Abstract
Genetic susceptibility to cutaneous melanoma has been investigated in Italian high-risk melanoma patients from different geographical regions. CDKN2A, CDK4, and MC1R genes have been screened in most studies, MITF and POT1 were screened in only one study, and none analyzed the TERT promoter. We carried out a mutational analysis of CDKN2A, CDK4 exon 2, POT1 p.S270N, MITF exon 10, MC1R, and the TERT promoter in 106 high-risk patients with familial melanoma (FM) and sporadic multiple primary melanoma (spMPM) from Central Italy and evaluated mutations according to the clinicopathological characteristics of patients and lesions. In FM, CDKN2A mutations were detected in 8.3% of the families, including one undescribed exon 1β mutation (p.T31M), and their prevalence increased with the number of affected relatives within the family. MC1R variants were identified in 65% of the patients and the TERT rs2853669 promoter polymorphism was identified in 58% of the patients. A novel synonymous mutation detected in MITF exon 10 (c.861A>G, p.E287E), although predicted as a splice site mutation by computational tools, could not functionally be confirmed to alter splicing. For spMPM, 3% carried CDKN2A mutations, 79% carried MC1R variants, and 47% carried the TERT rs2853669 promoter polymorphism. MC1R variants were associated with fair skin type and light hair color both in FM and in spMPM, and with a reduction of age at diagnosis in FM patients. Mutations in CDK4 exon 2 and the POT1 p.S270N mutation were not detected. A low frequency of CDKN2A mutations and a high prevalence of MC1R variants characterize high-risk melanoma patients from Central Italy.
Keywords: CDK4, CDKN2A, familial melanoma, MC1R, MITF, multiple primary melanoma, POT1, TERT promoter
Introduction
Cutaneous melanoma is a complex disorder, with genetic and environmental factors contributing toward its pathogenesis. Approximately 8–12% of melanomas are diagnosed in individuals with a hereditary predisposition 1,2. Individuals with at least one first-degree relative with melanoma have an approximately two-fold increased risk of developing the disease, and the risk increases with the number of affected relatives 2. Multiple primary melanomas (MPM) develop in about 5% of sporadic melanoma patients and in up to 19% of melanoma patients with a positive family history 2.
CDKN2A (cyclin-dependent kinase inhibitor 2A; MIM: 600160) and CDK4 (cyclin-dependent kinase 4; MIM: 123829) are well-known high-risk melanoma susceptibility genes, identified about 20 years ago 3,4. Germline mutations of the CDKN2A gene are described in 20% of familial melanoma (FM) patients, with the frequency increasing to 40% in families with three or more affected members 5. Early age at diagnosis, presence of MPM, and cosegregation of pancreatic cancer within the family have shown a significant association with CDKN2A mutations, although the effects vary widely across continents 5. In sporadic MPM (spMPM) patients, CDKN2A mutations have been reported in 3–15% of the patients 6–8. Mutations of the CDK4 gene are rare, with only 17 families carrying the mutation worldwide 9.
More recently, the introduction of next-generation sequencing methodologies led to the identification of new melanoma susceptibility genes implicated in undiscovered pathways. The telomere-elomere-shelterin POT1 (protection of telomeres 1; MIM: 606478) gene was identified as a new high-penetrance susceptibility gene for FM 10,11 and the p.S270N germline variant has been reported as a founder mutation in Italy with a frequency comparable with that of CDKN2A mutations 11. Two additional genes encoding proteins of the shelterin complex, ACD (adrenocortical dysplasia homologue; MIM: 609377) and TERF2IP (telomeric repeat binding factor 2; MIM: 605061), have recently been associated with familial melanoma 12. A novel variant occurring in the promoter region of the TERT (telomerase reverse-transcriptase; MIM: 187270) gene was identified as a high-risk predisposition allele in two melanoma-dense families from Germany and UK 13,14.
In addition to high-risk susceptibility genes, the importance of intermediate-risk genes, such as MC1R (melanocortin-1 receptor; MIM: 155555), has been highlighted by candidate gene and genome-wide association studies. MC1R variants (D84E, R142H, R151C, R160W, and D294H and I155T) are classified as red hair color (RHC) or ‘R’ variants and non-RHC or ‘r’ alleles according to the strength of association with the RHC phenotype 15. MC1R variants have been associated with melanoma risk that appears to be stronger in RHC carriers and in darkly pigmented patients 16. More recently, the E318K substitution in the MITF-M (microphthalmia-associated transcription factor, M variant; MIM: 156845) gene has been reported as conferring an intermediate risk for melanoma and as a susceptibility gene for renal cell carcinoma 17,18. Development of MPM, increased nevus count, and nonblue eye color are clinical characteristics of MITF E318K mutation carriers 17–19.
Genetic susceptibility to cutaneous melanoma has been investigated in Italian high-risk melanoma patients from different geographical regions. CDKN2A6,20–30, CDK46,21,22,27,28,30, and MC1R6,24,25,28,31,32 genes have been screened in almost all published studies, MITF in only one study 19, and POT1 in the discovery manuscript 11; no studies have analyzed the promoter of the TERT gene. In the present study, we carried out a mutational analysis of the major known melanoma susceptibility genes (CDKN2A, CDK4 exon 2, POT1 p.S270N, MC1R, MITF exon 10, and the TERT promoter) in high-risk FM and spMPM patients from Central Italy and evaluated the association of mutational status with the clinicopathological characteristics of patients and tumors.
Patients and methods
Patients’ recruitment
Patients from melanoma families (FM patients) with at least two first-degree or second-degree relatives or with at least three documented cases of melanoma irrespective of the degree of relatedness and spMPM patients recruited between 2000 and 2012 at the Department of Dermatology, University of L’Aquila, Italy, were included in the study.
Basic demographic information and phenotypic characteristics of patients were determined through a standardized questionnaire (sex, education, lifetime residential history, medical history, family history of melanoma, and personal and family history of other cutaneous and visceral neoplasms). Skin examination was performed by a dermatologist, who evaluated skin type, hair color (red, blond, light brown, dark brown, black), eye color (blue, green, light brown, and dark brown), number of melanocytic nevi, and presence or absence of clinically atypical nevi.
The diagnosis of cutaneous melanoma was confirmed histopathologically for all cases included in the study. Detailed data on patient’ age at diagnosis, anatomical site of melanoma, and histopathological data including clinicopathologic variant [superficial spreading melanoma (SSM), nodular melanoma, acral lentiginous melanoma, lentigo maligna/lentigo maligna melanoma, others, spitzoid melanoma, occult melanoma] and Breslow thickness were collected.
Approval for this study was obtained from the local ethics committee and a written informed consent was obtained from the participants. The study was carried out according to the Helsinki declaration.
