Abstract
The TRPM (transient receptor potential melastatin) family belongs to the superfamily of TRP cation channels. The TRPM subfamily is composed of eight members that are involved in diverse biological functions such as temperature sensing, inflammation, insulin secretion, and redox sensing. Since the first cloning of TRPM1 in 1998, tremendous progress has been made uncovering the function, structure, and pharmacology of this family. Complete structures of TRPM2, TRPM4, and TRPM8, as well as a partial structure of TRPM7, have been determined by cryo-EM, providing insights into their channel assembly, ion permeation, gating mechanisms, and structural pharmacology. Here we summarize the current knowledge about channel structure, emphasizing general features and principles of the structure of TRPM channels discovered since 2017. We also discuss some of the key unresolved issues in the field, including the molecular mechanisms underlying voltage and temperature dependence, as well as the functions of their C-terminal domains.
1. Introduction
The TRPM (transient receptor potential melastatin) family is the largest and most diverse subfamily of the TRP superfamily [1]. In 1998, TRPM1 was the first gene of the TRPM family identified and cloned [2]. Since then, the family has been found to have eight members, TRPM1 to TRPM8. These molecules are widely expressed and contribute to cellular Ca2+ signaling by promoting Ca2+ entry into the cytosol in response to stimuli such as changes in the concentration of ions, small molecules, and lipids, some of which acting as intracellular second messengers. Ca2+ entry results either from the channels being Ca2+-permeable (TRPM1, TRPM2, TRPM3, TRPM6/7, TRPM8) or by their effects on other channels (such as voltage-gated Ca2+ channels, by modulating the membrane potential). A change in the cellular Ca2+/Mg2+ concentration or a change in membrane potential and electrical activity of the cell can then affect biological processes including the sensing of oxidative stress, the regulation of endothelial permeability and cell death, magnesium homeostasis, myogenic response, and the regulation of vascular tone [3–7]. The members of this family have thus attracted increasing attention in the past decade as promising drug targets for treatment of neurodegenerative disorders [8], cardiovascular diseases [9], type II diabetes [10], inflammation [11], and inflammatory pain [12].
The TRPM family members are diverse, not only in their physiological functions but also in their biophysical properties, including ion conduction and selectivity, gating mechanism, and ligand recognition. For instance, most of the TRPM channels are nonselective Ca2+-permeable cation channels; only TRPM4 and TRPM5 are impermeable to Ca2+ [13, 14]. TRPM4, TRPM5, and TRPM8 are voltage-sensitive, but they lack the classic voltage sensor of other voltage-gated ion channels, which consists of several positively charged residues in the S4 helix. A hallmark of the members of the TRPM family is their polymodal nature: they are regulated by stimuli including voltage, temperature, and the binding of ions, lipids, or other ligands. Pioneering work by McKemy et al. [15], and Peier et al. [16] led to the identification of TRPM8 as a sensor for cold temperatures and the cooling agent menthol. Later work showed that other members of the TRPM family are also sensitive to temperature. The TRPM subfamily covers a wide range of temperatures from cool (TRPM8 [15, 16]), to warm (TRPM4/5 [17] and TRPM2 [18]), to hot (TRPM3 [19]), yet the mechanism underlying their temperature sensitivity is still unclear.
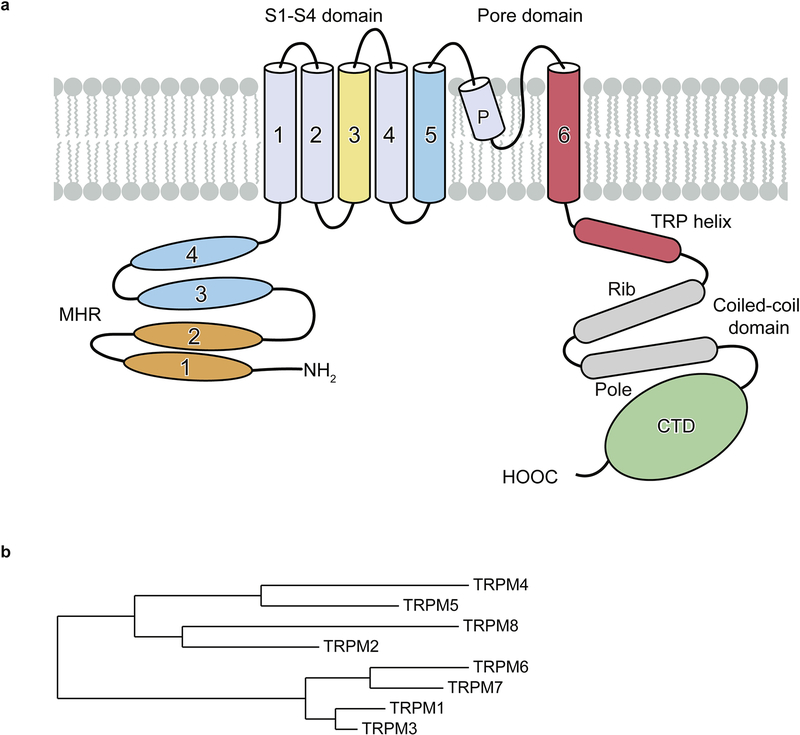
The TRPM channels have a large cytosolic domain of between 732 and 1611 amino acids for each subunit, making them the largest members of the TRP superfamily. They share a characteristic N-terminal TRPM homology region (MHR) domain, a transmembrane domain (TMD) consisting of six transmembrane helices, a TRP helix, a C-terminal coiled-coil domain, and a C-terminal domain (CTD) that differs among the members (Figure 1a). Despite some common structural features, the members of the TRPM subfamily are less conserved than the members of other subfamilies. They have been divided into four subgroups according to their sequence similarities (Figure 1b): 1) TRPM1 and TRPM3, 2) TRPM2 and TRPM8, 3) TRPM4 and TRPM5, and 4) TRPM6 and TRPM7. Their expression and functions are summarized in Table 1.
Figure 1: Family tree and domain organization of TRPM family.
a, Domain organization of a monomer of the human TRPM family; the C-terminal domain (CTD) differs among family members. The colors of TMD refer to Figure 6. b, The relatedness of the human TRPM family members.
Tabel 1:
Summary of expression, function, related diseases and ligands of TRPM channels.
| Name | Expression | Functions | Diseases | Ligands |
|---|---|---|---|---|
| TRPM1 | Retina ON bipolar cells; Skin melanocytes. | Depolarization of the bipolar cell; suppress melanoma metastasis. | Congenital stationary night blindness. |
Activation: pregnenolone sulphate Inhibition: Zn2+ |
| TRPM2 | Central nervous system; Immune cells; Pancreatic β cells | Core body temperature sensation; Oxidative sensation; Insulin secretion; Immune response. | Bipolar disorder; Ischaemia-reperfusion injury; Alzheimer’s disease. |
Activation: ADPR, cADPR, 2′-deoxy-ADPR, 2′-P-ADPR, 3′-P-ADPR, 2-F-ADPR, AMPCPR, Ca2+ Inhibition: econazole, clotrimazole, flufenamic acid, N-(p-amylcinnamoyl)anthranilic acid, 2-APB, Scalardial, 3-MFA, 8-Br-cADPR, 8-Br-ADPR, 8-Ph-ADPR, 8-Ph-2′-deoxy-ADPR, 8-(3-acetylphenyl)-ADPR, 8-thiophenyl-ADPR, and more Other: PIP2 |
| TRPM3 | Primary nociceptive neurons; Pancreatic beta-cell. Kidney | Glucose homeostasis; Heat sensation and inflammatory pain. | Visual epilepsy, retinal dystrophy. |
Activation: pregnenolone sulphate, CIM0216. Inhibition: primidone |
| TRPM4 | Heart, liver | Regulation of calcium oscillations after T cell activation, prevention of cardiac conduction disorders, Regulating smooth muscle contraction. | Brugada syndrome, Cardiac conduction defect. |
Activation: Ca2+ Inhibition: ATP, ADP, AMP, DVT, 9-Phenanthrol. |
| TRPM5 | Pancreatic beta-cells; tuft cells; solitary chemosensory cells. | Modulation of insulin secretion and sensory transduction in taste cells. | Beckwith-Wiedemann syndrome. |
Activation: Ca2+, PIP2, Steviol glycosides, Rutamarin. Inhibition: TPPO |
| TRPM6 | Kidney, intestine | Magnesium uptake and homeostasis in kidney and intestine. | Hypomagnesemia. |
Activation: Mg2+ Inhibition: Ruthenium red |
| TRPM7 | Ubiquitous. | Magnesium and Calcium homeostasis, cell viability. | Neuronal degenerative diseases. |
Activation: Mg2+-ATP, breakdown of PIP2, increase in cAMP concentrations Inhibition: Mg2+, spermine, 2-APB, MnTBAP |
| TRPM8 | Sensory neurons, prostate. | Cold sensation. | Inflammatory/neuropathic pain; prostate cancer. |
Activation: menthol, icilin. Inhibition: WS-12, CPS-369. |
The architecture of TRPM channels remained unknown until late 2017, when several TRPM4 structures and one TRPM8 structure were solved using single-particle cryo-EM and reported at about the same time [20–23]. The TRPM4 structures have been solved in different ligand-bound states by different labs and nicely complement each other. They include an apo state (Protein Data Bank IDs 6BCJ & 6BCL, 6BQR, and 6BWI; resolution 3.1 Å, 3.2 Å, and 3.7 Å, respectively); a Ca2+-bound state (PDB ID 6BQV, 3.1 Å); an ATP-bound state (PDB IDs 6BCO & 6BCQ, 2.9 Å); and a Ca2+/DVT bound state (PDB ID 5WP6, 3.8 Å) [20–22]. The TRPM8 structure from the collared flycatcher (Ficedula albicollis) was captured in apo state (faTRPM8, PDB ID 6BPQ, 4.1 Å) [23]. In 2018, an invertebrate TRPM2 structure from Nematostella vectensis (nvTRPM2, PDB ID 6CO7, 3.0 Å) in complex with Ca2+ and partial TRPM7 structures from mouse (Mus musculus) with truncation of the C-terminal enzymatic domain were reported (mmTRPM7, PDB IDs 5ZX5, 6BWD, 6BWF, 3.3, 3.7, and 4.1 Å) [24, 25]. At that time, all the TRPM structures had been captured in a closed conformation. Later, an active/open structure (PDB ID 6DRJ, 3.3 Å) and an apo/closed structure (PDB ID 6DRK, 3.8 Å) of the zebrafish (Danio rerio) full-length TRPM2 channel (drTRPM2) were reported; that open structure was the first of a TRPM channel [26].
