In this review, Szanto et al. summarize the metabolic regulatory roles of PARP enzymes and their associated pathologies.
Keywords: PARP, ARTD, adipocyte, adipogenesis, mitochondria, lipolysis, differentiation, white adipocytes, brown adipocytes, beige adipocytes, stem cell, PARylation, high fat diet, obesity, insulin resistance, AFLD, NAFLD, atherosclerosis
Abstract
Poly(ADP-ribose) polymerases (PARPs or ARTDs), originally described as DNA repair factors, have metabolic regulatory roles. PARP1, PARP2, PARP7, PARP10, and PARP14 regulate central and peripheral carbohydrate and lipid metabolism and often channel pathological disruptive metabolic signals. PARP1 and PARP2 are crucial for adipocyte differentiation, including the commitment toward white, brown, or beige adipose tissue lineages, as well as the regulation of lipid accumulation. Through regulating adipocyte function and organismal energy balance, PARPs play a role in obesity and the consequences of obesity. These findings can be translated into humans, as evidenced by studies on identical twins and SNPs affecting PARP activity.
Brief introduction to ADP-ribose metabolism
The field of poly(ADP-ribose) polymerases (PARPs or ARTDs) has come a long way since the discovery of a nuclear poly(ADP-ribosyl)ating (PARylating) enzyme in 1963 (Chambon et al. 1963). PARPs now constitute a superfamily of at least 17 members in human that share a conserved catalytic domain (Amé et al. 2004; Hottiger et al. 2010). ADP-ribosylation is a posttranslational modification, during which the ADP-ribosylation enzymes cleave NAD+ and attach the resulting ADP-ribose (ADPR) units to acceptor proteins. ADP-ribosylation is referred to as mono(ADP-ribosyl)ation (MARylation), oligo(ADP-ribosyl)ation, or poly(ADP-ribosyl)ation (PARylation), based on the number of the ADPR units added to the acceptor protein (Amé et al. 2004; Hottiger et al. 2010). Although all PARPs inherited the family name of the founding member, PARP-1, the PARP “polyenzymes” include only PARP-1, PARP-2, and the tankyrases (PARP-5a and PARP-5b) (Gibson and Kraus 2012). Other members perform only MARylation or oligo(ADP-ribosyl)ation, while PARP13 possesses no enzymatic activity (Hottiger et al. 2010). To our current understanding, the majority of PARP activity is attributed to PARP1 (80%–85%), while the rest is largely attributed to PARP2 (Amé et al. 1999; Schreiber et al. 2002; Szanto et al. 2011). In most cases, the major acceptor of PAR is PARP1 itself (termed auto-PARylation); nevertheless, with the use of state-of-the-art proteomics a large set of PARylated or ADP-ribosylated proteins were identified and this process is termed trans-PARylation (Chapman et al. 2013; Gibson et al. 2016; Abplanalp et al. 2018; Leslie Pedrioli et al. 2018; Palazzo et al. 2018) (for a comprehensive database of ADP-ribosylated proteins see Vivelo et al. (2017).
ADP-ribose unit(s) have rapid turnover and are removed by isoforms of poly(ADP-ribose) glycohydrolase (PARG) (O'Sullivan et al. 2019; Slade 2020), ADP-ribosyl hydrolase 3 (ARH3) (Oka et al. 2006; Rack et al. 2020), and ADP-ribosyl protein lyase (Kawaichi et al. 1983). PAR polymers can be recognized by a set of proteins that consequently localize to sites marked by PARP enzymes (Barkauskaite et al. 2013; Feijs et al. 2013). Karlberg et al. (2013) classified enzymes involved in ADPR metabolism and recognition as writers, readers, and erasers.
PARP1, PARP2, and PARP3 can be activated by DNA strand breaks and aberrant DNA forms (Menissier-de Murcia et al. 1989; Gradwohl et al. 1990; Kutuzov et al. 2013, 2015). Recently, other regulatory routes were described. PARP2 is activated by RNA forms (Léger et al. 2014); numerous signal transduction pathways, or the stability of PARP proteins were shown to modify the activity of PARP isoforms (Gagné et al. 2009; Cantó et al. 2013). PARPs, especially PARP1 and PARP2, are major NAD+ consumers in the cell (Bai et al. 2011a,b; Mohamed et al. 2014) and play a crucial role in regulating NAD+ availability and the nonredox functions of NAD+ (often referred to as the NAD+ node) (Houtkooper et al. 2010). On the other hand, PARP activity is dependent on NAD+ levels in cellular compartments and requires a continuous supply of NAD+. Nicotinamide mononucleotide adenylyl transferase (NMNAT) -1, -2, and -3 are NAD+ synthase enzymes that produce NAD+ from nicotinamide mononucleotide and ATP (Chiarugi et al. 2012; Cohen 2020). Thus, NMNATs can “feed” PARPs with their substrate and modulate PARP catalytic activity (Berger et al. 2007; Zhang et al. 2012; Ryu et al. 2018). There are pharmacological inhibitors available for the study of PARP biology, as well as for clinical use. Clinically available PARP inhibitors include ABT-888 (Veliparib from Abbott/Abbvie) rucaparib (Rubraca from Agouron/Pfizer/Clovis), talazoparib (Talzenna from Lead/Biomarin/Medivation/Pfizer), olaparib (Lynparza from KuDos Pharmaceuticals/AstraZeneca+Merck), and niraparib (Zejula from Merck/Tesaro/GSK) (for detailed review, see Slade 2020; Curtin and Szabo 2013). Although, none of the current PARP inhibitors seem to discriminate between PARP enzymes (Wahlberg et al. 2012), enzyme-specific inhibition of mono-PARP enzymes may be possible (Venkannagari et al. 2016; Upton et al. 2017; Holechek et al. 2018).
PARP enzymes have widespread biological functions ranging from DNA repair and chromatin structure (Javle and Curtin 2011; De Vos et al. 2012; Dantzer and Santoro 2013), RNA transcription, protein translation, and degradation (Kraus and Hottiger 2013; Bai 2015), cell division, tumor biology (Curtin and Szabo 2013), immune processes (Fehr et al. 2020) metabolism, and mitochondrial biology (Bai and Cantó 2012; Bai et al. 2015), oxidative stress biology, and cell death and differentiation, and aging (Mangerich et al. 2010; Burkle and Virag 2013; Fatokun et al. 2014). In this review, we focus on the metabolic properties of PARP enzymes.
PARP enzymes in metabolism
PARP enzymes impact metabolism at multiple points, exerting regulatory functions on higher order organismal and basic cellular processes. From another perspective, PARPs impact both central and peripheral metabolic regulation. Frequently, PARP activation represent pathological disruptive metabolic signals. Here, we briefly review PARP-mediated pathways in metabolic regulation. Metabolic pathologies associated with PARP activation are listed in Table 1.
Table 1.