Molecular analysis
Genomic DNA was extracted from whole blood using a QIAamp DNA-blood midi kit (Qiagen, Hilden, Germany). Mutational screening of exons 1α, 1β, 2, and 3, including the exon–intron boundaries of CDKN2A, exon 2 of CDK4, promoter region of the TERT gene (from −497 bp to the ATG start site), exon 10 of MITF, and the entire open reading frame of MC1R, was performed by PCR and direct sequencing on a 3500 Genetic-Analyzer (Thermo-Fisher, Foster City, California, USA). In detail, PCR amplification of the regions of interest was performed in a Gene-Amp PCR-System 9700 (Thermo-Fisher) using the primers listed in Table S1. PCR experiments were conducted using 1.25 U of AmpliTaq Gold-360 (Thermo-Fisher) in a 50-μl volume, containing the ×1 reaction buffer provided by the manufacturer, 1.6 mmol/l of MgCl2, 200 μmol/l of each deoxynucleoside triphosphate, 0.2 μmol/l of each primer, and 100 ng genomic DNA template. Five per cent dimethyl sulfoxide was added to the reaction solution. PCR amplification conditions were as follows: 95°C for 7 min, 35 cycles of 94°C for 1 min, Tm (°C) (Table S1) for 1 min, and 72°C for 1 min, followed by a final extension step at 72°C for 7 min.
Genotyping assay of the g.124493086C>T (p.S270N) mutation of the POT1 gene was carried out using TaqMan SNP Genotyping Assays (Thermo-Fisher). PCRs containing 30 ng of DNA, ×1 TaqMan Genotyping Master Mix; ×1 TaqMan genotyping assay mix (Thermo-Fisher), and water to reach the final volume of 10 μl were performed in 96-well plates using the standard TaqMan protocol on a 7500 Fast Real Time-PCR System (Thermo-Fisher). Water controls and positive controls were run in parallel with patients’ DNA samples.
The disease-causing potential of the splice site variant c.861A>G in the MITF gene was evaluated on RNA extracted from whole blood of the carrier patient and of a healthy control using the QIAamp RNA-blood mini kit (Qiagen). Total RNA was treated with DNase I (Qiagen) to avoid residual genomic DNA contamination. First-strand cDNA was generated from 1 μg of RNA with the ThermoScript RT-PCR System (Thermo-Fisher) according to the manufacturers’ protocols. The entire MITF-M transcript (reference number NM_00248.3) was amplified using primer pairs exon-spanning (Table S1) and sequenced by the Sanger method on a 3500 Genetic-Analyzer (Thermo-Fisher).
All mutations were confirmed by an analysis of a second independent blood sample.
Computational prediction analysis
Nine computational tools were run to predict the effect of the p.T31M change on p14ARF function: SIFT, PROVEAN, SNAP, Polyphen-2, Panther, MutationTaster, CONDEL, SNP&GO, and CADD. They are based on phylogenetic and structural information and yield a score indicating how amino acid substitutions could alter the protein structure. Pholyphen-2 was performed with two different training sets: HumDiv and HumVar in version 2.2.2. PROVEAN was used in version 1.1 with a score cut-off of 2.5. SIFT was run with a prediction cut-off of 0.05 and CADD with a deleteriousness cut-off of 15 for the scaled scores. The NetPhos 2.0 bioinformatic tool (http://www.cbs.dtu.dk/services/NetPhos/) was used to predict structural phosphorylation change on p14ARF because of the p.T31M substitution.
In silico splicing prediction tests for the p.E287E (c.861A>G) MITF-M variant were performed using the MutationTaster (http://www.mutationtaster.org/), Human Splicing Finder (http://www.umd.be/HSF/), and Ex-skip (http://ex-skip.img.cas.cz/) bioinformatics tools.
For prediction analysis, Ensembl transcript ID ENST00000579755 for p14ARF and ENST00000394351 for MITF-M were used.
Statistical analysis
χ2-test or Fisher’s exact test was used, as appropriate, to test for the significance of the mutation frequency according to the clinical characteristics of melanoma patients and the clinicopathological features of melanoma lesions. For analysis, CDKN2A mutational status was categorized as wild type or mutated. The unknown change p.T31M in exon 1 β, the p.A148T polymorphism in exon 2, and the *500C>G and *540C>T polymorphisms in the 3′UTR uncoding region were defined as wild type in the overall frequencies. For MC1R, the D84E, R142H, R151C, R160W, and D294H, I155T, insC_537 variants were considered as RHC variants, whereas all the others were termed non-RHC. Synonymous variants were considered wild type.
For statistical analysis, variables were categorized as follows: median age at melanoma diagnosis (≤40 years or>40 years), number of primary melanomas (single or multiple), number of affected relatives in a family (two or more than two), history of nonmelanoma skin cancers (presence or absence), number of melanocytic nevi (≤50 or>50), histopathological subtype (SSM or other subtypes), melanoma thickness (in situ or invasive; Breslow thickness ≤1 or> 1 mm), and melanoma anatomical site (axial, including head and trunk, or extremities; head or trunk, or extremities or palms and soles). For MC1R, we also considered phenotypical characteristics such as skin type (I/II, III/IV), hair color (red/blond, light brown, dark brown/black), and eye color (blue/green, light brown, dark brown).
Semiquantitative data (age at diagnosis, Breslow thickness) were analyzed using Student’s t-test or by medians using the Mann–Whitney test or a non parametric two-tailed Wilcoxon test, as appropriate.
Data were analyzed using the GraphPad Prism statistical package, version 5.03. The statistical significance was considered at P less than 0.05.
Results
Patients’ characteristics
A total of 106 high-risk melanoma patients, including 72 FM patients (40 women and 32 men) from 48 melanoma-prone families and 34 spMPM patients (15 women and 19 men), were enrolled in this study. The demographic and clinical characteristics of the patients and related tumors are listed in Table 1.
Table 1.
Demographic and clinical characteristics of melanoma patients and histopathological features of tumors
|
n (%) |
|||
|---|---|---|---|
| FM | spMPM | P | |
| Characteristics of patients | N = 72 | N = 34 | |
| Sex | |||
| Males | 32 (44.4) | 19 (55.9) | 0.30 |
| Females | 40 (55.6) | 15 (44.1) | |
| Age at diagnosis | |||
| Age (years) [mean (range)] | 49 (15–81) | 41 (19–80) | 0.09 |
| ≤ 40 | 22 (30.6) | 16 (47.1) | 0.13 |
| > 40 | 50 (69.4) | 18 (52.9) | |
| Nevus count | |||
| ≤ 50 | 41 (56.9) | 12 (35.3) | 0.08 |
| > 50 | 28 (38.9) | 19 (55.9) | |
| Clinically atypical nevi | |||
| Yes | 18 (25.0) | 7 (20.6) | 1.00 |
| No | 49 (68.1) | 20 (58.8) | |
| Skin type | |||
| I | 7 (9.7) | 4 (11.8) | 0.68 |
| II | 40 (55.6) | 16 (47.1) | |
| III | 23 (31.9) | 13 (38.2) | |
| IV | 2 (2.8) | 1 (2.9) | |
| Hair color | |||
| Red/blond | 15 (20.8) | 11 (32.3) | 0.48 |
| Light brown | 31 (43.1) | 13 (38.2) | |
| Dark brown/black | 24 (33.3) | 10 (29.4) | |
| Eye color | |||
| Blue/green | 36 (50.0) | 15 (44.1) | 0.49 |
| Light brown | 24 (33.3) | 11 (32.3) | |
| Dark brown/black | 10 (13.9) | 8 (23.5) | |
| History of NMSC | |||
| Yes | 9 (12.5) | 1 (2.9) | 0.16 |
| No | 59 (81.9) | 32 (94.1) | |
| Number of primary melanomas | |||
| Single | 58 (80.6) | NA | NA |
| Multiple | 14 (19.4) | 34 (100) | 1.00 |
| Two melanomas | 13 (18.1) | 30 (88.2) | |
| Three melanomas | 1 (9.7) | 2 (5.9) | |
| Four melanomas | 0 (0) | 1 (2.9) | |
| Five melanomas | 0 (0) | 1 (2.9) | |
| Characteristics of tumors | N = 87 | N = 75 | |
| Breslow thickness | |||
| In situ | 32 (36.8) | 37 (49.3) | 0.20 |
| Invasive | 50 (57.5) | 37 (49.3) | |
| ≤ 1 mm | 70 (80.5) | 64 (85.3) | 1.00 |
| > 1 mm | 12 (13.8) | 10 (13.3) | |
| Median value (range) | 0.57 (0.22–5.00) | 0.60 (0.17–2.66) | 0.24 |
| Histopathological subtype | |||
| SSM | 74 (85.1) | 72 (96.0)* | NA* |
| NM | 0 (0) | 2 (2.6) | |
| LM/LMM | 6 (6.9) | 1 (1.3) | |
| ALM | 3 (3.4) | 0 (0) | |
| Others | 3 (3.4) | 0 (0) | |
| Anatomical site of melanoma | |||
| Head | 7 (8.0) | 2 (2.6) | < 0.01 |
| Trunk | 33 (37.9) | 47 (62.7) | |
| Extremities | 42 (48.3) | 26 (34.7) | |
| Palms or soles | 3 (3.4) | 0(0) | |
| Axial | 40 (46.0) | 49 (65.3) | 0.02 |
| Extremities | 45 (51.7) | 26 (34.7) | |
Numbers do not always add up to the total because of missing data.
