Abstract
Sudden unexpected death in epilepsy (SUDEP) is the leading cause of epilepsy-related mortality, but the precise cellular substrates involved remain elusive. Epilepsy-associated ion channel genes with co-expression in brain and heart have been proposed as SUDEP candidate genes since they provide a singular unifying link between seizures and lethal cardiac arrhythmias. Here, we generated a conditional knockout (cKO) mouse with neuron-specific deletion of Kcna1, a SUDEP-associated gene with brain-heart co-expression, to test whether seizure-evoked cardiac arrhythmias and SUDEP require the absence of Kv1.1 in both brain and heart or whether ablation in neurons is sufficient. To obtain cKO mice, we developed a floxed Kcna1 mouse which we crossed to mice with the Synapsin1-Cre transgene, which selectively deletes Kcna1 in most neurons. Molecular analyses confirmed neuron-specific Kcna1 deletion in cKO mice and corresponding loss of Kv1.1 except in cerebellum where Synapsin1-Cre is not highly expressed. Survival studies and electroencephalography, electrocardiography, and plethysmography recordings showed that cKO mice exhibit premature death, epilepsy, and cardiorespiratory dysregulation but to a lesser degree than global knockouts. Heart rate variability (HRV) was increased in cKO mice with peaks during daytime suggesting disturbed diurnal HRV patterns as a SUDEP biomarker. Residual Kv1.1 expression in cKO cerebellum suggests it may play an unexpected role in regulating ictal cardiorespiratory dysfunction and SUDEP risk. This work demonstrates the principle that channelopathies with brain-heart expression patterns can increase death risk by brain-driven mechanisms alone without a functionally compromised heart, reinforcing seizure control as a primary clinical strategy for SUDEP prevention.
INTRODUCTION
Sudden unexpected death in epilepsy (SUDEP) is the leading cause of epilepsy-related mortality, claiming the lives of ~7% of people with epilepsy by age 40 and causing the second most years of potential life lost of any neurological disease, second only to stroke (Sillanpää and Shinnar, 2010; Thurman et al., 2014). SUDEP is generally defined as the unpredictable and unanticipated death of a reasonably healthy person with epilepsy, where no cause of death can be found (Dlouhy et al., 2016). The primary suspected mechanism is that seizures propagate to cardiorespiratory centers in the brain disturbing cardiac and/or respiratory physiology resulting in death (Dlouhy et al., 2016). However, the precise underlying causes and cellular substrates of SUDEP remain elusive.
The genetic architecture of SUDEP is complex and heterogeneous with multiple and diverse genes contributing to risk susceptibility (Leu et al., 2015; Bagnall et al., 2016). Exome-based analyses of rare variants in SUDEP patients indicate that ≥40% of cases are associated with pathogenic mutations in epilepsy (brain) or cardiac arrhythmia (heart) genes; however, no single gene rises to genome-wide significance (Bagnall et al., 2016). Epilepsy-associated ion channel genes with expression in both brain and heart have been proposed as candidate genes for SUDEP susceptibility since they provide a singular molecular link between epilepsy and lethal cardiac arrhythmias (Glasscock, 2014; Massey et al., 2014; Goldman et al., 2016). Conceptually, these brain-heart genes can influence cardiac function intrinsically or extrinsically via the autonomic nervous system increasing risk of SUDEP (Glasscock, 2014).
One such brain-heart ion channel gene that is associated with SUDEP risk in mice and humans is the human epilepsy gene KCNA1, which encodes Kv1.1 voltage-gated potassium channel α-subunits (Zuberi et al., 1999; Glasscock et al., 2010; Klassen et al., 2014). Kv1.1 subunits are best known for their functional role in neurons where they act to dampen membrane excitability and regulate action potential firing properties (Jan and Jan, 2012). Mice lacking Kv1.1 due to global Kcna1 gene knockout (i.e., Kcna1−/−) have been used extensively to study the pathophysiology of SUDEP by brain-driven mechanisms. Similar to observations in patients, Kcna1−/− mice exhibit seizure-related sudden death with postictal electrocerebral shutdown and bradyarrhythmias which culminate in cardiorespiratory arrest (Glasscock et al., 2010; Lhatoo et al., 2010; Ryvlin et al., 2013; Moore et al., 2014; Aiba and Noebels, 2015; Dhaibar et al., 2019). Autonomic dysregulation appears to contribute to the deleterious cardiac dysfunction in Kcna1−/− mice, as suggested by numerous findings in these animals including an increased frequency of atropine-sensitive atrioventricular (AV) conduction blocks; peri-ictal bradycardia; vagal axonal hyperexcitability; increased heart rate variability (HRV); prolonged lifespan with vagotomy; and abnormal brain-heart interaction dynamics (Glasscock et al., 2010, 2012; Moore et al., 2014; Mishra et al., 2017; Glasscock, 2019 p.1).
Although traditionally Kv1.1 has been thought to be absent in the heart, recent discoveries have demonstrated that Kv1.1 subunits are indeed present in cardiomyocytes where they control action potential morphology and contribute to arrhythmia susceptibility (Glasscock et al., 2015; Si et al., 2018). Therefore, Kv1.1 deficiency in the heart may contribute to the cardiac abnormalities in Kcna1−/− mice by rendering the heart a vulnerable substrate for cardiac dysfunction triggered by excessive neural activity (i.e., seizures or vagal hypertonia). In this study, we explore the anatomical and cellular origins of neurocardiac dysfunction and SUDEP due to Kv1.1 deficiency by generating neuron-specific Kcna1 conditional knockout (cKO) mice via a newly developed floxed Kcna1 allele. Using neuron-specific cKO mice, this work addresses the question of whether cardiac abnormalities and premature death in Kcna1−/− mice require the absence of Kv1.1 in both the heart and the brain or whether neuron-specific Kv1.1 deficiency is sufficient for manifestation of neurocardiac phenotypes. Furthermore, this work examines the broader question of whether intrinsic cardiac dysfunction contributes to SUDEP risk when mutations occur in epilepsy genes with brain-heart expression.
MATERIALS AND METHODS
Animals
Neuron-specific Kcna1 conditional knockout (cKO) mice (i.e., Synapsin1-Cre+/−; Kcna1del/− were generated by crossing heterozygous Kcna1 floxed (fl) mice (Kcna1fl/+) with heterozygous Kcna1 global knockout (KO) mice (Kcna1+/−) carrying one copy of the Synapsin1-Cre transgene (i.e., Synapsin1-Cre+/−, Kcna1+/−). Synapsin1-Cre+/−, Kcna1+/− mice were generated by crossing hemizygous transgenic Synapsin1-Cre+/− mice with heterozygous Kcna1+/− mice. Synapsin1-Cre (Syn1-Cre) mice were purchased from Jackson Labs (Bangor, MN) where they are cataloged as B6.Cg-Tg(Syn1-cre)671Jxm/J (JAX 003966). Kcna1+/− mice, which are maintained on a Tac:N:NIHS-BC genetic background, carry a null (KO) allele of the Kcna1 gene due to targeted deletion of the entire open reading frame (Smart et al., 1998). Generation of the Kcna1fl/+ mice, which are maintained on a C57BL/6N background, is described below. Mice were housed at 22°C, fed ad libitum, and submitted to a 12-h light/dark cycle. All experiments were performed in accordance with National Institutes of Health (NIH) guidelines with approval from the Institutional Animal Care and Use Committee of the Louisiana State University Health Sciences Center-Shreveport.
Generation of floxed Kcna1 mice
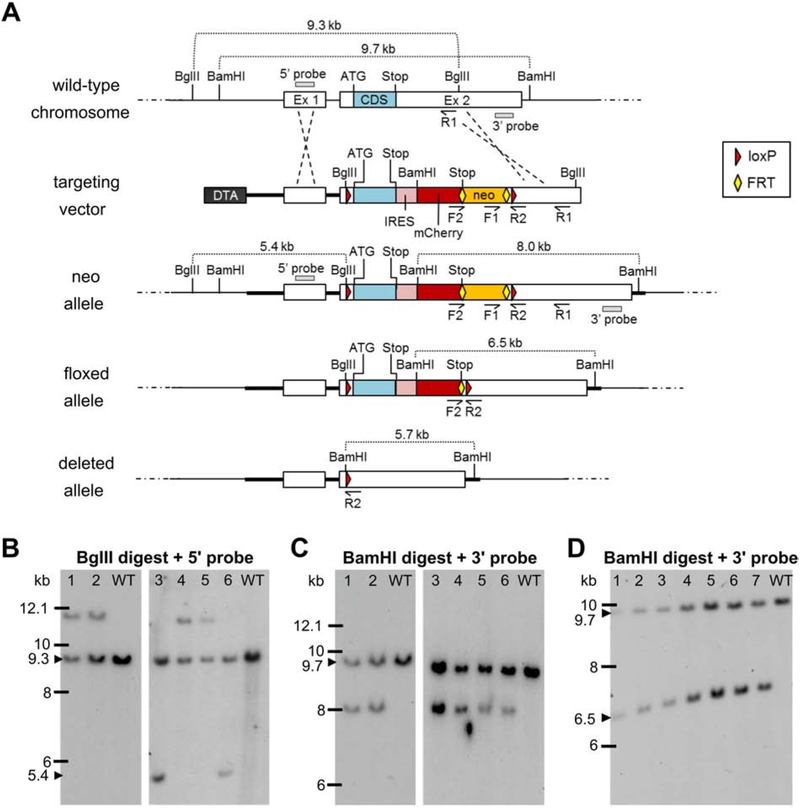
Mice carrying floxed alleles of Kcna1 (Kcna1fl) were developed in collaboration with the company genOway (Lyon, France) using a homologous recombination targeting strategy in mouse embryonic stem (ES) cells (summarized in Fig. 1). In brief, the mouse Kcna1 gene, located on chromosome 6F1-F3, is composed of 2 exons spanning ~9.3 kb of genomic DNA The entire 495-amino acid protein coding sequence (CDS) is contained in the 5’ portion of exon 2. Three DNA sequences spanning upstream of exon 1 to the 3’ untrans lated region (UTR) of exon 2 were PCR amplified from mouse genomic C57BL/6 ES cell DNA to generate the long (4.8 kb) and short (3 kb) homology arms necessary for assembly of the targeting vector. The resulting PCR products were subcloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA) via TA-cloning and the subclones sequenced to confirm the absence of mutations in the regions of interest before selection for the targeting vector construction. Utilizing the subcloned genomic Kcna1 sequences, the targeting vector plasmid (13.9 kb) was then generated by insertion of: (1) two loxP sites flanking the exon 2 CDS (one in the 5’ UTR and the other between the end of the CDS and the beginning of the 3’ UTR); (2) a Diphtheria Toxin A (DTA) marker; (3) an FRT-flanked neomycin gene cassette; and (4) an IRES (internal ribosome entry site)-mCherry reporter. The presence of the DTA cassette acted as a negative selection marker reducing the isolation of non-homologous recombined ES cell clones and enhancing the chance of isolating ES cell clones harboring the distal loxP site. The FRT-flanked neo cassette acted as a positive ES cell selection marker and was removed in vivo with Flp recombinase-expressing deleter mice. mCherry was cloned using the pmCherry vector #632522 from Clontech Laboratories, Inc (Mountain View, CA). The IRES-mCherry cassette was designed to act as a fluorescent reporter approximating expression of the floxed, non-deleted Kcna1 gene.