At the end of 2018, the structures of human (Homo sapiens) TRPM2 (hsTRPM2) in the apo, ADPR-bound, and ADPR/Ca2+-bound states were published, showing a different relationship of the C-terminal domain to the rest of the protein relative to the drTRPM2 structures, although the resolution was relatively low (PDB IDs 6MIX, 6MIZ, and 6MJ2; 3.6, 6.1, and 6.4 Å, respectively) [27]. Another paper describing drTRPM2 showed interesting intermediate states having twofold symmetry and illustrated the changes in TRPM2 from the closed to the open conformation (PDB ID 6D73, 3.8 Å) [28]. A recent paper by Huang et al. reported four structures of human TRPM2 in the apo/resting; single agonist occupied; preopen; and antagonist-bound states at higher resolution (PDB IDs 6PUO, 6PUR, 6PUS, and 6PUU; 3.3, 4.4, 3.7, and 3.7 Å, respectively) [29]. It concluded that the MHR1/2 domain serves as a conserved orthostatic ligand-binding site for TRPM2 activation and inhibition across species. In 2019, two more TRPM8 structure papers have been published, revealing several important ligand binding sites for TRPM8 antagonists, agonists, and modulators (Yin et al., PDB IDs 6NR2, 6NR3, and 6NR4, resolution 4.0, 3.4, and 4.3 Å, respectively; Diver et al., PDB IDs 6O6R, and 6O72, resolution 3.2, and 3.0 Å, respectively) [30, 31]. The work by Diver et al. revealed a well-defined selectivity filter and outer pore loop of TRPM8, as well as the molecular mechanism underlying TRPM8 channel desensitization and antagonism [31]. All the available structures of TRPM channels are summarized in Table 2.
Table 2:
Summary of available TRPM structures
| Name | Species | Ligand condition & Functional States | PDB | EMDB | Resolution | Reference |
|---|---|---|---|---|---|---|
| TRPM2 | Nematostella vectensis | Ca2+, closed | 6CO7 | EMD-7542 | 3.0 Å | Zhang et al. [24] |
| Danio rerio | Apo, closed | 6DRK | EMD-8901 | 3.8 Å | Huang et al. [26] | |
| Danio rerio | Ca2+/ADPR, open | 6DRJ | EMD-7999 | 3.3 Å | Huang et al. [26] | |
| Danio rerio | Apo, C4 symmetry, closed | 6PKV | EMD-20367 | 4.3 Å | Yin et al. [28] | |
| Danio rerio | Apo, C2 symmetry, closed | 6PKW | EMD-20368 | 4.5 Å | Yin et al. [28] | |
| Danio rerio | Ca2+, closed | 6D73 | EMD-7822 | 3.8 Å | Yin et al. [28] | |
| Danio rerio | Ca2+/ADPR, intermediate | 6PKX | EMD-20369 | 4.2 Å | Yin et al. [28] | |
| Homo sapiens | Apo, closed | 6MIX | EMD-9132 | 3.6 Å | Wang et al. [27] | |
| Homo sapiens | ADPR, closed | 6MIZ | EMD-9133 | 6.1 Å | Wang et al. [27] | |
| Homo sapiens | Ca2+/ADPR | 6MJ2 | EMD-9134 | 6.4 Å | Wang et al. [27] | |
| Homo sapiens | Apo, closed | 6PUO | EMD-20478 | 3.3 Å | Huang et al. [29] | |
| Homo sapiens | ADPR, closed | PUR | EMD-20479 | 4.4 Å | Huang et al. [29] | |
| Homo sapiens | Ca2+/ADPR, preopen/closed | 6PUS | EMD-20480 | 3.7 Å | Huang et al. [29] | |
| Homo sapiens | Ca2+/8-Br-cADPR, closed | 6PUU | EMD-20482 | 3.7 Å | Huang et al. [29] | |
| TRPM4 | Homo sapiens | Ca2+/DVT, closed | 5WP6 | EMD-8871 | 3.8 Å | Winker et al. [20] |
| Homo sapiens | Apo, closed | 6BQR | EMD-7132 | 3.2 Å | Autzen et al. [22] | |
| Homo sapiens | Ca2+, closed | 6BQV | EMD-7133 | 3.1 Å | Autzen et al. [22] | |
| Mus musculus | Apo (short coiled coil), closed | 6BCJ | EMD-7081 | 3.1 Å | Guo et al. [21] | |
| Mus musculus | Apo (long coiled coil), closed | 6BCL | EMD-7082 | 3.5 Å | Guo et al. [21] | |
| Mus musculus | ATP (short coiled coil), closed | 6BCO | EMD-7083 | 2.9 Å | Guo et al. [21] | |
| Mus musculus | ATP (long coiled coil), closed | 6BCQ | EMD-7085 | 3.3 Å | Guo et al. [21] | |
| Homo sapiens | Apo, closed | 6BWI | EMD-7299 | 3.7 Å | Duan et al. [55] | |
| TRPM7 (truncated, no CTD) | Mus musculus | EDTA, closed | 5ZX5 | EMD-6975 | 3.3 Å | Duan et al. [25] |
| Mus musculus | Mg2+, closed | 6BWD | EMD-7297 | 3.7 Å | Duan et al. [25] | |
| Mus musculus | Mg2+-unbound, closed | 6BWF | EMD-7298 | 4.1 Å | Duan et al. [25] | |
| TRPM8 | Ficedula albicollis | Apo, closed | 6BPQ | EMD-7127 | 4.1 Å | Yin et al. [23] |
| Ficedula albicollis | Menthol analog WS-12 and PIP2, closed | 6NR2 | EMD-0487 | 4.0 Å | Yin et al. [30] | |
| Ficedula albicollis | Ca2+, icilin (high occupancy) and PIP2, closed | 6NR3 | EMD-0488 | 3.4 Å | Yin et al. [30] | |
| Ficedula albicollis | Ca2+, icilin (low occupancy) and PIP2, closed | 6NR4 | EMD-0489 | 4.3 Å | Yin et al. [30] | |
| Parus major | Apo, closed | 6O6A | EMD-0631 | 3.6 Å | Diver et al. [31] | |
| Parus major | AMTB, closed | 6O6R | EMD-0636 | 3.2 Å | Diver et al. [31] | |
| Parus major | TC-I 2014, closed | 6O72 | EMD-0638 | 3.0 Å | Diver et al. [31] | |
| Parus major | Ca2+, desensitized/closed | 6O77 | EMD-0639 | 3.2 Å | Diver et al. [31] | |
2. Channel architecture and domain organization
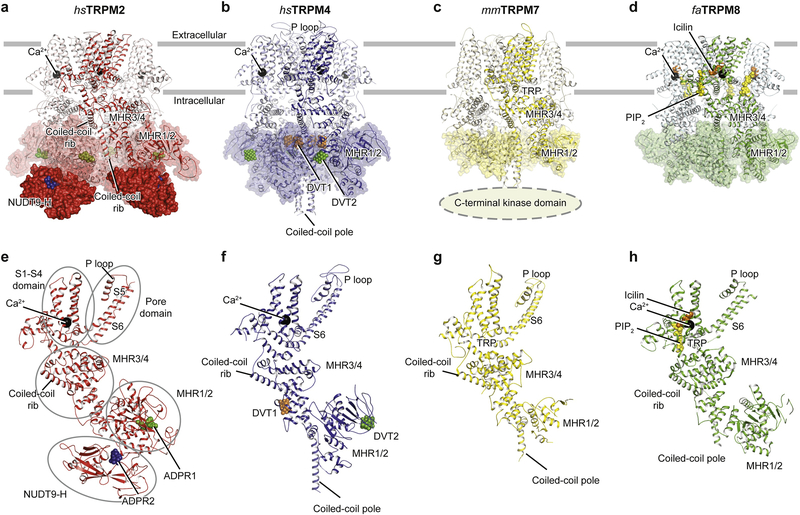
Most of the available TRPM channel structures—including TRPM4, the non-full-length TRPM7, and TRPM8—share a similar three-layered assembly that has the TMD, MHR3/4, and MHR1/2 domains from top to bottom (Figure 1a, Figure 2b–d, 2f–h). TRPM2 includes a fourth layer underneath MHR1/2 that harbors the unique C-terminal NUDT9-H domain (Figure 1a, Figure 2a, e), a domain with high homology to the mitochondrial ADPR pyrophosphatase NUDT9 [32]. The domain organization of the TMD is comparable with that of other TRP subfamilies, as well as with that of voltage-gated Ca2+ [33], K+ [34], and Na+ [35] channels. The transmembrane helixes S5 and S6 plus the p loop form the ion-conducting pore domain, which is surrounded by the S1-S4 domain (Figure 1a). The pore domain and the S1-S4 domain are connected through the S4-S5 linker (Figure 1a), which is believed to play an important role in the gating of TRP channels [26]. The S1-S4 domain of TRPM channels contains binding sites for agonist Ca2+ and other ligands, while in voltage-gated Ca2+, K+ and Na+ channels the S1-S4 domain contains a voltage sensor.