PARP-mediated metabolic diseases
PARPs in regulating central and peripheral organismal metabolic homeostasis
PARP enzymes are widely expressed in almost all tissues and cells of the human organism, including metabolic tissues and organs, such as the liver, skeletal muscle, hormone glands, adipose tissue (white, brown, and beige), and the nerve system (Bai 2015). Central metabolic regulation encompasses the coordinated regulatory activity of the central nervous system and the hormonal system, which allows the organism to adjust to environmental and internal metabolic challenges. Such signals are integrated into the nuclei of the ventromedial hypothalamus, which serve as a central orchestrator and zeitgeber for other organs through hypothalamic neurohormonal changes (Cedernaes et al. 2019). Whole body genetic deletion of PARP1 alters feeding entrainment in mice and changes spontaneous locomotor activity (Bai et al. 2011b), suggesting a role for PARP1 in the circadian phase of entrainment. PARP1 expression and PARP1 activity show circadian changes in murine models and humans that contribute to circadian entrainment of transcriptional programs in skeletal muscle, the liver, and in the cells of the immune system (Mocchegiani et al. 2003, 2004; Asher et al. 2010; Zhao et al. 2015). PARP1 can achieve circadian regulation of gene transcription through the following actions: (1) interacting with 11-zinc-finger protein or CCCTC-binding factor (CTCF) and converting parts of the chromatin to heterochromatin in a time-dependent fashion (Zhao et al. 2015) and (2) interacting with and ADP-ribosylating Clock protein (Asher et al. 2010). Yet-uncovered pathways may also be active. PARP1 activation seems to be vital for sensing or mediating NAD+/NADH levels to be integrated into cellular energy sensing and signaling. Although, the aforementioned pathways were described in nonneuronal models, PARPs are abundantly expressed and active in the nervous system (Komjati et al. 2004; Fatokun et al. 2014) and feeding and locomotion behavior changes in the PARP1 knockout mice (Bai et al. 2011b), making it likely that these processes are active in neurons and other cellular elements of the nervous system. It is important to note that disrupting circadian entrainment increases the risk for obesity and the consequences of obesity (Kettner et al. 2015); however, this has not been studied in the context of PARP activation.
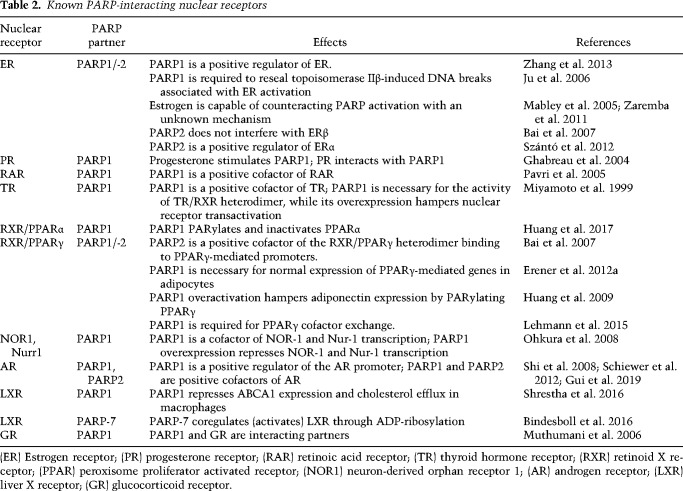
PARPs interfere with hormonal signaling at various points. PARPs regulate hormone levels, including intramuscular androgen production (Marton et al. 2018b). Fasting serum insulin levels were lower in PARP2 knockout mice (Bai et al. 2011a), weak PARP inhibitors were shown to restore insulin expression (Ye et al. 2006) and the deletion of Tankyrase 1 (PARP5a, TNK1) induced serum insulin levels (Yeh et al. 2009). Pharmacological inhibition or genetic deletion of PARP1 protects against streptozotocin-induced β-cell death that impairs insulin production (Burkart et al. 1999). Interestingly, the deletion of PARP2 impairs β-cell function and proliferation through blocking pdx-1 (Bai et al. 2011a). PARP1 and PARP2 were shown to modulate adipokine expression (Bai et al. 2007; Yeh et al. 2009; Erener et al. 2012a,b; Lehmann et al. 2015).
The sensing of hormones is also regulated by PARPs. Nuclear hormone receptors use PARPs as cofactors (Table 2). Therefore, nuclear hormone receptor activation is PARP-dependent. Insulin-like growth factor (IGF)-1 signaling is potentiated by PARP inhibition (Amin et al. 2015). Furthermore, PARP1 interferes with GLP-1 signaling that may interfere with insulin secretion from β cells (Liu et al. 2011). PARP1 and PARP2 activation were shown to be a key step in the development of insulin resistance (for review, see Bai and Cantó 2012).
Table 2.
Known PARP-interacting nuclear receptors
Hormones, such as insulin (Horvath et al. 2008), estrogens (Mabley et al. 2005; Jog and Caricchio 2013; Joshi et al. 2014), androgens (Shimizu et al. 2013), progesterone (Ghabreau et al. 2004), artificial steroids, and vitamin D (Marton et al. 2018b) can modulate the expression and activity of PARP1 and PARP2. Endocrine disruptors were also shown to modulate PARP activity (Chen et al. 2013; Guerriero et al. 2018). These observations suggest feedback loops where PARPs interfere with hormonal signaling and hormones regulate PARP availability and activity.
PARPs interplay with energy sensor systems in cells (for review, see Bai et al. 2015). These systems assess the energy charge of cells (NAD+/NADH or ATP/(ADP + AMP) ratio) and the availability of nutrients (amino acids, oxygen, etc.) and shape cellular metabolism to meet these challenges.
PARPs in carbohydrate metabolism
PARPs regulate points in glycolysis (Hopp et al. 2019), the core pathway of glucose catabolism. PARP1 activation hampers glycolytic flux, inducing metabolic dysfunction (Ying et al. 2002, 2003; Devalaraja-Narashimha and Padanilam 2009; Módis et al. 2012; Robaszkiewicz et al. 2014). Tankyrase 1 and Tankyrase 2 (TNK1, TNK2) regulate glucose transporter 4 (Glut4) translocation to the cytoplasmic surface in an ADP-ribosylation-dependent manner and, thus play a vital role in regulating glucose (and glutamine) availability and glycolytic flux (Yeh et al. 2007). The next step in glucose catabolism is the phosphorylation of glucose by hexokinase to form glucose-6-phosphate, which represents a commitment to glycolysis. Hexokinase is localized to the mitochondrial surface to help synchronize glycolytic flux and mitochondrial oxidation (Andrabi et al. 2014). PARP1 activation disrupts this synchronized function, reducing glycolytic influx (Andrabi et al. 2014; Fouquerel et al. 2014). This observation is further underlined by the observation that the supplementation of pyruvate, the end product of glycolysis, can alleviate cellular dysfunction and cell death upon PARP1 activation (Ying et al. 2002, 2003; Suh et al. 2005; Zeng et al. 2007). In agreement with these observations, the down-regulation of PARP1 supports glycolysis (Regdon et al. 2019). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is an NAD+-dependent enzyme in glycolysis. PARP1 can PARylate and hence inhibit GAPDH (Du et al. 2003). Furthermore, since GAPDH is NAD+-dependent, NAD+ breakdown by cytoplasmic PARPs can limit GAPDH activity and, consequently, glycolytic flux (Hopp et al. 2019). These results were confirmed by the observation that PARP1 knockout mice have higher respiratory quotient, suggesting a shift toward glucose oxidation (Bai et al. 2011b). Although pyruvate dehydrogenase complex is not considered as a member of the glycolytic enzymatic machinery, it is important to note that three subunits of the complex (PDPR, PDHA1, and PDHX) are subject to poly-ADP-ribosylation, which may regulate the fate of pyruvate, whether it can enter the TCA cycle, convert to lactate, or undergo gluconeogenesis (Hopp et al. 2019).
PARP10 and PARP14 are two poorly characterized members of the PARP family. Nevertheless, they seem to be connected to carbohydrate metabolism. Silencing of PARP10 induces glycolysis and mitochondrial oxidation, rendering cells hypermetabolic (Márton et al. 2018a). PARP14 can support glycolysis in lymphoma cells, although the molecular mechanism has not been elucidated (Cho et al. 2011). Another interesting feature of PARP14 is its physical interaction with phosphoglucose isomerase, an enzyme that enables the entry of fructose into glycolysis (Yanagawa et al. 2007). The actual consequence of this interaction is unknown.
When considering carbohydrate metabolism, the regulatory mechanisms should also be mentioned. PARPs interact with HIFs, GSK3b, and AMPK, sensors that regulate glycolytic flux and the coupling of glycolysis to mitochondrial oxidation. These pathways are reviewed in Bai et al. (2015). A high-glucose or high-fructose diet can induce the expression of PARP1 (Choi et al. 2017; Huang et al. 2019). The interplay between carbohydrate metabolism and PARPs was extensively reviewed in Hopp et al. (2019).
