In this study, Questa et al. studied natural variation in Arabidopsis response to cold and warm temperatures and identify an additional phase in the Polycomb silencing mechanism, downstream from H3K27me3 spreading. Their findings reveal an additional phase in the Polycomb silencing mechanism needed for subsequent reactivation and provides avenues to dissect broader evolutionary changes at FLC.
Keywords: FLOWERING LOCUS C, Polycomb, epigenetics, mathematical modeling, natural variation, vernalization
Abstract
In Arabidopsis thaliana, the cold-induced epigenetic regulation of FLOWERING LOCUS C (FLC) involves distinct phases of Polycomb repressive complex 2 (PRC2) silencing. During cold, a PHD–PRC2 complex metastably and digitally nucleates H3K27me3 within FLC. On return to warm, PHD–PRC2 spreads across the locus delivering H3K27me3 to maintain long-term silencing. Here, we studied natural variation in this process in Arabidopsis accessions, exploring Lov-1, which shows FLC reactivation on return to warm, a feature characteristic of FLC in perennial Brassicaceae. This analysis identifies an additional phase in this Polycomb silencing mechanism downstream from H3K27me3 spreading. In this long-term silencing (perpetuated) phase, the PHD proteins are lost from the nucleation region and silencing is likely maintained by the read-write feedbacks associated with H3K27me3. A combination of noncoding SNPs in the nucleation region mediates instability in this long-term silencing phase with the result that Lov-1 FLC frequently digitally reactivates in individual cells, with a probability that diminishes with increasing cold duration. We propose that this decrease in reactivation probability is due to reduced DNA replication after flowering. Overall, this work defines an additional phase in the Polycomb mechanism instrumental in natural variation of silencing, and provides avenues to dissect broader evolutionary changes at FLC.
In annual Arabidopsis thaliana, timing of the transition to flowering is controlled by a range of environmental and endogenous signals. The prolonged cold aligns flowering with spring in a process known as vernalization. An important factor in vernalization is the MADS-box transcription factor gene FLOWERING LOCUS C (FLC). FLC encodes a floral repressor that directly represses many genes including FLOWERING LOCUS T (FT) (Searle et al. 2006). Winter cold silences FLC expression, and this repression is epigenetically maintained providing memory of having been exposed to low temperatures and allowing the plant to respond to the long days of spring through activation of FT, which moves via the phloem to the shoot apical meristem to activate the floral transition (Corbesier et al. 2007; Jaeger and Wigge 2007).
Vernalization relies on the activity of a Polycomb repressive complex 2 (PHD–PRC2), containing the core components VERNALIZATION2 [VRN2/Su(z)12], MSI1, FIE/EED, and the E(z) homologs SWINGER (SWN) and CURLY LEAF (CLF) (Gendall et al. 2001; Wood et al. 2006; Finnegan and Dennis 2007; De Lucia et al. 2008; Angel et al. 2011), and the PHD proteins VERNALIZATION5 (VRN5), VERNALIZATION5/VIN3-LIKE 1 (VEL1), and the cold-inducible VERNALIZATION INSENSITIVE3 (VIN3) (Mylne et al. 2004; Sung and Amasino 2004; Greb et al. 2007). The silencing is through a cis-based chromatin switching mechanism at FLC (Berry et al. 2015), which occurs in distinct phases: (1) specific nucleation of H3K27me3 close to the FLC transcription start site during cold exposure, and (2) subsequent spreading of H3K27me3 to cover the entire FLC locus when plants are returned to the warm (Finnegan and Dennis 2007; De Lucia et al. 2008). Nucleation confers reversible metastable epigenetic memory, while the H3K27me3 spread state is widely associated with long-term epigenetic silencing of FLC (Yang et al. 2017). Increasing cold exposure leads to a higher proportion of FLC loci switched to the Polycomb silenced state, reflected in increasing average H3K27me3 levels in the plant (Angel et al. 2011).
Dynamics of FLC silencing vary in Arabidopsis accessions collected from different locations (Shindo et al. 2006; Coustham et al. 2012; Li et al. 2014; Duncan et al. 2015). In a Northern Swedish accession, Lov-1, FLC silencing requires many more weeks of cold exposure and this was found to be the result of cis polymorphisms (SNPs) that quantitatively modulate the silencing (Coustham et al. 2012). Similarly, cis variation between annual and perennial Arabis alpina FLC homologs has been found to modulate FLC silencing (Kiefer et al. 2017) and this influences stability of H3K27me3 levels over FLC in both the A. alpina and Arabidopsis halleri (Wang et al. 2009; Nishio et al. 2016). How and at what stage these noncoding SNPs affect Polycomb silencing of FLC is unknown.
Here, we provide an in-depth experimental and theoretical analysis of the influence of the Lov-1 noncoding SNPs on FLC epigenetic silencing. Our analysis reveals a further distinct phase in the sequence of events in Polycomb silencing, with the SNPs causing instability in epigenetic memory even after full spreading of H3K27me3 across the locus. We analyzed the SNPs and show how their epistatic combination mediates this instability. Our work defines an additional mechanistic phase in Polycomb-based epigenetic memory and provides findings likely to have implications for flowering time across plants with a broad range of reproductive strategies.
Results
Lov-1 FLC is never completely stably repressed by vernalization
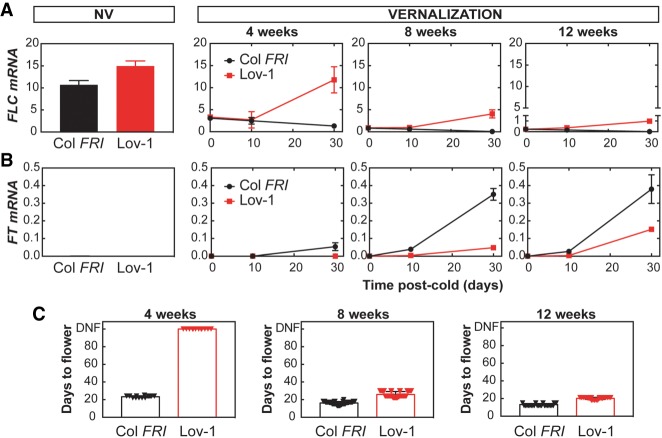
A detailed analysis of FLC silencing after different lengths of cold treatment revealed that Lov-1 FLC is never completely stably repressed (Fig. 1A). Cold exposure (5°C) caused down-regulation of FLC mRNA that was maintained at low levels for 10 d after transfer to warm temperatures (22°C) (Fig. 1A), but not after 30 d. Even after a 12-wk cold treatment, Lov-1 FLC expression reactivated to some extent (Fig. 1A), unlike the rapid vernalizing genotype Col FRI. Reduced levels of FLC allow activation of FT mRNA, therefore, to examine the consequences of FLC reactivation we measured the levels of FT after vernalization. Some degree of FT activation was observed in Col FRI plants after any cold treatment (Fig. 1B). However, in the conditions tested, FT activation was not detected after 4 wk of cold in Lov-1 (Fig. 1B). Longer cold exposure was required in Lov-1 to activate FT, which then correlated with flowering time. In Col FRI, all tested cold exposures effectively accelerated flowering upon return to warm temperatures (Fig. 1C). As found previously (Shindo et al. 2006), Lov-1 did not flower after 4-wk vernalization, but required longer cold treatments to accelerate flowering as demonstrated by the 8- and 12-wk cold treatments (Fig. 1C).
Figure 1.
FLC in Lov-1 reactivates to different levels after different periods of cold. (A) Expression of FLC mRNA before the cold and after 4, 8, and 12 wk of vernalization. (NV) Nonvernalized. (B) Expression of FT mRNA in the same samples. (A,B) Values are means ± SD of three biological replicates. (C) Flowering time was recorded as days to flower after vernalization after the indicated period of cold. Triangles indicate individual measurements (n = 12–48). (DNF) Did not flower.
H3K27me3 can be lost from Lov-1 FLC even after fully spreading across the locus
H3K27me3 accumulation at FLC differs significantly between rapid and slower vernalizing A. thaliana accessions (Shindo et al. 2006; Coustham et al. 2012; Duncan et al. 2015). To provide a more detailed view of H3K27me3 dynamics over the different phases of vernalization, we carried out ChIP analysis, comparing Col FRI and Lov-1 FLC, including a higher density of primers and more time points during and after cold treatment (Fig. 2). At the cell population level, H3K27me3 gradually accumulated during the cold at the nucleation region of FLC in Col FRI (Fig. 2), consistent with the PHD–PRC2 complex accumulating at the same region during cold exposure (De Lucia et al. 2008; Yang et al. 2017). In Lov-1, as in previous observations, the population level H3K27me3 at FLC is significantly lower in NV conditions (P-value < 0.0001), and after 2 wk (P-value < 0.0001) and 4 wk (P-value = 0.0018) of cold (Fig. 2; Supplemental Fig. S1). Lov-1 required at least 8 wk of cold for H3K27me3 to attain similar levels as Col FRI (Fig. 2B,C; Supplemental Fig. S1).
Figure 2.
