Abstract
Background
Fusions are increasingly pursued as oncology therapeutic targets. This study evaluated differences in outcomes for fusion vs. non-fusion targets.
Methods
Outcome was compared for patients with fusions versus other alterations for Food and Drug Administration (FDA)-approved single-agents (from package inserts) and for patients treated at the University of California San Diego.
Results
Twenty-eight FDA-approved drugs (N = 6,189 patients) were included in the analysis. Median response rate was 68% vs. 50% for fusions vs. non-fusion matches (odds ratio of 1.67 (p<0.0001)); solid tumor therapies had an odds ratio of 2.07 (p<0.0001) and hematologic therapies, an odds ratio of 3.35 (p<0.0001) for fusion vs. non-fusion targets. The UCSD analysis included 79 patients with fusions treated of 2,455 screened. Patients matched to fusions had longer median [95% confidence interval] PFS (11.6 (4.0 – 35.4) months) as compared to those unmatched to fusions (4.9 (3.5–8.8) months) (p=0.034). Patients with fusions matched to other alterations present in the tumor had median PFS indistinguishable from those patients with fusions given unmatched therapy (4.0 versus 5.0 months (p = 0.75)).
Conclusions
Significantly higher response rates and longer PFS were seen by targeting fusions vs. non-fusions. Our observations suggest that fusions are important targets and that additional studies are needed to confirm that optimized therapy may require targeting fusions even in the presence of other alterations.
Keywords: fusions, precision medicine, matched therapy, molecular alteration
Precis
Higher response rates and longer progression free survivals were seen by targeting fusions vs. non-fusions. Fusions are important drivers and optimized therapy may require targeting fusions even in the presence of other alterations.
INTRODUCTION
With the advent of next generation sequencing (NGS) and molecular profiling, new information about molecular pathways and targets is being unveiled in oncology. Targeted therapies have been developed to interfere with specific aberrant targets that are involved in the growth, progression, and spread of cancer 1. Potential targets include proteins that are either more abundant in cancer cells or are mutated to drive cancer progression. The majority of targeted therapies are antibodies that impact extracellular and cell-surface proteins or small molecule inhibitors that can also suppress protein pathways inside the cell. Several targeted therapies have been Food and Drug Administration (FDA) approved as single agents to be given in the presence of a specific molecular alteration (usually a mutation, amplification or fusion) and clinical trial designs have evolved to allow for selection of patients with specific molecular alterations 2.
Fusions are increasingly being pursued as therapeutic targets 3, 4. They arise as a result of genomic rearrangements that include chromosomal inversion, interstitial deletions, duplications, and translocations. The fusion gene leads to a fusion protein that is often a strong oncogenic driver. Some fusions result in a constitutively active tyrosine kinase enzyme. Examples include BCR-ABL, which was the first fusion identified and has led to dramatic improvements in outcomes for chronic myelogenous leukemia (CML) 5–7. Other well-studied genes involved in fusions include ROS1 8, ALK 9, PML-RARA 10, and NTRK 11, 12, each of which has a unique mechanism of action and has resulted in breakthrough drugs for the affected cancer. Beyond fusions, other drugs target mutations (e.g., KIT 13, BRAF 14, EGFR 15) or amplifications (e.g. ERBB2 16). In some cases, targeting fusions appears to result in remarkably high response rates. Perhaps one of the most striking examples is the NTRK fusion-targeting drug larotrectinib, with response rates of approximately 75% 11.
In order to determine if there are overall differences in response rates when drugs were approved for targeting a fusion versus a mutation or amplification, we examined all drugs approved by the FDA as single agents for a specific genomic alteration. In addition, we analyzed a group of 2,455 patients at the University of California San Diego Moores Cancer Center for Personalized Cancer Therapy in order to determine progression-free survival (PFS) when patients with fusions accompanied by additional genomic alterations were matched to drugs based on the fusions or on the non-fusion alterations. Our results indicate that targeting fusions versus mutations or amplifications results in significantly better outcomes, suggesting that fusions may be important targets for optimized precision medicine therapeutic approaches.