Figure 2: Comparison of the tetrameric architecture and single subunits of representative TRPM channels.
a-d, Overall structures of (a) hsTRPM2, (b) hsTRPM4, (c) mmTRPM7, and (d) faTRPM8 viewed parallel to the membrane. The mmTRPM7 structure is not full length; the kinase domain is truncated. (e-h) Single subunits viewed parallel to the membrane plane, with secondary structure elements labeled in panel e. Ligands are shown as spheres: ADPR1 in green; ADPR2 in blue; Ca2+ in black; DVT1 in orange; DVT2 in green; icilin in orange; and PIP2 in yellow.
The shape of the intracellular MHR domain is distinct among TRPM members, primarily because of different conformations of the MHR1/2 domain. Despite the differences, however, the MHR domains in TRPM channels all form a large hollow penetrated by a part of C-terminal coiled-coil domains (a coiled-coil “pole”) vertically, and by a second part of the coiled-coil domain (helical “ribs”) horizontally (Figure 2e–h). The C-terminal rib and pole constitute a unique umbrella-like shape that plays an important role in subunit assembly (Figure 2e–h, see Winkler et al. for the umbrella-like shape due to limited space) [20, 25, 29, 30] and also provides sites for ligand binding which will be discussed in the following section [20]. The rib helix connects through a flexible linker to the TRP helix, which is a hallmark of TRP channels and connects to the gating helix S6 (see Figure 1a). It is therefore reasonable to speculate that the rib helix and the TRP helix may form a complicated communication network for transferring signals from the MHR domains to the S6 helix in order to modulate ion channel gating [26].
In TRPM4, TRPM7, and TRPM8, the intersubunit interface is primarily formed between adjacent MHR domains (Figure 2b–d, 2f–h). In TRPM2, the intersubunit interface is mainly formed between the NUDT9-H domain and MHR1/2 domain (Figure 2a, e). They are mediated through an extensive interface between NUDT9-H domain and the cognate MHR2 domain in both drTRPM2 and hsTRPM2 (Figure 2a, e) [27, 29], while an additional intersubunit interface has been observed in hsTRPM2, formed between the adjacent NUDT9-H and MHR domains (refer to Huang et al. eLife due to limited figure numbers) [27, 29]. As a result, the NUDT9-H domain in hsTRPM2 is markedly better defined than in the drTRPM2. Whether the C-terminal kinase domain of TRPM7 and TRPM6 is also involved in intersubunit interaction still needs to be determined.
2.1. Ligand recognition and binding sites
The TRPM family members are affected by a wide variety of stimuli including small molecules, lipids, proteins, voltage, and temperature. The binding sites of several ligands have been unambiguously identified by structural and functional studies. Four ligands have been shown to bind to the TMD:
Ca2+ in nvTRPM2 [24], drTRPM2 [26] and hsTRPM2 [29] (PDB IDs 6CO7, 6DRJ, 6PUS, and 6PUU); the hsTRPM4 [22] (PDB ID 6BQV); the faTRPM8 [30] (PDB IDs 6NR3, 6NR4); and the great tit (Parus major) pmTRPM8 [31] (PDB ID 6O77) (Figure 3a–c, 3d–f)
PIP2 (phosphatidylinositol 4,5-bisphosphate) in faTRPM8 (Figure 3c, o) [24, 30] (PDB ID 6NR3);
icilin and the menthol analog WS-12 in faTRPM8 [30] (PDB IDs 6NR4, 6NR2) (Figure 3c, i, l); and
AMTB(N-(3-aminopropyl)-2-[(3-methylphenyl)methoxy]-N-(2-thienylmethyl)benzamide hydrochloride) and TC-I 2014 (3-[7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1Hbenzimidazol-2-yl]-1-oxa-2-azaspiro[4.5]dec-2-ene) in pmTRPM8 [31] (PDB IDs 6O6R and 6O72).
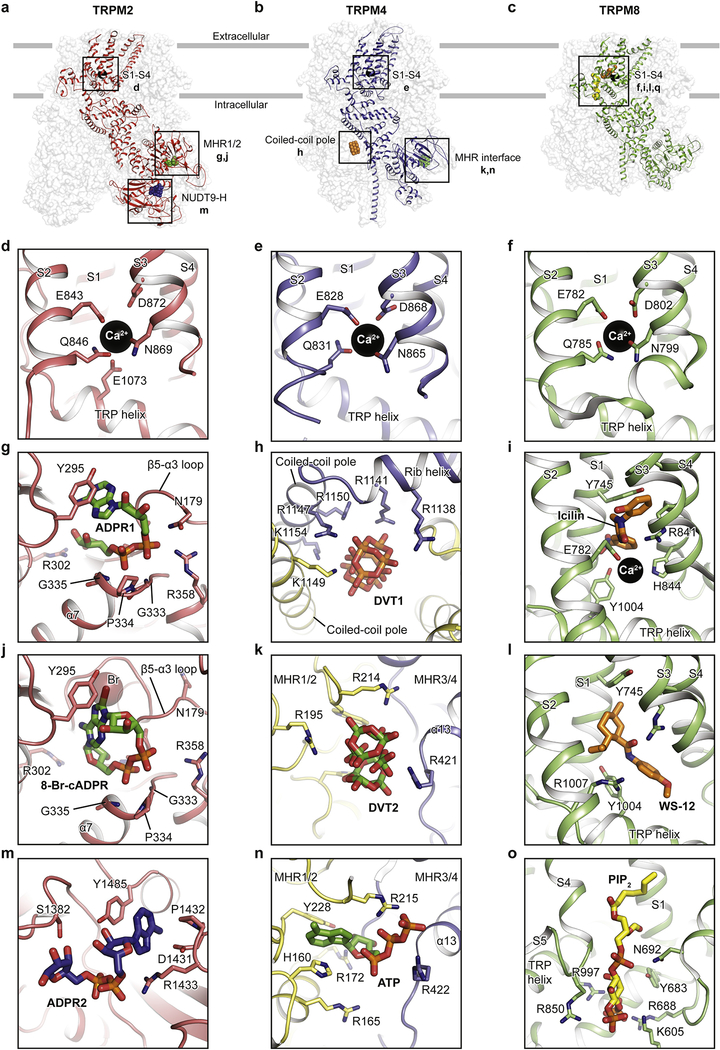
Figure 3: Ligand-binding sites of hsTRPM2, hsTRPM4, and faTRPM8.
The locations of available ligand-binding sites are boxed. Ca2+ is shown as black spheres, while other ligands and key residues involved in ligand binding are shown as sticks. The binding site of Ca2+ in (d) hsTRPM2, (e) hsTRPM4 and (f) faTRPM8. The hsTRPM2 binding sites of (g) ADPR1 in the MHR1/2 domain, (j) 8-Br-cADPR in the MHR1/2 domain, and (m) ADPR2 in the NUDT9-H domain. The binding sites in hsTRPM4 of (h) DVT1 at the kink of the rib helix and coiled-coil pole and of (k) DVT2 and (n) ATP at the interface of MHR1/2 and MHR3/4. The faTRPM8 binding sites of (i) icilin, (l) the menthol analog WS-12, and (o) PIP2 in the TMD. Please see the report by Diver et al. for the AMTB and TC-I 2014 binding sites; the figures in this review were prepared before that report was out.
Another four ligands are found in the cytosolic domain:
adenosine diphosphate ribose (ADPR) and 8-bromo-cyclic ADPR in hsTRPM2 (Figure 3a, g, j, m) [29] (PDB IDs 6PUR, 6PUS, and 6PUU); and
DVT (decavanadate) (Figure 3b, h, k) and ATP (Figure 3b, n) in TRPM4 [20, 21] (PDB IDs 6BCO, 6BCQ, and 5WP6).
Among these ligands, ADPR, Ca2+, PIP2, and ATP are physiological agents, while icilin, WS-12, 8-Br-cADPR, AMTB, TC-I 2014, and DVT are pharmacological compounds.