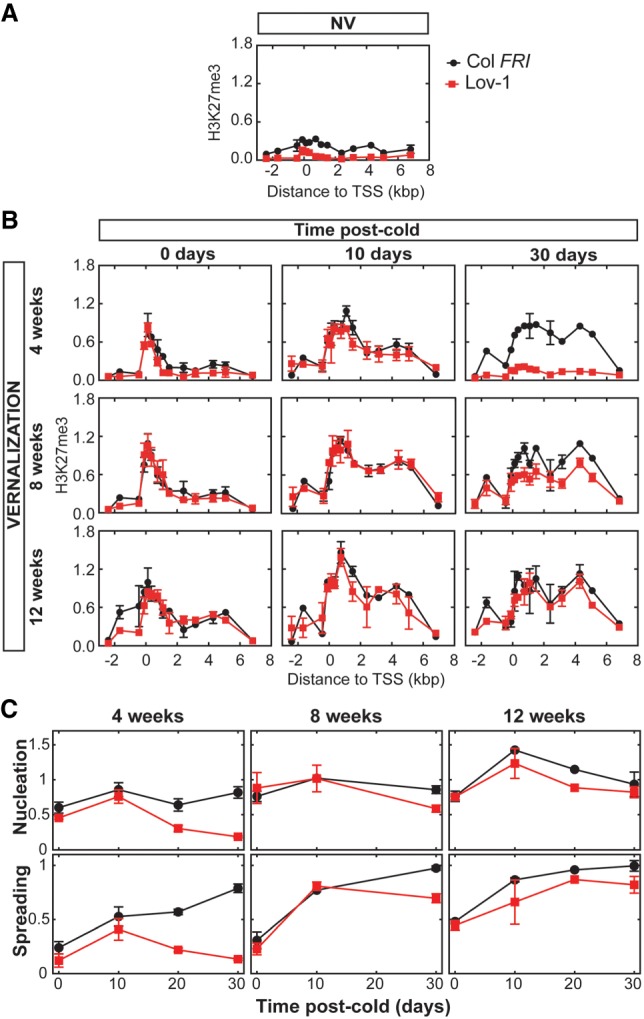
Cell population level H3K27me3 initially accumulates at Lov-1 FLC, but can subsequently be lost. (A) Comparison of Col FRI and Lov-1 H3K27me3 levels along the FLC locus using ChIP qPCR with primers in the locations indicated after 10 d of growth at 22°C. (B) As for A, showing samples following the indicated vernalization and postcold treatment. (C) The data of B (with additional time points 20 d warm after 4 and 12 wk of cold) shown as the average value of all primers in the nucleation region (fifth through eighth primers of Supplemental Table S1) and body region (denoted as “spreading”) (12th–13th primers of Supplemental Table S1). (A,B) Values are means ± SEM of two to four biological replicates. (C) Error bars indicate SEM.
In Col FRI, spreading of H3K27me3 across the whole FLC locus occurred after the plants were moved to warm temperatures, though some spreading occurred in the cold with longer treatments (Fig. 2B,C). Similarly, H3K27me3 spread across the FLC locus in Lov-1 (Fig. 2B,C), after 10 d of growth in warm temperatures regardless of the duration of the cold treatment (Fig. 2B, middle column). In all cases, after 10-d warm the gene body H3K27me3 levels were proportional to the duration of cold. In contrast, the H3K27me3 that had accumulated in the cold and spread across the gene body was lost from the Lov-1 FLC chromatin after 30 d in the warm (Fig. 2B, right column). Such a phenomenon has previously been reported for FLC and its orthologs in perennial species (Wang et al. 2009; Nishio et al. 2016). Reduction of H3K27me3 and therefore lack of maintenance of stable FLC repression had also been reported in mature leaves of Col FRI, when 10-wk-old vegetative plants were vernalized for 4 wk (Finnegan and Dennis 2007). However, in the latter case it is not clear whether H3K27me3 had fully spread along the locus before FLC reactivation. It is worth noting that in young seedlings and under the conditions tested in the current work, we never detected postcold FLC reactivation in Col FRI plants (Figs. 1A, 3B–D, 5A,C, 6B; Supplemental Figs. S3, S9A).
Figure 3.
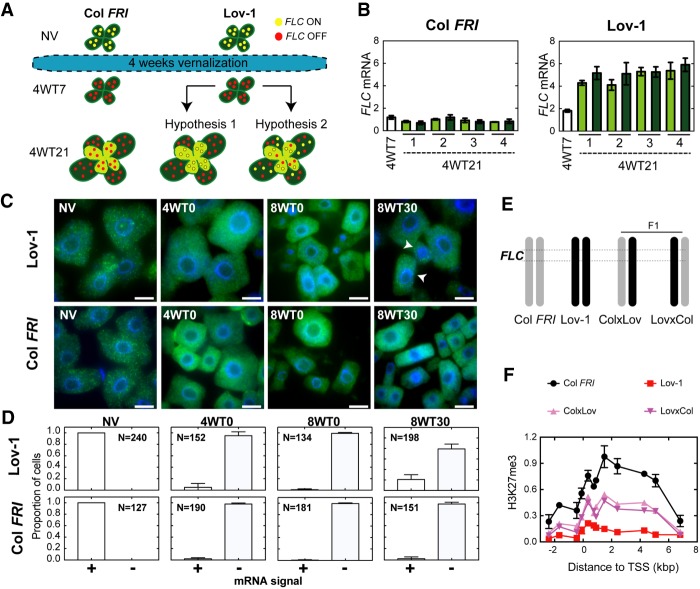
Stochastic and cell autonomous reactivation of Lov-1 FLC. (A) Schematic depicting the experimental design. Col FRI and Lov-1 plants were pregrown for 7 d (NV) and then vernalized for 4 wk. Cells with active (on) or inactive (off) FLC expression are represented with yellow and red circles, respectively. After vernalization, plants were moved to warm conditions. At 7 d after cold, vernalized plants were divided into two groups: (1) Leaf samples were collected (4WT7) for RNA extraction, and (2) all leaves were marked with a pen and the plants were allowed to grow for two additional weeks. At the end of this period (4WT21), leaves that were present at 4WT7 (old leaves, dark green) and leaves that arose after 4WT7 (new leaves, pale green) were sampled separately. xWTy refers to y days of warm following x weeks of cold treatment. (B) Expression of FLC mRNA in Col FRI and Lov-1 plants at 7 d (4WT7) and 21 d (4WT21) after cold. White bars represent 4WT7 data. Dark-green and pale-green bars represent FLC levels in old and new leaves at 4WT21 (dashed line), respectively. Numbers on X-axes (1–4) depict the four individual plants tested at 4WT21. One-tailed, paired Student's t-test results: P-value = 0.9578 testing whether new leaves have higher FLC than old leaves at 4WT21 in Lov-1; P-value = 0.4939 for the same in Col FRI. One-tailed, two-sample t-test results: P-value =3.572 × 10−6 testing whether pooled 4WT21 leaves have higher FLC than 4WT7 in Lov-1; P-value = 0.9916 for the same in Col FRI. (C) smRNA FISH showing FLC mRNA in isolated root meristematic cells from Lov-1 and Col FRI plants, either nonvernalized (NV) or treated with 4 (4WT0) and 8 (8WT0) wk of cold, and then 30 d (8WT30) in postcold warm. Cells showing stochastic reactivation (arrowheads). Scale bars, 10 μm. (D) Quantification of the number of cells with (+) and without (−) FLC mRNA signal at the time points depicted in C. N = number of cells analyzed. (E) Schematic structure of chromosome 5 of Col FRI (gray), Lov-1 (black) and F1 plants generated from reciprocal crosses between Col FRI and Lov-1. Dashed lines depict the FLC region. (F) ChIP-qPCR profiles of H3K27me3 at 30 d following a 4-wk vernalization treatment (4WT30) in Col FRI, Lov-1, and crosses. Lov-1 and Col FRI data are means ± SEM of two biological replicates. Data for one biological replicate is shown for each type of F1. Leaves of 24 individual plants were sampled for each biological replicate.
In parallel, we tested the behavior of H3K36me3 (Supplemental Fig. S2), a chromatin modification that antagonizes H3K27me3 at the FLC nucleation region during vernalization (Yang et al. 2014). A high peak of H3K36me3 was found at the FLC transcriptional start site in NV conditions both in Col FRI and Lov-1 (Supplemental Fig. S2A). Cell population H3K36me3 levels decreased during cold in both genotypes (Supplemental Fig. S2B) and the reduced levels were maintained 10 d after cold. Together with Lov-1 FLC transcript reactivation (Fig. 1A), H3K36me3 levels rose at 30 d after 4 and 8 wk of cold in Lov-1 FLC, whereas H3K36me3 levels remained low in the stably silenced Col FRI FLC (Supplemental Fig. S2B). Only after 12 wk of cold were no differences observed in H3K36me3 levels between the two genotypes.
We next considered what might underlie these differences in Lov-1 FLC regulation. Lov-1 FLC effectively nucleates H3K27me3 in the cold, thereby differing from the behavior of FLC in the PHD–PRC2 mutants vin3, vrn5 (Supplemental Fig. S3), and vrn2 (Chandler et al. 1996; Gendall et al. 2001; Sung and Amasino 2004; Greb et al. 2007), all of which block nucleation of H3K27me3. Although similar in terms of FLC mRNA reactivation, H3K27me3 initially spreads across the gene body in Lov-1, unlike in lhp1 and clf mutants, which nucleate but cannot spread H3K27me3 (Fig. 2; Supplemental Fig. S3; Mylne et al. 2006; Sung et al. 2006; De Lucia et al. 2008; Yang et al. 2017). These data suggest that the components required for transcriptional down-regulation and Polycomb silencing during the cold are functional and not affected by the cis-polymorphisms present in the Lov-1 FLC allele. It is only later, in the long-term maintenance phase, that these elements mediate the reactivation phenotype of the Lov-1 accession. Previous models have proposed that spreading of H3K27me3 along the FLC locus is sufficient to maintain the FLC repressed state through subsequent cell divisions (Angel et al. 2011). For Lov-1 FLC, this is true for only a limited number of days following transfer to warm temperatures.
Reactivation is the loss of silencing at individual FLC loci
Vernalization is a cell-autonomous process with each copy of FLC in individual cells switching from an epigenetically on to off state during cold exposure (Angel et al. 2011; Berry et al. 2015). In the analysis described above, FLC expression measurements and ChIP experiments were undertaken on whole plant samples. Thus, the Lov-1 FLC reactivation and H3K27me3 profiles reflect cell population averages and do not directly reveal FLC reactivation dynamics at individual loci. We reasoned that, following vernalization, the higher postcold FLC transcript levels detected in Lov-1 could be achieved in two different ways. One possibility was that the increased levels of FLC are the result of a new population of cells that never experienced cold temperatures, but instead arise from the division of “FLC-active” cells after cold. A second option would be that cells that had epigenetically silenced FLC to off during the cold could then revert to the on state, a more similar scenario to the meristem reversion observed in some perennial species (Aikawa et al. 2010).