METHODS
FDA-single agent approved oncology targeted therapeutics
All FDA-approved oncology targeted therapeutics with an approval for a specific molecular alteration were included in the analysis. Drugs were identified though review of the FDA website 17 and a review article on FDA-approved cancer therapies 18 through 12/31/2018. Package inserts were reviewed to determine the molecular alteration that was approved for drug use. Only drugs that were approved as single agents and were approved for a specific genomic alteration—mutation, amplification or fusion–-were used in the analysis. Agents only approved as adjuvant therapy or maintenance were excluded. Response rates were determined from efficacy studies listed in the package inserts. For drugs with several efficacy studies listed in the package insert, the response rate was an average of the studies. For agents with more than one alteration as a target, each was listed separately for the analysis. For solid tumors and lymphoma, response rate included complete and partial responses. For acute myeloid leukemia (AML) therapeutics, response rate included complete response with combined full or partial hematologic recovery. For CML therapeutics, response rate was the complete cytogenetic response rate for chronic phase. Therapeutics were segregated by fusions (e.g., ALK, ROS1, NTRK) and non-fusions (other alterations). Response rates were summarized for all patients, solid tumors, and hematologic malignancies. The odds ratio for likelihood of response with fusion as compared to non-fusion was calculated and compared using the chi square test of association.
UCSD patient population
Patients with FoundationOne reports who were enrolled in the University of California San Diego Study of Personalized Cancer Therapy to Determine Response and Toxicity (UCSD-PREDICT ), which encompasses an institutional review board (IRB)-approved observational cohort study at UCSD were screened for the study (August 2012-September 2018). FoundationOne testing evaluated between 182 and 406 genes. The current study was performed in accordance with the UCSD IRB guidelines for data analysis and for any investigational treatments for which patients gave consent. Patients with at least one fusion mutation who were either treated exclusively at UC San Diego or who had sufficient information in the electronic medical record to evaluate progression-free survival (PFS) were included in the analysis. Patients with hematologic malignancies were excluded. PFS was calculated from the start date of therapy to the date of progression. Date of progression was determined from review of imaging (progression by RECIST criteria) or date of clinical progression as documented in physician notes. Patients were censored if the regimen was changed due to toxicity at the start date of the new therapy, for death prior to progression, or at the date of last follow-up for those lost to follow-up.
The log-rank test was used to compare PFS for therapy matched to a fusion alteration vs. other therapies and for pairwise comparisons of: therapy matched to a fusion alteration, therapy matched to a non-fusion alteration, and unmatched therapies. All statistical comparisons were completed using SAS v. 9.4 and p-values of less than or equal to 0.05 were considered significant.
RESULTS
FDA-approved cancer monotherapy
A total of 28 FDA approved single-agent oncology targeted therapeutics were included in the analysis with response rates available for 6,189 patients. Of the 28 drugs used the in the analysis, 11 were approved for fusion targets and 16 were approved for non-fusion targets. Imatinib had two separate FDA-approved indications: targeting the BCR-ABL fusion in CML and KIT a non-fusion for GIST. The fusions included ALK, ROS1, NTRK, PML-RARA, and BCR-ABL. Non-fusion targets included EGFR, BRAF, KIT, BRCA, HER2, FLT3, IDH1, IDH2.
Response rates ranged 53%−79% for solid tumor fusions, 14%−75% for solid tumor non-fusions, 31%−84% for hematology fusions, and 21%−55% for hematology non-fusions. The median response rate was 68% vs. 50% for fusions (N = 2,654 patients) vs. non-fusions (N = 3,535 patients) with an odds ratio of 1.67 (p<0.0001) (Tables 1 and 2). For solid tumor therapies, median response rate was 74% vs. 51% for fusions vs. non-fusions with an odds ratio of 2.07 (p<0.0001). For hematology therapies median response rate was 55% vs. 28% (fusions versus non-fusions) with an odds ratio of 3.35 (p<0.0001). Thus, overall fusion targets gave significantly higher response rates than non-fusion targets.