2.1.1. Binding sites in the TMD
The Ca2+ binding site was first described to being close to the S3 helix in hsTRPM4, with the Ca2+ ion coordinated by Glu828, Gln831, Asn865, and Asp868 (Figure 3e) [22]. Ca2+ has since been found to bind to the same site in TRPM2 and TRPM8 (Figure 3d, f) [26, 29, 30]. Sequence alignments show that this binding site is highly conserved across the family members [24, 26, 30, 31]. Mutagenesis studies using electrophysiology have provided strong evidence that this Ca2+ binding site plays a major role in channel gating [24].
The binding site for PIP2 is thought to play an important role in channel modulation in several TRPM members. For example, it is required for the activation of TRPM8 [36] and TRPM2 [37]. A phospholipid density was first observed in the nvTRPM2 structure near the Ca2+ binding site [24]. The binding site for PIP2 in TRPM8 was defined by Yin et al. [30] (Figure 3o). It is in a similar site as the phospholipid density in nvTRPM2 in the TMD, embraced by the pre-S1 domain, the TRP domain, the S4-S5 linker, and the adjacent MHR4 domain. This is a key position involved in channel activation. In the same paper, the binding sites of two widely known cooling agents, icilin and menthol, were also located near the Ca2+-binding site, surrounded by the S1, S3, and S4 helixes and the TRP domain (Figure 3i, l) [30]. The authors showed an allosteric coupling between PIP2 and the cooling agents [30]. A recent paper reported the binding sites of two antagonists, AMTB and TC-I 2014, in a membrane-embedded cleft formed by the lower half of the S1-S4 domain (not illustrated due to limited space, refer to Diver et al.) [31].
2.1.2. Binding sites in the cytosolic domain
The large cytosolic domain provides numerous possible binding sites for small molecules that modulate channel function. The binding sites in the cytosolic domain are known for the following molecules: 1) ADPR1 and 8-Br-cADPR are in the MHR1/2 domain of TRPM2 (Figure 3g, j); 2) ATP and DVT2 at the intrasubunit interface of MHR1/2 and MHR3/4 in TRPM4 (Figure 3k, n); 3) DVT1, at the kink of the C-terminal rib and pole helices in TRPM4 (Figure 3h); and 4) ADPR2, in the CTD of TRPM2 (Figure 3m).
2.1.2.1. Within the MHR1/2 domain
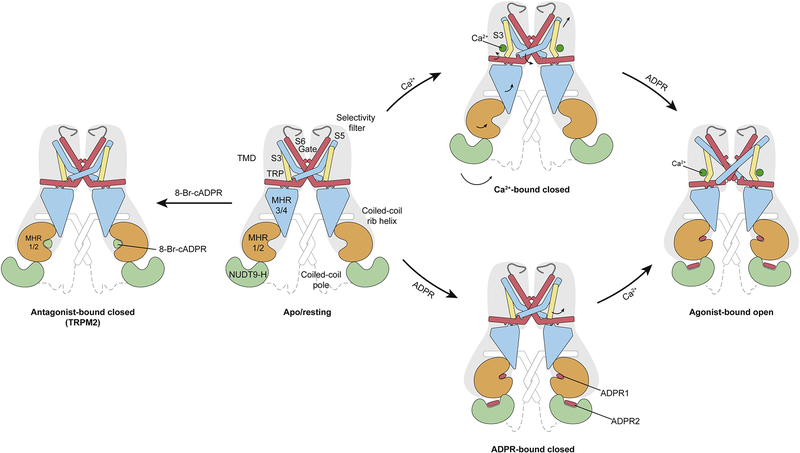
TRPM2 is activated by ADPR in the presence of Ca2+ [38–41]. Despite the consensus view that ADPR is bound in the C-terminal NUDT9-H domain of TRPM2, an ADPR molecule having a U-shape was unexpectedly found within the clamshell-like MHR1/2 domain of drTRPM2 [26]. We suggested that this is a universal ADPR binding site across all TRPM2 orthologues, not only because the key binding residues are highly conserved through evolution, but also because mutation of these residues reduces or abolishes the Ca2+/ADPR-induced channel activation in both hsTRPM2 and drTRPM2 [26, 29]. However, Wang et al. [27] reported that the MHR1/2 binding site does not exist in hsTRPM2. While the resolution of the structure was not sufficient to detect ADPR in the MHR1/2 domain, their conclusion was based on their observation that mutation of the residues in the MHR1/2 domain of hsTRPM2 had no major effect on activation of the channel downstream of hydrogen peroxide or activation by Ca2+/ADPR [27]. They thus suggested that the activation of hsTRPM2 involves a different mechanism than that of drTRPM2; specifically, that hsTRPM2 uses the NUDT9-H domain for channel gating, while drTRPM2 uses MHR1/2.
We recently resolved this contradiction by showing that ADPR clearly binds to both the MHR1/2 domain of hsTRPM2 (at ADPR1, in a U shape) and the NUDT9-H domain (at ADPR2, in an extended shape) (Figure 3a, g, m), and both binding sites are indispensable for channel activation [29]. We conclude that the MHR1/2 binding site is a universal binding site for ADPR across TRPM2 from different species and plays a key role in channel activation. Interestingly, the antagonist 8-Br-cADPR binds only in the MHR1/2 domain (Figure 3a, j). Given that the NUDT9-H domain is not required for the activation of invertebrate TRPM2 channels [42] and the MHR1/2 binding site is more conserved across TRPM2 than the NUDT9-H domain is, we suggest that MHR1/2 acts as a primary ligand-binding site in the TRPM2 channels. This binding site is not conserved in other TRPM family members, explaining why only TRPM2 recognizes ADPR.
2.1.2.2. At the intrasubunit interface of MHR1/2 and MHR3/4
Nucleotides such as ATP inhibit Ca2+-induced currents in TRPM4, but they do not affect its closest homolog, TRPM5 [43]. The molecular basis for this difference was unclear until Guo et al. [21] identified the ATP binding site in TRPM4, which is at the interface between the MHR1/2 domain and the adjacent MHR3 domain (Figure 3b, n). The adenosine group of ATP is surrounded by aromatic residues (His160 and Phe228), while the triphosphate group protrudes into the interface and is surrounded by several basic residues that are absent in TRPM5, which explains its lack of inhibition by ATP [21].
The ATP-bound TRPM4 structure is nearly identical to the apo structure except for the region near the ATP binding site, suggesting that ATP may inhibit TRPM4 by stabilizing the channel in an apo-like conformation. This is reminiscent of the inhibitory mechanism of hsTRPM2 by the antagonist 8-Br-cADPR [29, 44]. At a similar position to where ATP was found, Winkler et. al. identified one of the two DVT molecules in the TRPM4 structure interacting with three positively charged arginine residues (Figure 3b, k) [20]. DVT is a negatively charged metal cluster that modulates the voltage dependence of TRPM4 and modulates the Ca2+-activated current amplitude in a Ca2+-concentration-dependent manner [20]. A sequence alignment between TRPM4 and TRPM5 showed that all three arginine residues are absent in TRPM5, explaining TRPM5’s insensitivity to DVT [20].
2.1.2.3. At the kink of the C-terminal rib and pole
In the TRPM structures, the hollow-shaped MHR domains embrace the C-terminal coiled-coil domain, creating plenty of space in between that supports the binding of small molecules (Figure 3b). Until now, only one ligand, DVT1 in TRPM4, has been found at this location (Figure 3b, h) [20].
2.1.2.4. In the CTD
Although the CTD exists in TRPM1, TRPM2, TRPM3, TRPM6, and TRPM7 and is considered to contain ligand-binding sites for nucleotides and/or interaction partners for G proteins, so far the CTD (NUDT9-H domain) has only been observed in the drTRPM2 and hsTRPM2 structures [26–29]. To date, just one ligand-binding site, ADPR2 in the NUDT9-H domain of hsTRPM2 (Figure 3a, m), has been defined in a study by Huang et al. [29]. This was the first time that ADPR bound to the NUDT9-H domain of TRPM2 had been observed, although it has been generally accepted that the NUDT9-H domain of TRPM2 binds ADPR [38, 45, 46]. ADPR2 nestles in an extended shape in the cleft of the NUDT9-H domain, with the adenine and terminal ribose moieties far apart (Figure 3m). Its adenine moiety stacks between Tyr1485 and Asp1431, while the α-phosphate group interacts with Arg1433. The NUDT9-H domain in hsTRPM2 and drTRPM2 seems to be indispensable for channel function, based on the observation that mutations of these key residues markedly affected the channel function [47, 48].