Figure 1.
Targeting strategy for generation and verification of the floxed Kcna1 allele. A, Utilizing subcloned genomic sequences from the endogenous wild-type Kcna1 chromosomal region, a targeting vector was generated containing two loxP sites (red triangles) flanking the exon 2 (Ex 2) coding sequence (CDS); a Diphtheria Toxin A (DTA) negative selection marker; a neomycin (neo) cassette positive-selection marker flanked by two FRT sites (yellow diamonds); and an internal ribosomal entry site (IRES)-mCherry fluorescent reporter. The targeting construct was introduced into embryonic stem (ES) cells to obtain the neo allele by homologous recombination. Recombined ES cell clones were then used to generate chimeric males which were mated to females with Flp recombinase to excise the neo cassette in the germline, eventually yielding mice with the floxed allele following another round of breeding. In the presence of Cre recombinase, the CDS and IRES-mCherry reporter of the floxed allele are removed to yield the deleted allele. The F1/R1 primer pair was used to screen ES cells by PCR for 3’ homologous recombination. The F2/R2 primer pair was used to identify by PCR heterozygous floxed mice with Flp-mediated excision of the neo cassette. Restriction enzyme sites for BgIII and BamHI are indicated, as well as the locations of the 5’ and 3’ probes used for Southern blotting. B-C, Southern blots of ES cell DNA digested with BgIII and BamHI to test for 5’ and 3’ homologous recombination, respectively, to yield the neo allele. Clone numbers 3 and 6 exhibited restriction fragments indicative of successful 5’ and 3’ recombination events. D, Southern blot of BamHI-digested genomic DNA from seven heterozygous floxed mice verifying Flp-mediated excision of the neo cassette.
The linearized targeting vector was transfected into ES cells from C57BL/6 mice using electroporation and 430 G418-resistant clones were recovered. To test for 3’ homologous recombination, PCR screening was performed using primers to F1 and R1 revealing 18 positive clones. Six of these recombined clones were further analyzed by Southern blot using probes targeted at the 5’ homology arm (5’ probe) and downstream of the targeting vector 3’ homology sequence (3’ probe). Southern blotting revealed two clones, numbers 3 and 6, with 5’ (Fig. 1B) and 3’ (Fig. 1C) fragments indicative of recombination, suggesting a targeting efficiency of about 0.5%. These two recombined ES cell clones were injected into C57BL/6J-Tyr blastocysts and re-implanted into OF1 pseudo-pregnant females, yielding 7 male mice with a high degree of chimerism between 50-100%. These highly chimeric males were mated to C57BL/6 Flp-deleter females to allow germline excision of the neomycin cassette. This breeding resulted in 4 pups with the non-excised recombined (neo) allele, which were then further mated to C576BL/6 Flp-deleter mice to excise the neomycin cassette yielding 7 animals heterozygous for the desired floxed Kcna1 allele, which was verified by PCR with the F2/R2 primer pair and Southern blotting with the 3’ probe (Fig. 1D).
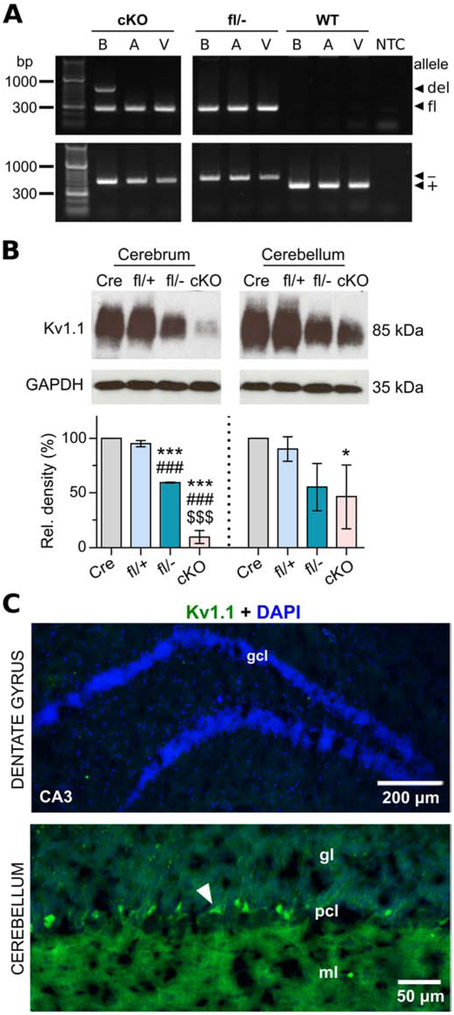
Figure 3.
Molecular and immunohistochemical characterization of neuron-specific Kcna1 cKO mice. A, PCR detection of the Kcna1 deletion (del; 679 bp), floxed (fl; 260 bp), null (−; 337 bp), and WT (+; 475 bp) alleles from genomic DNA isolated from brain (B), atrium (A), and ventricle (V) of cKO mice and Kcna1fl/− and WT controls. NTC, no template control. B, Representative Western blots for Kv1.1 and GAPDH loading control from the cerebrum and cerebellum of hemizygous Syn-Cre (Cre), heterozygous Kcna1 floxed (fl/+), compound heterozygous Kcna1 floxed/null (fl/−), and neuron-specific Kcna1 cKO (cKO) mice (1-2 months old) with corresponding quantification of Kv1.1 protein levels (n=3/genotype). C, Immunohistochemistry and fluorescence imaging in cKO mice reveals an absence of anti-Kv1.1 immunoreactivity (green) in the dentate gyrus but persistent expression in the cerebellum. However, the intensity of staining in basket cell terminals (white arrowhead) appeared less intense than Kcna1fl/+ mice. CA3, Cornu Ammonis of the hippocampus; DAPI, 4′,6-diamidino-2-phenylindole; gcl, granule cell layer; gl, granular layer; ml, molecular layer; pcl, Purkinje cell layer. *P < 0.05 vs. Cre; ***P < 0.0001 vs. Cre; ###P < 0.0001 vs. fl/+; $$$P < 0.0001 vs. fl/− (1-way ANOVA; post-hoc Tukey test).
Genotyping
Genomic DNA was isolated by enzymatic digestion of tail clips using Direct-PCR Lysis Reagent (Viagen Biotech, Los Angeles, CA, USA). Genotypes were determined by performing PCR amplification of genomic DNA using allele-specific primers. For detection of the Kcna1 global KO allele, the following primer sequences were used to yield amplicons of 337 bp for the wild-type (WT) allele and 475 bp for the KO allele: a WT-specific primer (5'-GCCTCTGACAGTGACCTCAGC-3'); a KO-specific primer (5'-CCTTCTATCGCCTTCTTGACG-3'); and a common primer (5'-GCTTCAGGTTCGCCACTCCCC-3'). For detection of the Syn1-Cre transgene, the following primer sequences were used to yield an amplicon of 450 bp: 5’-CTCAGCGCTGCCTCAGTCT-3’ and 5’-GCATCGACCGGTAATGCA-3’. As an internal positive control, the Syn1-Cre reaction was multiplexed with a primer pair that yields a 381 bp amplicon of the WT allele of the Kcnq1 gene: 5’-GCCTCACTATCCTGGTAGGC-3’ and 5’-GCCGCTTCTGTGAAGTACC-3’. For detection of the Kcna1fl allele, the following primer sequences were used to yield amplicons of 197 bp for the WT allele and 260 bp for the floxed allele: a WT-specific primer (5’-GCTCCTCTACTATCAGCAAGTCTGAGTACATGG-3’); a floxed-specific primer (5’-ATCAAGTTGGACATCACCTCCCACAAC-3’); and a common primer (5’-AAGGGGTTTGTTTGGGGCTTTTGTT-3’). For detection of the deleted Kcna1 floxed allele (i.e., Kcna1del) resulting from Cre-mediated recombination, the following primers were used to yield an amplicon of 679 bp for the deleted allele: the common primer in the Kcna1fl reaction above and 5’-CTCCAGTTTTACGAAGTTGTAAACAGATCGG-3’.
Western blotting
Age-matched mice of both sexes (1-3 months old) were euthanized by isoflurane overdose and their brains quickly removed and dissected on ice to separate cerebrum and cerebellum. Tissues were then homogenized with a mechanical shearer in ten volumes of ice-cold RIPA buffer (pH 7.4) containing EDTA and a cocktail of protease inhibitors (Thermo Scientific, Waltham, MA). Nuclei and cellular debris were removed from tissue homogenates by centrifuging at 9,300 relative centrifugal force (RCF) for 10 min at 4°C. The supernatants containing the crude membrane fractions were collected and stored at −80°C until used. Bradford reagent (Bio-Rad Laboratories, Hercules, CA) was used to determine protein concentrations of the brain homogenates and equal amounts of protein were resolved on 7.5% SDS-polyacrylamide gels by electrophoresis. The resolved proteins were then transferred onto nitrocellulose membranes by wet transfer at 4°C. The membranes were treated with blocking buffer made of phosphate buffered saline with Tween (PBST; 0.1% v/v) and milk protein (5% w/v) for 1 h at room temperature (RT; ~22°C) and then incubated overnight with mild shaking at 4°C in primary antibody solution prepared in blocking buffer. The primary antibodies used were mouse monoclonal anti-Kv1.1 (1:500; K20/78; Antibodies Incorporated; Davis, CA) and goat polyclonal anti-GAPDH conjugated to HRP (1:500; V-18; Santa Cruz Biotechnology, Dallas, TX). Following overnight incubation, the membranes were washed 3 times for 5 min with PBST and then the membranes treated with anti-Kv1.1 primary antibodies were further incubated for 1 h at RT with goat anti-mouse IgG-HRP secondary antibody (1:2000; Santa Cruz Biotechnology, Dallas, TX) in blocking buffer. After a final wash in PBST, immunoreactive bands were visualized by an enhanced chemiluminiscence (ECL) detection kit (GE Healthcare, Pittsburgh, PA) and developed on Amersham hyperfilm. Band intensity was quantified by spot densitometry analysis using ImageJ software (National Institutes of Health; Bethesda, MD), normalized to GAPDH levels, and reported as relative intensity of the control.