We designed an experiment to distinguish between these two hypotheses (Fig. 3A). We vernalized Col FRI and Lov-1 plants for 4 wk (Fig. 3A). Seven days after the cold treatment (4WT7), we collected all rosette leaves from a pool of four Col FRI and Lov-1 plants to determine FLC expression levels (Fig. 3A,B). In parallel, we marked the leaves of four individual Col FRI and Lov-1 plants to identify the ones present already at that time and allowed them to grow for an additional 14 d in the warm. At 4WT21, we sampled separately those leaves that were present at 4WT7 (the marked leaves) (dark-green leaves in Fig. 3A) and the newly formed leaves (the unmarked leaves) (pale-green leaves in Fig. 3A) for RNA extraction. If hypothesis 1 is correct, only the postcold developed leaves would produce FLC mRNA. Thus, we should observe a significant difference in FLC expression levels between the old and the newly formed leaves. In contrast, a more even FLC expression in both leaf types would favor hypothesis 2. Although we cannot exclude the possibility that the pool of “new leaves” could also contain some leaves that arose from leaf primordia that developed during the cold, the “old leaves” pool certainly will not contain leaves formed after the cold, and so under hypothesis 1 they will not reactivate. Therefore, this experimental design should still allow us to distinguish between the two hypotheses, depending on whether we observed differences in FLC expression levels between the two types of pool (“old leaves” present during cold vs. “new leaves” mixture of cold and postcold leaves). As expected, we observed that postcold stable repression of FLC occurs in Col FRI (Fig. 3B). In Lov-1, although FLC levels increase from 4WT7 to 4WT21, there is no evidence that the FLC levels at 4WT21 differ significantly between the two different types of leaves tested (Fig. 3B, P-value = 0.9578 comparing old and new leaves), suggesting that the reactivation observed is more likely the result of the postcold loss of chromatin silencing at individual FLC loci (hypothesis 2).
We were able to confirm this result by performing single-molecule RNA FISH (smRNA FISH) in root meristematic cells (Duncan et al. 2016). We designed probes to specifically visualize single FLC mRNA molecules in individual cells of Arabidopsis root meristematic tissue during and after cold treatment. Lov-1 and Col FRI exhibit high levels of FLC mRNA expression before the cold (NV in Fig. 3C), and thus all cells analyzed showed FLC mRNA signal. As expected, the cold-induced FLC repression detected in whole plants (Fig. 1A) is also observed at the level of individual cells (4WT0 and 8WT0, Fig. 3C), with Lov-1 and Col FRI cells losing FLC mRNA signal during cold. Thirty days after the cold (8WT30), Col FRI FLC is stably silenced with no cells exhibiting FLC mRNA expression (Fig. 3C). In contrast, we observed individual Lov-1 root meristematic cells showing FLC reactivation (arrowheads, Fig. 3C). Lov-1 FLC mRNA reactivation may also be observed in a single cell out of many cells in a tissue section (Supplemental Fig. S4A–D), again favoring hypothesis 2 stated above. In addition, we also detected sectors of FLC mRNA reactivation within other tissue samples (Supplemental Fig. S4E). We reasoned that these sectors might be the result of clonal division of cells that had reactivated FLC expression. To show that the behavior described above is specific to FLC, we also tested expression of PP2A mRNA, which was evenly expressed at 9WT30 both in Col FRI and Lov-1 cells (Supplemental Fig. S4F). Thus, our results strongly suggest that Lov-1 FLC reactivation is an on/off (digital), cell-autonomous process.
So far, we have provided evidence that Lov-1 FLC reactivation occurs stochastically in individual cells. We next asked whether the loss of H3K27me3 occurs at individual copies of the FLC gene. To this end, we performed reciprocal crosses between Lov-1 and Col FRI (Fig. 3E), and analyzed the H3K27me3 ChIP profiles of F1 plants after vernalization (4WT30). The intermediate accumulation of H3K27me3 in F1 plants (Fig. 3F) suggests that while the Col FRI allele is covered by H3K27me3, the Lov-1 allele has lost this histone modification. Tracking two FLC copies with different fluorescent tags previously showed that one copy of FLC may be switched off, while the other remains switched on, when located in different chromosomes within the same cell (Berry et al. 2015). Here, our data support the hypothesis that memory of FLC activity after vernalization can be stored locally in the chromatin environment of the FLC gene itself, even when the two copies are located in allelic positions.
Modeling FLC reactivation
The Lov-1 FLC allele remains silenced for ∼10 d after cold after 4 wk of vernalization (Fig. 1A). After that time, cell-autonomous stochastic reactivation occurs (Fig. 3). This delay in reactivation is not straightforward to explain, since the temperature is constant for the whole postcold period. Furthermore, the reactivation rate after this delay is relatively high, as FLC levels rise back to NV levels after 30-d warm following 4-wk cold (Fig. 1A).
Our previous work had revealed that local nucleation of PHD–PRC2 (without subsequent spreading) gave a metastable memory state, which transitions to a long-term epigenetically silenced state associated with H3K27me3 spread across the whole locus (Yang et al. 2017). To explain the Lov-1 results, we propose that there is an additional, later phase in the silencing mechanism, which we term the perpetuated state (Fig. 4). In this state, the PHD proteins are lost from the nucleation region, with silencing maintained potentially by the positive read–write feedbacks associated with H3K27me3 (Margueron et al. 2009). The SNPs in Lov-1 FLC would then lead to instability of the perpetuated state, explaining the observed delay in reactivation, as multiple transitions back to the high expression H state must occur before expression is reactivated. The spread and perpetuated states are also observable in Col FRI FLC, but the changes in H3K27me3 are more subtle in that case. In our detailed ChIP profile of Figure 2, we observed a reduction of H3K27me3 in the nucleation region after 30 d after cold in Col FRI, with the levels coming down to those of the body region. However, the perpetuated state in Col FRI remains stably silenced because the Lov-1 SNPs are absent. This result suggests that the same processes are occurring in both Lov-1 and Col FRI. Consistently, the levels of the metastable memory protein VRN5 are also decreased at the FLC locus after cold in Col FRI (Yang et al. 2017).
Figure 4.
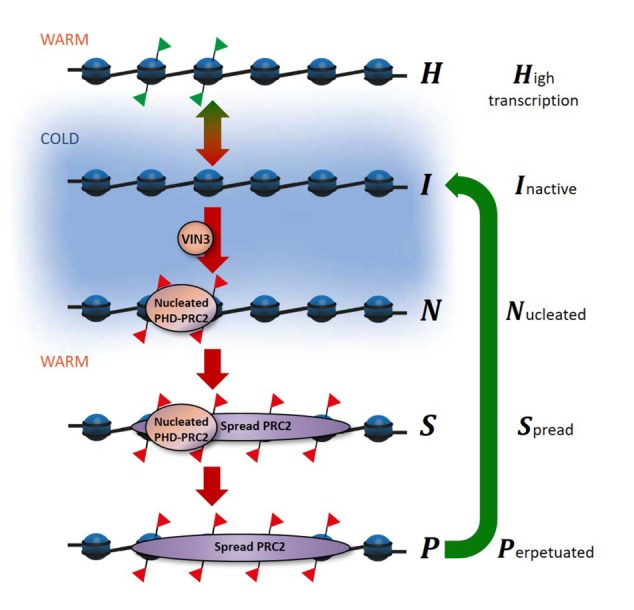
Model of FLC reactivation in Lov-1. The proposed phases of FLC silencing and reactivation. Schematic of the FLC gene, showing the histone modifications in different epigenetic states. (Red flags) H3K27me3; (green flags) H3K36me3. VIN3 is necessary for the nucleation of PHD–PRC2, which is associated with H3K27me3 in the nucleation region of FLC. Spreading of the PRC2 results in H3K27me3 modifications along the whole gene. The green arrow at the right shows the Lov-1-specific reactivation step. Transitions between these states are simulated in the mathematical model. The blue background indicates the cold-dependent transitions.
We developed a mathematical model incorporating the epigenetic phases shown in Figure 4 (Supplemental Fig. S5), in an effort to reproduce the reactivation of Lov-1 FLC after 4 wk of vernalization. This model is an extension of the vernalization model presented in Antoniou-Kourounioti et al. (2018), adapted to Lov-1 and fitted to H3K27me3 and H3K36me3 ChIP data, as well as FLC expression data. This model broadly reproduced all these data, except for the continued reduction of the FLC levels after the cold in Col FRI (Supplemental Fig. S6).
We also explored why the extent of Lov-1 FLC reactivation is different depending on the duration of cold. We found that the model introduced above actually gives a higher rate of reactivation after longer cold, contrary to the data. To illustrate this effect, we constructed a simpler model, ignoring for simplicity the delay in reactivation. This model describes a single transition from off to on in the warm, and can be written using a single ordinary differential equation (see the Materials and Methods for details). In such a simple model with a constant probability for any off cell to reactivate, the reactivation rate is proportional to the fraction of off cells at the end of the cold. Therefore, the reactivation rate will always be higher for the 8- and 12-wk cold-treated compared with the 4-wk cold-treated because more copies are available to reactivate at those times. This is the opposite of what we observed. We conclude that the reactivation probability for off cells must change with time to explain this discrepancy. Below we will discuss reasons for this effect and incorporate it into the model.
FT is not responsible for the change in the reactivation probability
We sought to determine why the Lov-1 FLC reactivation probability diminishes with increasing duration of cold treatments. This observation suggested that a potential factor involved in FLC postcold stable repression also changes its expression after longer vernalization treatments. An obvious candidate factor is FT, a direct target of FLC, given its postcold expression profile. FT mRNA expression was not activated in Lov-1 after a 4-wk vernalization treatment (Fig. 1B); instead, longer cold exposures of 8 and 12 wk were required for FT activation. In addition, it had previously been reported that FT feeds back to repress FLC at both vegetative and reproductive stages (Chen et al. 2014; Chen and Penfield 2018; Luo et al. 2019). Therefore, we wondered whether the lack of FT activation in Lov-1 after 4 wk of cold could explain FLC reactivation in that case and, more generally, the differences in the reactivation probability (Fig. 1A,B).