PARPs in lipid metabolism
There is an ever-growing body of evidence for the involvement of PARPs in lipid metabolism. As a prime example, PARP2 was found to be connected to cholesterol and triglyceride metabolism in a genome-wide association study (Manunza et al. 2014).
Cellular and organismal fatty acid homeostasis are regulated by PARPs. Erener et al. (2012b) reported hypercholesterolaemia in PARP1 knockout mice. The pattern of polyunsaturated fatty acid metabolites is dysregulated in PARP1 knockout mice (Kiss et al. 2015) and there seems to be a correlation between PARP1 activity and erythrocyte membrane composition (Bianchi et al. 2016). Furthermore, the composition of membrane-constituent lipids was altered upon the deletion of PARP2 (Marton et al. 2018b).
Fatty acid absorption and fatty acid biosynthesis had not been studied in the context of PARP enzymes and poly-ADP-ribosylation; however, the involvement of PARPs is likely, as suggested by scattered data in the literature. For example, the deletion of PARP2 reduces the expression of fatty acid synthase in the white adipose tissue (Bai et al. 2007). The expression of the fatty acid transporters, FABP7, FABP3, CD36, and aP2 (FABP4), are regulated by PARP1, PARP2, and tankyrases (Bai et al. 2007; Yeh et al. 2009; Erener et al. 2012a; Kiss et al. 2015). The deletion of PARP1, PARP2, or PARP10 induces mitochondrial fatty acid oxidation (Bai et al. 2011a,b; Márton et al. 2018a). Upon the genetic deletion of PARP2, the respiratory quotient decreases, suggesting a preference for fatty acid oxidation both in the active and in the sleeping period of the daily cycle (Bai et al. 2011a). Acylation of histone proteins by fatty acids may serve as epigenetic marks, a recent study suggested the PARP-sirtuin interplay may be a key factor in regulating acyl epigenetic marks (Faraone-Mennella et al. 2019).
Certain fatty acid-type lipid species can regulate the expression of PARPs. Serum deprivation of a plethora of lipid species (Sun et al. 2019) can inhibit PARP2 expression, similar to lipoic acid (Zhang et al. 2014). Caloric restriction reduces, while a high-fat diet induces the expression of PARP1 (Bai et al. 2011b; Salomone et al. 2017; Huang et al. 2019). In a similar fashion, fatty acid synthase activation or overexpression can also induce PARP1 expression (Wu et al. 2016).
Another arch of lipid metabolism is cholesterol homeostasis and the metabolism of cholesterol derivatives. The central organ for cholesterol biosynthesis is the liver, although other organs, such as skeletal muscle, also possess functional enzymatic machinery for cholesterol biosynthesis. Dietary cholesterol is taken up from the intestines and is then transported to the liver by chylomicrons. Excess cholesterol is excreted in the bile that is subsequently emptied into the intestines. Collectively, this is called the enterohepatic circulation of cholesterol. The liver can excrete cholesterol into low-density lipoprotein (LDL) that are then sent to the periphery to supply cholesterol to cells. Peripheral cholesterol is returned to the liver by high-density lipoproteins (HDL). This is the peripheral circulation of cholesterol in humans. Mice have little HDL, therefore, LDL performs the functions of HDL in mice. Cholesterol is a starting compound for the synthesis of steroid hormones, vitamin D, and bile acids.
PARP2 negatively regulates de novo cholesterol biosynthesis through suppression of sterol-regulatory element-binding protein expression. The deletion of PARP2 induces increased cholesterol biosynthesis in the liver and skeletal muscle (Szántó et al. 2014; Marton et al. 2018b). A fraction of excess cholesterol seems to be incorporated into biomembranes (Marton et al. 2018b). The deletion of PARP2 does not affect the enterohepatic circulation of cholesterol. However, PARP2 deletion reduces the expression of hepatic ATP-binding cassette subfamily A member 1 (ABCA1), a major transporter of cholesterol to lipoproteins (Szántó et al. 2014). In line with this, serum HDL levels are lower in PARP2 knockout mice (Szántó et al. 2014). However, it is not easy to translate this finding into the human situation.
PARP1 expression and activity correlate negatively with ABCA1 expression (Shrestha et al. 2016). In addition, PARP1 regulates the expression of microsomal epoxide hydrolase (mEH), a key sodium-dependent bile acid transporter in hepatocytes (Peng et al. 2015). Furthermore, a lipid-activated enzyme, acyl-CoA-binding domain containing 3, activates PARP1 activity (Chen et al. 2015). Knockout and pharmacological inhibitor studies show that PARP1 inhibition improves HDL/LDL levels in mice (Diestel et al. 2003; Kiss et al. 2006; Oumouna-Benachour et al. 2007; Hans et al. 2008; von Lukowicz et al. 2008; Zerfaoui et al. 2008; Hans et al. 2009a, b; Xu et al. 2014). In humans, an SNP that renders PARP1 less active correlates with decreases total cholesterol levels, increases in HDL and decreased risk for coronary artery disease (Wang et al. 2017).
Lipids can be stored physiologically or pathophysiologically in multiple organs, where excess lipids cause damage to the tissue. Lipid-mediated activation of PARP1 may have a crucial role in organ or cellular damage (Diestel et al. 2003; Kiss et al. 2006; Hans et al. 2008; Bai and Csóka 2015; Chen et al. 2015). Ectopic lipid deposition to the walls of arteries happens in atherosclerosis. PARP inhibition or genetic deletion of PARP1 alleviates the symptoms of atherosclerosis by reducing plaque area, lipid deposition, inflammation, and the HDL/LDL ratio (Martinet et al. 2002; Kiss et al. 2006; Ambrose et al. 2009; Liu et al. 2011; Sunderland et al. 2011; Shen et al. 2012; Wei et al. 2013; Xu et al. 2014).
The liver, although it has limited lipid storage, is also a site for abnormal lipid deposition in alcoholic and nonalcoholic fatty liver disease (AFLD and NAFLD, respectively). Alcohol consumption induces PARylation (Nomura et al. 2001). Logically, pharmacological PARP inhibition confers protection against steatosis, inflammation, and liver tissue injury in AFLD (Mukhopadhyay et al. 2017). While the genetic deletion of PARP2 is protective against nonalcoholic hepatic lipid accumulation (Bai et al. 2011a), there is apparent ambiguity in the literature on the role of PARP1 concerning whether the genetic ablation of PARP1 exacerbates NAFLD (Erener et al. 2012b) or pharmacological PARP inhibition protects against steatosis, inflammation, and liver tissue injury in NAFLD (Bai et al. 2011b; Gariani et al. 2017; Mukhopadhyay et al. 2017; Huang et al. 2018). The differences have not been elucidated yet.
General outline of adipogenesis
“Professional” lipid storage cells in mammals are adipocytes classified as white, brown, and beige adipocytes.
Brown or multilocular (referring to the numerous intracellular lipid droplets) adipocytes are localized to specific regions, including the interscapular and perirenal regions and lining the large arteries (Cannon and Nedergaard 2004). Brown adipocytes are characterized by high mitochondrial content and high uncoupling protein-1 (UCP1) expression (Kajimura 2015). This tissue is vital in human newborns and in rodents for maintaining core body temperature through uncoupled respiration and through that, in maintaining organismal energy balance, regulating fatty acid and glucose oxidation, and preventing or alleviating obesity and its consequences (Cannon and Nedergaard 2004).