ALM, acral lentiginous melanoma; FM, familial melanoma; LM/LMM, lentigo maligna/lentigo maligna melanoma; NA, not applicable; NM, nodular melanoma; NMSC, nonmelanoma skin cancer; spMPM, sporadic multiple primary melanoma; SSM, superficial spreading melanoma.
Significant difference in frequency distribution between FM and spMPM (P = 0.03) on comparing SSM versus others (NM, LM/LMM, ALM, spitzoid, occult).
In terms of the number of affected individuals within families, 38/48 (79%) families had two affected members and the remaining 10 (10/48, 21%) had more than two affected patients (five families with three affected members, three with four members, one with five members, and one with six members). As for the degree of relatedness, 33/48 (69%) families had first-degree affected relatives and 15/48 (31%) families had second-degree affected relatives. Among the FM patients, 58/72 (81%) presented a single melanoma, whereas 14/72 (19%) were diagnosed with MPM (13 patients with two melanomas and one with three melanomas) for a total of 87 melanoma lesions (Table 1). The median age of melanoma onset was 49 (range: 15–81) years. Age at melanoma diagnosis was significantly younger in FM patients with MPM (40 years) than in FM patients with a single melanoma (50 years, P=0.04) (data not shown).
As for spMPM patients, 30/34 (88%) were diagnosed with two melanomas, 2/34 (6%) with three melanomas, 1/34 (3%) with four, and 1/34 (3%) with five melanomas, for a total of 75 melanoma lesions (Table 1). The median age at first melanoma diagnosis in spMPM patients was 41 (19–80) years. spMPM patients developed almost exclusively melanomas of the SSM subtype compared with FM patients (P=0.03). Melanomas were more frequently located on the extremities in FM patients and on the trunk in spMPM patients (P<0.01) (Table 1).
Mutational analysis
Familial melanoma
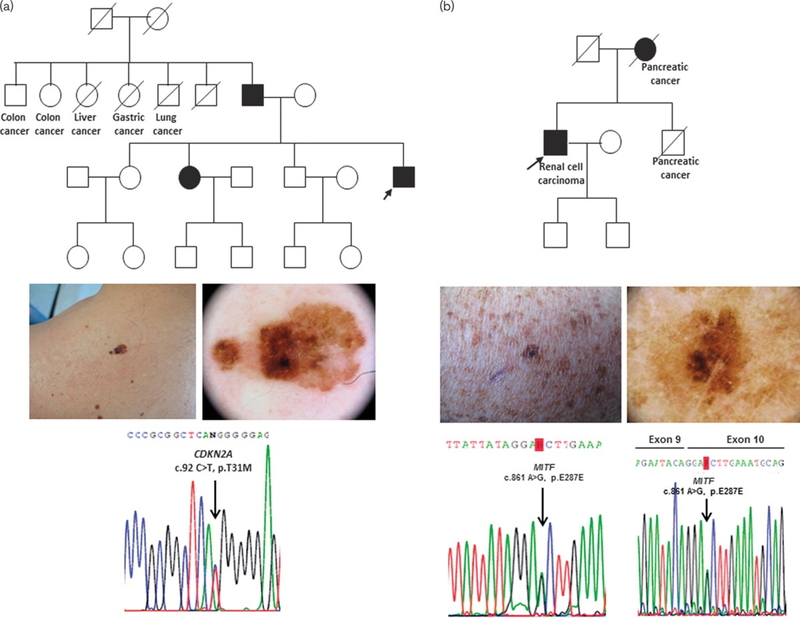
Overall, we detected 4 CDKN2A germline missense mutations (Table 2) in exon 2, p.V59A (c.222T>C), p.N71I (c.212A>T), p.H83Q (c.295C>A), and p.114L (c.387C>T), in six FM patients from four families, all with first-degree affected relatives and one novel nucleotide change with unknown function in exon 1β, p.T31M (c.92C>T).
Table 2.
CDKN2A mutations, the TERT polymorphism, and MC1R variants in familial melanoma and sporadic multiple primary melanoma patients
| FM (N = 72) [n (%)] |
spMPM (N = 34) [n (%)] |
||||
|---|---|---|---|---|---|
| CDKN2A | |||||
| Genomic region | Nucleotide change | Effect on p16INK4A | Effect on p14ARF | ||
| Exon 1β | c.92C > T | None | p.T31M | 1 | |
| Exon 2 | c.214G > T | p.S56I | p.Q70H | 1 | |
| Exon 2 | C.222T > C | p.V59A | none | 2 | |
| Exon 2 | c.212A > T | p.N71I | p.Q85H | 1 | |
| Exon 2 | c.295C > A | p.H83Q | none | 1 | |
| Exon 2 | c.387C > T | p.P114L | A128V | 2 | |
| Exon 2 | C.442G > A | p.A148T | None | 12 (16.7) | 3 (8.8) |
| 3′UTR | *500C > G | NA | NA | 31 (43.1) | 10 (29.4) |
| 3′UTR | *540C > T | NA | NA | 8 (11.1) | 9 (26.5) |
| TERT | |||||
| rs2853669 | c.-245T > C | 42 (58.3) | 16 (47) | ||
| MC1R | |||||
| Wild type | 25 (34.7) | 7 (20.6) | |||
| Any variant | 47 (65.3) | 27 (79.4) | |||
| One variant | 23 (31.9) | 15 (44.1) | |||
| Two variants | 24 (33.3) | 12 (35.3) | |||
| Any RHC variants | 30 (41.7) | 15 (44.1) | |||
| Any NRHC variants | 17 (23.6) | 12 (35.3) |
FM, familial melanoma; NA, not applicable; NRHC, nonred hair color; RHC, red hair color; spMPM, sporadic multiple primary melanoma.