Table 1:
Single agent targeted therapies approved for genomically altered cancers (mutation, amplification or fusion) and response rates*
| Solid tumors: Fusion target | |||||
| Drug | Disease Indication | Line of Therapy | Aberrant gene | Number of studies** | Response Rate (%)** |
| Alectinib | NSCLC | 2+ | ALK | 1 | 79% |
| Brigatinib | NSCLC | 2+ | ALK | 1 | 53% |
| Ceritinib | NSCLC | 2+ | ALK | 1 | 73% |
| Crizotinib | NSCLC | 1 | ALK/ROS1 | 2 | 56% |
| Entrectinib | Solid tumors | 1+ | NTRK/ROS1/ALK | 1 | 78% |
| Larotrectinib | Solid tumors | 1+ | NTRK | 1 | 75% |
| Solid tumor: Non-fusion target | |||||
| Drug | Disease Indication | Line of Therapy | Aberrant gene | Number of studies** | Response Rate (%) |
| Afatinib | NSCLC | 1+ | EGFR | 1 | 50% |
| Dabrafenib | Melanoma | 1+ | BRAF | 1 | 54% |
| Dacomitinib | NSCLC | 1+ | EGFR | 1 | 75% |
| Erlotinib | NSCLC | 1+ | EGFR | 1 | 65% |
| Gefitinib | NSCLC | 1 | EGFR | 1 | 70% |
| Imatinib | GIST | 1 | KIT | 1 | 51% |
| Olaparib | Ovarian | 4+ | BRCA | 1 | 34% |
| Osimertinib | NSCLC | 1+ | EGFR | 1 | 51% |
| Rucaparib | Ovarian | 3+ | BRCA | 1 | 54% |
| Talazoparib | Breast | 2+ | BRCA | 1 | 50% |
| TDM-1 | Breast | 2+ | HER2 | 1 | 44% |
| Trastuzumab | Breast | 2+ | HER2 | 1 | 14% |
| Vemurafenib | Melanoma | 1+ | BRAF | 1 | 48% |
| Hematologic malignancies: Fusion Target | |||||
| Drug | Disease Indication | Line of Therapy | Aberrant gene | Number of studies** | Response Rate (%) |
| All trans-retinoic acid | APL | 1+ | PML-RARA | 1 | 72% |
| Bosutinib | CML | 2+ | BCR-ABL | 2 | 31% |
| Dasatinib | CML | 1+ | BCR-ABL | 2 | 63% |
| Imatinib | CML | 1 | BCR-ABL | 1 | 73% |
| Nilotinib | CML | 1+ | BCR-ABL | 1 | 84% |
| Ponatinib | CML | 2+ | BCR-ABL | 1 | 46% |
| Hematologic malignancies: Target non-fusion | |||||
| Drug | Disease Indication | Line of Therapy | Aberrant gene | Number of studies** | Response Rate (%) |
| Enasidenib | AML | 2+ | IDH2 | 1 | 23% |
| Gilteritinib | AML | 2+ | FLT3 | 1 | 21% |
| Ivosidenib | AML | 2+ | IDH1 | 1 | 33% |
| Vemurafenib | ECD | 1+ | BRAF | 1 | 55% |
Approved by the Food and Drug Administration (FDA)
Refers to number of studies provided in FDA package insert; Response rate is per package insert. If more than one study listed, the mean is given