Beige adipocytes are localized within white adipose tissue depots mixed with white adipocytes (Wu et al. 2012). Beige cells share the morphological characteristics of white adipocytes; nevertheless, beige cells respond to adrenergic stimuli by mitochondrial biogenesis, induction of UCP1 expression, fatty acid breakdown, and heat generation. Beige adipocytes are characterized by a futile creatine cycle (Kristóf et al. 2016; Bertholet et al. 2017; Kazak et al. 2017) that is not present in brown cells and is vital for heat generation. Importantly, a mutation in the fto gene was associated with impaired beige adipogenesis and, consequently, impaired mitochondrial biogenesis and organismal energy balance (Claussnitzer et al. 2015).
White adipocytes are cells specialized for fat storage. Morphologically, these cells are unilocular and when stimulated respond with triglyceride breakdown through hormone-sensitive lipase (HSL). There are multiple adipose tissue depots in the body and their metabolic behavior is quite different in terms of lipid mobilizing capacity or heat generation (Garaulet et al. 2006; Roca-Rivada et al. 2011; Sacks et al. 2013; Luche et al. 2015). The switching on of beige adipocytes in white adipose depots or the transdifferentiation of white adipocytes to brown or beige cells is termed “browning” (Kajimura 2015).
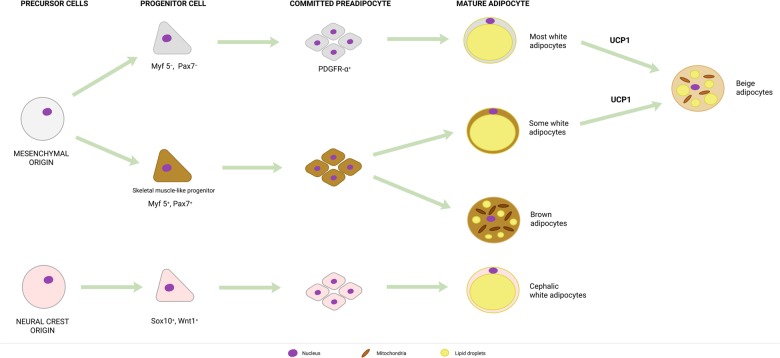
According to the classical scheme of adipocyte differentiation, Pax7+ Myf5+ brown cell precursors segregate from the dermatomyotome, while Pax7− Myf5− stem cells differentiate to white and beige adipocytes (Rosen and Spiegelman 2014). This picture is, in fact, more complex (Fig. 1). Lineage tracing studies revealed that there are multiple lineages giving rise to white adipocytes. The majority of these are of mesenchymal origin; nevertheless, depots in the head region stem from the neural crest (Sox10+, Wnt1+ precursors) (Billon et al. 2007; Sanchez-Gurmaches and Guertin 2014a). Mesenchymal precursors can be Myf5+ or Myf5−. The proportion of white adipocytes derived from Myf5+ or Myf5− precursors vary between the adipose tissue depots (Sanchez-Gurmaches and Guertin 2014a). Beige adipocytes can differentiate from the same precursors as the white adipocytes, except for neural crest-derived precursors (Sanchez-Gurmaches and Guertin 2014a). Finally, brown adipocytes differentiate from Pax7+ Myf5+ dermatomyotomal precursors (Sanchez-Gurmaches and Guertin 2014a).
Figure 1.
The general scheme of adipose tissue lineage differentiation. Abbreviations are defined in the text.
The in vitro models of (human) adipose tissue-derived stem cells (hADMSCs), (embryonic) fibroblasts, or immortalized cell lines (e.g., 3T3-L1, 3T3-F442A, etc.) (Ruiz-Ojeda et al. 2016) are useful tools in understanding transcriptional control over adipogenesis. The differentiation protocol usually involves a complete stop of proliferation by growing cells at confluency, followed by the induction of differentiation by a cocktail of hormones including insulin, a synthetic glucocorticoid, dexamethasone, and 3-isobutyl-1-methylxanthine (IBMX), a phosphodiesterase inhibitor. After the induction of differentiation, cells undergo commitment and committed cells undergo a few rounds of cellular division, called mitotic clonal expansion (Fig. 2). It is not known whether clonal expansion also characterizes the in vivo differentiation of adipocytes. After clonal expansion, cells begin accumulating lipids in lipid droplets (in vitro differentiated adipocytes are multilocular), termed terminal differentiation (Fig. 2; Ruiz-Ojeda et al. 2016; Mota de Sa et al. 2017).
Figure 2.
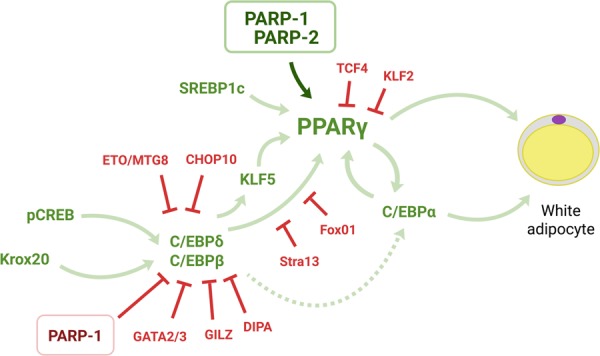
The involvement of PARP enzymes in the transcriptional control of white adipogenesis. Abbreviations are defined in the text.
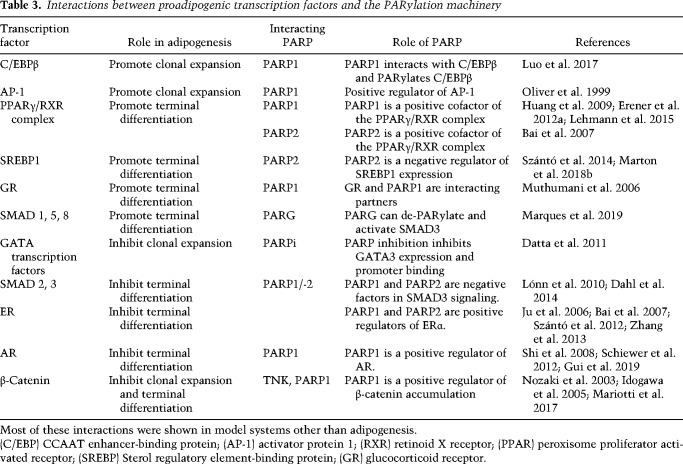
Concerted action of a large set of transcription factors is needed to guide adipogenic differentiation ( (Fig. 2; Mota de Sa et al. 2017). Adipogenic transcription factors interacting with PARPs are listed in Table 3. Classically, clonal expansion of white adipocytes was shown to be mediated by the self-amplifying activation of C/EBPδ and C/EBPβ that subsequently induces the expression of C/EBPα and, finally, the expression of peroxisome proliferator activated receptor (PPAR) γ1 and PPARγ2 expression (Fajas et al. 1998).
Table 3.
Interactions between proadipogenic transcription factors and the PARylation machinery
PPARγ1 and PPARγ2 belong to the family of nuclear receptors and are crucial in driving adipogenesis and adipocyte function through supporting the expression of major adipogenic genes (Fajas et al. 1998). PPARγ-dependent genes include lipoprotein lipase (LPL), fatty acid transporters (CD36 and aP2), TG storage proteins (e.g., perilipin), and adipokines (e.g., leptin, adiponectin) (Auwerx et al. 2003). While PPARγ1 is expressed ubiquitously, PPARγ2 expression is restricted to adipocytes and macrophages (Fajas et al. 1997; Nagy et al. 1998). Both PPARγ isoforms are lipid activated, suggesting an intricate modulation of PPARγ activity by lipid species (Nagy et al. 1998). The ligand-mediated activation of PPARγ involves the exchange of repressor cofactors (e.g., NCoR-1) to coactivator factors (e.g., p300) that facilitate chromatin relaxation and the initiation of transcription (Gelman et al. 1999; Coste et al. 2008).
