 1 A. J Neurol Neurosurg Psychiatry. 2017. November;88(11):941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giambonini-Brugnoli G, Buchstaller J, Sommer L, Suter U, Mantei N. Distinct disease mechanisms in peripheral neuropathies due to altered peripheral myelin protein 22 gene dosage or a Pmp22 point mutation. Neurobiol Dis. 2005. April;18(3):656–68. [DOI] [PubMed] [Google Scholar]
- 61.Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N. Statistical significance of quantitative PCR. BMC Bioinformatics. 2007. April;8:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J Caveats in the Established Understanding of CMT1A. Ann Clin Transl Neurol. 2017. 08;4(8):601–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taniuchi M, Clark HB, Johnson EM. Induction of nerve growth factor receptor in Schwann cells after axotomy. Proc Natl Acad Sci U S A. 1986. June;83(11):4094–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klein D, Groh J, Wettmarshausen J, Martini R. Nonuniform molecular features of myelinating Schwann cells in models for CMT1: distinct disease patterns are associated with NCAM and c-Jun upregulation. Glia. 2014. May;62(5):736–50. [DOI] [PubMed] [Google Scholar]
- 65.Hutton EJ, Carty L, Laurá M, et al. c-Jun expression in human neuropathies: a pilot study. J Peripher Nerv Syst. 2011. Dec;16(4):295–303. [DOI] [PubMed] [Google Scholar]
- 66.Hantke J, Carty L, Wagstaff LJ, et al. c-Jun activation in Schwann cells protects against loss of sensory axons in inherited neuropathy. Brain. 2014. November;137(Pt 11):2922–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krajewski KM, Lewis RA, Fuerst DR, et al. Neurological dysfunction and axonal degeneration in Charcot-Marie-Tooth disease type 1 A. Brain. 2000. July;123 ( Pt 7):1516–27. [DOI] [PubMed] [Google Scholar]
- 68.Warner LE, Svaren J, Milbrandt J, Lupski JR. Functional consequences of mutations in the early growth response 2 gene (EGR2) correlate with severity of human myelinopathies. Hum Mol Genet. 1999;8(7):1245–51. [DOI] [PubMed] [Google Scholar]
- 69.Baets J, Deconinck T, De Vriendt E, et al. Genetic spectrum of hereditary neuropathies with onset in the first year of life. Brain. 2011. September;134(Pt 9):2664–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Decker L, Desmarquet-Trin-Dinh C, Taillebourg E, Ghislain J, Vallat JM, Charnay P. Peripheral myelin maintenance is a dynamic process requiring constant Krox20 expression. J Neurosci. 2006. September 20;26(38):9771–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fledrich R, Schlotter-Weigel B, Schnizer TJ, et al. A rat model of Charcot-Marie-Tooth disease 1A recapitulates disease variability and supplies biomarkers of axonal loss in patients. Brain. 2012. January;135(Pt 1):72–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.