We tested this hypothesis by transferring Col FRI and Lov-1 plants to the warm in either long-day (LD) or short-day (SD) photoperiods, conditions that either promote or prevent FT activation, respectively (Fig. 5). After 5 wk of vernalization, further FLC reduction was observed in Col FRI at 10 and 20 d after cold, independent of the photoperiod (Fig. 5A,C). As expected, FT activation only occurred in LD (Fig. 5D) but not in SD photoperiods (Fig. 5B). A similar result was found for FT in Lov-1, although these plants required a longer treatment of 10 wk of cold to induce FT (Fig. 5D). Nevertheless, FLC reactivation was detected after 10 wk in Lov-1 at 20 d after cold under both SD and LD conditions, and therefore regardless of the absence or presence of FT transcripts, respectively (Fig. 5A,C). Indeed, in LD, where there was more FT, the reactivation was higher, unlike what we would expect if FT was stabilizing FLC repression. Thus, our data indicate that FT levels do not influence FLC epigenetic stability in Lov-1. Another possibility is that the Lov-1 polymorphisms could disrupt either or both of the previously reported FT-binding sites at FLC (Chen and Penfield 2018; Luo et al. 2019). However, if FT was necessary for stable silencing of FLC, we would expect reactivation also in Col FRI in SD conditions where there is no FT. The lack of such FLC postcold reactivation (Fig. 5A) argues against any role of FT in stabilizing FLC after vernalization. Consistently, it has been observed that the FT feedback repression of FLC can be overcome by vernalization (Chen and Penfield 2018).
Figure 5.
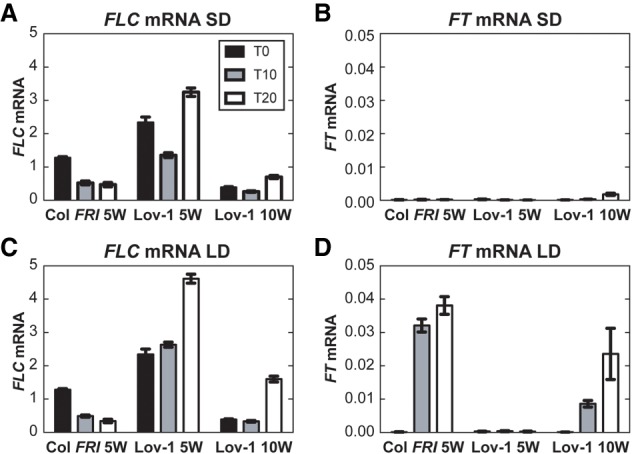
FT is not required for FLC stable repression after vernalization. (A–D) Expression of FLC mRNA (A,C) and FT mRNA (B,D) in Col FRI and Lov-1 at 0 d (black bars), 10 d (gray bars), and 20 d (white bars) after 5 and 10 wk (only for Lov-1) of vernalization treatment. After vernalization, plants were moved to 22°C at either short day (SD; A,B) or long day (LD; C,D) photoperiods. Values are means ± SEM of three biological replicates.
Earlier flowering after longer cold is correlated with less reactivation
Given the lack of a role for FT, we continued to explore what might influence the FLC reactivation rate, focusing on a role for flowering itself. To further probe this aspect, Lov-1 plants were grown for longer after cold (beyond 30 d) in SD conditions, where flowering is suppressed. Our results show that, in the absence of floral induction, Lov-1 plants continued to reactivate their FLC even after 12 wk of vernalization (Supplemental Fig. S7A). This is consistent with results from perennials that showed more reactivation in meristems that continued vegetative growth compared with flowering meristems (Lazaro et al. 2018).
We then considered that the transition to flowering suppressed postcold production of leaves and that this was the factor that influenced the level of FLC reactivation. Continued vegetative growth after cold involves continued cell division/DNA replication. DNA replication represents the major challenge to maintenance of chromatin states, by disrupting histone modifications (Alabert and Groth 2012). Fewer replication events therefore mean a reduced need for the Polycomb apparatus that rebuilds the silenced state, especially after DNA replication. Indeed, we see a reduction in the levels of the core PRC2 methyltransferase components SWN and CLF at the FLC locus when the plants had flowered and the rosette had stopped growing (Supplemental Fig. S7B,C), as assayed by ChIP experiments in a Col FRI background at T30.
Plants that had been vernalized for longer, flowered sooner, thus adding fewer new rosette leaves (Supplemental Fig. S7D,E). In contrast, after short vernalization treatments, more leaves are produced before flowering, so there is more cell division/DNA replication. In the case of Lov-1, this could be the cause of reactivation of FLC and therefore delay flowering even further, such that most Lov-1 plants never flower after a short 4-wk vernalization (Fig. 1C). The continued leaf production and the many rounds of cell division/DNA replication eventually cause FLC reactivation to almost NV starting levels (Fig. 1A).
Incorporating this hypothesis into our full model, we were able to reproduce a reduction in the reactivation rate of Lov-1 FLC dependent on the duration of cold exposure (Supplemental Fig. S8). The model captured all the qualitative features of the FLC mRNA, H3K27me3 and H3K36me3 levels in the cold and subsequent warm for various cold durations, except for the continued reduction of the FLC levels after the cold in Col FRI, as previously. The reduced growth of the rosette in response to flowering occurs in both Lov-1 and Col FRI. However, after many rounds of replication, only Lov-1 FLC reactivates, while Col FRI FLC remains stably silent, suggesting that FLC silencing in Col FRI is intrinsically more stable and therefore it can withstand the continued challenge of replication in high growth conditions. We next addressed the differences between Lov-1 and Col FRI FLC that could confer this difference.
Lov-1 FLC polymorphisms influence stability of the perpetuated silenced state
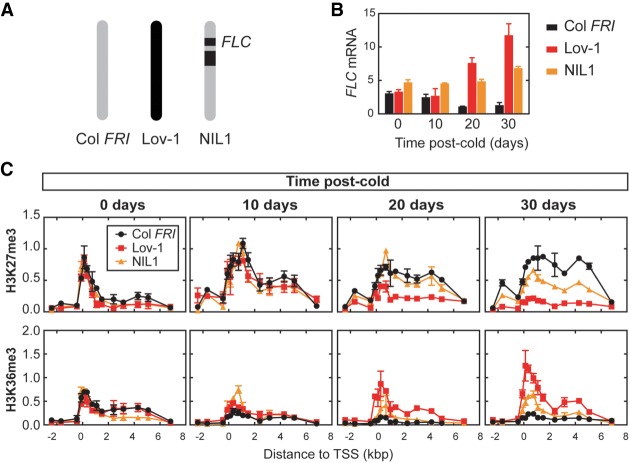
Analysis of chimeric FLC fusions had previously shown that cis, noncoding polymorphisms quantitatively modulate chromatin silencing of Lov-1 FLC (Coustham et al. 2012). To test whether these polymorphisms are responsible for the instability of the perpetuated state in Lov-1 FLC, we performed ChIP experiments using near isogenic lines (NIL1) (Fig. 6A; Coustham et al. 2012). Similar to Lov-1, FLC expression is reactivated in NIL1 plants after 4 wk of cold, although this reactivation is not as strong as that observed in Lov-1 (Fig. 6B). Nevertheless, NIL1 FLC reactivation occurred even after 8 wk of cold and postcold FT activation was delayed in both Lov-1 and NIL1 compared with Col FRI (Supplemental Fig. S9). Nucleation and spreading of H3K27me3 occurred in all the plants tested (Fig. 6C), but Lov-1 and NIL1 showed a significant loss of H3K27me3 at 30 d after cold and accumulation of H3K36me3 (Fig. 6C), with the result for H3K27me3 confirmed in a second NIL line (NIL2) (Supplemental Fig. S10; Duncan et al. 2015). The intermediate phenotype of the NILs between Col FRI and Lov-1 may reflect factors other than the cis-localized SNPs contributing to instability in FLC silencing.
Figure 6.
Lov-1 cis, noncoding polymorphisms lead to instability of perpetuated silenced state at Lov-1 FLC. (A) Schematic structure of chromosome 5 of Col FRI (gray), Lov-1 (black) and the nearly isogenic line NIL1 (Coustham et al. 2012). Black boxes represent the introgressed Lov-1 genomic segments in NIL1. (B) Expression of FLC mRNA in Col FRI, Lov-1 and NIL1 at 0, 10, 20, and 30 d of growth in the warm after a 4-wk vernalization treatment. Values are means ± SEM of three to six biological replicates. (C) Accumulation of H3K27me3 and H3K36me3 in the same samples and time points as described in B. Values are means ± SEM of two to four biological replicates.
Among all the noncoding polymorphisms between the Col-0 and Lov-1 FLC alleles, four single-nucleotide changes (Lov-1 key SNPs) at the 5′ end of the gene are linked to the differential FLC silencing of Lov-1 and Col FRI (Coustham et al. 2012). These Lov-1 SNPs are located at positions −121, −56, +326, and +598 bp from the transcriptional start site (TSS) of the gene (Fig. 7A). We sought to determine the contribution of these four SNPs to the disruption of the long-term epigenetic memory observed in Lov-1 FLC after the cold. To address this question, we started by comparing the vernalization response of Lov-1 with another accession, Edi-0. The reason for choosing Edi-0 was that it shares three of the key SNPs with Lov-1 (−121, −56, and +326) (Fig. 7A; Li et al. 2014), even though it is stably silenced after relatively short cold periods, with a rapid vernalization (RV) response type equivalent to Col FRI (Shindo et al. 2006; Li et al. 2014). We compared the accumulation of H3K27me3 in Lov-1 and Edi-0 before (Fig. 7B) and after 4 wk of cold treatment (Fig. 7C). Edi-0 FLC maintains high levels of H3K27me3 after 4 wk of cold exposure (Fig. 7C), suggesting that the +598 SNP is the causative SNP for the Lov-1 phenotype.
Figure 7.