The novel substitution c.92C>T, resulting in the p.T31M substitution of p14ARF protein, was identified in the index patient of a family with three first-degree affected members (Fig. 1a). To assess the pathogenic role of this variant in p14ARF, we used nine computational effect prediction tools, obtaining an overall accuracy of 62%. Six out of nine prediction tools estimated the mutation as neutral, not affecting p14ARF function, whereas the activity predictor SIFT, the MAPP tool, and the latest and currently best sensible CADD tool attributed a damaging effect to this variant with low scores. We also carried out a structural in silico analysis to analyze the potential involvement of the T31 site in p14ARF phosphorylation. NetPhos 2.0 results indicated that p14ARF contains three serines and one threonine as potential phosphorylation sites. Interestingly, T31 is included in the prediction, with a high score of 0.921, yielding a very likely phosphorylation site and suggesting a potential influence of the T31M alteration in p14ARF post-translational modification.
Fig. 1:
p14ARFp.T31M and MITF p.E287E mutations in Italian familial melanoma (FM) patients. (a) c.92C>T, p.T31M in exon 1β of the CDKN2A gene. Upper panel: pedigree of the FM patient carrying the mutation. Middle panel: clinical and dermoscopic images of melanoma. Bottom panel: electropherogram of 1β mutated sequence; (b) c.861A>G, p.E287E in exon 10 of the MITF gene. Upper panel: pedigree of the FM patient carrying the mutation. Middle panel: clinical and dermoscopic images of melanoma. Bottom panel: electropherogram of MITF exon 10 mutation, DNA sequence (left); electropherogram of MITF exon 9/exon 10 junction, cDNA sequence (right). In the pedigrees, index cases are indicated by an arrow, strike-through symbols indicate deceased individuals, and solid symbols represent melanoma-affected patients.
CDKN2A mutations were identified more frequently in members of families with more than two affected members than in individuals of families with two affected members (P=0.04) (Table S2). A significant association for the presence of CDKN2A mutations and occurrence of NMSC, which were all diagnosed as basal cell carcinoma, was observed (P=0.03) (Table S2).
Moreover, we found three known CDKN2A polymorphisms (Table 2). The p.A148T variant in exon 2 was detected in 12/72 (17%) FM cases and the 3′UTR region polymorphisms, *500C>G and *540C>T, in 34/72 (47%) FM cases (*500C>G in 26 patients, *540C>T in three and both *500C>G and *540C>T in five) (Table 2). 3′UTR polymorphisms were significantly more frequent in patients with melanoma of the trunk (P=0.02) (Table S2).
Mutational screening of the TERT promoter identified the rs2853669 polymorphism (G>C substitution at −245 bp) in 42 of 72 (58%) FM patients (Table 2). No statistical differences were found between carriers and noncarriers of the polymorphism according to the characteristics of patients or the clinicopathological features of melanomas.
None of our FM patients carried mutations in exon 2 of the CDK4 gene and none harbored the p.S270N mutation in the POT1 gene.
We detected a synonymous mutation, p.E287E (c.861A>G), at the third base of the first codon of exon 10 of the MITF gene in a FM patient with a personal history of renal cell carcinoma and familial occurrence of pancreatic cancer in two first-degree relatives (mother and brother) (Fig. 1b). The patient was negative for mutations in the other screened high-penetrance genes and carried the TERT rs2853669 polymorphism, the 3′UTR *500C>G variant, and the V60L variant in the MC1R gene. In silico splicing prediction tests for the p.E287E (c.861A>G) MITF variant, with MutationTaster, Human Splicing Finder, and Ex-skip tools showed that this substitution could potentially alter an exonic splicing enhancer site, likely disturbing normal splicing. An in-vitro analysis of the splicing site variant was carried out by amplifying and sequencing the cDNA of the MITF-M variant obtained from the carrier patient and a healthy control. No splicing variants differing in size from the mutated transcript and the wild-type form were detected. Therefore, we subjected the PCR products to Sanger sequencing, but no splicing alteration in exon 10 occurred in the patient (Fig. 1b).
In the MC1R analysis, we detected 10 nonsynonymous variants in 47/72 (65%) FM patients, with the V60L change being the most prevalent variant (25/72, 34%), followed by R151C (15/72, 21%) (Table 2 and S3). Twenty-three of 47 (49%) patients carried one MC1R allelic variant and 24/47 (51%) carried two allelic variants (six homozygotes and 18 compound heterozygotes) (Table 2). In terms of clinical characteristics, MC1R variants, mainly RHC substitutions, were associated with a light phenotype (skin type and hair color, P=0.02 and <0.001, respectively) and with a reduction of age at diagnosis (P=0.01) compared with noncarriers (Table 3). In addition, a reduction of age at diagnosis was also observed in four FM patients harboring both a CDKN2A mutation and MC1R variants compared with patients carrying only a CDKN2A mutation (median age 39 vs. 58 years, respectively), although the low number of the mutation carriers did not allow us to perform consistent statistical tests.
Table 3.
Frequency of MC1R variants according to the clinical and histopathological characteristics of familial melanoma and sporadic multiple primary melanoma patients
| FM |