Immunohistochemistry
Mice of both sexes (1-4 months old) were euthanized by isoflurane overdose and serially intracardially perfused with phosphate buffered saline (PBS), 4% paraformaldehyde, and 2.5% paraformaldehyde with 4% sucrose. The brains were then removed and cryoprotected for 3 days by placing them in solutions with progressively increasing concentrations of sucrose for 1 day each (10%, 20% and 30% sucrose in PBS). Next, the brains were embedded in optimum cutting temperature (OCT) embedding medium and cut into 10 μm sections using a cryostat maintained at −20°C. Sections were directly mounted on slides and allowed to thaw at room temperature for ~30 minutes immediately before processing. Sections were blocked and permeabilized for 1 h in antibody vehicle (10% BSA, 0.3% Triton X-100 in PBS), followed by incubation in mouse monoclonal anti-Kv1.1 primary antibody (1:1000 dilution in vehicle; clone K20/78; Antibodies Incorporated; Davis, CA;) for 15-20 h at room temperature. Subsequently, sections were washed three times in vehicle and incubated for 1 h in Alexa Fluor 488 goat anti-mouse IgG1 secondary antibody (1:1000 dilution in vehicle; Invitrogen; Carlsbad, CA). Sections were then slide-mounted using ProLong Glass anti-fade reagent (Invitrogen, Carlsbad, CA). Images were captured using a Nikon A1R confocal microscope (Nikon Instruments Inc., Melville, NY) with Nikon NIS-Elements C software.
Cold swim seizure susceptibility
Individual 3-month old mice were placed in the middle of a tank of 17°C water to swim for 1 min. After swimming, the mice were placed in a dry and clean mouse cage without bedding at room temperature (~22°C) for observation.
Flurothyl seizure susceptibility
Mice (30-31 d old) were placed in an air-tight Plexiglas chamber (18.4 x 15.0 x 34.5 cm) and allowed to acclimate for five minutes before exposure to the liquid convulsant flurothyl (2,2,2-trifluoroethyl ether; SynQuest Laboratories, Alachua, FL). Flurothyl was infused (20 μL/min) with a syringe pump onto Whatman filter paper suspended at the top of the chamber from which it vaporized. Two observers recorded the latencies (in seconds) from the first drip of flurothyl to the onset of fully generalized (tonic-clonic) seizure, which typically consisted of about 3-5 s of falling down and/or running/bouncing. The latencies to these events served as surrogate measures of epileptogenicity (i.e., seizure susceptibility/threshold). Immediately following generalized seizure onset, the mice were quickly removed to fresh air and placed in a cage for observation and recovery. Each mouse was tested individually and received only one exposure to flurothyl. The testing chamber was cleaned and aerated between each trial.
Video-electroencephalography-electrocardiography-plethysmography (EEG-ECG-pleth) recordings
Mice (4-6 weeks old) of both sexes were anesthetized with either an anesthetic cocktail or avertin and then surgically implanted with bilateral silver wire EEG and ECG electrodes (0.005-inch diameter) attached to a microminiature connector (Omnetics Connector Corporation, Minneapolis, MN) for recording in a tethered configuration. The anesthetic cocktail contained ketamine (80-100 mg/kg), xylazine (6-10 mg/kg), and acepromazine (1-2 mg/kg) and was administered by intraperitoneal (i.p.) injection. Avertin (0.02 mL/g, i.p.) was prepared by mixing 250 mg 2,2,2-tribromoethanol (Sigma-Aldrich) with 155 μL 2-methyl-2-butanol (Sigma-Aldrich) dissolved in 19.75 mL double-distilled water heated to 40°C, followed by filter-sterilization prior to use. When anesthetic cocktail was used, the reversal agent Antisedan (1 mg/kg) was administered by subcutaneous (s.c.) injection immediately following completion of the surgical procedure. Carprofen (4-5 mg/kg, s.c.) was given the day of the surgery and 24 hours postsurgery for analgesia. EEG wires were inserted into the subdural space through cranial burr holes overlying parietotemporal cortex for the recording electrodes and above frontal cortex for the ground and reference electrodes, as described previously (Mishra et al., 2018). For ECG, two wires were tunneled subcutaneously on both sides of the thorax and sutured in place to record cardiac activity, as described previously (Mishra et al., 2018). Mice were allowed to recover from surgery for 24-48 h before recording simultaneous video and EEG-ECG for ≥24 h continuously while the animals were housed in a plexiglass container (40-cm length x 20-cm width x 23-cm height). Biosignals were band-pass filtered by applying 0.3-Hz high-pass and 200-Hz low-pass filters for EEG and a 3.0-Hz high-pass filter for ECG. Sampling rates were set to 500 Hz for EEG and 2 kHz for ECG.
For recording respiratory waveforms in combination with EEG-ECG, a subset of mice of both sexes were placed in an unrestrained whole-body plethysmography (pleth) chamber (Data Sciences International, St. Paul, MN, USA) with a lid that was modified to accommodate wires for recording EEG-ECG in a tethered configuration, as done previously (Dhaibar et al., 2019). Following a 45-min acclimatization period in the chamber, video and EEG-ECG-pleth were simultaneously recorded in 6-8 h sessions during the light phase of the day (i.e., between 6:00 am and 6:00 pm) using Ponemah data acquisition and analysis software (Data Sciences International, St. Paul, MN, USA). Respiratory signals were acquired at a 500-Hz sampling rate.
Analysis of video-EEG, ECG, and pleth recordings
Seizures were identified by visual inspection of the EEG and defined as high-amplitude, rhythmic electrographic discharges lasting ≥ 5 s. Seizure frequency was calculated as the number of seizures per hour of recording, which was then expressed as seizures per day. The seizure duration was defined as the time from the onset of electrographic seizure until the cessation of spiking.
The RR intervals of the ECG waveforms were used to estimate heart rate (HR) and heart rate variability (HRV) for the first 24-h recording period overall as a whole and for the 12-h day- (6:00 AM to 6:00 PM) and 12-h night-phase (6:00 PM to 6:00 AM) portions. For each ECG recording, six separate RR interval series were derived by sampling 2-min ECG segments every four hours to provide three light phase and three dark phase measurements. RR interval series for each segment were automatically generated and manually confirmed using Ponemah software (Data Sciences International, St. Paul, MN). RR intervals were only sampled during times when the mouse was stationary. To further improve the quality of the data, the derived RR intervals were edited to delete abnormal RR intervals caused by occasional physiological artifacts, such as skipped or ectopic beats, as described previously (Peltola, 2012). HRV was measured in the time domain using the standard deviation of the RR intervals (SDNN); the coefficient of variance (CV; defined as the SDNN/RRmean x100); the root mean square of successive differences between RR intervals (RMSSD); and the percentage of consecutive RR intervals differing by >6 ms (pNN6). The HR and HRV values for each animal represent an average of the six total segments or an average of the three daytime or three nightime segments, as indicated in the text.
Skipped heart beats (e.g., AV conduction blocks or sinus pauses) were identified from the ECG waveforms over the entire 24-h recording period and defined as a prolongation of the RR interval equaling ≥ 1.5 times the previous RR interval.
Respiratory rate (breaths per min; BPM); tidal volume (Tv); and minute ventilation (Mv) were measured using Ponemah analysis software (Data Sciences International, St. Paul, MN). Average respiratory characteristics for each mouse were calculated by sampling 50 consecutive breaths every hour for 5 h for a total of 250 breaths, as done previously (Dhaibar et al., 2019). Measurements were only obtained during periods when animals were stationary, as verified by video monitoring. Respiratory variability was calculated as the coefficient of variance (CV) using the formula: CV=σ/μ, where σ is standard deviation of breath intervals and μ is mean of breath intervals. Sighs and apneas were also manually quantified. Sighs were defined as deep augmented breaths with a ≥25% higher amplitude and ≥125% higher tidal volume compared to the preceding 10 s, whereas apneas were identified as cessations of plethysmographic signals for ≥2 respiratory cycles, or 0.8-s, as done previously (Dhaibar et al., 2019).
Statistical analysis
Data are presented as mean ± SEM. Prism 6 for Windows (GraphPad Software Inc, La Jolla, CA, USA) was used for statistical analyses. Survival curves were evaluated using the Kaplan-Meier log-rank (Mantel-Cox) test. For comparisons involving only two groups, unpaired two-tailed Student’s t tests were employed. For comparisons involving more than two groups, oneway analysis of variance (ANOVA) was used followed by Tukey post-hoc tests. Outliers were identified using the ROUT method with Q = 1% and excluded from analyses. Results were considered significant if P < 0.05.