The induction of the expression of PPARγ isoforms is a common denominator of beige and brown adipogenesis, similar to white adipogenesis. Mitochondrial biogenesis is a key factor for the differentiation of beige and brown adipocytes. The concerted action of the energy stress sensor web is vital for the induction of mitochondrial biogenesis, including the activation of AMPK or SIRT1 (Qiang et al. 2012; Shan et al. 2013; Wang et al. 2015; Abdul-Rahman et al. 2016; Nagy et al. 2019).
The role of PARP enzymes in adipogenesis
The first observation that PARPs modulate adipogenesis came in 1995 by Smulson et al. (1995) using the 3T3-L1 model system and 3AB, a rather unspecific PARP inhibitor. This study showed that pharmacological PARP inhibition hampers 3T3-L1 differentiation (Smulson et al. 1995). Indeed, PARPs play a role in the regulation of adipogenesis and adipose tissue function. Since this first observation, much data has emerged along with numerous controversial issues.
Early commitment and clonal expansion
PARP1, PARP2, and PARP7 have pivotal roles in decision making between retaining stem cell properties and differentiation in nonadipogenic models (Yélamos et al. 2006; Farrés et al. 2013, 2015; Nozaki et al. 2013; Roper et al. 2014; Vida et al. 2016). Therefore, PARPs may be crucial in the early commitment of cells toward preadipocytes and adipose lineages (Fig. 1). To date, no studies have been published concerning the role of PARPs in commitment to adipocyte lineages in an in vivo setting (e.g., as in Sanchez-Gurmaches and Guertin 2014b). However, PARP1 has a crucial role in preadipocyte commitment to white adipocyte differentiation in in vitro systems (Luo et al. 2017; Ryu et al. 2018).
In the in vitro differentiation of 3T3-L1 preadipocytes, a characteristic PARylation pattern was detected (Luo et al. 2017). In confluency (growth arrest), PARP1 auto-PARylation dominates cells, after which the PARylation signal is low in the clonal expansion phase and boosts again in terminal differentiation (Luo et al. 2017). In terminal differentiation, PARP1 auto-PARylation returns, nevertheless, lower molecular weight PARylation signals are also detected (Luo et al. 2017).
As noted in the previous chapter, the clonal expansion phase is dominated by the self-intensifying loop between C/EBPβ and C/EBPδ. This loop is vital for the subsequent transcription of C/EBPα and PPARγ transcription factors that then transcribe the “executors” of lipogenesis. PARP1 can PARylate C/EBPβ on K133, E135, and E139 residues, resulting in decreased binding of C/EBPβ to the promoters of C/EBPα or PPARγ2. Hence, genetic or pharmacological inactivation of PARP1 supports adipocyte differentiation (Luo et al. 2017). The deletion of these PARylation sites enhance C/EBPβ binding to target promoters and renders C/EBPβ resistant to PARP inhibitors. These findings provide a physiological explanation for reduced PARylation during the clonal expansion phase.
Another mechanism for the regulation of PARP1 activity and clonal expansion is the compartment-specific NAD+ biosynthesis through NMNAT enzymes. Ryu et al. (2018) showed that blocking nuclear NMNAT-1 induces adipocyte differentiation through limiting nuclear NAD+ for PARP1. In other words, PARP1 activation and fueling PARP1 activation by NMNAT-1 can keep preadipocytes undifferentiated. The cytosolic NMNAT-2 is induced early in adipocyte differentiation (4 h after induction) and shifts nuclear NAD+ biosynthesis to the cytosol to support glycolysis (Ryu et al. 2018). As a “side effect,” nuclear PARylation is reduced, supporting white adipocyte differentiation (Ryu et al. 2018).
Adipocyte terminal differentiation
Adipocyte terminal differentiation in in vitro models is characterized by increasing C/EBPα and PPARγ protein expression and lipid accumulation. This phase of terminal differentiation is associated with the accumulation of PARP1 and PAR formation (Erener et al. 2012a; Luo et al. 2017). In the studies of Erener et al. (2012a,b), pharmacological and genetic PARP inhibition blocked the differentiation of 3T3-L1 cells. When PARP1 was blocked in the course of 3T3-L1 differentiation, a major reduction in the expression of C/EBPα and PPARγ2 and a set of PPARγ-dependent transcripts was observed, in stark contrast to the previously discussed studies (Luo et al. 2017; Ryu et al. 2018).
Lower adipocyte differentiation was linked to a slower resolution of transcription-coupled topoisomerase II-inflicted double strand breaks and the consequent slower initiation of RNA polymerase II-mediated transcription in the absence of PARP activity (Pavri et al. 2005; Erener et al. 2012a; Lehmann et al. 2015). Furthermore, PARP inhibition supported the binding of NCoR-1 (an inhibitory cofactor of PPARγ), while decreasing the binding of p300 (an activating cofactor of PPARγ) (Lehmann et al. 2015). In a cardiomyocyte model, pharmacological, and genetic PARP1 inhibition led to increased PPARγ activity (Huang et al. 2009), in contrast to the observations detailed above.
There is apparent contradiction between the results showing that PARP1 and NAD+ biosynthesis during the commitment phase blocks (Luo et al. 2017; Ryu et al. 2018), while during terminal differentiation PARP1 supports adipocyte differentiation (Erener et al. 2012a,b; Lehmann et al. 2015). To date, no explanation is given to the discrepancies that is backed by experimental proof. Nevertheless, the visibly contradictory results may be both true. The contradictory reports do observe PARP auto-PARylation in confluent and in terminally differentiated cells (Erener et al. 2012a; Luo et al. 2017) suggesting that similar processes may take place in all cases; however, the dependence of the cells on early commitment may be different. In our hands different clones of the 3T3-L1 cells have different behavior in differentiation and response to PARP inhibitors (unpublished data).
The genetic silencing of PARP2 led to lipodystophy in chow diet-fed mice, which was mirrored when primary fibroblasts were differentiated to mature adipocytes (Bai et al. 2007). Decreased adipocytic differentiation was a result of blunted PPARγ activation. PARP2 binds to PPARγ-mediated promoters (e.g., aP2) and supports mRNA transcription. Reduced expression of the PPARγ-dependent genes in the PARP2 knockout mice points toward hampered PPARγ activation in the absence of PARP2 (Bai et al. 2007).
In the above-mentioned studies (Bai et al. 2007; Huang et al. 2009; Erener et al. 2012a,eb; Lehmann et al. 2015; Luo et al. 2017; Ryu et al. 2018), PARP inhibition or the genetic deletion of PARP1 or PARP2 modulated genes involved in fatty acid uptake (lipoprotein lipase [LPL], fatty acid binding protein 4 [FABP4, aP2], and CD36), lipid storage (perilipin), fatty acid biosynthesis (fatty acid synthase [FAS]), and adipokines (leptin, adiponectin, and resistin) in white adipocyte differentiation models. The deletion of tankyrase-1 induced leptin and adiponectin expression and secretion from white adipose tissue (Yeh et al. 2009). These genes are PPARγ-dependent and encompass all processes needed for triglyceride uptake and storage. To date, no studies have reported fatty acid release disorders in relation to the modulation of PARP1 or PARP2 activity (Bai et al. 2007; Erener et al. 2012b).
Switch between white, brown, or beige adipogenesis
PARPs may have a role in selecting between the differentiation to white, brown, and beige adipocytes. PARP1 and PARP2 were shown to modulate skeletal muscle myoblast differentiation and health (Butler and Ordahl 1999; Vyas et al. 2001; Hu et al. 2013; Chacon-Cabrera et al. 2015). Therefore, it is also likely that PARPs can influence white/brown/beige diversion. This hypothesis is further supported by the widespread interactions between energy stress sensors, mitochondrial biogenesis regulators, and PARPs (Bai et al. 2015).