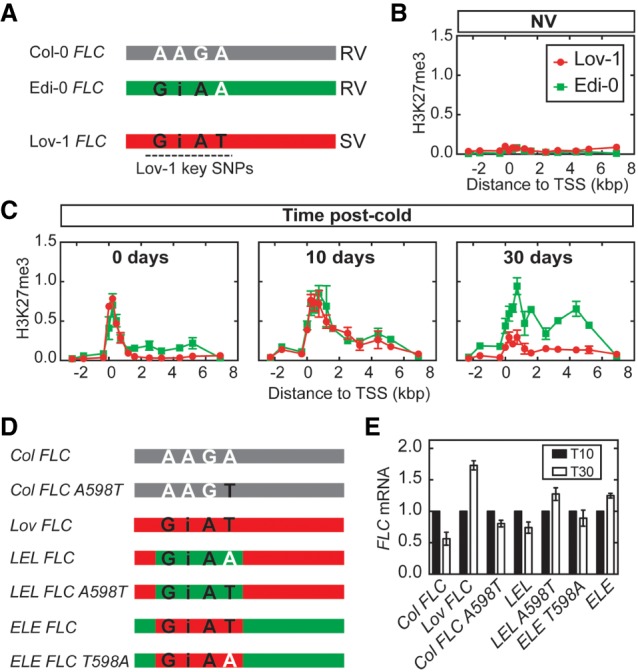
Lov-1 key SNPs prevent long-term epigenetic memory at FLC. (A) Schematic representation of FLC genomic structures (not to scale) of the A. thaliana accessions Col-0 (gray), Edi-0 (green), and Lov-1 (red). The region of the Lov-1 key SNPs (Coustham et al. 2012) is depicted with a dotted black line. (Black letters) Lov-1 SNPs; (white letters) Col-0 SNPs; (i) indel; (RV) rapid vernalization response; (SV) slow vernalization response. (B,C) Accumulation of H3K27me3 in Lov-1 (red) and Edi-0 (green) before (NV; B) and at 0, 10, and 30 d in the warm after a 4-wk vernalization treatment (C). Values are means ± SEM of two (Lov-1) and three (Edi-0) biological replicates. (D) Schematic of different transgenic lines carrying wild type and mutated Col-0 FLC transgenes (gray), and Lov-1 (red) and chimeric Lov-1/Edi-0 (red/green) FLC transgenes. (Black letters) Lov-1 SNPs; (white letters) Col-0 SNPs. (E) Expression of FLC mRNA at 10 (black bars) or 30 (white bars) days in the warm after a 4-wk vernalization treatment in the different transgenic lines from D. Data are shown normalized to T10. Values are means ± SEM of four biological replicates.
To further explore whether +598 prevents stable FLC silencing, we generated the following transgenic lines in a common Col FRI background (Fig. 7D): (1) Lov FLC lines that carry a Lov-1 FLC transgene, and (2) Col FLC A598T lines that carry a Col-0 type FLC construct in which we introduced the A-to-T change at the position +598 (A598T). To account for the variation that may arise due to variable transgene copy number and insertion sites in the new transgenic lines generated, the experiments were carried out using pools of lines. One rosette leaf of each independent transgenic line (36 Lov FLC and 24 Col FLC A598T) was collected and pooled for RNA extraction to test FLC expression in plants exposed to 4 wk of cold (Fig. 7E). FLC mRNA levels of Lov FLC and Col FLC A598T were compared with those of a previously characterized transgenic line that carries the Col-0 FLC transgene (Col FLC) (Csorba et al. 2014). As expected, FLC was reactivated in the Lov FLC lines (Fig. 7E), reinforcing that Lov-1 cis polymorphisms are needed for FLC unstable silencing. However, FLC was stably repressed in both Col FLC and Col FLC A598T lines (Fig. 7E) indicating that introducing the +598 SNP alone in the Col-0 FLC background is not enough to disrupt FLC silencing after cold.
Previous work from our laboratory had demonstrated that swapping the FLC region that contains the four Lov-1 SNPs reciprocally between Lov-1 and Edi-0 FLC alleles can revert the vernalization response of these two Arabidopsis accessions (Li et al. 2014). Transgenic lines carrying the chimeric Lov-1/Edi-0/Lov-1 (LEL) FLC allele (LEL FLC, a pool of 77 independent transgenic lines) in a Col FRI background (Fig. 7D) showed stable FLC silencing after 4 wk of cold (Fig. 7E; Li et al. 2014). In contrast, lines carrying the Edi-0/Lov-1/Edi-0 (ELE) FLC construct (ELE FLC, a pool of 61 independent transgenic lines) in a Col FRI background (Fig. 7D) showed FLC reactivation (Fig. 7E; Li et al. 2014), which results in a slow vernalization response similar to the Lov FLC line. Thus, the ELE FLC vernalization response matches that of the Lov-1 accession rather than the Edi-0 accession, while the LEL FLC chimera shows a vernalization response similar to Edi-0 and Col FRI.
To further test the effect of the +598 SNP in FLC silencing, we generated two additional transgenic constructs in a Col FRI background (Fig. 7D): (1) LEL FLC A598T that carries a LEL FLC chimera in which we substituted the A to T at position +598, and (2) ELE FLC T598A that carries an ELE FLC with T mutated to A at position +598. Interestingly, introducing the +598 SNP into the LEL chimera results in FLC reactivation after 4 wk of cold similar to ELE, Lov FLC, and the Lov-1 accession (Fig. 7E). In contrast, removing the Lov-1 +598 T of the ELE FLC results in stable silencing similar to LEL, Col FLC, Col FRI, and Edi-0 (Fig. 7E). In these cases, 60 and 74 independent transgenic lines were pooled for LEL FLC A598T and ELE FLC T598A, respectively. These results show that SNP +598 has a strong effect on Lov-1 FLC reactivation, but only when combined with the other three Lov-1 SNPs. Thus, the noncoding SNPs interact epistatically to influence the stability of the perpetuated silenced state at FLC. The transgenic analysis presented in this study was performed in the common genetic background Col FRI to specifically test how the Lov-1 noncoding SNPs interfere with the FLC perpetuated silencing following vernalization. However, we cannot exclude that other trans-factors in addition to the noncoding SNPs contribute to Lov-1 FLC instability (Fig. 6B,C; Supplemental Fig. S10).
The alignment of the FLC region covering the four Lov-1 SNPs (Supplemental Fig. S11) shows that SNPs −121, −56, and +598 are variable among all the taxa evaluated (A. thaliana, A. halleri, and A. alpina), with no clear evolutionary conservation between long-term stability (Col-0 and Edi-0 FLCs) or instability (Lov-1 FLC, A. halleri FLC, and A. alpina PEP1) of epigenetic silencing of the FLC orthologs. As suggested previously by Kiefer et al. (2017), the Lov-1 SNP +326 seems to be ancestral, showing conservation among Lov-1 and Edi-0 accessions of A. thaliana, A. halleri, and A. alpina, only being different in the FLC allele of A. thaliana Col-0. However, as it is conserved both in Edi-0 and Lov-1, it does not correlate with unstable FLC silencing after vernalization. Therefore, our data suggest that different specific combinations of noncoding SNPs at FLC have evolved to influence the stability of the long-term PRC2 silenced state.
Discussion
Our work on the mechanistic dissection of natural variation in the vernalization response of A. thaliana accessions has enabled us to elaborate the phases of PRC2 silencing, an understanding that may be widely applicable. Previous models have proposed a separate nucleation state that confers metastable memory, followed by spreading of PRC2 and H3K27me3 across the locus for long-term silencing (Yang et al. 2017). We now add a further phase where the metastable memory element is lost and silencing may be maintained predominantly by the histone modification feedbacks, in a configuration we term the perpetuated state.
Analysis of H3K27me3 dynamics using Lov-1 NILs allowed us to demonstrate that A. thaliana long-term epigenetic memory is coordinated in cis by noncoding SNPs along the FLC locus. Furthermore, using a transgenic approach, we showed that the +598 Lov-1 SNP contributes to postcold FLC instability, but only when in combination with the other three Lov-1 SNPs. These SNPs are within the nucleation site, yet specifically affect the long-term memory state. The nucleation region could therefore be viewed as a Polycomb Response Element (PRE) by analogy with flies, where a PRE is required to effectively propagate H3K27me3 once silencing has become established (Coleman and Struhl 2017; Laprell et al. 2017). At FLC, the nucleation region (PRE) is sufficient to nucleate PHD–PRC2 and confer metastable silencing, and our work here has shown that this process is not affected by the Lov-1 FLC SNPs. The Lov-1 SNPs do, however, affect long-term epigenetic silencing of the perpetuated state, in a process that is potentially mediated by a histone read–write mechanism. In contrast, polymorphisms at the FLC locus in Col FRI might confer greater stability to this perpetuated state. The instability of the perpetuated FLC silencing state caused by Lov-1 SNPs is likely equivalent to the PRC2-maintained state in Drosophila that cannot hold memory through DNA replication after excision of the PRE (Coleman and Struhl 2017; Laprell et al. 2017). In mammalian cells, the JARID2/MTF2–PRC2 nucleated state may be comparable with the PHD–PRC 2 metastable state (Oksuz et al. 2018; Perino et al. 2018).
The absence of PRC trans-factors can result in loss of epigenetic silencing through cell division (Gaydos et al. 2014; Audergon et al. 2015; Ragunathan et al. 2015; Coleman and Struhl 2017; Laprell et al. 2017). However, it is unlikely that the Lov-1 SNPs differentially affect binding of the PHD proteins VIN3 and VRN5, as they disassociate from the FLC locus within 10 d following transfer from cold (Yang et al. 2017), and both the nucleation and spreading processes are unaffected by the SNPs. Potential candidates to influence maintenance of the perpetuated state could be the H3K27me3 methyltransferases SWN and CLF, as we demonstrated that they transiently bind to FLC after cold (Supplemental Fig. S7B,C). Similarly, it remains to be investigated whether the SNPs modulate LHP1 activity at FLC. Another possibility is that H3K27me3 is actively removed from the FLC locus and a candidate for such a role would be the H3K27 demethylase EARLY FLOWERING 6 (ELF6) (Crevillén et al. 2014). This could potentially also explain the difference in starting FLC mRNA levels between Lov-1 and Col FRI (Fig. 1A). However, the ELF6 binding profile is not limited to the area covered by the Lov-1 SNPs (Yang et al. 2014). Stronger association of a trans-factor that promotes transcription could also explain the observed phenotype as transcription opposes Polycomb silencing (Berry et al. 2017). In all these cases, the perpetuated silent state would become more unstable in Lov-1 compared with Col FRI, and therefore more susceptible to switching due to the challenge of replication.