Abbreviations: AML=acute myelogenous leukemia; CML=chronic myelogenous leukemia; ECD=Erdheim-Chester disease; NHL=non-Hodgkin’s lymphoma; NSCLC=non-small cell lung cancer
Table 2:
Summary of response rates to FDA-approved drugs directed at fusions versus non-fusion (mutations or amplifications) alterations (see also Table 1)
| Fusions | Non-fusions | |||||||
|---|---|---|---|---|---|---|---|---|
| Number of Drugs | Number of patients | Median response rate (IQR 25–75 percentile) | Number of drugs | Number of patients | Median response rate (IQR 25–75 percentile) | Odds ratio (95% CI) (response rate of fusions versus non-fusions) | Chi-square test for association p-value | |
| All Patients | 12 | 2654 | 68% (51%– 74%) | 17 | 3535 | 50% (34%–54%) | 1.67 (1.51–1.85) | <0.0001 |
| Solid Tumors | 6 | 784 | 74% (60%–77%) | 13 | 3002 | 51% (48%–54%) | 2.07 (1.75–2.44) | <0.0001 |
| Hematology | 6 | 1870 | 55% (42%–70%) | 4 | 533 | 28% (23%–38%) | 3.35 (2.71–4.14) | <0.0001 |
Abbreviations: CI = confidence interval; IQR = interquartile range
UCSD patient population
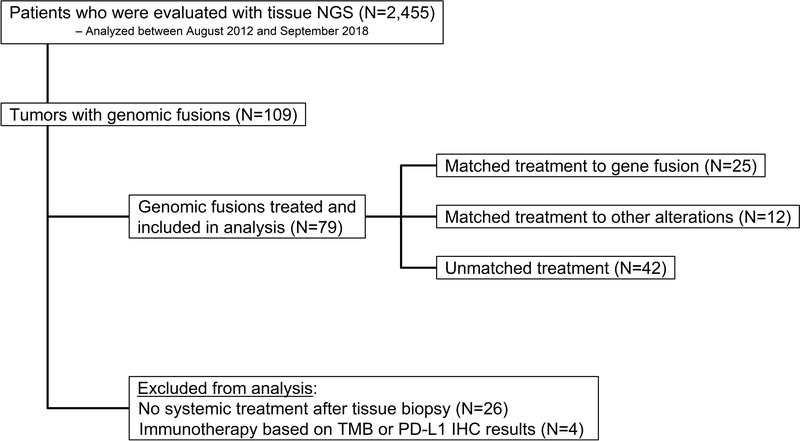
A total of 2,455 patients with next generation sequencing were screened for the study and 109 tumors with genomic fusions were identified (4.4% of samples). The consort diagram is depicted in Figure 1. Of the 79 patients with fusions who were treated and met eligibility, 25 received a therapy matched to a gene fusion, 12 received a therapy matched to another alteration, and 42 received unmatched treatment. Of these patients, 34 (43%) had fusions that were predicted to result in oncogenic tyrosine kinase activity. Patients treated with immunotherapy alone based on tumor mutational burden of PD-L1 immunohistochemistry were excluded from the analysis (N=4). A total of 26 patients did not have documented systemic treatment and were excluded from the analysis. Additional details on the patients and therapies administered are provided in Supplemental Tables 1A–C.
Figure 1.
Consort diagram for the study. Overall, genomic fusions were found in 4.4% of all samples (109/2,455).
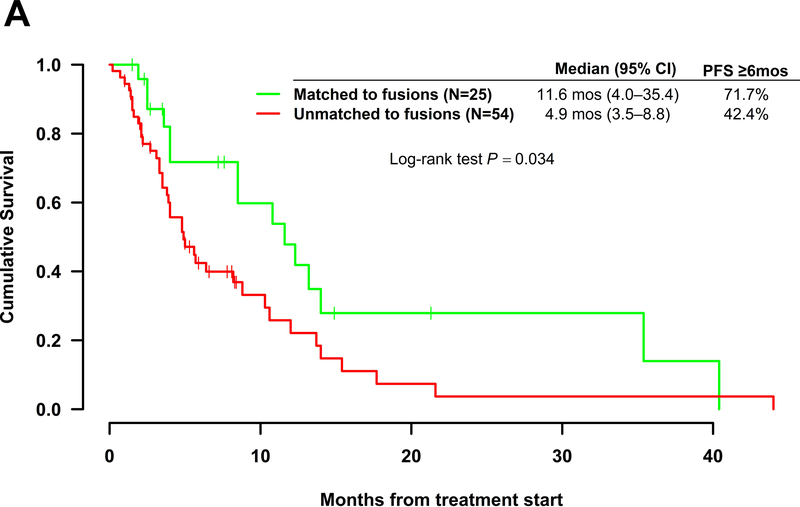