spMPM |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MC1R Wild-type | Any MC1R variant | Pa | RHC | Pb | NRHC | P c | MC1R wild-type | Any MC1R variant | Pa | RHC | Pb | NRHC | Pc | |
| Characteristics of patients | ||||||||||||||
| Total (N) | 72 | 34 | ||||||||||||
| n | 25 | 47 | 30 | 17 | 7 | 27 | 15 | 12 | ||||||
| Age at diagnosis | ||||||||||||||
| ≤ 40 | 4 (16.0) | 18 (38.3) | 0.06 | 12 (40.0) | 0.07 | 6 (35.3) | 0.13 | 3 (42.9) | 13 (48.2) | 1.00 | 8 (53.3) | 1.00 | 5 (41.7) | 0.58 |
| > 40 | 21 (84.0) | 29 (61.7) | 18 (60.0) | 11 (64.7) | 4 (57.1) | 14 (51.8) | 7 (46.7) | 7 (58.3) | ||||||
| Median value (range) | 53 (20–81) | 45 (15–80) | 0.03 | 44 (15–77) | 0.01 | 48 (26–80) | 0.04 | 44 (19–78) | 41 (19–80) | 0.64 | 39 (19–80) | 0.57 | 42 (28–67) | 0.70 |
| Skin type | ||||||||||||||
| I/II | 11 (44.0) | 35 (74.5) | 0.08 | 25 (83.3) | 0.02 | 10 (58.8) | < 0.01 | 2 (28.6) | 18 (66.7) | 0.42 | 12 (80.0) | 0.06 | 6 (50.0) | 0.02 |
| III/IV | 12 (48.0) | 12 (25.5) | 5 (16.7) | 7 (41.2) | 5 (71.4) | 9 (33.3) | 3 (20.0) | 6 (50.0) | ||||||
| Hair color | ||||||||||||||
| Red-blond | 0 (0) | 13 (27.7) | < 0.01 | 12 (40.0) | < 0.01 | 1 (5.9) | < 0.01 | 1 (14.3) | 10 (37.0) | < 0.01 | 7 (46.7) | < 0.01 | 3 (25.0) | < 0.01 |
| Medium | 8 (32.0) | 23 (48.9) | 12 (40.0) | 11 (64.7) | – | 13 (48.2) | 6 (40.0) | 7 (58.3) | ||||||
| Dark | 13 (52.0) | 11 (23.4) | 6 (20.0) | 5 (29.4) | 6 (85.7) | 4 (14.8) | 2 (13.3) | 2 (16.7) | ||||||
| Eye color | ||||||||||||||
| Light | 11 (44.0) | 25 (53.2) | 0.46 | 15 (50.0) | 0.46 | 10 (58.8) | 0.72 | 2 (28.6) | 13 (48.2) | 0.06 | 8 (53.3) | 0.22 | 5 (41.7) | 0.12 |
| Medium | 7 (28.0) | 17 (36.2) | 12 (40.0) | 5 (29.4) | 1 (14.3) | 10 (37.0) | 4 (26.7) | 6 (50.0) | ||||||
| Dark | 5 (20) | 5 (10.6) | 3 (10.0) | 2 (11.8) | 4 (57.1) | 4 (14.8) | 3 (20.0) | 1 (8.3) | ||||||
| Nevus count | ||||||||||||||
| ≤ 50 | 17 (68.0) | 24 (51.1) | 0.12 | 16 (53.0) | 0.25 | 8 (47.1) | 0.25 | 4 (57.1) | 8 (29.6) | 0.17 | 5 (33.3) | 0.34 | 3 (25.0) | 0.31 |
| > 50 | 6 (24.0) | 21 (44.7) | 12 (40.0) | 9 (52.9) | 2 (28.6) | 17 (63.0) | 9 (60.0) | 8 (66.7) | ||||||
| Clinically atypical nevi | ||||||||||||||
| Yes | 4 (16.0) | 14 (29.8) | 0.25 | 8 (26.7) | 0.51 | 6 (35.3) | 0.40 | 1 (14.3) | 6 (22.2) | 1.00 | 6 (40.0) | 0.35 | 0 (0) | 0.11 |
| No | 19 (76.0) | 30 (63.8) | 20 (66.7) | 10 (58.8) | 5 (71.4) | 15 (55.6) | 8 (53.3) | 7 (58.3) | ||||||
| Number of family members | ||||||||||||||
| 2 | 20 (80.0) | 33 (70.2) | 0.41 | 21 (70.0) | 0.53 | 12 (70.6) | 0.41 | NA | NA | NA | NA | |||
| > 2 | 5 (20.0) | 14 (29.8) | 9 (30.0) | 5 (29.4) | NA | NA | NA | NA | ||||||
| Number of primary melanomas | ||||||||||||||
| Single | 21 (84.0) | 37 (78.7) | 0.75 | 22 (73.3) | 0.51 | 15 (88.2) | 0.30 | NA | NA | NA | NA | |||
| Multiple | 4 (16.0) | 10 (21.3) | 8 (26.7) | 2 (11.8) | NA | NA | NA | NA | ||||||
| History of NMSC | ||||||||||||||
| Yes | 4 (16.0) | 5 (10.6) | 0.47 | 2 (6.7) | 0.39 | 3 (17.6). | 0.25 | 0 (0) | 1 (3.7) | 0.62 | 1 (6.7) | 1.00 | 0 (0) | 0.30 |
| No | 19 (76.0) | 40 (85.1) | 27 (90.0) | 13 (76.5) | 7 (100) | 25 (92.6) | 13 (86.7) | 12 (100) | ||||||
| Characteristics of melanomas | ||||||||||||||
| Total (N) | 87 | 75 | ||||||||||||
| n | 29 | 58 | 38 | 20 | 16 | 59 | 35 | 24 | ||||||
| Breslow thickness | ||||||||||||||
| In situ | 12 (41.4) | 20 (34.5) | 0.63 | 10 (26.3) | 0.29 | 10 (50.0) | 0.18 | 8 (50.0) | 29 (49.1) | 1.00 | 12 (34.3) | 0.37 | 17 (70.8) | 0.14 |
| Invasive | 15 (51.7) | 35(60.3) | 25 (65.8) | 10 (50.0) | 8 (50.0) | 29 (49.1) | 22 (62.9) | 7 (29.2) | ||||||
| ≤ 1 mm | 24 (82.7) | 46 (79.3) | 1.00 | 29 (76.3) | 1.00 | 17 (85.0) | 0.82 | 13 (81.2) | 51 (86.4) | 0.65 | 29 (82.9) | 0.64 | 22 (91.7) | 0.42 |
| > 1mm | 3 (10.3) | 9 (15.5) | 6 (15.8) | 3 (15.0) | 3 (18.8) | 7 (11.9) | 5 (14.3) | 2 (8.3) | ||||||
| Median value (range) | 0.43 (0.22–4.55) | 0.68 (0.24–0.50) | 0.27 | 0.65 (0.24–3.15) | 0.45 | 0.85 (0.27–5.0) | 0.37 | 0.77 (0.27–2.20) | 0.60 (0.17–2.66) | 0.91 | 0.57 (0.17–2.66) | 0.71 | 0.92 (0.5–1.45) | 0.46 |
| Anatomical site | ||||||||||||||
| Head | 3 (10.3) | 4 (6.9) | 0.42 | 1 (2.6) | 0.35 | 3 (15.0) | 0.28 | 0(0) | 2 (3.4) | 0.03 | 1 (2.9) | 0.02 | 1 (4.2) | 0.09 |
| Trunk | 10 (34.5) | 23 (39.6) | 15 (39.5) | 8 (40.0) | 6 (37.5) | 41 (69.5) | 26 (74.3) | 15 (62.5) | ||||||
| Extremities | 16 (55.2) | 26 (44.8) | 20 (52.6) | 6 (30.0) | 10 (62.5) | 16 (27.1) | 8(22.9) | 8 (33.3) | ||||||
| Palm or soles | 0 (0) | 3 (5.2) | 1(2.6) | 2 (10.0) | 0 (0) | 0(0) | 0(0) | 0(0) | ||||||
| Axial | 13 (44.8) | 27 (46.5) | 0.77 | 16 (42.1) | 0.40 | 11 (55.0) | 0.68 | 6 (37.5) | 43 (72.9) | < 0.01 | 27 (77.1) | 0.02 | 16 (66.7) | < 0.01 |
| Extremities | 16 (55.2) | 29 (50.0) | 21 (55.3) | 8 (40.0) | 10 (62.5) | 16 (27.1) | 8 (22.9) | 8 (33.3) | ||||||
| Histopathological subtype | ||||||||||||||
| SMM | 25 (86.2) | 49 (84.5) | 0.74 | 35 (92.1) | 0.69 | 14 (70.0) | 0.61 | 16 (100) | 56 (94.9) | 1.00 | 32 (91.4) | 0.54 | 24 (100) | 0.09 |
| Other | 3 (10.3) | 9 (15.5) | 3 (7.9) | 6 (30.0) | 0 (0) | 3 (5.1) | 3 (8.6) | 0 (0) | ||||||
Numbers do not always add up to the total because of missing data.
FM, familial melanoma; NA, not applicable; NMSC, nonmelanoma skin cancer; NRHC, nonred hair color; RHC, red hair color; spMPM, sporadic multiple melanoma; SSM, superficial spreading melanoma.
P value calculated comparing MC1R-mutated versus MC1R wild-type patents.
P value calculated comparing RHC carriers versus MC1R wild-type patients.
Test for linear trend across categories (wild type, NRHC, RHC).