RESULTS
Characterization of floxed Kcna1 mice
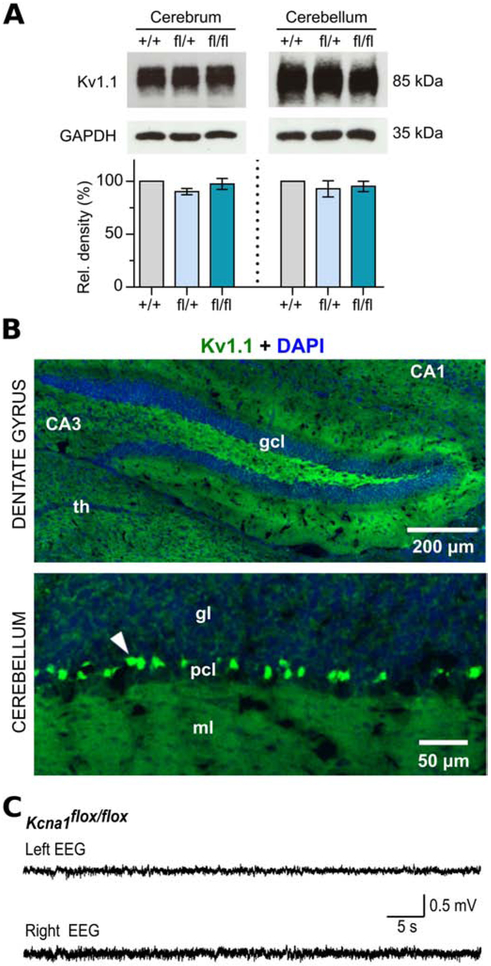
Kcna1fl/fl and Kcna1fl/+ mice are viable with no obvious abnormalities in general health or gross behavior. However, mice from the Kcna1fl strain are difficult to breed, probably due to their C57BL/6N genetic background, which in some studies has been associated with increased pup mortality due to strain-specific differences in maternal behaviors (Brown et al., 1999; Gaskill et al., 2013). Although litters of Kcna1fl/+ breeding pairs are normal sized (about 6-8 pups per litter), the number of pups that survive to weaning is often only about 2-3 animals per litter due to maternal cannibalism or neglect. Immunoblots of brain tissue from floxed mice demonstrated that the allele does not significantly alter Kv1.1 protein levels (Fig 2A). In addition, immunostaining showed the normal expression pattern of Kv1.1 protein in the brains of floxed mice (Fig 2B). In electroencephalography (EEG) recordings, Kcna1fl/fl mice exhibited normal brain activity with no evidence of seizures (Fig 2C). When forced to swim in cold (17°C) water, Kcna1fl/fl mice (n=4) did not show cold-swim induced neuromyotonia, characterized by violent tremors with closed eyes and flickering whiskers, which is a consistent response to cold swims in Kcna1 global knockout mice (i.e., Kcna1−/− (Zhou et al., 1998). Thus, the Kcna1fl allele is comparable to the normal wild-type (WT) allele of Kcna1 and does not appear to act as a partial loss-of-function mutation.
Figure 2.
Molecular, immunohistochemical, and electrophysiological characterization of floxed Kcna1 mice. A, Representative Western blots for Kv1.1 and GAPDH loading control from the cerebrum and cerebellum of WT (+/+), heterozygous Kcna1 floxed (fl/+), and homozygous Kcna1 floxed (fl/fl) mice (~3 months old) with corresponding quantification of Kv1.1 protein levels (n=3/genotype). B, Immunohistochemistry and fluorescence imaging of dentate gyrus and cerebellum in Kcna1fl/+ mice reveals anti-Kv1.1 immunoreactivity (green) in the expected WT pattern for Kv1.1, such as in basket cell terminals in the cerebellum (white arrowhead). CA1 and CA3, Cornu Ammonis regions of the hippocampus; DAPI, 4′,6-diamidino-2-phenylindole; gcl, granule cell layer; gl, granular layer; ml, molecular layer; pcl, Purkinje cell layer; th, thalamus. C, EEG recording from a Kcna1fl/fl mouse showing normal brain activity.
Generation of neuron-specific Kcna1 conditional knockout (cKO) mice
To determine the underlying neural contribution of Kv1.1 deficiency to cardiac dysfunction and SUDEP, Kcna1fl/− mice were crossed to transgenic Synapsin1-Cre (Syn1-Cre) mice to generate Kcna1 conditional knockout (cKO) animals (i.e., Syn1-Cre+/−; Kcna1del/−) with the Kcna1fl allele deleted (Kcna1del) selectively in neurons. The Syn1-Cre transgene has been used successfully to induce neuronal gene deletion in other studies of neurogenic SUDEP (Yuskaitis et al., 2018, 2019). The Syn1-Cre driver exhibits approximately pan-neuronal Cre expression, targeting most differentiated neurons in the brain (including brainstem) and spinal cord; however, expression is lower in cerebellum and absent in astroglia (Zhu et al., 2001; Yu et al., 2011; Ferguson et al., 2012; Ruisu et al., 2013; Yu and Lieberman, 2013). Kcna1fl/− mice were used as breeders to generate cKOs instead of Kcna1fl/fl mice for several reasons: 1) usage of Kcna1fl/− mice prevented the potential of unwanted germline recombination (Rempe et al., 2006) since parental germ cells carrying both the Cre and floxed alleles were never generated; (2) the presence of only a single floxed allele has the potential advantage of higher efficiency Cre recombination; and (3) breeding difficulties associated with Kcna1fl/fl mice limited their availability. Importantly, despite 50% reduction in Kcna1 gene dosage, Kcna1+/− heterozygotes are electrophysiologically indistinguishable from WT at the levels of EEG and nerve activity (Smart et al., 1998).
PCR, immunoblots, and immunohistochemistry confirmed tissue-specific deletion of Kcna1 in the neurons of cKO mice. PCR amplification of regionally-isolated genomic DNA revealed the presence of the Kcna1del allele in the brains of cKO mice but not in atrial or ventricular tissues of the heart, confirming neuron-specific gene deletion (Fig. 3A). In addition to the Kcna1del allele, the Kcna1fl allele was also detectable by PCR in the brains of cKO mice probably due primarily to the presence of other Kv1.1-positive non-neuronal cell types in which Syn1-Cre is not expressed. Western blot experiments showed a nearly complete absence of Kv1.1 protein in the cerebrum of cKO mice compared to WT controls (Fig. 3B). The small amount of residual Kv1.1 expression in cKO cerebrum may be due in part to glia where Kv1.1 has been observed at low levels (Smart et al., 1997; Hallows and Tempel, 1998) or a lack of recombination of the floxed allele in a small fraction of neurons (May et al., 2004; Ren et al., 2013). The cerebrum of Kcna1fl/− mice exhibited about a 50% reduction in Kv1.1 protein levels, which was expected since they carry one null allele. In accordance with previous reports that the Syn1-Cre driver does not express Cre at a high level in the cerebellum (Zhu et al., 2001), immunoblotting revealed high levels of Kv1.1 protein remaining in cKO cerebellum which were indistinguishable from Kcna1fl/− mice (Fig. 3B). Immunostaining further confirmed the absence of Kv1.1 protein in the neurons of cKO brain, except in the cerebellum where Kv1.1 immunoreactivity persisted (Fig. 3C).
Premature death and epilepsy in neuron-specific Kcna1 cKO mice
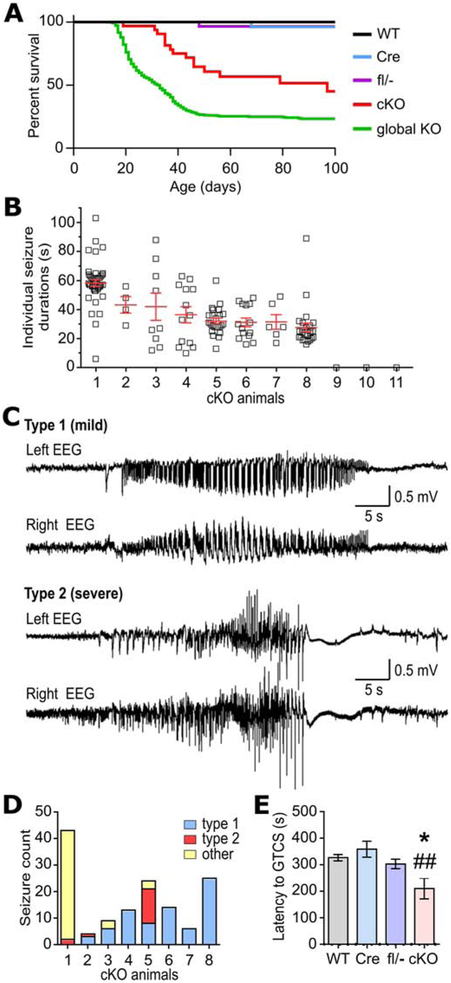
Kcna1 cKO mice began to exhibit premature death beginning between the fourth and fifth postnatal weeks (Fig. 4A). cKO animals had a median survival of 97 d with about 45% living to 100 d. In contrast, global Kcna1 KO animals begin dying about two weeks earlier than cKO mice (between postnatal weeks two and three) and at a higher rate (Fig. 4A). In global KO mice, median survival is 33 d and only 23% live to 100 d. Except for a few rare sporadic deaths, no control genotypes exhibited early lethality (Fig. 4A). Although no deaths were directly observed in cKO mice, their early lethality was most likely seizure-induced since dead animals were found in outstretched positions indicative of tonic extension.
Figure 4.
Premature death and seizure phenotypes in neuron-specific Kcna1 cKO mice. A, Kaplan-Meier survival curves for WT (n=50), hemizgous Syn1-Cre (Cre; n=36), compound heterozygous Kcna1 floxed/null (fl/−; n=32), neuron-specific Kcna1 cKO (cKO; n=32), and global Kcna1 knockout (global KO; n=553) mice. In cKO mice, survival was significantly decreased compared to Cre, fl/−, and WT controls (P < 0.0001, Log-rank test) but significantly increased compared to global KO mice (P = 0.0002, Log-rank test). B, Plot of individual seizure durations for every seizure recorded in each cKO animal in the study. C, Representative EEG traces from the left and right recording electrodes in cKO mice showing the two main types of seizure activity based on electrographic pattern. Type 1 seizures were generally associated with mild, non-convulsive behaviors, whereas type 2 seizures were associated with more severe, tonic-clonic behaviors. D, Total number of seizures segregated by type for each seizure-positive cKO mouse. Seizures classified as “other” exhibited a mixture of EEG patterns that could not be categorized as solely type 1 or 2. E, Quantification of the time latency to generalized tonic-clonic seizure (GTCS) upon exposure to flurothyl in WT (n=5), Cre (n=6), fl/− (n=5), and cKO mice (n=4). *P=0.04; ##P=0.006 (1-way ANOVA; post-hoc Tukey test).
To definitively test for the occurrence of spontaneous seizures and cardiac dysfunction, continuous 24-hour recordings of simultaneous video EEG-ECG were performed. Spontaneous seizures were observed in 8 of 11 (73%) cKO mice but none in controls (Fig. 4B). The absence of seizures in some cKO mice was probably due to variability in seizure frequencies between animals, as has been reported previously in global KOs (Mishra et al., 2017), which prevented detection during the limited sampling window, rather than incomplete penetrance of the epilepsy phenotype. cKO mice exhibited an average seizure frequency of 11.3 ± 4.1/d with an average seizure duration per animal of 38 ± 3 s for the seizure-positive mice. Of the 138 individual seizures recorded in cKO mice, 17% (23/138) lasted > 60 s (Fig. 4B).