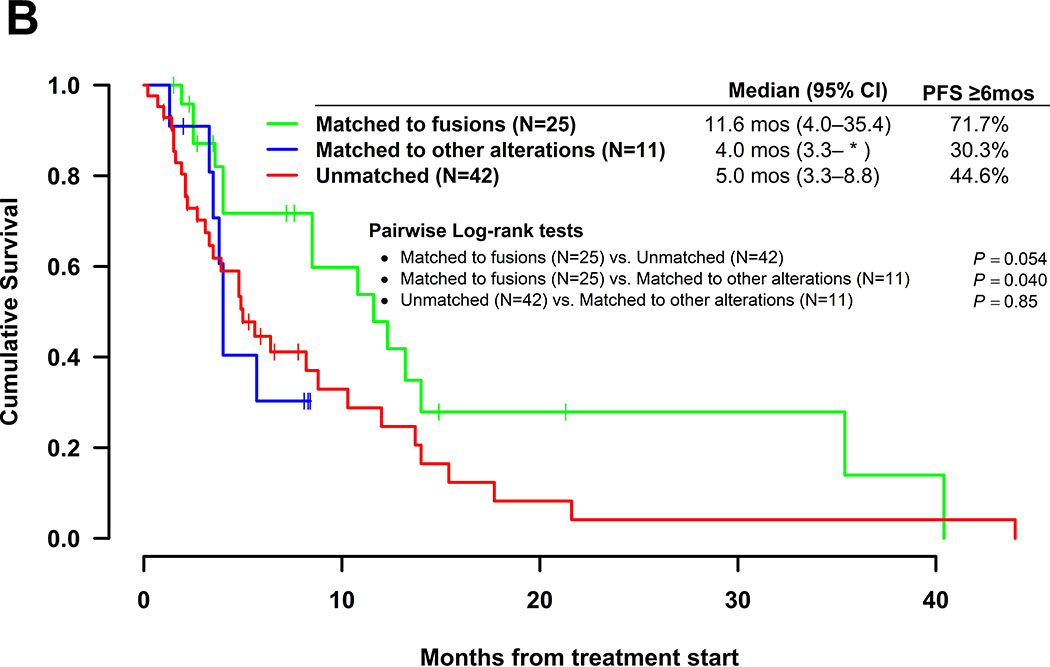
Kaplan Meir plots for PFS are shown for the 79 patients with fusions in Figure 2. Patients matched to fusions had longer median [95% confidence interval] PFS (11.6 (4.0 – 35.4) months) as compared to those unmatched to fusions (4.9 (3.5–8.8) months) (includes matched to other alterations or unmatched), which was significant by the log rank test (p=0.034). Pairwise comparisons revealed no significant differences in median PFS for any of the groups (matched to fusions, matched to other alterations, and unmatched). However, patients on therapies matched to fusions had PFS trending toward significance (median = 11.6 versus 4 months) when compared to those patients with fusions who were matched to other alterations (p=0.098). Importantly amongst patients with fusions, those matched to another alteration had no difference in median PFS as compared to those that were unmatched (4.0 versus 5.0 months; p = 0.75).
Figure 2.
Kaplan Meir curves for progression-free survival. All patients harbored tumors bearing fusion mutations and received systemic therapies (N=79). A. Therapy matched to fusion alteration (N=25) vs. therapy unmatched to fusion alteration (N=54). B. Pairwise comparisons for patients on therapies matched to fusion alteration (N=25) vs. therapies matched to a non-fusion alteration (N=11) vs. unmatched therapies (N=42). One patient in the matched group had an unclear match and was excluded from the pairwise comparison analysis. *Upper limit of 95% confidence interval was not estimable.
DISCUSSION
Metastatic cancers have complex molecular landscapes 19 with biological heterogeneity existing between histologies and individual patients 20–24. Thus, selecting therapies based on the molecular profile of an individual tumor is important for treating advanced cancer, and some (but not all) recent clinical trials of matching patients to therapies based on the molecular alterations found in their tumors have resulted in improved response rates and outcomes 25–27. Many malignancies will have several targetable mutations and identifying which are the most important for tumor growth is important for a precision medicine approach to cancer therapy.