Our work has not only helped to dissect the fundamental Polycomb silencing mechanism, but has also enabled a broader dissection of the evolutionary changes at FLC. Instability in silencing of an FLC homolog was first observed in the perennial species A. alpina (Wang et al. 2009). Based on the parallels with Lov-1 FLC behavior, it is possible that instability of a perpetuated state may be the basis of this FLC reactivation in perennial plants. The molecular analysis of FLC behavior in perennial plants shows many phases in parallel to A. thaliana FLC. For example, analysis of A. halleri AhgFLC expression showed three distinct phases in a natural environment: (1) quantitative down-regulation during winter, (2) maintenance at low levels for ∼8 wk, and (3) reactivation to NV levels to reset AhgFLC for the following flowering season (Aikawa et al. 2010). The latter profile suggests that nucleation and spreading of H3K27me3 at AhgFLC chromatin transiently maintain low levels of the floral repressor following winter, generating a time window for flowering. The subsequent loss of the repressive modification would trigger AhgFLC reactivation, which coincides with meristem reversion. It remains to be investigated whether noncoding SNPs at AhgFLC are responsible for its seasonal expression variation, but for A. alpina, replacing the endogenous PEP1 with the FLC from its annual relative was sufficient to give stable silencing and an annual flowering response (Hyun et al. 2019). Furthermore, increasing evidence supports the hypothesis that cis-acting sequence variation influences postcold FLC stability in the Brassicaceae (Irwin et al. 2016; Kiefer et al. 2017). We envisage that the molecular understanding described here, together with the mathematical model, will help unpick the relative contributions of SNPs, a perpetuated silenced state, and DNA replication-dependent reactivation, on the behavior of FLC orthologs in plants with a range of reproductive strategies.
Materials and methods
Plant material
All mutants and transgenic lines were in Col FRIsf2 background (referred to as Col FRI throughout this work) (Lee et al. 1994). Mutant alleles of vin3 (vin3-4) (Bond et al. 2009), vrn5 (vrn5-8) (Greb et al. 2007), clf (clf-81) (Kim et al. 1998), lhp1 (lhp1-3) (Larsson et al. 1998) in Col FRI background were generated previously (Yang et al. 2017). Arabidopsis thaliana natural accessions Lov-1 and Edi-0 (Shindo et al. 2006) as well as the Lov-1 introgressed lines NIL1 (Coustham et al. 2012) and NIL2 (Duncan et al. 2015) were described elsewhere.
Col FLC (Csorba et al. 2014) and Lov FLC (Coustham et al. 2012) transgenic lines were described previously. Briefly, a SacI fragment (∼12 kb) that comprises ∼6 kb of the genomic sequence of the FLC (Col or Lov-1) gene with its flanking natural 5′ (∼3.5 kb) and 3′ (∼2.6 kb) sequences were cloned into pBLUESCRIPT (pBluescript-FLCprom::FLC), subcloned into a pENTRY by SacI/XhoI digestion (pENTRY-FLCprom::FLC), and subsequently subcloned into a pSLJ-DEST vector (a modified version of the pSLJ series) (Jones et al. 1992) using Gateway Cloning technology (Thermo Fisher Scientific). The pENTRY-FLCprom::FLC was mutagenized to generate the Col FLC A598T construct, following the Megaprimer method of site-directed mutagenesis (Sarkar and Sommer 1990), using the primers detailed in Supplemental Table S2. The previously reported ELE FLC and LEL FLC constructs (referred to as RSR and SRS, respectively, in Li et al. 2014) were mutagenized using overlap extension PCR cloning to obtain the ELE FLC T598A and LEL FLC A598T lines, using the primers detailed in Supplemental Table S2. The single Col FLC transgenic line used in the current work had been selected previously (Csorba et al. 2014) for its similarity with Col FRI regarding FLC mRNA expression and flowering time. All the other constructs (Lov FLC, Col FLC A598T, ELE FLC T598A, and LEL FLC A598T) were transformed into flc-2 FRI plants (Michaels and Amasino 1999). Multiple independent transgenic lines were used for RNA expression analysis (36 for Lov FLC, 24 for Col FLC A598T, 74 for ELE FLC T598A, 60 for LEL FLC A598T, 61 for ELE FLC, and 77 for LEL FLC) (Li et al. 2014). All of them complemented the early flowering phenotype of flc-2 FRI. SWN-YFP (Wang et al. 2006) and 35S::GFP-CLF/clf-28 (Schubert et al. 2006) were both described previously.
Growth conditions
Seeds were surface sterilized and sown on MS medium plates without glucose and stratified for 48 h at 4°C. For nonvernalized (NV) conditions, plants were grown for 10 d in long-day conditions (LD; 16 h light at 22°C, 8 h darkness at 18°C). For vernalization experiments, plants were pregrown for 7 d, and then transferred to 5°C (unless indicated otherwise) under short-day conditions (SD; 8 h light, 16 h darkness) for different durations of cold before being returned to either LD (16 h light at 22°C, 8 h darkness at 18°C) or SD (8 h light, 16 h darkness, at 20°C). For flowering time measurements as well as for ChIP and RNA expression experiments at 20 and 30 d after cold, plants were transferred from plates to soil 10 d after vernalization and grown in controlled environment chambers in long-day conditions (16 h light at 22°C, 8 h darkness at 20°C).
Flowering was counted as days to flower after the cold treatment, from moving plants to warm until bolting, but did not include pregrowth or time spent in the cold. Bolting was scored when stems reached 2 cm in height.
For single molecule RNA FISH (smRNA FISH) imaging, plants were grown and vernalized in vertically oriented Petri dishes containing MS medium. Nonvernalized plants were grown for 7 d at 22°C with a 16-h photoperiod and vernalized plants were grown under the conditions mentioned above for different lengths of time. Following vernalization, plants were transferred to larger plates (one plant per plate) containing fresh MS medium and grown horizontally for an additional 30 d at warm conditions (LD, 16 h light at 22°C).
RNA preparation and qPCR
RNA was extracted as described (Box et al. 2011), using phenol equilibrated to pH8, followed by lithium chloride precipitation. RNA was DNase-treated with Turbo DNA-Free DNase (Life Technologies). cDNA was synthesized with SuperScript III reverse transcriptase (Life Technologies) using either gene-specific primers or oligo dT (12–18). qPCR was performed using SYBR Green master mix II on a LightCycler 480 II (both Roche). Primer sequences are listed in Supplemental Table S2. UBC was used as the normalization gene control.
Chromatin immunoprecipitation
Nuclei were extracted using Honda buffer as described previously (Sun et al. 2013), using 3 g of crosslinked plant material for histone ChIP and at least 5 g for SWN-YFP/CLF-GFP ChIP. In all histone ChIP reactions, sonication, immunoprecipitation, DNA recovery, and purification were performed as previously described (Angel et al. 2011). The antibodies used were anti-H3 (Abcam, ab1791), anti-H3K27me3 (Millipore 07-449), and anti-H3K36me3 (Abcam ab9050). All ChIP experiments were quantified by quantitative PCR (qPCR) with appropriate primers (Supplemental Table S1). For H3K27me3 analysis, SHOOT MERISTEMLESS (STM) was used as the internal control and data are represented as the ratio of (H3K27me3 FLC/H3 FLC) to (H3K27me3 STM/H3 STM). In the case of H3K36me3, ACTIN was used as the internal control and the data are represented as the following: ratio of (H3K36me3 at FLC/H3 FLC) to (H3K36me3 ACT/H3 ACT).
For SWN-YFP/CLF-GFP ChIP, purified nuclei were resuspended in RIPA buffer (1X PBS, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, Roche Complete tablets, 1mM PMSF). Two rounds of sonication were performed in a Bioruptor (Diagenode) at medium setting. Sonication I: four times during 5 min (30 sec on/30 sec off); sonication II: twice for 5 min (30 sec on/30 sec off). After each round of sonication, samples were centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatant was collected in a fresh tube for the subsequent step. Protein A agarose/salmon sperm DNA (Millipore) and anti-GFP (Abcam, ab290) antibodies were used for immunoprecipitation. The data are represented as IP to input.
Single-molecule RNA FISH (smRNA FISH)
smRNA FISH was carried out as described by Duncan et al. (2017). Briefly, roots were fixed for 30 min in 4% paraformaldehyde. Samples were prepared on slides by squashing root tissue under coverslips, ethanol permeabilized, washed, and incubated overnight at 37°C with RNA FISH probes. smRNA FISH probes (Supplemental Tables S3–S5) were used at a final concentration of 25 nM. A Zeiss Elyra PS1 inverted microscope was used for imaging. A x100 oil-immersion objective (1.46 NA) and cooled EM-CCD Andor iXon 897 camera (512512 QE > 90%) were used to obtain all images. Quasar 570 and 670 probes were excited by 561- and 642-nm laser lines, with signals detected between 570 and 640 nm and between 655 and 710 nm, respectively. DAPI signal was acquired with a 405-nm laser excitation and signal detection between 420 and 480 nm. Maximum projections and analysis of three-dimensional pictures were performed using Fiji (an implementation of ImageJ, a public domain program by W. Rasband available from http://rsb.info.nih.gov/ij). Z-stacks of Supplemental Figure S4 were deconvolved using AutoQuant X2 (Media Cybernetics).
Statistical analysis
Comparing H3K27me3 levels in Col FRI and Lov-1
A two-tailed paired Student's t-test was applied (paired t-test function in Graphpad) to compare whether the levels of H3K27me3 were significantly different between Lov-1 and Col FRI under each cold treatment (see Supplemental Fig. S1). The primer pairs are “paired” according to position between the genotypes to remove the effect of the primer position on the H3K27me3 levels and instead only look at the effect of the genotype. Five comparisons were performed, one for each time point in Supplemental Figure S1, and so we performed the Bonferroni correction to adjust the significance level to α = 0.01.
Comparing reactivation of Lov-1 FLC in old and new leaves
A one-tailed paired t-test (function ttest from Matlab R2018b) was used to compare the reactivation in old and new leaves in each plant, with the null hypothesis that in new leaves the reactivation is not greater than in old leaves (Fig. 3). Additionally, the 4WT21 samples were pooled and compared against the 4WT7 results for Col FRI and Lov-1, using a one-tailed, two-sample t-test (function ttest2 from Matlab R2018b).
Alignment of FLC orthologs
Genomic sequences of FLC orthologs were aligned using ClustalW.
Modeling
Simple FLC expression model showing that the reactivation probability decreases with duration of time in the cold
We introduced a simple model for FLC reactivation. In the warm, we had the transition M → A, where M represents the fraction of FLC off gene copies and A represents the fraction of FLC on gene copies. We assumed that this transition happens with a constant probability a.