Two main types of EEG patterns were observed for spontaneous seizures in cKO mice (Fig. 4C). The most common kind of seizure pattern (type 1) occurred 54% of the time (75/138) and was characterized on EEG by sharp waves with a polyspike pattern. The second most common kind of seizure pattern (type 2) occurred 12% of the time (16/138) and consisted of runs of polyspike activity with progressively increasing amplitude followed by postictal flattening. The remainder of the seizures (other; 47/138) exhibited various combinations of the EEG features of type 1 and 2 seizures, which prevented easy classification. Phenotypically, type 1 seizures were associated with relatively mild non-convulsive behaviors consisting of behavioral arrest that was sometimes followed by facial clonus, head nodding, straub tail, and occasional small whole-body jerks. In contrast, type 2 seizures were associated with more severe convulsive behaviors consisting of initial behavioral arrest, head nodding, and straub tail (like type 1 seizures) which progressed to more severe phenotypes such as forelimb clonus, walking backwards, falling to one side, generalized tonic-clonic activity, and bouts of running and bouncing. Other seizures that did not fall into the type 1 and 2 categories exhibited behaviors that were a mixture of those two seizure types. Examination of the distribution of seizure types across cKO mice showed that most seizure-positive animals (6/8) exhibited mild type 1 seizures as their predominant or exclusive seizure type (Fig. 4D). In addition to spontaneous seizures, cKO mice also exhibited cold swim-induced convulsions, as previously reported in global KO mice (Zhou et al., 1998).
To test whether neuron-specific Kcna1 deletion influences seizure threshold, animals were exposed to the volatile convulsant flurothyl (2,2,2-trifluoroethyl ether), and the time latency to generalized tonic-clonic seizure was measured. Kcna1 cKO mice (n=4) exhibited significantly reduced latencies to seizure (211 ± 39 s) compared to WT controls (327 ± 12 s, n=5; F=5.408, P=0.0092, 1-way ANOVA). In contrast, Kcna1fl/− and SynCre+/− control mice showed latencies that were similar to WT animals (Fig. 4E). Thus, selective Kv1.1 deficiency in neurons leads to premature death, spontaneous seizures, and decreased seizure threshold.
Cardiac dysregulation in neuron-specific Kcna1 cKO mice
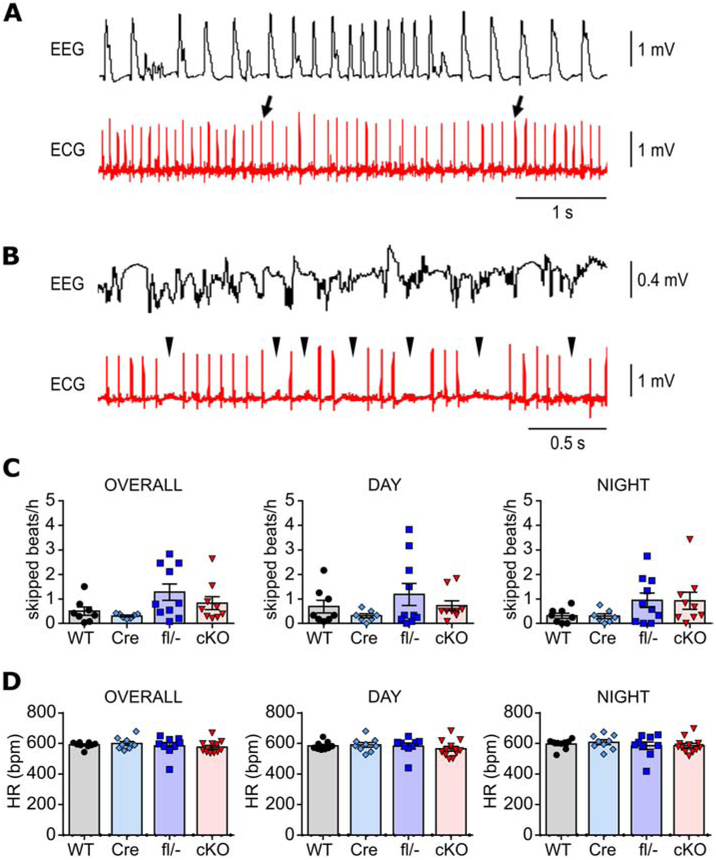
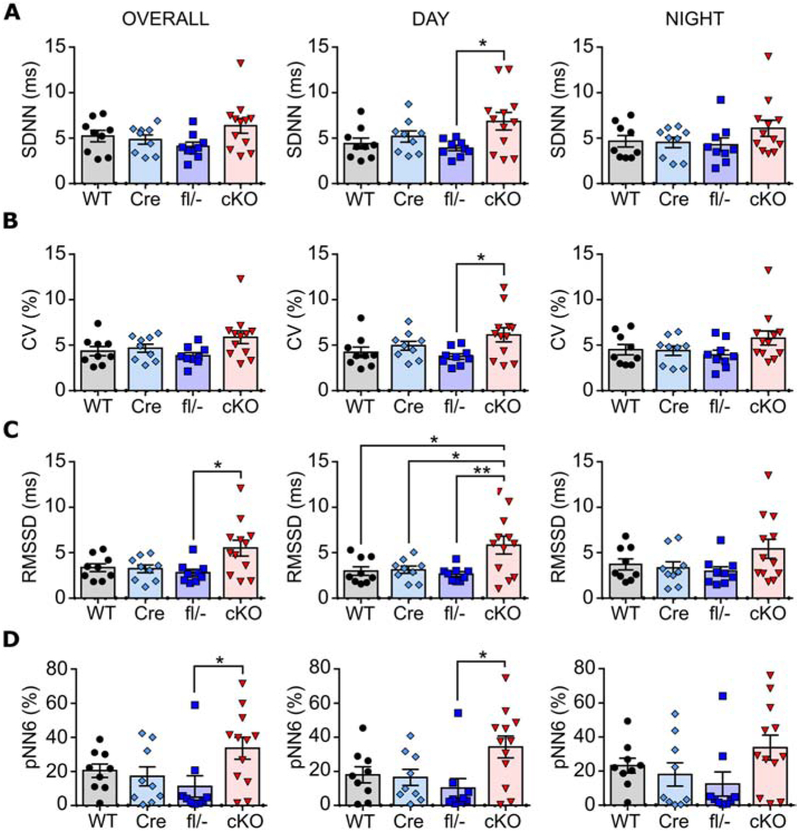
To determine whether neuron-specific deletion of Kcna1 is sufficient to cause cardiac dysfunction, ECGs were analyzed during ictal and interictal periods in cKO mice. Spontaneous seizures in cKO mice evoked cardiac abnormalities including bradycardia (Fig. 5A) and skipped heart beats (such as sinus exit and AV blocks; Fig. 5B), but such events were rare, occurring in only 7% of all seizures (9/138) and in only five out of eight animals with seizures. Previously, Kcna1−/− global knockouts have been shown to exhibit interictal cardiac abnormalities indicative of neurogenic origin, including an increase in skipped heart beats (e.g., AV conduction blocks) and increased heart rate variability (HRV) measured as the root mean square of successive beat-to-beat differences (RMSSD) (Glasscock et al., 2010; Mishra et al., 2017; Vanhoof-Villalba et al., 2018). The AV blocks in Kcna1−/− mice can be alleviated by parasympathetic inhibition with atropine, whereas RMSSD serves as an index of parasympathetic tone. During interictal periods, cKO mice did not exhibit significant differences in the frequency of skipped heart beats compared to controls (Fig. 5C). Although mean heart rates were similar across genotypes (Fig. 5D), cKO mice showed increased HRV that was most pronounced during the day, which corresponds to the time when mice normally tend to sleep more (Fig. 6). SDNN and CV, HRV measures of net autonomic activity, were similar between genotypes overall; however, cKO mice exhibited significantly higher values compared to Kcna1fl/− mice during the day (SDNN: F=3.389, P=0.029, 1-way ANOVA; CV: F=3.109, P=0.039, 1-way ANOVA; Fig. 6A,B) Overall RMSSD was also significantly increased in cKO mice compared to Kcna1fl/− animals (F=4.187, P=0.12, 1-way ANOVA; Fig. 6C). During the day, RMSSD in cKO animals was about two- to three-fold higher than control genotypes (F=5.352, P=0.0039; 1-way ANOVA), but during the night, RMSSD was not significantly different (Fig. 6C). pNN6, another HRV index of parasympathetic activity, was also significantly increased in cKO mice overall and during the day compared to Kcna1fl/− mice (F=3.659, P=0.022; 1-way ANOVA; Fig. 6D). Thus, neuron-specific Kv1.1 deficiency is sufficient for ictal cardiac dysfunction and increased HRV, especially during the day.
Figure 5.
Ictal and interictal cardiac abnormalities in neuron-specific Kcna1 cKO mice. A, Simultaneous EEG-ECG recording showing bradycardia (between the two arrows) during a seizure in a cKO mouse. B, Simultaneous EEG-ECG recording showing multiple AV blocks (arrowheads), which appear as skipped heart beats, during a seizure in a cKO mouse. C, Quantification of the frequency of skipped heart beats (e.g., AV blocks or sinus pauses) during the day and night and overall (day and night combined) in WT, hemizygous Syn1-Cre (Cre), compound heterozygous Kcna1 floxed/null (fl/−), and neuron-specific Kcna1 cKO mice (cKO). D, Quantification of mean heart rate by genotype during the day and night and overall (day and night combined).
Figure 6.
Heart rate variability (HRV) analysis in neuron-specific Kcna1 cKO mice. Quantification of HRV during the day and night and overall (day and night combined) using the time domain measures of total autonomic activity, SDNN (A) and CV (B), and of parasympathetic tone, RMSSD (C) and pNN6 (D). *P<0.05; **P<0.01 (1-way ANOVA; post-hoc Tukey test). WT, wild-type; Cre, hemizygous Syn1-Cre; fl/−, compound heterozygous Kcna1 floxed/null; cKO, neuron-specific Kcna1 cKO.