The current study evaluates the importance of matching therapies to fusions. The analysis of FDA-approved single-agent targeted therapies (N = 6,189) found significantly higher response rates in patients matched to drugs based on their fusions as compared to non-fusion matches; these differences held for subset comparisons of hematologic and solid tumor therapeutics.
We also reviewed our UCSD patient population for patients with fusions and compared PFS of patients with fusions on therapies matched to fusions vs. those with fusions but matched to other alterations or unmatched. We found that, in patients with fusions, treatments regimens matched to fusions gave significantly improved PFS compared to those not matched to fusions (median = 11.6 versus 4.9 months; p=0.034). Furthermore, patients with fusions matched to other alterations had indistinguishable PFS as compared to those patients with fusions that were unmatched (median 4 versus 5 months; p = 0.75). On the other hand, patients with fusions matched to drugs based on the fusions showed a trend towards longer PFS than patients with fusions matched to other alterations (median = 11.6 versus 4 months; p=0.098). While the current data set is limited, this suggests that there may be a lack of benefit in matching patients to drugs on the basis of other alterations in the presence of a fusion. This observation merits investigation with a larger sample size.
A prior study evaluated 59 patients treated with matched targeted therapies for fusions in the phase 1 clinic at MD Anderson and found superior median PFS (7.1 months) and overall survival (OS) (19.6 months) as compared to patients historically matched to mutations (PFS = 5.2, OS = 13.4 months) or unmatched (PFS = 2.2, OS = 9 months) 28.
A prior study of 7000 tumors from The Cancer Genome Atlas project found the 3.0% of tumors contained a likely oncogenic kinase fusion. Thyroid cancer was found to have the highest frequency of fusions at 13%, while other tumors such as renal cell carcinoma had much lower frequencies 29. Other studies found <1% of fusions in the majority of cancers, but higher numbers of fusions in endocrine system tumors (35%), bone tumors (15%), soft tissue tumors (20%), and male genital organ tumors (80%) 30. Thus, fusions are not common events in most solid tumors; however they represent important therapeutic targets when they are identified. Many hematologic malignancies harbored higher frequencies of fusions; acute myelogenous leukemia (20% of patients); acute lymphoblastic leukemia (30%); B-cell neoplasms (30%); T-cell neoplasms (15%), and chronic myelogenous leukemia (100%) 30.
Thus, fusions are present in a wide variety of solid tumors in addition to hematologic malignancies. Many fusions lead to a state of oncogenic addiction which makes them ideal targets for anticancer therapeutics. The discovery of BCR-ABL and a therapy targeted to this fusion transformed CML into a chronic condition 5–7. The NTRK inhibitor larotrectinib has an unprecedented 75% response rate for metastatic solid tumors 11. The presence of a fusion may provide additional targets to the fusion itself. Indeed, fusions have been suggested to be important for neoantigens which generate cytotoxic T-cell responses and can help mediate responses to immunotherapy 31.
The current study was limited by small sample size in the UCSD patient population, but we were still found a significant difference between therapies targeted to fusions and those not targeted to fusions. Differences in the specific fusions found between patients in the matched to other alterations vs. those matched to fusions may also have contributed to the difference seen between these groups. The analysis of FDA-approved single-agent targeted therapies had large numbers of patients but was limited to studies listed on the package inserts. Significant differences between fusion and non-fusion targets were found for all groups. The analysis only focused on FDA-approved fusion and non-fusion targeted therapeutics and thus only represents successful, effective therapies. While targeting these fusions led to remarkable clinical outcomes, drug development for other fusions has been challenging. Oxaliplatin and cisplatin were considered targeted therapies for ATM and BRCA alterations, however it is important to note that these agents work by inducing DNA damage and taking advantage of the defective DNA repair mechanisms in cancer cells rather than directly targeting DNA mutations themselves.
In conclusion, we demonstrated improved response rates and PFS in targeting fusions vs. non-fusions. In our patients, targeting non-fusions in patients with fusions gave results identical to those with unmatched therapies. However, this observation was made in a small heterogeneous group of patients and therefore needs to be confirmed in larger studies. When taking the FDA approval results in concert with the UCSD patient database, the observations suggest that targeting fusions is important for both hematologic and solid malignances when they are present.