2.2. The ion-conducting pore and selectivity
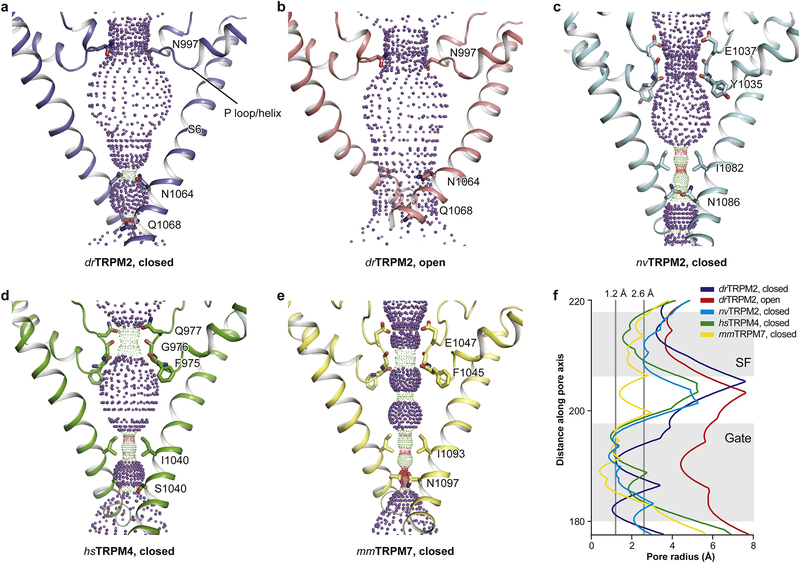
Like other TRP channels, TRPM channels have two restriction sites: a selectivity filter formed by the P loop near the extracellular side and a gate shaped by S6 near the intracellular side. Most of the TRPM channels show a triangular-shaped vestibule (Figure 4a, c, d, e), similar to that reported for TRPV [49–54]. The structures having a triangle-shaped vestibule are EDTA-drTRPM2 [26], hsTRPM4 [20–22, 55], nvTRPM2 [24], mmTRPM7 [25], faTRPM8 (not shown in the figure 4 due to less well-defined pore region) [23, 30], and pmTRPM8 [31] (the pmTRPM8 structure was not known when the figure in this review was created). They have a restriction site with a radius of 1.0 Å or smaller formed by the cytosolic ends of the S6 helices, which prevents the entrance of ions and thus is assigned as the closed state (Figure 4f).
Figure 4: Comparison of ion-conducting pores.
The shape and size of the ion-conducting pore of (a) EDTA-drTRPM2, (b) ADPR/Ca2+-drTRPM2, (c) Ca2+-nvTRPM2, (d) Ca2+/DVT-hsTRPM4, and (e) mmTRPM7 (not full length). The side chains of restriction residues are shown as sticks. Purple, green, and red spheres define pore radii of > 2.3 Å, 1.2–2.3 Å, and < 1.2 Å, respectively. (f), Pore radius as a function of distance along the pore axis.
The hsTRPM2 structure in complex with Ca2+ and ADPR has been reported as an open state by Wang et al., but that interpretation is ambiguous [27]. The reasons are a) that most of the transmembrane domain was defined at 6.4 Å overall resolution and without visualizing any pore-lining side chains, and b) that the key flipping of the S4-S5 linker from one side of the TRP helix to the other for channel opening as described in both Huang et. al. and Yin et. al [26, 28] didn’t happen. This will be discussed in the following section. The ADPR/Ca2+-hsTRPM2 structure of Huang et al. at 3.7 Å clearly showed a closed pore [29]. In contrast, ADPR/Ca2+-drTRPM2 has a bowl-shaped vestibule and an elevated gate (Figure 4b), large enough to pass hydrated Ca2+ and Na+ ions; it is thus the only open structure determined in the TRPM family so far (Figure 4f). Despite a closed pore in the desensitized pmTRPM8 structure, remarkable conformational changes were observed in the TMD and TRP domains, revealing a molecular mechanism underlying TRPM channel desensitization [31].
TRPM4 and TRPM5 are the only known TRP channels that are impermeable to Ca2+ but permeable to monovalent Na+ or K+. Nilius et al. have proposed that negatively charged residues in the pore loop of TRPM4 and TRPM5 determine the permeation of monovalent cations [56], and Topala et al. reported that the acidic residues in the selectivity filter are crucially involved in the divalent cation permeation of TRPM6 [57]. It was not clear why TRPM2 is permeable to Ca2+, because the amino acids that form its selectivity filter are highly conserved when compared with those of TRPM4 and TRPM5. Also, TRPM2 lacks the determinant acidic residue for calcium permeability found in TRPM1, TRPM3, TRPM6, and TRPM7. Indeed, replacing Gln981 in hsTRPM2 (corresponding to Asn997 in the drTRPM2) by a Glu substantially increased calcium permeability [58].
The differences in ion selectivity within this family were finally understood when several TRPM structures allowed a comparison of their selectivity filters. drTRPM2 has an unusually short selectivity filter, the single neutral residue Asn997, giving rise to a long and flat P helix–P loop hinge (Figure 4a, b) [26]. The other TRPM channels, including the invertebrate nvTRPM2, have a longer filter of three residues as a result of a shorter P helix–P loop hinge (Figure 4c–e) [20, 24, 25]. We thus speculate that a shorter selectivity filter leads to a reduced ability to differentiate between ions. The nvTRPM2 has a selectivity filter with long configuration similar to that of hsTRPM4, but it is Ca2+-permeable because it has a Glu instead of a neutral Gln (Figure 4c, d) [20, 24]. Furthermore, although both TRPM7 and TRPM4 share a similar three-residue selectivity filter (Figure 4d–e), that of TRPM4 is narrower, explaining why TRPM7 is permeable to divalent ions and TRPM4 is not.
2.3. Gating mechanism of TRPM channels
Currently, the only TRPM channel for which we have both open and closed structures is drTRPM2 [26] (Figure 4a, b; Figure 5a, b). Despite lacking an open state of TRPM8, the pmTRPM8 structures in a desensitized state and an apo state reveal remarkable conformational changes [31]. These structures provide deep insight into the mechanism of ligand-mediated gating of TRPM channels. In this section, we summarize the activation and inhibition mechanism of TRPM2 and desensitization and inhibition mechanism of TRPM8, respectively.
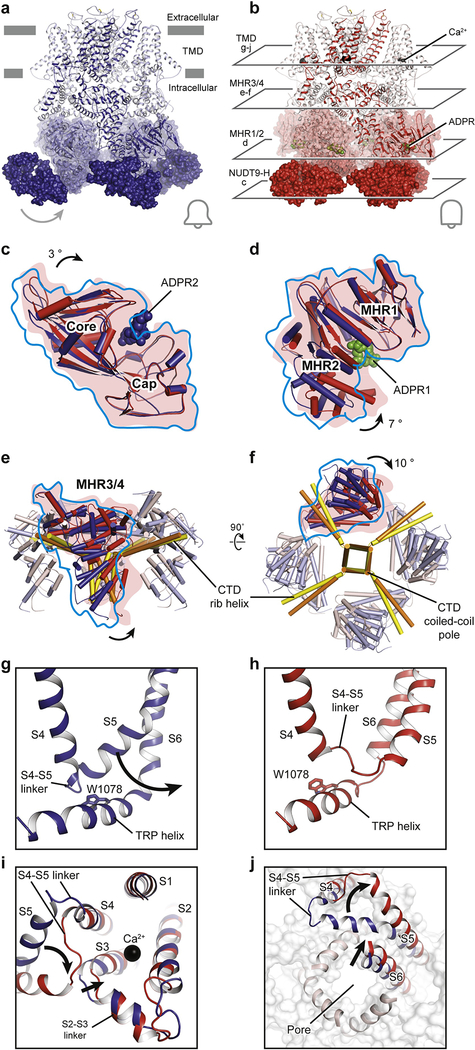
Figure 5: Gating mechanism of the voltage-independent TRPM2 channel.
Structures of the (a) EDTA-drTRPM2 and (b) ADPR/Ca2+-drTRPM2. Comparison of the (c) NUDT9-H domains and the (d) MHR1/2 domains of EDTA-hsTRPM2 and ADPR/Ca2+-hsTRPM2 by superimposition of the cap regions or the MHR1 domains. ADPR1 and ADPR2 are shown as green and blue spheres. The domain closure induced by ADPR binding in NUDT9-H and MHR1/2 is indicated. (e, f), Superimposition of the linker layers of EDTA-hsTRPM2 (blue) and ADPR/Ca2+-hsTRPM2 (red) by aligning the CTD coiled-coil poles. (g, h), Detailed view of the interaction between MHR4 and the TRP helix. The movement of the S4-S5 linker is indicated and the Trp1078 of TRP helix is shown in sticks. (i, j), Superimposition of EDTA-drTRPM2 (blue) and ADPR/Ca2+-drTRPM2 (red) structures by aligning the S1-S4 domains. (i), The conformational changes of S2-S3 linker and S3 upon Ca2+ binding are indicated. (j), The flipping of S4-S5 linker upon channel opening, viewed from the intracellular side, is indicated.
In the drTRPM2 structures, the binding of ADPR induces the closure of the bi-lobed MHR1/2 domain in the ligand-sensing layer, accompanied by a swing of NUDT9-H and the adjacent MHR1/2 (Figure 5c, d). The motion is translated to the linker layer, with MHR3/4 tilting up toward the TMD, leading to a repositioning of the TRP helix (Figure 5e–h). We thus defined MHR3/4 as a signal-transducing domain (STD). Its motion unlocks the restriction of the S4-S5 linker by the TRP helix and enables S5 to relocate from one side of the TRP helix to the other, changing from a flexed to a straightened conformation (Figure 5g–h). We believe this flipping is the key element for channel activation. A stepwise flipping of the S5 helix has also been observed in Yin et al. drTRPM2 with two-fold symmetry [28]. In the drTRPM2 structure in Huang et al., the binding of Ca2+ within the S1-S4 domain repositions S3, which frees space for the relocation of the S4-S5 linker (Figure 5i, j). That relocation promotes an outward tilting of the S6 helix, ultimately resulting in channel opening (Figure 5g–j; Figure 6).