Respiratory dysregulation in neuron-specific Kcna1 cKO mice
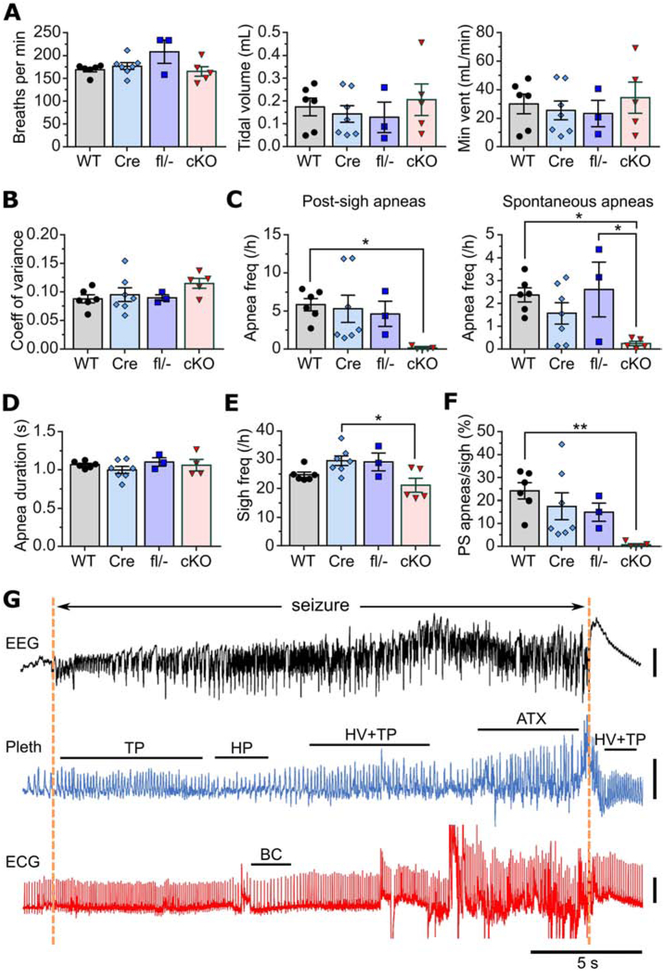
Since brain-driven respiratory dysfunction has been hypothesized to contribute to ictal cardiac abnormalities and SUDEP in Kcna1−/− global knockouts (Dhaibar et al., 2019), simultaneous whole-body plethysmography (pleth), EEG, and ECG recordings were performed in cKO mice to evaluate breathing, brain, and heart activity, respectively. Baseline measurements of interictal respiratory rate, tidal volume, and minute ventilation showed no significant differences between cKO mice and controls (Fig. 7A). Unlike global knockouts which have greatly increased respiratory variability (Dhaibar et al., 2019), the inter-breath time intervals in cKO mice were relatively stable with a degree of variation similar to controls (Fig. 7B). However, like global knockouts (Dhaibar et al., 2019), cKO mice also exhibited a nearly complete absence of interictal apneas (Fig. 7C), including reductions of 98% in post-sigh apnea frequency (F=3.618, P=0.035; 1-way ANOVA) and of 90% in spontaneous apnea frequency (F=4.345, P=0.019; 1-way ANOVA) (Dhaibar et al., 2019). When cKO mice did experience interictal apneas, they were of similar durations to those seen in control animals (Fig. 7D). Sigh frequency in cKO mice was significantly reduced by 29% relative to Cre controls (F=4.463, P=0.017; 1-way ANOVA), but not compared to the other control groups (Fig. 7E). Thus, an alteration in sigh frequency in cKO mice does not appear to account for their drastic reduction in apnea occurrence. In addition, as observed in global knockouts, the percentage of sighs that evoked an apnea was significantly reduced in cKO mice (F=4.737, P=0.014; 1-way ANOVA; Fig. 7F), suggesting impaired ability of a sigh to generate an apnea by the normal reflex mechanisms (Dhaibar et al., 2019).
Figure 7.
Ictal and interictal respiratory abnormalities in neuron-specific Kcna1 cKO mice. A, Baseline measurements of interictal respiratory rate (breaths per min), tidal volume, and minute ventilation (vent) in WT, hemizygous Syn1-Cre (Cre), compound heterozygous Kcna1 floxed/null (fl/−), and neuron-specific Kcna1 cKO mice (cKO). B, Comparison between genotypes of respiratory variability, measured as the coefficient of variance of the inter-breath intervals. C-F, Quantification across genotypes of post-sigh apnea frequency (C), spontaneous apnea frequency (D), sigh frequency per hour (E), and percentage ratio of post-sigh apneas to total number of sighs (F). G, Representative recording of simultaneous EEG-Pleth-ECG activity during a seizure in a cKO mouse. Onset and termination of the seizure are indicated by dotted orange lines. Tachypnea (TP) occurs at seizure onset followed by hypopnea (HP) and hyperventilation (HV) with tachypnea, which develops into irregular ataxic (ATX) breathing that terminates at the end of the seizure. Hyperventilation with tachypnea is evident during the postictal period. Bradycardia (BC) occurs during the seizure but subsequent to tachypnea and hypopnea, suggesting the cardiac slowing may be triggered by brain-driven respiratory dysfunction. *P<0.05; **P<0.01 (1-way ANOVA; post-hoc Tukey test). ECG, electrocardiography; EEG, electroencephalography; Pleth, plethysmography. Vertical scale bars: 0.5 mV for EEG; 0.5 mL for pleth; 1 mV for ECG.
During the 53 seizures captured during EEG-ECG-pleth recordings, cKO mice exhibited numerous types of respiratory abnormalities (Fig. 7G). Similar to our previous observations in global KO mice (Dhaibar et al., 2019), the cKO animals showed hyperventilation in 94% of seizures, which usually coincided with ictal onset. Tachypnea and ataxic breathing were also commonly observed during about 92% and 42% of seizures, respectively. Less common ictal respiratory events included hypopnea (28% of seizures), bradypnea (17% of seizures), and sighs (8% of seizures). Apnea was only observed during one seizure and it was of relatively short duration, lasting only about 0.9 s. In addition, although ataxic breathing occurred with a similar frequency during seizures in global and cKO mice, the duration appeared noticeably shorter in cKO seizures. To compare the average duration of ataxic breathing in cKO mice to global KO mice, the data from our previous study of global KO mice (Dhaibar et al., 2019) was reanalyzed. The average bout of ataxic breathing in cKO seizures was 7.6 ± 0.9 s (n=22), which was significantly shorter than global KO mice, which exhibited average bouts of 15.6 ί 1.5 s (n=33; P=0.0002, two-tailed unpaired t-test). Finally, when cardiac dysfunction was present during a seizure in cKO mice, it was always preceded by breathing abnormalities, suggesting a link between brain-driven respiratory mechanisms and ictal cardiac events (Fig. 7G).
DISCUSSION
A primary unanswered question in the field of epilepsy research is whether neural or cardiac mechanisms are more important for SUDEP pathophysiology. To test this, a floxed Kcna1 allele was developed to allow determination of the tissue-specific origins of neurocardiac phenotypes in the Kcna1−/− SUDEP model. The primary finding of this study was that neuron-specific Kcna1 deletion is capable of partially recapitulating many of the salient phenotypes observed in Kcna1 global KO mice including epilepsy, premature death, and cardiorespiratory dysregulation, indicating that these deficits are primarily brain-driven. However, the restriction of Kv1.1 deficiency to neurons ameliorated the severity of these phenotypes compared to global KO mice which lack Kv1.1 in both neurons and cardiomyocytes. Thus, although Kcna1 deletion in heart is not necessary for premature death and cardiorespiratory dysregulation, a Kv1.1-deficient heart may synergistically interact with the brain to increase risk of mortality and the prevalence cardiac abnormalities.
The premature death, seizure, and cardiorespiratory phenotypes of cKO mice were overall less severe than those in global Kcna1 KO mice. About twice as many cKO mice lived to 100 d compared to global KO mice, and cKO median survival was prolonged by about 2 months. Although cKO mice exhibited a seizure frequency close to global KO mice, the cKO seizure phenotype appeared less severe overall as measured by seizure duration and cardiorespiratory effects. Only 17% of seizures exceeded 60 s duration in cKO mice, which was about half the percentage observed in previous studies of global KOs which reported between 29-35% (Glasscock et al., 2010; Mishra et al., 2017). Only 7% of cKO seizures in EEG-ECG recordings evoked cardiac dysfunction compared to at least 30% in a previous study of global KO animals (Glasscock et al., 2010). In addition, the frequency of interictal skipped heart beats in cKO mice was lower than previous observations in global KO mice (about 1/h versus 5/h), suggesting less impaired cardiac dysfunction at baseline (Glasscock et al., 2010). Although cKO animals were similar to global KOs in that they exhibited a nearly complete absence of interictal apneas, their respiratory variability was not statistically different from WT, unlike global KO mice which exhibit abnormally variable breathing (Dhaibar et al., 2019). Finally, bouts of ictal ataxic breathing, an indicator of more deleterious respiratory dysfunction which often preceded cardiac dysfunction in global KO mice (Dhaibar et al., 2019), was significantly abbreviated in cKO mice by about 50%. Thus, the overall picture that emerges is that neuro-cardio-respiratory phenotypes in cKO mice are generally similar to global KO animals but are of lesser severity. One explanation for the milder phenotype of cKO mice is the presence of intact Kv1.1 subunits in the heart which render it more resistant to the deleterious effects of seizures. In addition, the predominantly non-convulsive seizures in cKO mice may put them at less risk of seizure-related death due to cardiorespiratory dysfunction, which in global KO mice tends to be a more consistent feature of severe tonic-clonic seizures (Dhaibar et al., 2019).