Therefore, the reactivation rate is proportional to the fraction of off copies M at all times. Because more FLC copies are silenced with longer cold durations, this rate will become greater with more vernalization. This is the opposite of what was observed in our experimental results (Fig. 1A), leading us to reject our assumption of a constant probability of reactivation.
Full FLC model
Our model of FLC reactivation (Fig. 4; Supplemental Fig. S5) built on previous work (Antoniou-Kourounioti et al. 2018), which modeled a sequence of cell-autonomous switches between digital states. Here, we kept the first two states the same as before—H (high transcription) and I (inactive)—but we separated the epigenetic state E (epigenetically silenced) into N (nucleated), S (spread), and the newly proposed state P (perpetuated). These new states allowed us to compare the model output against the ChIP data, as well as with the FLC expression data. Switches between these states included the VIN3-independent pathway switch (H ↔ I) as in the previous work, as well as the VIN3-dependent nucleation switch (now the I → N and H → N switches). The newly added switches were spreading (N → S), loss of nucleated PHD–PRC2 (S → P), and reactivation (P → I), the latter being specific to Lov-1.
A further complication that must be considered in this model is that an active cell cycle is required for spreading (see below and Yang et al. 2017) and presumably also for reactivation. Because only a restricted number of cells are progressing through the cell cycle (and so are dividing) in plants, in the model we must consider two populations of FLC gene copies, those in dividing cells and those in nondividing cells. The normalized number of gene copies in each of the FLC states in dividing cells was represented by the variables Hs, Is, Ns, Ss, and Ps, and for nondividing cells, the variables were H, I, N, S, and P, respectively. In both cases, these numbers were normalized to the initial number of total gene copies in dividing cells, so that the dividing copy variables could be defined as fractions (as the total number of these cells does not change). This normalization was for simplicity and did not affect our results.
A simplified model for replication was implemented: The division of a dividing cell was assumed to lead to a new dividing cell that replaces the old, and a constant number (dn) (defined in Supplemental Table S6) of nondividing cells that all appear instantaneously. This structure was based on the fact that nonstem cells only divide a fixed number of times (approximately five). Our estimate ignored endoreduplicating cells for simplicity, but including this population would give a limited number of further events, and we found that even a large change in the dn parameter did not affect our results. In dividing cells, the gene copies undergo switching between FLC states without their numbers being changed after a cycle of replication and division, and where we assumed that the state does not affect the likelihood of division. Therefore, the number of total gene copies in dividing cells was constant. Gene copies in nondividing cells were generated by the division of dividing cells and they can also undergo transitions according to Supplemental Figure S5.
The spreading (N → S) switch is known to depend on an active cell cycle (Yang et al. 2017). The loss of nucleated PHD–PRC2 (S → P) and reactivation (P → I) switches are also assumed to depend on an active cell cycle, presumably replication, based on evidence showing that, in the absence of spreading, the metastable memory is lost only in cycling cells (Yang et al. 2017). This is because replication is the major challenge to the stability of silencing, presumably due to loss of histone modifications and other factors defining the silenced state. To incorporate these aspects in our model, the N → S, S → P, and P → I transitions could only occur in dividing cells. Furthermore, at every division of a nucleated gene copy (Ns), all offspring were assumed to have spread (Ss and S) and so the spreading rate was the same as the division rate. For the other transitions (S → P and P → I), only a proportion of the dividing cells switches during division (Supplemental Fig. S5). Ordinary differential equations were used to describe the dynamics of the system:
and
where s1, r1, s2, r2, s3, and g are functions that determine the rates of the transitions and are explained in detail in the following sections. The rate g is the division rate as well as the spreading rate. Rates ps and dn are constants given in Supplemental Table S6.
Temperature input
The temperature conditions [T(t)] used in the model simulation matched the experimental conditions with the exception of the pregrowth duration (parameter tg in Supplemental Table S6). The function T(t) takes the form
tg was fitted to the data since the growth rate in the first few days is not consistent with the later growth rate. This parameter, together with the growth rate and temperature dependence of growth, also affected the rate of spreading after cold. Since only the dividing cells are capable of spreading, the apparent rate of spreading is diluted by the nondividing cell population whose number is dependent on the duration of pregrowth.
Early transcriptional shutdown and VIN3-dependent silencing
The “VIN3-independent pathway” (Hepworth et al. 2018) and the “VIN3-dependent” nucleation switch (including the dynamics of VIN3) were modeled as described in Antoniou-Kourounioti et al. (2018). The same temperature sensitivity was also used, though in this study only 5°C (cold) and 22°C (warm) were relevant.
Therefore, in the model here, the VIN3-independent transition rates for H ↔ I were
and
where ps1, Tr1, Tr2, and pr1 are parameters defined in Supplemental Table S6, and Tn is the mean nighttime temperature, where night was defined previously as the time from 6 h before midnight to 6 h after midnight (Antoniou-Kourounioti et al. 2018). In the present work, the temperature was constant throughout the day–night cycle, and therefore Tn was simply the temperature of the treatment.
The VIN3-dependent transition rate (for I → N) is
where T is temperature, V is the VIN3 concentration, calculated according to the LSCD model of Antoniou-Kourounioti et al. (2018), and T1, T2, and ps2 are parameters defined in Supplemental Table S6.
The newly defined transitions N → S, S → P, and P → I only occur in dividing cells and therefore have rates that depend on the temperature-dependent growth rate
and
where pg1, pg2, ps3, and pr2 are parameters defined in Supplemental Table S6.
Initial conditions
The initial conditions of our system were chosen to match the observed initial conditions of the ChIP data, with the additional constraint that, in Col FRI, the relative numbers of gene copies in each of the FLC states, as well as the normalized FLC mRNA concentration ([FLC], defined in the next section), are at steady state in the warm, with the latter equal to 1. Therefore, at t = 0,
Ns = 0, Ss = 0, Ps = P0, and [FLC] = 1, where P0 describes the relative number of gene copies that are off before vernalization and it is defined in Supplemental Table S6. This parameter value was fitted to the levels of H3K27me3 in the body region at the NV time point. Also important is that we defined the starting levels of the gene copies in nondividing cells to be equal to 0 (H = I = N = S = P = 0), so that their relative amounts before the cold match the relative amounts of the corresponding dividing cell variables that produce them.
Model of FLC concentration
The FLC concentration is defined as the amount of FLC divided by the volume, which is approximated here as being proportional to the total number of gene copies (n), where the constant of proportionality is absorbed into the other parameters. n is a monotonically increasing variable calculated by n = Hs + Is + Ns + Ss + Ps + H + I + N + S + P = 1 + H + I + N + S + P.
We modeled the FLC amount according to previous work (Antoniou-Kourounioti et al. 2018), but adjusting for the separate dividing and nondividing cells as described here. FLC is transcribed only in the H state of the FLC gene, so the FLC mRNA amount (NFLC) obeys
where pf1 and pf are the transcription and degradation rates of NFLC, respectively. The FLC mRNA concentration (F) is
and therefore
For simplicity, we ignored the effect of dilution on FLC mRNA (dn/dt)/n, since the dynamics of the mRNA due to its relatively fast degradation (half-life of ∼6 h) (Ietswaart et al. 2017) will be much faster than dilution due to growth of the plant [pf ≫ (dn/dt)/n]. In fact, while the plants are very small (and so n is small), the two rates are comparable. However, the dilution rate decreases very quickly with size so that by the end of the pregrowth period (t = 0) we can approximate
and the equation for dF/dt becomes
We chose the initial value of F to be at steady state:
The FLC concentration normalized to this initial level ([FLC] = F/F0) therefore obeys
With our choice of initial conditions and with the dynamical transitions permitted in the warm in Col FRI, the FLC concentration then remains at steady state in the initial warm period.
The previous equation reduces to the equation for FLC mRNA concentration previously used (Antoniou-Kourounioti et al. 2018), but where the H in that work is replaced by (Hs + H)/n to account for the different populations of cells.
Comparison of numerical model output with data
For comparison of the model output with the ChIP data, we chose parameters pA, pn, and pN (defined in Supplemental Table S6) and compared each of the histone modification measurements with the corresponding value, as indicated.
H3K36me3 in nucleation region:
H3K27me3 in nucleation region:
H3K27me3 in body region:
Matlab version R2018b was used to solve the models numerically, with solver ode15s (Shampine and Reichelt 1997).
Reactivation probability decreasing through stalled growth and flowering
Flowering stalls growth in the rosette and therefore also stalls the cell cycle. As an active cell cycle is necessary for spreading and reactivation in the model, we might expect flowering to have an effect on the reactivation rate. Since the FLC concentration affects flowering time, FLC can effectively feedback on itself (Supplemental Fig. S8E).
In Supplemental Figure S8, we propose a decreasing probability for reactivation related to flowering time after the cold. We implemented this decreasing reactivation probability by changing the growth rate function so that it depends on the history of the FLC concentration. Flowering time is also controlled by the current temperature the plant is experiencing, so this was also included in the changing growth rate function. Specifically, the growth rate becomes
where r is the ratio of the current growth rate to the initial growth rate and is given by
At t = 0, r0 = 1 and r decays exponentially in permissive conditions determined by the function
where pk−1 defines the timescale over which the growth rate decays. pFLC is the FLC threshold below which the flowering transition may begin (defined in Supplemental Table S6). Therefore, while the temperature conditions are inductive (warm: T > T2) and the FLC concentration is permissive of flowering, the plant flowering is accelerated and therefore on average the growth rate is decreasing exponentially in time.
During the postcold warm, and while the FLC is below the threshold, r will be decreasing while simultaneously FLC will be reactivating. For short cold treatments, FLC will be above the threshold so r will not decrease and reactivation will not slow down (Supplemental Fig. S8F). Conversely, for longer cold treatments, where the FLC levels will be lower at the start of the warm, r will be decreasing while FLC remains under the threshold, leading to less reactivation (Supplemental Fig. S8G).
A more quantitative comparison of the slowing growth rate and the change in reactivation will require more accurate measurements of growth and division/endoreduplication at a cellular level. Indeed, assuming that it reflects replication, the difference between leaf addition rates observed in Supplemental Figure S7D,E can only explain the difference in reactivation rate at a qualitative not quantitative level.