The deletion of PARP1 or PARP2, as well as the pharmacological inhibition of PARP, supports mitochondrial biogenesis (Virag et al. 1998a; Bai et al. 2011a,b,2015; Szanto et al. 2011; Mohamed et al. 2014) via the preservation of cellular NAD+ pools and the subsequent activation of the SIRT1–PGC1α axis (Cantó et al. 2013; Bai et al. 2015). In agreement with this, Nagy et al. (2019) found that in vitro treatment of hADMSC cells, differentiated to white adipocytes, with olaparib induced browning of the cells, marked by mitochondrial biogenesis and UCP1 induction. In the olaparib-treated cells, beige cell markers were not induced, suggesting browning induced transdifferentiation to brown adipocytes. In good agreement with that observation, in PARP1 knockout mice, we detected more active brown adipose tissue (lower lipid deposition, induction of UCPs, increased fatty acid oxidation, and higher mitochondrial content), increased energy expenditure, and improved capacity to withstand cold exposure (Bai et al. 2011b). We detected increased cellular NAD+ content and SIRT1 activity in both models (Bai et al. 2011b; Nagy et al. 2019). Interestingly, the brown adipose tissue of the PARP2 knockout mice was not more active (Bai et al. 2011a). To date, no thorough studies were performed to assess the contribution of PARPs to beige and brown adipocyte differentiation. These findings are in agreement with the observations that better NAD+ availability (Yamaguchi et al. 2019) or SIRT1 activation supports brown and beige differentiation (Qiang et al. 2012; Khanh et al. 2018).
Lipid accumulation, obesity, insulin sensitivity
A role for PARP enzymes in obesity has been reported. In a study of monozygotic twins, higher PARP activity was found in the subcutaneous white adipose tissue of the heavier cotwin (Jukarainen et al. 2016). Furthermore, in weight loss adipocytic PARP activity is reduced, while SIRT1 activity is up-regulated (Rappou et al. 2016). In murine studies, PARP1, PARP2, and tankyrase-1 were shown to be involved in modulating energy balance and obesity. Similar to the ambiguity in the role of PARP1 in adipocyte differentiation, the studies on the organismal role of PARP1 in obesity and its consequences are also contradictory. In our studies, PARP1 knockout mice were leaner when kept on chow diet that was accentuated on high-fat feeding (Bai et al. 2011b). This study was backed by a study from another laboratory. PARP1 knockout mice had lower body weight and white adipose tissue mass when on a high-fat diet (Erener et al. 2012b). Furthermore, treatment of mice with an orally administered PARP inhibitor, MRLB-45696, (PARP1 is responsible for 80%–85% of total cellular PARP activity) (Schreiber et al. 2002; Szanto et al. 2011) prevented weight gain on a high-fat diet (Lehmann et al. 2015). In contrast to these studies, a report by Devalaraja-Narashimha and Padanilam (2010) reported a complete opposite phenotype; the PARP1 knockout mice became seriously obese as compared with their wild-type counter partners upon high-fat feeding. In all studies, a hypercaloric high-fat diet was used.
Obesity is a complex pathology and cannot be solely attributed to the dysfunction of white adipocytes; a complex deregulation of organismal energy homeostasis is involved (Rosen and Spiegelman 2014). In the above-mentioned studies that reported a lean phenotype, an energy expenditure phenotype was described due to mitochondrial biogenesis in the brown adipose tissue and the skeletal muscle, attributed mainly to the activation of the NAD+–SIRT1 axis (Bai et al. 2011b; Pirinen et al. 2014; Lehmann et al. 2015). The improved metabolic fitness yielded improved glucose tolerance and insulin sensitivity, with skeletal muscle being responsible for glucose clearance both in chow-fed and high-fat-fed mice (Bai et al. 2011b). In the monozygotic twin study, the activation of the NAD+–SIRT1 axis and the consequently lower PARP activity was associated with a leaner, metabolically healthier phenotype (Jukarainen et al. 2016). The contradictory study (Devalaraja-Narashimha and Padanilam 2010) reported an opposing rearrangement of energy homeostasis characterized by lower oxygen consumption, energy deliberation, worsened glucose clearance, and insulin resistance.
These are again opposing results without good experimental explanation. A root cause for the disagreement between the studies could be that these studies were conducted on two different knockout PARP1 mouse strains. One of the strains was generated by Wang et al. (1995) and deposited at Jackson Laboratories; the other strain was generated in the laboratory of de Murcia et al. (1997). The mice generated by Wang et al. (1995) were on an SV129 background, while the mice generated by de Murcia et al. (1997) were on a C57/Bl6J background. The metabolic behavior of the two different backgrounds is profoundly different (Andrikopoulos et al. 2005; Berglund et al. 2008) and might be the explanation for the differing results. A solution for these issues could be the use of a transgenic PARP1loxP mouse strain that will bypass developmental issues and enable the study of interorgan interactions (JAX 2019).
Induction of mitochondrial biogenesis by enhancing the NAD+–SIRT1 axis in the skeletal muscle after the genetic deletion of PARP2 brought about a lean phenotype (Bai et al. 2011a; Mohamed et al. 2014). Interestingly, the brown adipose tissue of the PARP2 knockout mice was not involved in the energy expenditure phenotype, in contrast to the PARP1 knockout mice (Bai et al. 2011a,b). In chow-fed mice, the deletion of PARP2 improved insulin sensitivity and glucose clearance. While on a high-fat diet, the ablation of PARP2 improved insulin sensitivity, but insulin secretion and glucose clearance were blunted due to inhibition of compensatory β-cell proliferation (Bai et al. 2011a).
Tankyrase expression is among the highest in the white adipose tissue and the brain (Yeh et al. 2009). White adipose tissue and energy homeostasis changes were observed in tankyrase knockout mice (Yeh et al. 2007, 2009). Interestingly, tankyrase expression may also affect brown adipose tissue (Yeh et al. 2009). However, the involvement of tankyrase in brown adipose tissue function was not investigated yet. Tankyrase knockdown was shown to impair Glut4 translocation and hence insulin-stimulated glucose uptake, resulting in down-regulation of glucose metabolism in differentiated 3T3-L1 adipocytes (Yeh et al. 2007). These effects were dependent on tankyrase activity (Yeh et al. 2007). In tankyrase knockout mice, the relative mass of the epididymal white adipose tissue decreased in parallel to enhanced energy expenditure marked by increased oxygen consumption (Yeh et al. 2009).
Future directions
PARP enzymes and PARP inhibition interfere with adipose tissue biology at multiple points. There are obesity-associated processes (e.g., inflammation) that are also PARP regulated, but their interplay had not been assessed. We give a brief overview of these processes below.
Inflammation plays diverse roles in obesity and adipose tissue homeostasis. Obesity is associated with inflammation and fibrosis of the adipose tissue (Reilly and Saltiel 2017). Preventing adipose tissue inflammation is a key step toward the “metabolically healthy” obese phenotype (Vishvanath and Gupta 2019). Furthermore, inflammatory signaling seems to be a player in diverting toward the beige lineage (Sun et al. 2018). PARP enzymes are involved in the regulation of inflammation; usually, the absence of PARP1 or PARP2 or pharmacological PARP inhibition is anti-inflammatory (Fehr et al. 2020), except for Th17-mediated processes (Kiss et al. 2019). Furthermore, increases in SIRT1 activity, which can be elicited by PARP inhibition, can suppress adipose tissue inflammation, and hence support its function (Gillum et al. 2011; Chalkiadaki and Guarente 2012). Importantly, there is evidence that the results of murine PARP inhibitor studies are likely translatable to humans (Morrow et al. 2009). PARP1 and PARP inhibition regulate IL6 (Lehmann et al. 2015), IL12m, IL13ra, SAA3, pu1, and MPEG1 (Erener et al. 2012b) expression. In the adipose tissue of PARP2 knockout mice, signs of inflammation were detected, including F4/80 positive cells and dilated capillaries, that were absent in their wild-type counter partners (Bai et al. 2007). Whether inflammatory processes are the cause or consequence of the distortion of adipose tissue function is unknown.