Figure 6: Schematic of ligand sensing and the activation mechanism of TRPM2.
Conformational changes of TRPM2 upon ligand binding are shown by arrows. 8-Br-cADPR binds only to the MHR1/2 domain and inhibits the TRPM2 channel by stabilizing the MHR1/2 domain in apo-like conformation. Channel activation requires both Ca2+ and ADPR; the binding of Ca2+ or ADPRs lone is not sufficient to open the channel. The simultaneous binding of two ADPRs and Ca2+ in three indispensable binding sites open the TRPM2 channel. ADPR1 bound in the MHR1/2 domain shows a “U” shape, while ADPR2 bound in the NUDT9-H domain is in an extended shape.
Even though an open structure of hsTRPM2 is currently lacking, we suggest that it shares a mostly conserved gating mechanism with drTRPM2, because the major conformational changes throughout the protein are consistent upon binding of ADPR and Ca2+. However, their NUDT9-H domains may contribute differently to channel gating, which will be discussed in the following section.
A comparison of the pmTRPM8 structures in the desensitized and apo states provides a mechanism underlying channel desensitization (Refer to the figure 6 in Diver et al. due to limited space) [31]. This involves 1) a rigid-body tilt of the S1-S4 domain away from the central axis, accompanied by large shifts of S5, the pore helix, and S6; 2) a shift of the register of the lower gate at S6, and a local α-to-π-helical transition of S6; 3) a large tilting of the TRP domain; and 4) stabilization of the outer pore loop. Although the flipping did not occur in the pmTRPM8 structures, the large movement of TRP helix further supports that idea that the position of the TRP helix relative to S1-S4 is closely associated with channel activation or inhibition [31].
2.4. Conclusion and Perspective
2.4.1. The function of the C-terminal domain
TRPM1, TRPM2, TRPM3, TRPM6, and TRPM7 have a C-terminal domain after the coiled-coil pole. The CTDs differ remarkably in amino acid sequence and domain architecture. The most well-known CTDs are the serine/threonine kinase domains of TRPM6 and TRPM7 and the NUDT9-H domain of TRPM2; these three channels have been termed “chanzymes”, because they have dual functions as both a channel and an enzyme [59]. In TRPM6 and TRPM7, the functional coupling between the kinase domain and the channel is not clear, and we don’t know their full-length structures. Experimental data shows that truncation of the kinase domains doesn’t affect channel function, indicating that the kinase domain and the channel are functionally independent [60, 61]. In 2014, Krapivinsky et al. reported exciting data that the kinase domain is proteolytically cleaved from the channel and translocates to the nucleus, where it binds components of chromatin-remodeling complexes and phosphorylates specific serines or threonines of histones. These actions result in epigenetic chromatin remodeling that affects gene expression [62]. It will be crucial to solve the full-length structure of TRPM6 or TRPM7 in order to help us understand the coupling between the kinase domain and the ion channel.
The NUDT9-H domain of TRPM2 has been studied intensively over the past few years in terms of both structure and function [26, 27, 47, 63, 64]. Ever since Perraud et al. first identified ADPR as a TRPM2 ligand [38], the NUDT9-H domain has been considered to be the only ADPR binding site and to play an important role in channel activation in the presence of Ca2+. This view was supported by two observations. First, in hsTRPM2, deletion of the NUDT9-H domain strongly decreases TRPM2 expression in the plasma membrane and abolishes channel gating [65]. Second, the mutation of residues presumably involved in ADPR binding in the NUDT9-H domain decrease or abolish channel activation [45–47, 66].
Over the years, several studies have challenged that traditional view. Burroughs et al. [67] identified by comparative genomic analysis a superfamily of proteins having a SLOG domain that features an atypical nucleotide binding pocket, and they showed that the members of the TRPM family have such a SLOG domain in their extended N-terminus. Kühn et al. found a splice variant of hsTRPM2 that lacks a small part of the N-terminus containing a CaM-binding IQ motif. The splice variant could no longer be activated, but a point mutation in the IQ motif did not interfere with activation by ADPR [68]. The same group also characterized nvTRPM2 and observed that removal of its NUDT9-H domain did not interfere with activation of the channel by ADPR [42].
In hsTRPM2, the NUDT9-H domain has been rendered catalytically inactive during evolution by a replacement of a Glu residue crucial for catalysis by an Ile [69]. In contrast, the NUDT9-H domain of the nvTRPM2 is an active ADPRase [42, 63], indicating that the NUDT9-H domain plays different roles in vertebrate and invertebrate TRPM2. This idea is supported by the differences observed among hsTRPM2, drTRPM2, and nvTRPM2 in the interactions between the NUDT9-H domain and the rest of the channel. In nvTRPM2, the NUDT9-H domain is completely invisible despite high resolution of the rest of the protein, likely due to the lack of interactions [24]. The NUDT9-H domain in drTRPM2 is visible but poorly defined, with a single major interface between the cognate MHR1/2 and NUDT9-H domain [26]. It is still unclear whether the NUDT9-H domain in drTRPM2 binds ADPR. The NUDT9-H domain in hsTRPM2 is extraordinarily well-defined, probably thanks to two major interfaces with the rest of the protein [29]. The tighter interaction between the NUDT9-H domain and the rest of the protein in more advanced species supports the view that the NUDT9-H domain gained functions during the evolution from invertebrates to mammals.
An important step toward a deeper understanding of the gating of hsTRPM2 will be to see whether its two ADPR binding sites are functionally equivalent. Until now, mutagenesis studies have indicated that both sites need to be occupied for the activation of the channel [29, 45–47, 70]. The conformation of ADPR bound to the sites differs significantly, with ADPR1 in the MHR1/2 domain having a horseshoe-like conformation and ADPR2 in the NUDT9-H domain having an extended conformation (see Figure 3g, m). This difference makes it more likely that nucleotide agonists of TRPM2, as well as synthetic ADPR analogues, will bind differently at the two sites.
Two recent observations are of great interest in this regard. The first is that two ADPR analogues, 8-(thiophen-3-yl)-ADPR and 8-(3-acetylphenyl)-ADPR, which were previously identified as antagonists of hsTRPM2 [71], act instead as agonists of both nvTRPM2 [72] and a variant of it that lacks the NUDT9-H domain. IDPR is one of two known alternative substrates (the other one being 2′-P-ADPR) of hsTRPM2 NUDT9-H domain [72–74], and is a weak agonist of hsTRPM2. It can increase the activity of nvTRPM2 by inhibiting the enzymatic activity of its NUDT9-H domain), but it does not activate the nvTRPM2 variant lacking the NUDT9-H domain [72].
The second observation is that 2′-deoxy-ADPR, an endogenous nucleotide, behaves as a superagonist of hsTRPM2, producing 10-fold higher currents than ADPR [75]. These observations suggest there might be important functional differences in the two ADPR binding sites in hsTRPM2. Developing a specific antagonist for either binding site might help elucidate their roles in the physiological function of the channel.
Recently, the CTDs in TRPM1 and TRPM3 have been shown to be directly inhibited by Gβγ subunits, indicating a potential regulation by GPCRs [76–79]. It would be interesting to solve the full-length structure of TRPM1 and TRPM3, as well as their complex with Gβγ subunits, in order to investigate the GPCR-modulated gating mechanism of TRPM channels.
2.4.2. The mechanism underlying temperature sensitivity
Temperature-sensitive TRP channels have been comprehensively reviewed elsewhere [80–82], so we will only briefly discuss several thermosensitive TRPM channels. A unique feature of the TRPM family is that more than half of the members are sensitive to a wide range of temperatures from cool to hot. The first member of the TRPM family shown to be thermosensitive was TRPM8, which is the primary cold sensor in the periphery [15, 16]. TRPM4 and TRPM5 are activated between 15 and 35 °C [17]. TRPM3 is a hot sensor in peripheral sensory neurons, sensing temperature of about 52 °C [19]. Togashi et al. first described the activation of TRPM2 by temperatures above 35°C in 2006 [18]. In 2016, exciting discoveries showing that TRPM2 is a key warmth sensor in peripheral sensory neurons and in the preoptic area of the hypothalamus were made by Tan et al. and Song et al., respectively. In peripheral neurons, TRPM2 is involved in sensing non-noxious warmth and contributes to thermotaxis, whereas in the hypothalamus it measures deep brain temperature and regulates core body temperature [83, 84]. Despite extensive studies on the mechanism underlying temperature sensation, many questions remain unanswered, including how these thermosensitive ion channels respond to different temperatures, the molecular nature of the temperature sensor, and its location in the channel.
2.4.3. Voltage-dependent gating mechanism
TRPM4, TRPM5, and TRPM8 are voltage-sensitive, but the structural mechanism of voltage sensitivity of these channels is not understood. Thus far, none of the voltage-sensitive TRPM channels have been captured in an open state, despite being detergent-solubilized, incorporated in nanodiscs, or isolated in the presence of PIP2 or of the positive modulator DVT. The capture of these channels in an open state might be helped by reconstituting the protein into liposomes; using mutations or antibodies that stabilize the open state; or using more-potent positive modulators that can shift the voltage dependence.