Sporadic MPM
CDKN2A was mutated in only one of 34 (3%) spMPM patients (Table 2), diagnosed with two melanomas. Three of 34 (9%) spMPM patients carried the A148T polymorphism, 17 of 34 (50%) carried at least one 3′UTR polymorphism (eight carried *500C>G, seven carried *540C>T, and two carried the 500C>G and *540C>T polymorphisms), and 16 of 34 (47%) patients carried the rs2853669 polymorphism in the TERT promoter (Table 2). No significant differences were observed in the clinical phenotype of patients and melanoma characteristics between carriers and noncarriers of the 3′UTR or the TERT promoter polymorphisms (data not shown).
We did not detect any mutation in exon 2 of the CDK4 gene and in exon 10 of the MITF gene or the p.S270N mutation in the POT1 gene in our spMPM patients.
As for MC1R, we detected 16 MC1R allelic variants in 27/34 (79%) spMPM patients (Table S3), with 15/27 (56%) carrying a single allelic variant and 12/27 (44%) carrying two variants (one homozygote and 11 compound heterozygotes) (Table 2). The V60L substitution was the most prevalent variant and was detected in 16/34 (47%) patients. The presence of at least one MC1R variant, mainly RHC substitutions, was associated with a light phenotype (fair skin type and light hair color, P=0.01 and <0.01, respectively). Carriers of RHC changes were diagnosed more frequently with melanoma on the trunk compared with wild-type patients (P=0.02) (Table 3).
Discussion
We characterized the mutation profile of the major melanoma susceptibility genes in a group of high-risk patients from Central Italy. We identified CDKN2A mutations in 8.3% of melanoma families, and their prevalence increased with the number of affected relatives within the family (P=0.04). MC1R variants were identified in 65% of FM patients and the TERT rs2853669 promoter polymorphism was identified in 58%. A new synonymous mutation (c.861A>G, p.E287E), not affecting the MITF exon 10 splicing, was detected in one FM patient. Among patients with spMPM, 3% carried CDKN2A mutations, 79% carried MC1R variants, and 47% carried the TERT rs2853669 promoter polymorphism. None of the patients carried mutations in CDK4 exon 2 nor harbored the POT1 p.S270N mutation.
The frequency of CDKN2A mutations in melanoma families has been shown to vary considerably among different populations depending on baseline melanoma incidence, founder effects, and selection criteria of the study population 8. The 8.3% mutation rate detected in our FM patients (4/48 families) is similar to that reported in some studies 22,24,28, but lower than that described in other studies 21,23,25,26 analyzing Italian families. Our findings confirm that a high number of affected members within the family is a strong predictive factor for CDKN2A genetic screening. About 80% of our families were small families, with only two affected members, and only 2.6% (1/38) of them carried a CDKN2A mutation. This value is in line with the 4% incidence rate reported in families with two affected relatives in two large international studies 5,33. We did not find any significant association between clinical features of patients or tumors and CDKN2A mutations because of the low frequency of mutations identified.
Outside the familial context, the development of multiple primary tumors can be related either to germline de-novo mutations or mutations in low-penetrance predisposing genes. The 3% prevalence of CDKN2A mutations in spMPM patients in the present study is lower than the 21.4% reported previously in a different series of spMPM patients by our group, where CDKN2A mutations were detected in three of 14 patients (G101W mutation in one patient and an intronic variant IVS2+1G>T in two patients) 6. The higher prevalence of CDKN2A mutations in the previous study might be related to the counting of the intronic variant, whose functional significance is uncertain and that has not been identified in any of the patients in the present study. Low mutation rates were also found in a Spanish cohort of spMPM patients (8.2%) and in two large multicenter population-based studies (2.9 and 6.9%, respectively) 7,8.
Our findings indicate that CDKN2A mutations account for a small percentage of high-risk melanoma patients in Central Italy. The low frequency of CDKN2A mutations in our cases might be explained by the modifying effect of intermediate/low-risk melanoma susceptibility genes, by the absence of a founder mutation, or by the involvement of as yet unknown predisposition genes.
We first report a novel substitution in exon 1β of CDKN2A, p.T31M, affecting the p14ARF protein, but not p16INK4A, in the only screened patient of a family with three first-degree affected relatives. This mutation has not been published previously or included in gene mutation databases (http://www.hgmd.cf.ac.uk/ac/index.php; http://wwwncbinlmnihgov/SNP/; and http://wwwncbinlmnihgov/clinvar). Six of nine computational tools predicted this mutation to be neutral and the other three attributed a damaging effect to this variant with low scores. In silico analysis showed that T31 is very likely to be a phosphorylation site, suggesting a potential influence of the T31M alteration in p14ARF post-translational modification.
The association of CDKN2A 3′UTR polymorphisms with melanoma risk has been suggested in different case–control studies 34–36, but no definite correlation with clinical features of patients or histopathological aspects of melanoma has been found 37. We observed a high frequency of CDKN2A 3′UTR polymorphisms in FM (47%) and spMPM (50%) patients, significantly higher in FM patients with melanoma on the trunk compared with patients with melanoma on other anatomical sites (P<0.01).
rs2853669 is a common and potentially functional polymorphism of the TERT promoter acting as a positive regulator of the TERT gene and required for telomerase activation during tumor progression and cellular immortalization. This polymorphism has been identified recently as a risk factor for lung cancer 38 and seems to affect survival in acute myeloid leukemia 39, bladder carcinoma 40, renal cell carcinoma 41, and glioblastoma 42, but few data are available in melanoma. Horn et al.13 described this polymorphism to be in complete allelic linkage with the −57 bp melanoma-predisposing variant of the TERT promoter in one melanoma family and in 40% of metastatic melanoma cell lines. In our patients, a high prevalence of the rs2853669 polymorphism was observed in FM (58%) and in spMPM (47%), but none of them carried the predisposing variant at −57 bp.
An FM patient with a personal history of renal cell carcinoma and a family history of pancreatic carcinoma harbored a synonymous mutation of the MITF gene occurring in the early exonic positions of exon 10, the p.E287E (c.861A>G), reported as a very rare variant (MAF: G=0.004) in the Exome Aggregation Consortium database (http://exac.broadinstitute.org/). Segregation of the mutation with the disease was not tested as the only affected family member was the mother, who died decades ago. Although not expected to alter the function of the protein, the p.E287E mutation introduces a sequence variation at the intron–exon boundary, an essential region for splicing and mRNA transcription. It has indeed been shown that exonic single-base substitutions may affect splicing when occurring at binding sites for splicing regulatory elements 43. The three computational tools (MutationTaster, Human Splicing Finder and Ex-skip) used for splicing prediction suggested that the p.E287E (c.861A>G) variant could likely disturb normal splicing, altering the enhancer site, but in-vitro functional analysis carried out by RT-PCR and subsequent Sanger sequencing could not detect a splicing variant of MITF exon 10 caused by the mutation. These findings could be explained by the use of total RNA from whole blood and not from melanocytes, in which different environmental stresses and senescence stimuli could influence the expression of splicing variants. Therefore, we cannot definitely conclude whether the c.861A>G variant is a disease-causing mutation, but the patient’s personal and family history of multiple cancers suggests a strong genetic component.
Mutational screening of our patients showed a high prevalence of MC1R polymorphisms (65% in FM, 79% in spMPM) as observed in other Italian studies 6,22,25,31,32, with the V60L being the most frequent MC1R variant detected in FM (35%) and spMPM (47%). MC1R RHC variants were identified in over 40% of both FM and spMPM patients, confirming their high prevalence in melanoma patients. In our study, RHC variants were associated with light phenotypic complexion, supporting the role of MC1R as an important determinant of human pigmentation 44,45.