Recent studies showed that the loss of microbiome diversity hampers adipose tissue browning (Suárez-Zamorano et al. 2015; Li et al. 2019). Intriguingly, the genetic deletion of PARP1 enhances the diversity of the gut microbiome (Larmonier et al. 2016; Vida et al. 2018), suggesting a possible link between PARP1 and adipose tissue browning. Disruption of circadian entrainment of feeding can also contribute to obesity (Hatori et al. 2012; Zarrinpar et al. 2016; Chaix et al. 2019) and, as noted earlier, the disruption of PARP1 leads to changes in the diurnal cycle of feeding and metabolism (Asher et al. 2010; Bai et al. 2011b). PARP activation can be a go/no-go signal in cell death (Virág et al. 1998b; Fatokun et al. 2014; Dawson and Dawson 2017) and PARPs regulate cellular proliferation (Bai 2015), two vital steps to adipocyte differentiation and selection between beige, brown, or white lineages. Similarly, PARP1 and PARP10 were implicated in the regulation of autophagy and mitophagy (Muñoz-Gámez et al. 2009; Kleine et al. 2012), processes that shape adipocyte differentiation (Kim and Lee 2014). PARPs affect nuclear structure and the epigenetic code (Wacker et al. 2007; Krishnakumar et al. 2008; Hottiger 2015; Zhao et al. 2015). PARP1 deficiency was shown to modulate H3K9me3 and H3K4me3 methylation during adipogenic differentiation (Erener et al. 2012a). Nevertheless, large scale studies are missing. There are genes reported to be PARP-mediated (e.g., MDH1) (Hopp et al. 2019) that regulate adipocyte differentiation. Again, the involvement of these genes in adipogenesis in the context of PARylation had not been assessed.
All adipose tissue depots are characterized by secretion of bioactive compounds such as peptide hormones (adipokines), bioactive lipids (lipokines), and RNA molecules with local (paracrine) and systemic (endocrine) effects on multiple metabolic tissues and the cardiovascular system. These bioactive compounds are synthesized and secreted as a function of the energy status of adipose tissues, which in turn regulates appetite, thermogenesis, glucose, and lipid metabolism (Scheja and Heeren 2019). The role of PARPs had not been studied in this direction. Along the same lines, large-scale endocrine studies are also missing.
The role of PARPs in adipogenesis and metabolism will clearly have practical applications not only in the strict sense of metabolism and metabolic diseases, but also from the perspective of cancer and cancer cachexia (Chacon-Cabrera et al. 2015, 2017; Barreiro and Gea 2018; Doles et al. 2018). These outstanding issues warrant further studies in the future.
Acknowledgments
We are grateful to Dr. Karen Uray (University of Debrecen) for the critical revision of the manuscript.
Our work is supported by grants from the Nemzeti Kutatási, Fejlesztési és Innovációs Hivatal (NKFIH) (K123975, PD121138, and GINOP-2.3.2-15-2016-00006) and the Hungarian Academy of Sciences (NKM-26/2019). This study was financed by the Higher Education Institutional Excellence Program (NKFIH-1150-6/2019) of the Ministry of Innovation and Technology in Hungary, within the framework of the biotechnology thematic program of the University of Debrecen.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.334284.119.
Freely available online through the Genes & Development Open Access option.
References
- Abdul-Rahman O, Kristóf E, Doan-Xuan QM, Vida A, Nagy L, Horváth A, Simon J, Maros T, Szentkirályi I, Palotás L, et al. 2016. AMP-activated kinase (AMPK) activation by AICAR in human white adipocytes derived from pericardial white adipose tissue stem cells induces a partial beige-like phenotype. PLoS One 11: e0157644 10.1371/journal.pone.0157644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abplanalp J, Hopp AK, Hottiger MO. 2018. Mono-ADP-ribosylhydrolase assays. Methods Mol Biol 1813: 205–213. 10.1007/978-1-4939-8588-3_13 [DOI] [PubMed] [Google Scholar]
- Ambrose HE, Willimott S, Beswick RW, Dantzer F, de Murcia JM, Yelamos J, Wagner SD. 2009. Poly(ADP-ribose) polymerase-1 (Parp-1)-deficient mice demonstrate abnormal antibody responses. Immunology 127: 178–186. 10.1111/j.1365-2567.2008.02921.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amé JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Höger T, Menissier-de Murcia J, de Murcia G. 1999. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 274: 17860–17868. 10.1074/jbc.274.25.17860 [DOI] [PubMed] [Google Scholar]
- Amé JC, Spenlehauer C, de Murcia G. 2004. The PARP superfamily. Bioessays 26: 882–893. 10.1002/bies.20085 [DOI] [PubMed] [Google Scholar]
- Amin O, Beauchamp MC, Nader PA, Laskov I, Iqbal S, Philip CA, Yasmeen A, Gotlieb WH. 2015. Suppression of homologous recombination by insulin-like growth factor-1 inhibition sensitizes cancer cells to PARP inhibitors. BMC Cancer 15: 817 10.1186/s12885-015-1803-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppagounder SS, Gagne JP, Poirier GG, Dawson VL, Dawson TM. 2014. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci 111: 10209–10214. 10.1073/pnas.1405158111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrikopoulos S, Massa CM, Aston-Mourney K, Funkat A, Fam BC, Hull RL, Kahn SE, Proietto J. 2005. Differential effect of inbred mouse strain (C57BL/6, DBA/2, 129T2) on insulin secretory function in response to a high fat diet. J Endocrinol 187: 45–53. 10.1677/joe.1.06333 [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. 2010. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142: 943–953. 10.1016/j.cell.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Auwerx J, Cock TA, Knouff C. 2003. PPAR-γ: a thrifty transcription factor. Nucl Recept Signal 1: e006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P. 2015. Biology of poly(ADP-ribose) polymerases: the factotums of cell maintenance. Mol Cell 58: 947–958. 10.1016/j.molcel.2015.01.034 [DOI] [PubMed] [Google Scholar]
- Bai P, Cantó C. 2012. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab 16: 290–295. 10.1016/j.cmet.2012.06.016 [DOI] [PubMed] [Google Scholar]
- Bai P, Csóka B. 2015. New route for the activation of poly(ADP-ribose) polymerase-1: a passage that links poly(ADP-ribose) polymerase-1 to lipotoxicity? Biochem J 469: e9–e11. 10.1042/BJ20150598 [DOI] [PubMed] [Google Scholar]
- Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. 2007. Poly(ADP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-γ heterodimer. J Biol Chem 282: 37738–37746. 10.1074/jbc.M701021200 [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Brunyánszki A, Huber A, Szántó M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, et al. 2011a. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab 13: 450–460. 10.1016/j.cmet.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Cantó C, Oudart H, Brunyánszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. 2011b. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13: 461–468. 10.1016/j.cmet.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Nagy L, Fodor T, Liaudet L, Pacher P. 2015. Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol Metab 26: 75–83. 10.1016/j.tem.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G. 2013. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J 280: 3491–3507. 10.1111/febs.12358 [DOI] [PubMed] [Google Scholar]
- Barreiro E, Gea J. 2018. PARP-1 and PARP-2 activity in cancer-induced cachexia: potential therapeutic implications. Biol Chem 399: 179–186. 10.1515/hsz-2017-0158 [DOI] [PubMed] [Google Scholar]