Supplementary Material
Acknowledgments
We thank all members of the Dean and Howard groups for discussions. We also thank Scott Berry for comments and discussions at the early phases of this project, Giuseppe Facchetti for discussions on the modeling work, and Cecilia Lövkvist for critically reading the manuscript. The project was supported by European Research Council Advanced Investigator grant MEXTIM and UK Biotechnology and Biological Sciences Research Council Institute Strategic Programme grant GEN (BB/P013511/1).
Author contributions: J.I.Q. and R.L.A.-K. performed all experiments and analyzed the data. S.R. and S.D. performed smRNA FISH experiments. J.I.Q. and P.L. generated Arabidopsis transgenic lines, and C.W. contributed to the characterization of their phenotype. R.L.A.-K. and M.H. performed the modeling. J.I.Q., R.L.A.-K, M.H., and C.D. designed the experiments, interpreted the data, and wrote the manuscript with input from all the other authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.333245.119.
Freely available online through the Genes & Development Open Access option.
References
- Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H. 2010. Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proc Natl Acad Sci 107: 11632–11637. 10.1073/pnas.0914293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13: 153–167. 10.1038/nrm3288 [DOI] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M. 2011. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476: 105–108. 10.1038/nature10241 [DOI] [PubMed] [Google Scholar]
- Antoniou-Kourounioti RL, Hepworth J, Heckmann A, Duncan S, Qüesta J, Rosa S, Säll T, Holm S, Dean C, Howard M. 2018. Temperature sensing is distributed throughout the regulatory network that controls FLC epigenetic silencing in vernalization. Cell Syst 7: 643–655.e9. 10.1016/j.cels.2018.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audergon PN, Catania S, Kagansky A, Tong P, Shukla M, Pidoux AL, Allshire RC. 2015. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 348: 132–135. 10.1126/science.1260638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry S, Hartley M, Olsson TSG, Dean C, Howard M. 2015. Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. Elife 4: e07205 10.7554/eLife.07205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry S, Dean C, Howard M. 2017. Slow chromatin dynamics allow Polycomb target genes to filter fluctuations in transcription factor activity. Cell Syst 4: 445–457.e8. 10.1016/j.cels.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond DM, Wilson IW, Dennis ES, Pogson BJ, Jean Finnegan E. 2009. VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. Plant J 59: 576–587. 10.1111/j.1365-313X.2009.03891.x [DOI] [PubMed] [Google Scholar]
- Box MS, Coustham V, Dean C, Mylne JS. 2011. Protocol: a simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 7: 7 10.1186/1746-4811-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Wilson A, Dean C. 1996. Arabidopsis mutants showing an altered response to vernalization. Plant J 10: 637–644. 10.1046/j.1365-313X.1996.10040637.x [DOI] [PubMed] [Google Scholar]
- Chen M, Penfield S. 2018. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 360: 1014–1017. 10.1126/science.aar7361 [DOI] [PubMed] [Google Scholar]
- Chen M, MacGregor DR, Dave A, Florance H, Moore K, Paszkiewicz K, Smirnoff N, Graham IA, Penfield S. 2014. Maternal temperature history activates Flowering Locus T in fruits to control progeny dormancy according to time of year. Proc Natl Acad Sci 111: 18787–18792. 10.1073/pnas.1412274111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RT, Struhl G. 2017. Causal role for inheritance of H3K27me3 in maintaining the off state of a Drosophila HOX gene. Science 356: eaai8236 10.1126/science.aai8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. 2012. Quantitative modulation of Polycomb silencing underlies natural variation in vernalization. Science 337: 584–587. 10.1126/science.1221881 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C. 2014. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590. 10.1038/nature13722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C. 2014. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci 111: 16160–16165. 10.1073/pnas.1419030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. 2008. A PHD–Polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci 105: 16831–16836. 10.1073/pnas.0808687105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S, Holm S, Questa J, Irwin J, Grant A, Dean C. 2015. Seasonal shift in timing of vernalization as an adaptation to extreme winter. Elife 4: e06620 10.7554/eLife.06620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S, Olsson TSG, Hartley M, Dean C, Rosa S. 2016. A method for detecting single mRNA molecules in Arabidopsis thaliana. Plant Methods 12: 13 10.1186/s13007-016-0114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S, Olsson TSG, Hartley M, Dean C, Rosa S. 2017. Single molecule RNA FISH in Arabidopsis root cells. Bio Protoc 7: e2240 10.21769/BioProtoc.2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. 2007. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol 17: 1978–1983. 10.1016/j.cub.2007.10.026 [DOI] [PubMed] [Google Scholar]
- Gaydos LJ, Wang W, Strome S. 2014. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345: 1515–1518. 10.1126/science.1255023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C. 2001. The VERNALIZATION2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535. 10.1016/S0092-8674(01)00573-6 [DOI] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C. 2007. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17: 73–78. 10.1016/j.cub.2006.11.052 [DOI] [PubMed] [Google Scholar]
- Hepworth J, Antoniou-Kourounioti RL, Bloomer RH, Selga C, Berggren K, Cox D, Collier Harris BR, Irwin JA, Holm S, Säll T, et al. 2018. Absence of warmth permits epigenetic memory of winter in Arabidopsis. Nat Commun 9: 639 10.1038/s41467-018-03065-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Vincent C, Tilmes V, Bergonzi S, Kiefer C, Richter R, Martinez-Gallegos R, Severing E, Coupland G. 2019. A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363: 409–412. 10.1126/science.aau8197 [DOI] [PubMed] [Google Scholar]
- Ietswaart R, Rosa S, Wu Z, Dean C, Howard M. 2017. Cell-size-dependent transcription of FLC and its antisense long non-coding RNA COOLAIR explain cell-to-cell expression variation. Cell Syst 4: 622–635.e9. 10.1016/j.cels.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JA, Soumpourou E, Lister C, Ligthart J-D, Kennedy S, Dean C. 2016. Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J 87: 597–605. 10.1111/tpj.13221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. 2007. FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054. 10.1016/j.cub.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Jones JD, Shlumukov L, Carland F, English J, Scofield SR, Bishop GJ, Harrison K. 1992. Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res 1: 285–297. 10.1007/BF02525170 [DOI] [PubMed] [Google Scholar]
- Kiefer C, Severing E, Karl R, Bergonzi S, Koch M, Tresch A, Coupland G. 2017. Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues. Mol Ecol 26: 3437–3457. 10.1111/mec.14084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. 1998. The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206: 175–183. 10.1007/s004250050389 [DOI] [PubMed] [Google Scholar]
- Laprell F, Finkl K, Müller J. 2017. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 356: 85–88. 10.1126/science.aai8266 [DOI] [PubMed] [Google Scholar]
- Larsson AS, Landberg K, Meeks-Wagner DR. 1998. The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Obeng-Hinneh E, Albani MC. 2018. Extended vernalization regulates inflorescence fate in Arabis alpina by stably silencing PERPETUAL FLOWERING1. Plant Physiol 176: 2819–2833. 10.1104/pp.17.01754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM. 1994. The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J 6: 903–909. 10.1046/j.1365-313X.1994.6060903.x [DOI] [Google Scholar]
- Li P, Filiault D, Box MS, Kerdaffrec E, van Oosterhout C, Wilczek AM, Schmitt J, McMullan M, Bergelson J, Nordborg M, et al. 2014. Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev 28: 1635–1640. 10.1101/gad.245993.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Chen T, Zeng X, He D, He Y. 2019. Feedback regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5′ FLC promoter region in Arabidopsis. Mol Plant 12: 285–288. 10.1016/j.molp.2019.01.013 [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. 10.1038/nature08398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. 10.1105/tpc.11.5.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne J, Greb T, Lister C, Dean C. 2004. Epigenetic regulation in the control of flowering. Cold Spring Harb Symp Quant Biol 69: 457–464. 10.1101/sqb.2004.69.457 [DOI] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C. 2006. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci 103: 5012–5017. 10.1073/pnas.0507427103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio H, Buzas DM, Nagano AJ, Suzuki Y, Sugano S, Ito M, Morinaga S, Kudoh H. 2016. From the laboratory to the field: assaying histone methylation at FLOWERING LOCUS C in naturally growing Arabidopsis halleri. Genes Genet Syst 91: 15–26. 10.1266/ggs.15-00071 [DOI] [PubMed] [Google Scholar]
- Oksuz O, Narendra V, Lee CH, Descostes N, LeRoy G, Raviram R, Blumenberg L, Karch K, Rocha PP, Garcia BA, et al. 2018. Capturing the onset of PRC2-mediated repressive domain formation. Mol Cell 70: 1149–1162.e5. 10.1016/j.molcel.2018.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perino M, van Mierlo G, Karemaker ID, van Genesen S, Vermeulen M, Marks H, van Heeringen SJ, Veenstra GJC. 2018. MTF2 recruits Polycomb repressive complex 2 by helical-shape-selective DNA binding. Nature Genet 50: 1002–1010. 10.1038/s41588-018-0134-8 [DOI] [PubMed] [Google Scholar]
- Ragunathan K, Jih G, Moazed D. 2015. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348: 1258699 10.1126/science.1258699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G, Sommer SS. 1990. The ‘megaprimer’ method of site-directed mutagenesis. BioTechniques 8: 404–407. [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. 2006. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649. 10.1038/sj.emboj.7601311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampine LF, Reichelt MW. 1997. The Matlab ODE suite. SIAM J Sci Comput 18: 1–22. 10.1137/S1064827594276424 [DOI] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C. 2006. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev 20: 3079–3083. 10.1101/gad.405306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. 2013. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621. 10.1126/science.1234848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM. 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164. 10.1038/nature02195 [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. 2006. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38: 706–710. 10.1038/ng1795 [DOI] [PubMed] [Google Scholar]
- Wang D, Tyson MD, Jackson SS, Yadegari R. 2006. Partially redundant functions of two SET-domain Polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci 103: 13244–13249. 10.1073/pnas.0605551103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC. 2009. PEP1 regulates perennial flowering in Arabis alpina. Nature 459: 423–427. 10.1038/nature07988 [DOI] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. 2006. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci 103: 14631–14636. 10.1073/pnas.0606385103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Howard M, Dean C. 2014. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24: 1793–1797. 10.1016/j.cub.2014.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Berry S, Olsson TSG, Hartley M, Howard M, Dean C. 2017. Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 357: 1142–1145. 10.1126/science.aan1121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.