We found a significant reduction of age at diagnosis in FM patients carrying MC1R variants compared with wild-type patients (P=0.03) and the reduction became stronger when we considered patients carrying RHC variants (P=0.01). This is a controversial issue as a significant reduction of age at diagnosis as the number or type of MC1R variants increased was reported in small studies including reports from Mediterranean populations 25,31,46,47, whereas no association was found in a large study on familial melanoma from three continents 5.
In CDKN2A-positive melanoma families, MC1R variants have been shown to significantly increase the penetrance of CDKN2A mutations, especially with respect to multiple MC1R variants and RHC variants 48. We observed the co-occurrence of CDKN2A and MC1R substitutions in four FM patients who presented a reduction of age at diagnosis compared with FM patients with only CDKN2A mutations (39 vs. 59 years).
In our spMPM cohort, MC1R variants, especially RHC changes, were prevalent in patients with melanoma on the trunk. A similar result was reported in Swedish spMPM and FM patients 49, whereas RHC variants were significantly associated with melanoma on the arms in a recent multicenter study by the GEM group 50. Differences in body-site grouping and categorization of MC1R variants make it difficult to compare our results with those of other studies 31,46.
The main limitation of our study was the small size of our sample populations, which did not provide us with adequate statistical power in some subgroup analysis. This was especially true when we analyzed the association between the presence of CDKN2A mutations and clinical aspects of patients or histopathological features of melanoma. In spMPM, we could not carry out any association analysis as only one patient had the CDKN2A mutation. In addition, the two novel mutations first identified in our study, the substitution in exon 1β of CDKN2A, p.T31M, and the synonymous change at the first codon of MITF exon 10, p.E287E, were analyzed by computational tools and in-vitro splicing analysis, respectively, but they need to be further characterized with in-depth functional studies.
In conclusion, a low frequency of CDKN2A mutations and a high prevalence of MC1R variants characterize high-risk melanoma patients from Central Italy. Our results support the role of CDKN2A mutations in familial melanoma with more than two affected members, and confirm the involvement of MC1R variants in both FM and spMPM patients, with a significant association with reduction of age at diagnosis in FM patients and the occurrence of melanoma on the trunk in spMPM patients. CDK4, POT1, and MITF genes and the TERT promoter were rarely mutated in our patients. The identification of predominant germline mutations in candidate susceptibility genes within a specific geographic area has particular relevance for prevention and early melanoma detection to refer patients and their families to clinical screening.
Acknowledgements
This study was supported by the Italian Ministry of the University and Scientific Research (PRIN-2012 grant 2012JJX494).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Zanetti R, Masini C, et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer 2005; 41:2040–2059. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 2.Psaty EL, Scope A, Halpern AC, Marghoob A. Defining the patient at high risk for melanoma. Int J Dermatol 2010; 49:362–376. NIH Library [Context Link] [DOI] [PubMed] [Google Scholar]
- 3.Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, et al. Germline p16 mutations in familial melanoma. Nat Genet 1994; 8:15–21. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 4.Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 1996; 12:97–99. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 5.Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007; 44:99–106. [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peris K, Fargnoli MC, Pacifico A, Surrenti T, Stolz W, Wolf P, et al. CDKN2A and MC1R mutations in patients with sporadic multiple primary melanoma. J Invest Dermatol 2004; 122:1327–1330. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 7.Berwick M, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, et al. The prevalence of CDKN2A germ-line mutations and relative risk for cutaneous malignant melanoma: an international population-based study. Cancer Epidemiol Biomarkers Prev 2006; 15:1520–1525. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 8.Helsing P, Nymoen DA, Ariansen S, Steine SJ, Maehle L, Aamdal S, et al. Population-based prevalence of CDKN2A and CDK4 mutations in patients with multiple primary melanomas. Genes Chromosomes Cancer 2008; 47:175–184. [Context Link] [DOI] [PubMed] [Google Scholar]
- 9.Puntervoll HE, Yang XR, Vetti HH, Bachmann IM, Avril MF, Benfodda M, et al. Melanoma prone families with CDK4 germline mutation: phenotypic profile and associations with MC1R variants. J Med Genet 2013; 50:264–270. NIH Library Bibliographic Links [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robles-Espinoza CD, Harland M, Ramsay AJ, Aoude LG, Quesada V, Ding Z, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet 2014; 46:478–481. NIH Library Bibliographic Links [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet 2014; 46:482–486. NIH Library Bibliographic Links [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoude LG, Pritchard AL, Robles-Espinoza CD, Wadt K, Harland M, Choi J, et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J Natl Cancer Inst 2014; 107:1–7. [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013; 339:959–961. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 14.Harland M, Petljak M, Robles-Espinoza CD, Ding Z, Gruis NA, van Doorn R, et al. Germline TERT promoter mutations are rare in familial melanoma. Fam Cancer 2016; 15:139–144. NIH Library [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaumont KA, Shekar SN, Newton RA, James MR, Stow JL, Duffy DL, et al. Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum Mol Genet 2007; 16:2249–2260. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 16.Pasquali E, Garcia-Borron JC, Fargnoli MC, Gandini S, Maisonneuve P, Bagnardi V, et al. MC1R variants increased the risk of sporadic cutaneous melanoma in darker-pigmented Caucasians: a pooled-analysis from the M-SKIP project. Int J Cancer 2015; 136:618–631. [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011; 480:99–103. NIH Library Bibliographic Links [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011; 480:94–98. [Context Link] [DOI] [PubMed] [Google Scholar]
- 19.Ghiorzo P, Pastorino L, Queirolo P, Bruno W, Tibiletti MG, Nasti S, et al. Prevalence of the E318K MITF germline mutation in Italian melanoma patients: associations with histological subtypes and family cancer history. Pigment Cell. Melanoma Res 2013; 26:259–262. NIH Library [Context Link] [DOI] [PubMed] [Google Scholar]
- 20.Fargnoli MC, Chimenti S, Keller G, Soyer HP, Dal Pozzo V, Hofler H, et al. CDKN2a/p16INK4a mutations and lack of p19ARF involvement in familial melanoma kindreds. J Invest Dermatol 1998; 111:1202–1206. [Context Link] [DOI] [PubMed] [Google Scholar]
- 21.Mantelli M, Barile M, Ciotti P, Ghiorzo P, Lantieri F, Pastorino L, et al. High prevalence of the G101W germline mutation in the CDKN2A (P16(ink4a)) gene in 62 Italian malignant melanoma families. Am J Med Genet 2002; 107:214–221. NIH Library Bibliographic Links [Context Link] [PubMed] [Google Scholar]