2.4.4. Pharmacology
TRPM channels are potential targets for the treatment of numerous diseases, including inflammatory pain (TRPM8), neurodegeneration (TRPM2), cardiovascular disease (TRPM4), and diabetes (TRPM2 and TRPM5). While several ligand-binding sites have been identified, numerous binding sites of agonists and antagonists await identification in order to better understand these important channels.
Acknowledgements
We thank D. Nadziejka for technical editing. We appreciate Du and Lü lab members including T. Walter, E. Haley, and Z. Ruan for proofreading. J.D. is supported by a McKnight Scholar Award, a Klingenstein-Simon Scholar Award, and a National Institutes of Health (NIH) grant (1R01NS111031-01). W.L. is supported by a NIH grant (1R56HL144929-01). Research in the Guse/Fliegert labs is supported by Deutsche Forschungs-gemeinschaft (SFB1328, projects A01, A05; Joachim-Herz-Foundation, Infectophysics consortium, project 4; and EU project INTEGRATA - DLV-813284).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Fleig A, Penner R, The TRPM ion channel subfamily: molecular, biophysical and functional features, Trends Pharmacol Sci. 25 (2004) 633–639. [DOI] [PubMed] [Google Scholar]
- [2].Duncan LM, Deeds J, Hunter J, et al. , Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis, Cancer Res. 58 (1998) 1515–1520. [PubMed] [Google Scholar]
- [3].Simon F, Varela D, Cabello-Verrugio C, Oxidative stress-modulated TRPM ion channels in cell dysfunction and pathological conditions in humans, Cell Signal. 25 (2013) 1614–1624. [DOI] [PubMed] [Google Scholar]
- [4].McNulty S, Fonfria E, The role of TRPM channels in cell death, Pflugers Arch. 451 (2005) 235–242. [DOI] [PubMed] [Google Scholar]
- [5].Schlingmann KP, Waldegger S, Konrad M, Chubanov V, Gudermann T, TRPM6 and TRPM-7-Gatekeepers of human magnesium metabolism, Biochim Biophys Acta. 1772 (2007) 813–821. [DOI] [PubMed] [Google Scholar]
- [6].Earley S, Waldron BJ, Brayden JE, Critical role for transient receptor potential channel TRPM4 in myogenic constriction of cerebral arteries, Circ Res. 95 (2004) 922–929. [DOI] [PubMed] [Google Scholar]
- [7].Zholos A, Johnson C, Burdyga T, Melanaphy D, TRPM channels in the vasculature, Adv Exp Med Biol. 704 (2011) 707–729. [DOI] [PubMed] [Google Scholar]
- [8].Sun Y, Sukumaran P, Schaar A, Singh BB, TRPM7 and its role in neurodegenerative diseases, Channels (Austin). 9 (2015) 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Abriel H, Syam N, Sottas V, Amarouch MY, Rougier JS, TRPM4 channels in the cardiovascular system: physiology, pathophysiology, and pharmacology, Biochem Pharmacol. 84 (2012) 873–881. [DOI] [PubMed] [Google Scholar]
- [10].Vennekens R, Mesuere M, Philippaert K, TRPM5 in the battle against diabetes and obesity, Acta Physiol (Oxf). 222 (2018). [DOI] [PubMed] [Google Scholar]
- [11].Zierler S, Hampe S, Nadolni W, TRPM channels as potential therapeutic targets against pro-inflammatory diseases, Cell Calcium. 67 (2017) 105–115. [DOI] [PubMed] [Google Scholar]
- [12].Held K, Voets T, Vriens J, TRPM3 in temperature sensing and beyond, Temperature (Austin). 2 (2015) 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Launay P, Fleig A, Perraud AL, Scharenberg AM, Penner R, Kinet JP, TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization, Cell. 109 (2002) 397–407. [DOI] [PubMed] [Google Scholar]
- [14].Hofmann T, Chubanov V, Gudermann T, Montell C, TRPM5 is a voltage-modulated and Ca(2+)-activated monovalent selective cation channel, Curr Biol. 13 (2003) 1153–1158. [DOI] [PubMed] [Google Scholar]
- [15].McKemy DD, Neuhausser WM, Julius D, Identification of a cold receptor reveals a general role for TRP channels in thermosensation, Nature. 416 (2002) 52–58. [DOI] [PubMed] [Google Scholar]
- [16].Peier AM, Moqrich A, Hergarden AC, et al. , A TRP channel that senses cold stimuli and menthol, Cell. 108 (2002) 705–715. [DOI] [PubMed] [Google Scholar]
- [17].Talavera K, Yasumatsu K, Voets T, et al. , Heat activation of TRPM5 underlies thermal sensitivity of sweet taste, Nature. 438 (2005) 1022–1025. [DOI] [PubMed] [Google Scholar]
- [18].Togashi K, Hara Y, Tominaga T, et al. , TRPM2 activation by cyclic ADP-ribose at body temperature is involved in insulin secretion, EMBO J. 25 (2006) 1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vriens J, Owsianik G, Hofmann T, et al. , TRPM3 is a nociceptor channel involved in the detection of noxious heat, Neuron. 70 (2011) 482–494. [DOI] [PubMed] [Google Scholar]
- [20].Winkler PA, Huang Y, Sun W, Du J, Lu W, Electron cryo-microscopy structure of a human TRPM4 channel, Nature. 552 (2017) 200–204. [DOI] [PubMed] [Google Scholar]
- [21].Guo J, She J, Zeng W, Chen Q, Bai XC, Jiang Y, Structures of the calcium-activated, nonselective cation channel TRPM4, Nature. 552 (2017) 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Autzen HE, Myasnikov AG, Campbell MG, Asarnow D, Julius D, Cheng Y, Structure of the human TRPM4 ion channel in a lipid nanodisc, Science. 359 (2018) 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yin Y, Wu M, Zubcevic L, Borschel WF, Lander GC, Lee SY, Structure of the cold- and menthol-sensing ion channel TRPM8, Science. 359 (2018) 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang Z, Toth B, Szollosi A, Chen J, Csanady L, Structure of a TRPM2 channel in complex with Ca(2+) explains unique gating regulation, Elife. 7 (2018) e36409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Duan J, Li Z, Li J, et al. , Structure of the mammalian TRPM7, a magnesium channel required during embryonic development, Proc Natl Acad Sci U S A. 115 (2018) E8201–E8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang Y, Winkler PA, Sun W, Lu W, Du J, Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium, Nature. 562 (2018) 145–149. [DOI] [PubMed] [Google Scholar]
- [27].Wang L, Fu TM, Zhou Y, Xia S, Greka A, Wu H, Structures and gating mechanism of human TRPM2, Science. 362 (2018) eaav4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yin Y, Wu M, Hsu AL, et al. , Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel, Nat Commun. 10 (2019) 3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Huang Y, Roth B, Lu W, Du J, Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel, Elife. 8 (2019) e50175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yin Y, Le SC, Hsu AL, Borgnia MJ, Yang H, Lee SY, Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel, Science. 363 (2019) eaav9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Diver MM, Cheng Y, Julius D, Structural insights into TRPM8 inhibition and desensitization, Science. (2019) 1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shen BW, Perraud AL, Scharenberg A, Stoddard BL, The crystal structure and mutational analysis of human NUDT9, J Mol Biol. 332 (2003) 385–398. [DOI] [PubMed] [Google Scholar]
- [33].Wu J, Yan Z, Li Z, et al. , Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 A resolution, Nature. 537 (2016) 191–196. [DOI] [PubMed] [Google Scholar]
- [34].Long SB, Campbell EB, Mackinnon R, Crystal structure of a mammalian voltage-dependent Shaker family K+ channel, Science. 309 (2005) 897–903. [DOI] [PubMed] [Google Scholar]
- [35].Pan X, Li Z, Zhou Q, et al. , Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1, Science. 362 (2018). [DOI] [PubMed] [Google Scholar]
- [36].Rohacs T, Lopes CM, Michailidis I, Logothetis DE, PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain, Nat Neurosci. 8 (2005) 626–634. [DOI] [PubMed] [Google Scholar]
- [37].Toth B, Csanady L, Pore collapse underlies irreversible inactivation of TRPM2 cation channel currents, Proc Natl Acad Sci U S A. 109 (2012) 13440–13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Perraud AL, Fleig A, Dunn CA, et al. , ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology, Nature. 411 (2001) 595–599. [DOI] [PubMed] [Google Scholar]
- [39].McHugh D, Flemming R, Xu SZ, Perraud AL, Beech DJ, Critical intracellular Ca2+ dependence of transient receptor potential melastatin 2 (TRPM2) cation channel activation, J Biol Chem. 278 (2003) 11002–11006. [DOI] [PubMed] [Google Scholar]
- [40].Starkus J, Beck A, Fleig A, Penner R, Regulation of TRPM2 by extra- and intracellular calcium, J Gen Physiol. 130 (2007) 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Csanady L, Torocsik B, Four Ca2+ ions activate TRPM2 channels by binding in deep crevices near the pore but intracellularly of the gate, J Gen Physiol. 133 (2009) 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kuhn FJ, Kuhn C, Winking M, Hoffmann DC, Luckhoff A, ADP-ribose activates the TRPM2 channel from the sea anemone Nematostella vectensis independently of the NUDT9H domain, PLoS One. 11 (2016) e0158060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ullrich ND, Voets T, Prenen J, et al. , Comparison of functional properties of the Ca2+-activated cation channels TRPM4 and TRPM5 from mice, Cell Calcium. 37 (2005) 267–278. [DOI] [PubMed] [Google Scholar]