Genetic background may partially contribute to the phenotypic differences between cKO and global KO mice, but it seems unlikely to account for the degree of differences observed. The cKO mice in this study were on a mixed Tac:N:NIHS-BC and C57 background, whereas the global KO mice in our previous studies were on a Tac:N:NIHS-BC background. Inbred mouse strains possess unique, strain-specific differences in seizure susceptibility, such as in seizure threshold tests using kainic acid or flurothyl (Löscher et al., 2017). However, our previous measurements of flurothyl-induced seizure latency in global KO mice on a Tac:N:NIHS-BC background and on a mixed Tac:N:NIHS-BC and C57BL/6 background gave nearly identical values, demonstrating the highly penetrant nature of the Kcna1 deletion mutation (Mishra et al., 2017; Vanhoof-Villalba et al., 2018). Additionally, in our two previous survival studies of global KO mice on mixed Tac:N:NIHS-BC and C57 backgrounds (Glasscock et al., 2007; Mishra et al., 2017), their median survival ranged between 35 to 50 d, which is about half that observed in this study (97 d). Furthermore, survival studies by other groups have shown that a C3H genetic background for global KO mice confers an even more severe SUDEP phenotype with a median survival of about 43 d and 100% mortality by about 65 d (Simeone et al., 2016). Thus, the drastically ameliorated mortality in cKO mice is probably not due to differences in genetic background alone since Kcna1 deletion exhibits a high incidence and early onset of mortality across multiple strains.
In addition to the heart, expression differences in glia and cerebellum could also potentially contribute to phenotypic differences between cKO and global KO mice. The Syn1-Cre transgene used to generate neuron-specific conditional Kcna1 gene deletion in this study is not significantly expressed in astrocytes and cerebellum (Zhu et al., 2001 p.1). Astrocytes are important regulators of neuronal excitability by controlling glutamate uptake, [K+]0 buffering, and brain water regulation, and a breakdown of these mechanisms can promote hyperexcitability and seizures (Murphy et al., 2017). Although Kv1.1 protein has been detected in cortical astrocytes, its expression level is relatively low and its role mostly unexplored (Smart et al., 1997). Therefore, it is unclear whether the absence of Kv1.1 in astrocytes of global KO mice impairs their function leading to enhanced neuronal excitability relative to cKO mice.
The lack of Kv1.1 deficiency in the cerebellum could contribute to the amelioration of seizure severity, premature death incidence, and respiratory dysregulation in cKO mice. Kv1.1 subunits are normally expressed at a high level in cerebellum, localizing to the pinceau terminal regions of basket cells and to axons in the granule cell layer (Wang et al., 1994). Cerebellar neuropathology is a common finding in sudden death syndromes, including both SUDEP and sudden infant death syndrome (SIDS) (Cruz-Sánchez et al., 1997; Lavezzi et al., 2006). In SUDEP, cerebellar atrophy has been reported in 16% of cases, as well as microscopic changes including Purkinje cell loss, Bergmann’s gliosis, and folial atrophy, which affect up to 61% of cases (Shields et al., 2002; Scorza et al., 2011). Although the cerebellum has not traditionally been considered to play a role in breathing, it is now recognized as a key brain structure for respiratory and cardiovascular regulation in response to extreme changes in CO2 levels or blood pressure (Xu and Frazier, 2002; Scorza et al., 2011). Thus, cerebellar abnormalities could increase SUDEP susceptibility by impairing normal compensatory responses that aid recovery from breathing or cardiovascular challenges (Scorza et al., 2011). Furthermore, the cerebellum can also act as a suppressor of seizure severity. In a mouse model of temporal lobe epilepsy, optogenetic stimulation of the cerebellum significantly reduced spontaneous seizure frequency and seizure duration (Krook-Magnuson et al., 2014). Therefore, a hypothesis to be tested in more detail in future studies is whether the preservation of Kv1.1 expression in the cerebellum of cKO mice acts to limit seizure duration and reduce SUDEP risk compared to global KO animals.
Parasympathetic indices of interictal HRV were elevated in cKO mice most significantly during the daytime hours, suggesting that disturbed diurnal HRV patterns could be a biomarker of SUDEP susceptibility. Patients with epilepsy generally tend to have lower HRV indicative of a shift in autonomic tone toward sympathetic dominance (Myers et al., 2018). Reduced HRV has also been correlated with increased SUDEP risk (DeGiorgio et al., 2010). However, individual cases also exist that show the opposite relationship, that sudden or progressive increases in HRV, indicative of shifts toward parasympathetic dominance, can be a leading prognosticator of terminal deterioration prior to SUDEP (Jeppesen et al., 2014; Myers et al., 2017). Time of day and arousal state may also affect HRV leading to windows of increased SUDEP susceptibility due to autonomic dysfunction. For example, drug-resistant epilepsy patients who inherently have higher SUDEP susceptibility exhibit circadian alterations in HRV that peak at night, suggesting autonomic imbalances that are maximal when patients are at rest (Yang et al., 2018). Furthermore, in a retrospective study of SUDEP victims, some patients had extremely high ratios of sleep:wake RMSSD due to abnormally high HRV during sleep and abnormally low HRV during wakefulness (Myers et al., 2018). This observation has led to an autonomic “second hit” hypothesis of SUDEP. This hypothesis proposes that people with epilepsy and severe autonomic dysfunction are closer to intolerable extremes of both parasympathetic and sympathetic tone depending on the time of day whereby an inciting event (e.g., a seizure) can push such an individual into a pathological zone of autonomic dysregulation leading to lethality (Myers et al., 2018). In the case of cKO mice, they exhibit abnormally high HRV that peaks during the daytime hours, suggesting brain-driven Kv1.1 deficiency leads to circadian alterations in cardiac control that may contribute to SUDEP susceptibility.
In summary, neuron-specific Kcna1 cKO mice demonstrate that channelopathies with brain-heart expression patterns are capable of increasing susceptibility to premature death by brain-driven mechanisms alone without the need for a functionally compromised heart. This finding further reinforces the clinical imperative of seizure control as a primary strategy for reducing SUDEP risk even when dual arrhythmogenic potential exists in the brain and heart. This work also raises the possibility that the cerebellum may play an unexpected role in regulating cardiorespiratory dysfunction in epilepsy and SUDEP probability. Finally, the elevation of daytime parasympathetic HRV measures in cKO mice reveals dysregulation of diurnal HRV patterns may be a new biomarker of augmented SUDEP risk.
HIGHLIGHTS.
A floxed Kcna1 allele was developed to generate neuron-specific Kv1.1 knockout mice
Neuron-specific Kv1.1 knockout (cKO) mice died early and had spontaneous seizures
Heart rate variability was increased in cKO mice, especially during daytime
During non-seizure periods, apneas were almost completely absent in cKO mice
Acknowledgements
This work was supported by grants from the National Institutes of Health (R01NS100954 and R01NS099188 to E.G.); a Malcolm Feist Predoctoral Fellowship (to K.T.) from Louisiana State University Health, Sciences Center- Shreveport (LSUHSC-S); an Ike Muslow Predoctoral Fellowship (to H.D.) from LSUHSC-S; and a Malcolm Feist Postdoctoral Fellowship (to V.M.) from LSUHSC-S. We would like to thank Oleg Chernyshev, M.D. (Department of Neurology at LSUHSC-S) for assistance with interpretation of EEG patterns. The address for Edward Glasscock is now the Department of Biological Sciences at Southern Methodist University in Dallas, TX.
Footnotes
Conflict of interest statement
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aiba I, Noebels JL (2015) Spreading depolarization in the brainstem mediates sudden cardiorespiratory arrest in mouse SUDEP models. Sci Transl Med 7:282ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall RD, Crompton DE, Petrovski S, Lam L, Cutmore C, Garry SI, Sadleir LG, Dibbens LM, Cairns A, Kivity S, Afawi Z, Regan BM, Duflou J, Berkovic SF, Scheffer IE, Semsarian C (2016) Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann Neurol 79:522–534. [DOI] [PubMed] [Google Scholar]
- Brown RE, Mathieson WB, Stapleton J, Neumann PE (1999) Maternal Behavior in Female C57BL/6J and DBA/2J Inbred Mice. Physiol Behav 67:599–605. [DOI] [PubMed] [Google Scholar]
- Cruz-Sánchez FF, Lucena J, Ascaso C,Tolosa E, Quintò L, Rossi ML (1997) Cerebellar cortex delayed maturation in sudden infant death syndrome. J Neuropathol Exp Neurol 56:340–346. [DOI] [PubMed] [Google Scholar]
- DeGiorgio CM, Miller P, Meymandi S, Chin A, Epps J, Gordon S, Gornbein J, Harper RM (2010) RMSSD, a measure of vagus-mediated heart rate variability, is associated with risk factors for SUDEP: the SUDEP-7 Inventory. Epilepsy Behav EB 19:78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaibar H, Gautier NM, Chernyshev OY, Dominic P, Glasscock E (2019) Cardiorespiratory profiling reveals primary breathing dysfunction in Kcna1-null mice: Implications for sudden unexpected death in epilepsy. Neurobiol Dis 127:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlouhy BJ, Gehlbach BK, Richerson GB (2016) Sudden unexpected death in epilepsy: basic mechanisms and clinical implications for prevention. J Neurol Neurosurg Psychiatry 87:402–413. [DOI] [PubMed] [Google Scholar]
- Ferguson CJ, Lenk GM, Jones JM, Grant AE, Winters JJ, Dowling JJ, Giger RJ, Meisler MH (2012) Neuronal expression of Fig4 is both necessary and sufficient to prevent spongiform neurodegeneration. Hum Mol Genet 21:3525–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill BN, Pritchett-Corning KR, Gordon CJ, Pajor EA, Lucas JR, Davis JK, Garner JP (2013) Energy Reallocation to Breeding Performance through Improved Nest Building in Laboratory Mice. PLOS ONE 8:e74153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasscock E (2014) Genomic biomarkers of SUDEP in brain and heart. Epilepsy Behav 38:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasscock E (2019) Kv1.1 channel subunits in the control of neurocardiac function. Channels Austin Tex 13:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasscock E, Qian J, Kole MJ, Noebels JL (2012) Transcompartmental reversal of single fibre hyperexcitability in juxtaparanodal Kv1.1-deficient vagus nerve axons by activation of nodal KCNQ channels.J Physiol 590:3913–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasscock E, Qian J, Yoo JW, Noebels JL (2007) Masking epilepsy by combining two epilepsy genes. Nat Neurosci 10:1554–1558. [DOI] [PubMed] [Google Scholar]