Supplementary Material
Acknowledgements/Funding
This work was supported by the Joan and Irwin Jacobs philanthropic fund and National Cancer Institute grant P30 CA023100.
Disclosures: Dr. Kurzrock receives consultant fees from Actuate Therapeutics, X-Biotech and NeoMed, as well as research funds from Incyte, Genentech, Pfizer, Foundation Medicine, Guardant, Sequenom, and Merck Serono, speaker fees from Roche, and has an ownership interest in Curematch Inc.
References
- 1.National Cancer Institute: Targeted Cancer Therapies. https://www.cancer.gov/about-cancer/treatment/types/targeted-therapies/targeted-therapies-fact-sheet Accessed: December 12, 2018.
- 2.Chae YK, Pan AP, Davis AA, et al. Path toward Precision Oncology: Review of Targeted Therapy Studies and Tools to Aid in Defining “Actionability” of a Molecular Lesion and Patient Management Support. Mol Cancer Ther. 2017;16: 2645–2655. [DOI] [PubMed] [Google Scholar]
- 3.Parker BC, Zhang W. Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment. Chin J Cancer. 2013;32: 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol. 2017;14: 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurzrock R, Shtalrid M, Romero P, et al. A novel c-abl protein product in Philadelphia-positive acute lymphoblastic leukaemia. Nature. 1987;325: 631–635. [DOI] [PubMed] [Google Scholar]
- 6.Kurzrock R, Gutterman JU, Talpaz M. The molecular genetics of Philadelphia chromosome-positive leukemias. N Engl J Med. 1988;319: 990–998. [DOI] [PubMed] [Google Scholar]
- 7.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344: 1031–1037. [DOI] [PubMed] [Google Scholar]
- 8.Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo-Coco F, Avvisati G, Vignetti M, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369: 111–121. [DOI] [PubMed] [Google Scholar]
- 11.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med. 2018;378: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis Oncol. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Huo X, Tang C, et al. Frequent KIT mutations in human gastrointestinal stromal tumors. Sci Rep. 2014;4: 5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417: 949–954. [DOI] [PubMed] [Google Scholar]
- 15.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res. 2015;5: 2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 16.Iqbal N, Iqbal N. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol Biol Int. 2014;2014: 852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.US Food and Drug Administration: Hematology/Oncology (Cancer) Approvals & Safety Notifications. https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm279174.htm. Accessed December 12, 2018.
- 18.Sun J, Wei Q, Zhou Y, Wang J, Liu Q, Xu H. A systematic analysis of FDA-approved anticancer drugs. BMC Syst Biol. 2017;11: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurzrock R, Giles FJ. Precision oncology for patients with advanced cancer: the challenges of malignant snowflakes. Cell Cycle. 2015;14: 2219–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheler JJ, Parker BA, Lee JJ, et al. Unique molecular signatures as a hallmark of patients with metastatic breast cancer: implications for current treatment paradigms. Oncotarget. 2014;5: 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wheler J, Lee JJ, Kurzrock R. Unique molecular landscapes in cancer: implications for individualized, curated drug combinations. Cancer Res. 2014;74: 7181–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18: 6373–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodon J, Soria JC, Berger R, et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med. 2019;25: 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sicklick JK, Kato S, Okamura R, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwaederle M, Parker BA, Schwab RB, et al. Precision Oncology: The UC San Diego Moores Cancer Center PREDICT Experience. Mol Cancer Ther. 2016;15: 743–752. [DOI] [PubMed] [Google Scholar]
- 28.Groisberg R, Hess KR, Hong DS, et al. Outcomes of patients with gene fusion driven cancers treated on early phase clinical trials American Society of Clinical Oncology. Chicago, Illinois; ., 2018. [Google Scholar]
- 29.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5: 4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7: 233–245. [DOI] [PubMed] [Google Scholar]
- 31.Yang W, Lee KW, Srivastava RM, et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat Med. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.