- [44].Kolisek M, Beck A, Fleig A, Penner R, Cyclic ADP-ribose and hydrogen peroxide synergize with ADP-ribose in the activation of TRPM2 channels, Mol Cell. 18 (2005) 61–69. [DOI] [PubMed] [Google Scholar]
- [45].Kuhn FJ, Luckhoff A, Sites of the NUDT9-H domain critical for ADP-ribose activation of the cation channel TRPM2, J Biol Chem. 279 (2004) 46431–46437. [DOI] [PubMed] [Google Scholar]
- [46].Yu P, Xue X, Zhang J, et al. , Identification of the ADPR binding pocket in the NUDT9 homology domain of TRPM2, J Gen Physiol. 149 (2017) 219–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fliegert R, Watt JM, Schobel A, et al. , Ligand-induced activation of human TRPM2 requires the terminal ribose of ADPR and involves Arg1433 and Tyr1349, Biochem J. 474 (2017) 2159–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yu P, Liu Z, Yu X, et al. , Direct Gating of the TRPM2 Channel by cADPR via Specific Interactions with the ADPR Binding Pocket, Cell Rep. 27 (2019) 3684–3695 e3684. [DOI] [PubMed] [Google Scholar]
- [49].Liao M, Cao E, Julius D, Cheng Y, Structure of the TRPV1 ion channel determined by electron cryo-microscopy, Nature. 504 (2013) 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zubcevic L, Herzik MA Jr., Chung BC, Liu Z, Lander GC, Lee SY, Cryo-electron microscopy structure of the TRPV2 ion channel, Nat Struct Mol Biol. 23 (2016) 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Singh AK, McGoldrick LL, Sobolevsky AI, Structure and gating mechanism of the transient receptor potential channel TRPV3, Nat Struct Mol Biol. 25 (2018) 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Deng Z, Paknejad N, Maksaev G, et al. , Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms, Nat Struct Mol Biol. 25 (2018) 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hughes TET, Lodowski DT, Huynh KW, et al. , Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM, Nat Struct Mol Biol. 25 (2018) 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Saotome K, Singh AK, Yelshanskaya MV, Sobolevsky AI, Crystal structure of the epithelial calcium channel TRPV6, Nature. 534 (2016) 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Duan J, Li Z, Li J, et al. , Structure of full-length human TRPM4, Proc Natl Acad Sci U S A. 115 (2018) 2377–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nilius B, Prenen J, Janssens A, et al. , The selectivity filter of the cation channel TRPM4, J Biol Chem. 280 (2005) 22899–22906. [DOI] [PubMed] [Google Scholar]
- [57].Topala CN, Groenestege WT, Thebault S, et al. , Molecular determinants of permeation through the cation channel TRPM6, Cell Calcium. 41 (2007) 513–523. [DOI] [PubMed] [Google Scholar]
- [58].Xia R, Mei ZZ, Mao HJ, et al. , Identification of pore residues engaged in determining divalent cationic permeation in transient receptor potential melastatin subtype channel 2, J Biol Chem. 283 (2008) 27426–27432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Scharenberg AM, TRPM2 and TRPM7: channel/enzyme fusions to generate novel intracellular sensors, Pflugers Arch. 451 (2005) 220–227. [DOI] [PubMed] [Google Scholar]
- [60].Schmitz C, Perraud AL, Johnson CO, et al. , Regulation of vertebrate cellular Mg2+ homeostasis by TRPM7, Cell. 114 (2003) 191–200. [DOI] [PubMed] [Google Scholar]
- [61].Matsushita M, Kozak JA, Shimizu Y, et al. , Channel function is dissociated from the intrinsic kinase activity and autophosphorylation of TRPM7/ChaK1, J Biol Chem. 280 (2005) 20793–20803. [DOI] [PubMed] [Google Scholar]
- [62].Krapivinsky G, Krapivinsky L, Manasian Y, Clapham DE, The TRPM7 chanzyme is cleaved to release a chromatin-modifying kinase, Cell. 157 (2014) 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Iordanov I, Toth B, Szollosi A, Csanady L, Enzyme activity and selectivity filter stability of ancient TRPM2 channels were simultaneously lost in early vertebrates, Elife. 8 (2019) e44556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kuhn F, Kuhn C, Luckhoff A, Different principles of ADP-ribose-mediated activation and opposite roles of the NUDT9 homology domain in the TRPM2 orthologs of man and sea anemone, Front Physiol. 8 (2017) 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Perraud AL, Schmitz C, Scharenberg AM, TRPM2 Ca2+ permeable cation channels: from gene to biological function, Cell Calcium. 33 (2003) 519–531. [DOI] [PubMed] [Google Scholar]
- [66].Perraud AL, Takanishi CL, Shen B, et al. , Accumulation of free ADP-ribose from mitochondria mediates oxidative stress-induced gating of TRPM2 cation channels, J Biol Chem. 280 (2005) 6138–6148. [DOI] [PubMed] [Google Scholar]
- [67].Burroughs AM, Zhang D, Schaffer DE, Iyer LM, Aravind L, Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling, Nucleic Acids Res. 43 (2015) 10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kuhn FJ, Kuhn C, Naziroglu M, Luckhoff A, Role of an N-terminal splice segment in the activation of the cation channel TRPM2 by ADP-ribose and hydrogen peroxide, Neurochem Res. 34 (2009) 227–233. [DOI] [PubMed] [Google Scholar]
- [69].Iordanov I, Mihalyi C, Toth B, Csanady L, The proposed channel-enzyme transient receptor potential melastatin 2 does not possess ADP ribose hydrolase activity, Elife. 5 (2016) e17600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Toth B, Iordanov I, Csanady L, Putative chanzyme activity of TRPM2 cation channel is unrelated to pore gating, Proc Natl Acad Sci U S A. 111 (2014) 16949–16954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Moreau C, Kirchberger T, Swarbrick JM, et al. , Structure-activity relationship of adenosine 5’-diphosphoribose at the transient receptor potential melastatin 2 (TRPM2) channel: rational design of antagonists, J Med Chem. 56 (2013) 10079–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kuhn FJP, Watt JM, Potter BVL, Luckhoff A, Different substrate specificities of the two ADPR binding sites in TRPM2 channels of Nematostella vectensis and the role of IDPR, Sci Rep. 9 (2019) 4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lin S, Gasmi L, Xie Y, et al. , Cloning, expression and characterisation of a human Nudix hydrolase specific for adenosine 5’-diphosphoribose (ADP-ribose), Biochim Biophys Acta. 1594 (2002) 127–135. [DOI] [PubMed] [Google Scholar]
- [74].Toth B, Iordanov I, Csanady L, Ruling out pyridine dinucleotides as true TRPM2 channel activators reveals novel direct agonist ADP-ribose-2’-phosphate, J Gen Physiol. 145 (2015) 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Fliegert R, Bauche A, Wolf Perez AM, et al. , 2’-Deoxyadenosine 5’-diphosphoribose is an endogenous TRPM2 superagonist, Nat Chem Biol. 13 (2017) 1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Shen Y, Rampino MA, Carroll RC, Nawy S, G-protein-mediated inhibition of the Trp channel TRPM1 requires the Gbetagamma dimer, Proc Natl Acad Sci U S A. 109 (2012) 8752–8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Quallo T, Alkhatib O, Gentry C, Andersson DA, Bevan S, G protein betagamma subunits inhibit TRPM3 ion channels in sensory neurons, Elife. 6 (2017) e26138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Dembla S, Behrendt M, Mohr F, et al. , Anti-nociceptive action of peripheral mu-opioid receptors by G-beta-gamma protein-mediated inhibition of TRPM3 channels, Elife. 6 (2017) e26280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Badheka D, Yudin Y, Borbiro I, et al. , Inhibition of Transient Receptor Potential Melastatin 3 ion channels by G-protein betagamma subunits, Elife. 6 (2017) e26147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Huang J, Zhang X, McNaughton PA, Modulation of temperature-sensitive TRP channels, Semin Cell Dev Biol. 17 (2006) 638–645. [DOI] [PubMed] [Google Scholar]
- [81].Tominaga M, The Role of TRP Channels in Thermosensation, in: Liedtke WB, Heller S (Eds.) TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades, Boca Raton (FL), 2007. [PubMed] [Google Scholar]
- [82].Wang H, Siemens J, TRP ion channels in thermosensation, thermoregulation and metabolism, Temperature (Austin). 2 (2015) 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Tan CH, McNaughton PA, The TRPM2 ion channel is required for sensitivity to warmth, Nature. 536 (2016) 460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Song K, Wang H, Kamm GB, et al. , The TRPM2 channel is a hypothalamic heat sensor that limits fever and can drive hypothermia, Science. 353 (2016) 1393–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]