- 22.Landi MT, Goldstein AM, Tsang S, Munroe D, Modi W, Ter-Minassian M, et al. Genetic susceptibility in familial melanoma from north eastern Italy. J Med Genet 2004; 41:557–566. NIH Library Bibliographic Links [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gensini F, Sestini R, Piazzini M, Vignoli M, Chiarugi A, Brandani P, et al. The p.G23S CDKN2A founder mutation in high-risk melanoma families from Central Italy. Melanoma Res 2007; 17:387–392. Ovid Full Text Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 24.Majore S, De Simone P, Crisim A, Eibenschutz L, Binni F, Antigoni I, et al. CDKN2A/CDK4 molecular study on 155 Italian subjects with familial and/or primary multiple melanoma. Pigment Cell Melanoma Res 2008; 21:209–211. NIH Library [Context Link] [DOI] [PubMed] [Google Scholar]
- 25.Pastorino L, Bonelli L, Ghiorzo P, Queirolo P, Battistuzzi L, Balleari E, et al. CDKN2A mutations and MC1R variants in Italian patients with single or multiple primary melanoma. Pigment Cell Melanoma Res 2008; 21:700–709. NIH Library [Context Link] [DOI] [PubMed] [Google Scholar]
- 26.Bruno W, Ghiorzo P, Battistuzzi L, Ascierto PA, Barile M, Gargiulo S, et al. Clinical genetic testing for familial melanoma in Italy: a cooperative study. J Am Acad Dermatol 2009; 61:775–782. NIH Library [Context Link] [DOI] [PubMed] [Google Scholar]
- 27.Binn F, Antigoni I, De Simone P, Majore S, Silipo V, Crisi A, et al. Novel and recurrent p14 mutations in Italian familial melanoma. Clin Genet 2010; 77:581–586. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 28.Menin C, Vecchiato A, Scaini MC, Elefanti L, Funari G, De Salvo GL, et al. Contribution of susceptibility gene variants to melanoma risk in families from the Veneto region of Italy. Pigment Cell Melanoma Res 2011; 24:728–730. NIH Library [Context Link] [DOI] [PubMed] [Google Scholar]
- 29.Pedace L, De Simone P, Castori M, Sperduti I, Silipo V, Eibenschutz L, et al. Clinical features predicting identification of CDKN2A mutations in Italian patients with familial cutaneous melanoma. Cancer Epidemiol 2011; 35:e116–e120. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 30.Di Lorenzo S, Fanale D, Corradino B, Calo V, Rinaldi G, Bazan V, et al. Absence of germline CDKN2A mutation in Sicilian patients with familial malignant melanoma: could it be a population-specific genetic signature? Cancer Biol Ther 2016; 17:83–90. NIH Library [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landi MT, Kanetsky PA, Tsang S, Gold B, Munroe D, Rebbeck T, et al. MC1R, ASIP, and DNA repair in sporadic and familial melanoma in a Mediterranean population. J Natl Cancer Inst 2005; 97:998–1007. NIH Library [Context Link] [DOI] [PubMed] [Google Scholar]
- 32.Fargnoli MC, Altobelli E, Keller G, Chimenti S, Hofler H, Peris K. Contribution of melanocortin-1 receptor gene variants to sporadic cutaneous melanoma risk in a population in central Italy: a case-control study. Melanoma Res 2006; 16:175–182. [Context Link] [DOI] [PubMed] [Google Scholar]
- 33.Begg CB, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, et al. Genes Environment and Melanoma Study Group. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst 2005; 97:1507–1515. [Context Link] [DOI] [PubMed] [Google Scholar]
- 34.Aitken J, Welch J, Duffy D, Milligan A, Green A, Martin N, et al. CDKN2A variants in a population-based sample of Queensland families with melanoma. J Natl Cancer Inst 1999; 91:446–452. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 35.Kumar R, Smeds J, Berggren P, Straume O, Rozell BL, Akslen LA, et al. A single nucleotide polymorphism in the 3’untranslated region of the CDKN2A gene is common in sporadic primary melanomas but mutations in the CDKN2B, CDKN2C, CDK4 and p53 genes are rare. Int J Cancer 2001; 95:388–393. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 36.Maccioni L, Rachakonda PS, Bermejo JL, Planelles D, Requena C, Hemminki K, et al. Variants at the 9p21 locus and melanoma risk. BMC Cancer 2013; 13:325 NIH Library [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamperska KM, Przybyla A, Kycler W, Mackiewicz A. The CDKN2a common variants: 148 Ala/Thr and 500 C/G in 3’ UTR, and their association with clinical course of melanoma. Acta Biochim Pol 2007; 54:119–124. NIH Library Bibliographic Links [Context Link] [PubMed] [Google Scholar]
- 38.Yoo SS, Do SK, Choi JE, Lee SY, Lee J, Cha S, et al. TERT polymorphism rs2853669 influences on lung cancer risk in the Korean population. J Korean Med Sci 2015; 30:1423–1428. NIH Library Bibliographic Links [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosrati MA, Willander K, Falk IJ, Hermanson M, Hoglund M, Stockelberg D, et al. Association between TERT promoter polymorphisms and acute myeloid leukemia risk and prognosis. Oncotarget 2015; 6:25109–25120. NIH Library Bibliographic Links [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hosen I, Rachakonda PS, Heidenreich B, de Verdier PJ, Ryk C, Steineck G, et al. Mutations in TERT promoter and FGFR3 and telomere length in bladder cancer. Int J Cancer 2015; 137:1621–1629. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 41.Hosen I, Rachakonda PS, Heidenreich B, Sitaram RT, Ljungberg B, Roos G, et al. TERT promoter mutations in clear cell renal cell carcinoma. Int J Cancer 2015; 136:2448–2452. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 42.Mosrati MA, Malmstrom A, Lysiak M, Krysztofiak A, Hallbeck M, Milos P, et al. TERT promoter mutations and polymorphisms as prognostic factors in primary glioblastoma. Oncotarget 2015; 6:16663–166673. NIH Library Bibliographic Links [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol 2009; 578:3–22. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 44.Rees JL. Genetics of hair and skin color. Annu Rev Genet 2003; 37:67–90. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 45.Naysmith L, Waterston K, Ha T, Flanagan N, Bisset Y, Ray A, et al. Quantitative measures of the effect of the melanocortin 1 receptor on human pigmentary status. J Invest Dermatol 2004; 122:423–428. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 46.Stratigos AJ, Dimisianos G, Nikolaou V, Poulou M, Sypsa V, Stefanaki I, et al. Melanocortin receptor-1 gene polymorphisms and the risk of cutaneous melanoma in a low-risk southern European population. J Invest Dermatol 2006; 126:1842–1849. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 47.de Torre C, Garcia-Casado Z, Martinez-Escribano JA, Botella-Estrada R, Banuls J, Oliver V, et al. Influence of loss of function MC1R variants in genetic susceptibility of familial melanoma in Spain. Melanoma Res 2010; 20:342–348. Ovid Full Text Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 48.Fargnoli MC, Gandini S, Peris K, Maisonneuve P, Raimondi S. MC1R variants increase melanoma risk in families with CDKN2A mutations: a meta-analysis. Eur J Cancer 2010; 46:1413–1420. [Context Link] [DOI] [PubMed] [Google Scholar]
- 49.Hoiom V, Tuominen R, Kaller M, Linden D, Ahmadian A, Mansson-Brahme E, et al. MC1R variation and melanoma risk in the Swedish population in relation to clinical and pathological parameters. Pigment Cell Melanoma Res 2009; 22:196–204. NIH Library Bibliographic Links [Context Link] [DOI] [PubMed] [Google Scholar]
- 50.Taylor NJ, Busam KJ, From L, Groben PA, Anton-Culver H, Cust AE, et al. Inherited variation at MC1R and histological characteristics of primary melanoma. PLoS One 2015; 10:e0119920 NIH Library [Context Link] [DOI] [PMC free article] [PubMed] [Google Scholar]