- Glasscock E, Voigt N, McCauley MD, Sun Q, Li N, Chiang DY, Zhou X-B, Molina CE, Thomas D, Schmidt C, Skapura DG, Noebels JL, Dobrev D, Wehrens XHT (2015) Expression and function of Kv1.1 potassium channels in human atria from patients with atrial fibrillation. Basic Res Cardiol 110:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasscock E, Yoo JW, Chen TT, Klassen TL, Noebels JL (2010) Kv1.1 potassium channel deficiency reveals brain-driven cardiac dysfunction as a candidate mechanism for sudden unexplained death in epilepsy. J Neurosci 30:5167–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman AM, Behr ER, Semsarian C, Bagnall RD, Sisodiya S, Cooper PN (2016) Sudden unexpected death in epilepsy genetics: Molecular diagnostics and prevention. Epilepsia 57 Suppl 1:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows JL,Tempel BL (1998) Expression of Kv1.1, a Shaker-like potassium channel, is temporally regulated in embryonic neurons and glia. J Neurosci 18:5682–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN (2012) Voltage-gated potassium channels and the diversity of electrical signalling. J Physiol 590:2591–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen J, Fuglsang-Frederiksen A, Brugada R, Pedersen B, Rubboli G, Johansen P, Beniczky S (2014) Heart rate variability analysis indicates preictal parasympathetic overdrive preceding seizure-induced cardiac dysrhythmias leading to sudden unexpected death in a patient with epilepsy. Epilepsia 55:e67–71. [DOI] [PubMed] [Google Scholar]
- Klassen TL, Bomben VC, Patel A, Drabek J, Chen TT, Gu W, Zhang F, Chapman K, Lupski JR, Noebels JL, Goldman AM (2014) High-resolution molecular genomic autopsy reveals complex sudden unexpected death in epilepsy risk profile. Epilepsia 55:e6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook-Magnuson E, Szabo GG, Armstrong C, Oijala M, Soltesz I (2014) Cerebellar Directed Optogenetic Intervention Inhibits Spontaneous Hippocampal Seizures in a Mouse Model of Temporal Lobe Epilepsy. eNeuro 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavezzi AM, Ottaviani G, Mauri M, Matturri L (2006) Alterations of biological features of the cerebellum in sudden perinatal and infant death. Curr Mol Med 6:429–435. [DOI] [PubMed] [Google Scholar]
- Leu C et al. (2015) Genome-wide Polygenic Burden of Rare Deleterious Variants in Sudden Unexpected Death in Epilepsy. EBioMedicine 2:1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhatoo SD, Faulkner HJ, Dembny K, Trippick K, Johnson C, Bird JM (2010) An electroclinical case-control study of sudden unexpected death in epilepsy. Ann Neurol 68:787–796. [DOI] [PubMed] [Google Scholar]
- Löscher W, Ferland RJ, Ferraro TN (2017) The relevance of inter-and intrastrain differences in mice and rats and their implications for models of seizures and epilepsy. Epilepsy Behav EB 73:214–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey CA, Sowers LP, Dlouhy BJ, Richerson GB (2014) Mechanisms of sudden unexpected death in epilepsy:the pathway to prevention. Nat Rev Neurol 10:271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Rohlmann A, Bock HH, Zurhove K, Marth JD, Schomburg ED, Noebels JL, Beffert U, Sweatt JD, Weeber EJ, Herz J (2004) Neuronal LRP1 functionally associates with postsynaptic proteins and is required for normal motor function in mice. Mol Cell Biol 24:8872–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V, Gautier NM, Glasscock E (2018) Simultaneous Video-EEG-ECG Monitoring to Identify Neurocardiac Dysfunction in Mouse Models of Epilepsy. J Vis Exp JoVE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V, Karumuri BK, Gautier NM, Liu R, Hutson TN, Vanhoof-Villalba SL, Vlachos I, Iasemidis L, Glasscock E (2017) Scn2a deletion improves survival and brain-heart dynamics in the Kcna1-null mouse model of sudden unexpected death in epilepsy (SUDEP). Hum Mol Genet 26:2091–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BM, Jerry Jou C, Tatalovic M, Kaufman ES, Kline DD, Kunze DL (2014) The Kv1.1 null mouse, a model of sudden unexpected death in epilepsy (SUDEP). Epilepsia 55:1808–1816. [DOI] [PubMed] [Google Scholar]
- Murphy TR, Binder DK, Fiacco TA (2017) Turning down the volume: Astrocyte volume change in the generation and termination of epileptic seizures. Neurobiol Dis 104:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, Bello-Espinosa LE, Symonds JD, Zuberi SM, Clegg R, Sadleir LG, Buchhalter J, Scheffer IE (2018) Heart rate variability in epilepsy: A potential biomarker of sudden unexpected death in epilepsy risk. Epilepsia 59:1372–1380. [DOI] [PubMed] [Google Scholar]
- Myers KA, McPherson RE, Clegg R, Buchhalter J (2017) Sudden Death After Febrile Seizure Case Report: Cerebral Suppression Precedes Severe Bradycardia. Pediatrics 140. [DOI] [PubMed] [Google Scholar]
- Peltola M (2012) Role of editing of R-R intervals in the analysis of heart rate variability. Front Physiol 3 Available at: https://www.frontiersin.org/articles/10.3389/fphys.2012.00148/full [Accessed May 9, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempe D, Vangeison G, Hamilton J, Li Y, Jepson M, Federoff HJ (2006) Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis 44:44–49. [DOI] [PubMed] [Google Scholar]
- Ren H, Plum-Morschel L, Gutierrez-Juarez R, Lu TY, Kim-Muller JY, Heinrich G, WardlawS L, Silver R, Accili D (2013) Blunted refeeding response and increased locomotor activity in mice lacking FoxO1 in synapsin-Cre-expressing neurons. Diabetes 62:3373–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruisu K, Kask K, Meier R, Saare M, Raid R, Veraksitš A, Karis A, Tõnissoo T, Pooga M (2013) Ablation of RIC8A function in mouse neurons leads to a severe neuromuscular phenotype and postnatal death. PloS One 8:e74031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvlin P et al. (2013) Incidence and mechanisms of cardiorespiratory arrests in epilepsy monitoring units (MORTEMUS): a retrospective study. Lancet Neurol 12:966–977. [DOI] [PubMed] [Google Scholar]
- Scorza FA, Terra VC, Arida RM, Sakamoto AC, Harper RM (2011) Sudden death in a child with epilepsy: potential cerebellar mechanisms? Arq Neuropsiquiatr 69:707–710. [DOI] [PubMed] [Google Scholar]
- Shields LBE, Hunsaker DM, Hunsaker JC, Parker JC (2002) Sudden unexpected death in epilepsy: neuropathologic findings. Am J Forensic Med Pathol 23:307–314. [DOI] [PubMed] [Google Scholar]
- Si M, Trosclair K, Hamilton KA, Glasscock E (2018) Genetic ablation or pharmacological inhibition of Kv1.1 potassium channel subunits impairs atrial repolarization in mice. Am J Physiol Cell Physiol 316:C154–C161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää M, Shinnar S (2010) Long-term mortality in childhood-onset epilepsy. N Engl J Med 363:2522–2529. [DOI] [PubMed] [Google Scholar]
- Simeone KA, Matthews SA, Rho JM, Simeone TA (2016) Ketogenic diet treatment increases longevity in Kcna1-null mice, a model of sudden unexpected death in epilepsy. Epilepsia 57:e178–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart SL, Bosma MM, Tempel BL (1997) Identification of the delayed rectifier potassium channel, Kv1.6, in cultured astrocytes. Glia 20:127–134. [DOI] [PubMed] [Google Scholar]
- Smart SL, Lopantsev V, Zhang CL, Robbins CA, Wang H, Chiu SY, Schwartzkroin PA, Messing A, Tempel BL (1998) Deletion of the K(V)1.1 potassium channel causes epilepsy in mice. Neuron 20:809–819. [DOI] [PubMed] [Google Scholar]
- Thurman DJ, Hesdorffer DC, French JA (2014) Sudden unexpected death in epilepsy: assessing the public health burden. Epilepsia 55:1479–1485. [DOI] [PubMed] [Google Scholar]
- Vanhoof-Villalba SL, Gautier NM, Mishra V, Glasscock E (2018) Pharmacogenetics of KCNQ channel activation in 2 potassium channelopathy mouse models of epilepsy. Epilepsia 59:358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kunkel DD, Schwartzkroin PA, Tempel BL (1994) Localization of Kv1.1 and Kv1.2, two K channel proteins, to synaptic terminals, somata, and dendrites in the mouse brain. J Neurosci 14:4588–4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Frazier DT (2002) Role of the cerebellar deep nuclei in respiratory modulation. Cerebellum Lond Engl 1:35–40. [DOI] [PubMed] [Google Scholar]
- Yang Z, Liu H, Meng F, Guan Y, Zhao M, Qu W, Hao H, Luan G, Zhang J, Li L (2018) The analysis of circadian rhythm of heart rate variability in patients with drug-resistant epilepsy. Epilepsy Res 146:151–159. [DOI] [PubMed] [Google Scholar]
- Yu T, Lieberman AP (2013) Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin. PLOS Genet 9:e1003462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Shakkottai VG, Chung C, Lieberman AP (2011) Temporal and cell-specific deletion establishes that neuronal Npc1 deficiency is sufficient to mediate neurodegeneration. Hum Mol Genet 20:4440–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuskaitis CJ, Jones BM, Wolfson RL, Super CE, Dhamne SC, Rotenberg A, Sabatini DM, Sahin M, Poduri A (2018) A mouse model of DEPDC5-related epilepsy: Neuronal loss of Depdc5 causes dysplastic and ectopic neurons, increased mTOR signaling, and seizure susceptibility. Neurobiol Dis 111:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuskaitis CJ, Rossitto L-A, Gurnani S, Bainbridge E, Poduri A, Sahin M (2019) Chronic mTORC1 inhibition rescues behavioral and biochemical deficits resulting from neuronal Depdc5 loss in mice. Hum Mol Genet 28:2952–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang CL, Messing A, Chiu SY (1998) Temperature-sensitive neuromuscular transmission in Kv1.1 null mice: role of potassium channels under the myelin sheath in young nerves. J Neurosci 18:7200–7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF (2001) Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev 15:859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi SM, Eunson LH, Spauschus A, De Silva R, Tolmie J, Wood NW, McWilliam RC, Stephenson JB, Stephenson JP, Kullmann DM, Hanna MG (1999) A novel mutation in the human voltage-gated potassium channel gene (Kv1.1) associates with episodic ataxia type 1 and sometimes with partial epilepsy. Brain 122 (Pt 5):817–825. [DOI] [PubMed] [Google Scholar]