- Berger F, Lau C, Ziegler M. 2007. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc Natl Acad Sci 104: 3765–3770. 10.1073/pnas.0609211104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund ED, Li CY, Poffenberger G, Ayala JE, Fueger PT, Willis SE, Jewell MM, Powers AC, Wasserman DH. 2008. Glucose metabolism in vivo in four commonly used inbred mouse strains. Diabetes 57: 1790–1799. 10.2337/db07-1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholet AM, Kazak L, Chouchani ET, Bogaczynska MG, Paranjpe I, Wainwright GL, Bétourne A, Kajimura S, Spiegelman BM, Kirichok Y. 2017. Mitochondrial patch clamp of beige adipocytes reveals UCP1-positive and UCP1-negative cells both exhibiting futile creatine cycling. Cell Metab 25: 811–822.e4. 10.1016/j.cmet.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi AR, Ferreri C, Ruggiero S, Deplano S, Sunda V, Galloro G, Formisano C, Mennella MR. 2016. Automodification of PARP and fatty acid-based membrane lipidome as a promising integrated biomarker panel in molecular medicine. Biomark Med 10: 229–242. 10.2217/bmm.16.3 [DOI] [PubMed] [Google Scholar]
- Billon N, Iannarelli P, Monteiro MC, Glavieux-Pardanaud C, Richardson WD, Kessaris N, Dani C, Dupin E. 2007. The generation of adipocytes by the neural crest. Development 134: 2283–2292. 10.1242/dev.002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindesboll C, Tan S, Bott D, Cho T, Tamblyn L, MacPherson L, Gronning-Wang L, Nebb HI, Matthews J. 2016. TCDD-inducible poly-ADP-ribose polymerase (TIPARP/PARP7) mono-ADP-ribosylates and co-activates liver X receptors. Biochem J 473: 899–910. 10.1042/BJ20151077 [DOI] [PubMed] [Google Scholar]
- Bodnár E, Bakondi E, Kovács K, Hegedűs C, Lakatos P, Robaszkiewicz A, Regdon Z, Virág L, Szabó E. 2018. Redox profiling reveals clear differences between molecular patterns of wound fluids from acute and chronic wounds. Oxid Med Cell Longev 2018: 1–12. 10.1155/2018/5286785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart V, Blaeser K, Kolb H. 1999. Potent β-cell protection in vitro by an isoquinolinone-derived PARP inhibitor. Horm Metab Res 31: 641–644. 10.1055/s-2007-978813 [DOI] [PubMed] [Google Scholar]
- Burkle A, Virag L. 2013. Poly(ADP-ribose): PARadigms and PARadoxes. Mol Aspects Med 2: 00157–00154. [DOI] [PubMed] [Google Scholar]
- Butler AJ, Ordahl CP. 1999. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol 19: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. 2004. Brown adipose tissue: function and physiological significance. Physiol Rev 84: 277–359. 10.1152/physrev.00015.2003 [DOI] [PubMed] [Google Scholar]
- Cantó C, Sauve A, Bai P. 2013. Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes. Mol Asp Med 34: 1168–1201. 10.1016/j.mam.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedernaes J, Waldeck N, Bass J. 2019. Neurogenetic basis for circadian regulation of metabolism by the hypothalamus. Genes Dev 33: 1136–1158. 10.1101/gad.328633.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon-Cabrera A, Fermoselle C, Salmela I, Yelamos J, Barreiro E. 2015. MicroRNA expression and protein acetylation pattern in respiratory and limb muscles of Parp-1−/− and Parp-2−/− mice with lung cancer cachexia. Biochim Biophys Acta 1850: 2530–2543. 10.1016/j.bbagen.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Chacon-Cabrera A, Mateu-Jimenez M, Langohr K, Fermoselle C, García-Arumi E, Andreu AL, Yelamos J, Barreiro E. 2017. Role of PARP activity in lung cancer-induced cachexia: effects on muscle oxidative stress, proteolysis, anabolic markers, and phenotype. J Cell Physiol 232: 3744–3761. 10.1002/jcp.25851 [DOI] [PubMed] [Google Scholar]
- Chaix A, Lin T, Le HD, Chang MW, Panda S. 2019. Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab 29: 303–319.e4. 10.1016/j.cmet.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A, Guarente L. 2012. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab 16: 180–188. 10.1016/j.cmet.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P, Weill JD, Mandel P. 1963. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun 11: 39–43. [DOI] [PubMed] [Google Scholar]
- Chapman JD, Gagné JP, Poirier GG, Goodlett DR. 2013. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J Proteome Res 12: 1868–1880. 10.1021/pr301219h [DOI] [PubMed] [Google Scholar]
- Chen ST, Lin CC, Liu YS, Lin C, Hung PT, Jao CW, Lin PH. 2013. Airborne particulate collected from central Taiwan induces DNA strand breaks, Poly(ADP-ribose) polymerase-1 activation, and estrogen-disrupting activity in human breast carcinoma cell lines. J Environ Sci Health A Tox Hazard Subst Environ Eng 48: 173–181. 10.1080/10934529.2012.717809 [DOI] [PubMed] [Google Scholar]
- Chen Y, Bang S, Park S, Shi H, Kim SF. 2015. Acyl-CoA-binding domain containing 3 modulates NAD+ metabolism through activating poly(ADP-ribose) polymerase 1. Biochem J 469: 189–198. 10.1042/BJ20141487 [DOI] [PubMed] [Google Scholar]
- Chiarugi A, Dolle C, Felici R, Ziegler M. 2012. The NAD metabolome - a key determinant of cancer cell biology. Nat Rev Cancer 2012: 13. [DOI] [PubMed] [Google Scholar]
- Cho SH, Ahn AK, Bhargava P, Lee CH, Eischen CM, McGuinness O, Boothby M. 2011. Glycolytic rate and lymphomagenesis depend on PARP14, an ADP ribosyltransferase of the B aggressive lymphoma (BAL) family. Proc Natl Acad Sci 108: 15972–15977. 10.1073/pnas.1017082108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Abdelmegeed MA, Song BJ. 2017. Diet high in fructose promotes liver steatosis and hepatocyte apoptosis in C57BL/6J female mice: role of disturbed lipid homeostasis and increased oxidative stress. Food Chem Toxicol 103: 111–121. 10.1016/j.fct.2017.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. 2015. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med 373: 895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS. 2020. Interplay between compartmentalized NAD+ synthesis and consumption: a focus on the PARP family. Genes Dev (this issue) 10.1101/gad.335109.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O'Malley BW, Auwerx J. 2008. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1α. Proc Natl Acad Sci 105: 17187–17192. 10.1073/pnas.0808207105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin N, Szabo C. 2013. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol Aspects Med 6: 1043–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl M, Maturi V, Lönn P, Papoutsoglou P, Zieba A, Vanlandewijck M, van der Heide LP, Watanabe Y, Söderberg O, Hottiger MO, et al. 2014. Fine-tuning of Smad protein function by poly(ADP-ribose) polymerases and poly(ADP-ribose) glycohydrolase during transforming growth factor β signaling. PLoS One 9: e103651 10.1371/journal.pone.0103651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer F, Santoro R. 2013. The expanding role of PARPs in the establishment and maintenance of heterochromatin. FEBS J 280: 3508–3518. 10.1111/febs.12368 [DOI] [PubMed] [Google Scholar]
- Datta R, Naura AS, Zerfaoui M, Errami Y, Oumouna M, Kim H, Ju J, Ronchi VP, Haas AL, Boulares AH. 2011. PARP-1 deficiency blocks IL-5 expression through calpain-dependent degradation of STAT-6 in a murine asthma model. Allergy 66: 853–861. 10.1111/j.1398-9995.2011.02549.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. 2017. Mitochondrial mechanisms of neuronal cell death: potential therapeutics. Annu Rev Pharmacol Toxicol 57: 437–454. 10.1146/annurev-pharmtox-010716-105001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. 1997. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci 94: 7303–7307. 10.1073/pnas.94.14.7303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalaraja-Narashimha K, Padanilam BJ. 2009. PARP-1 inhibits glycolysis in ischemic kidneys. J Am Soc Nephrol 20: 95–103. 10.1681/ASN.2008030325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalaraja-Narashimha K, Padanilam BJ. 2010. PARP1 deficiency exacerbates diet-induced obesity in mice. J Endocrinol 205: 243–252. 10.1677/JOE-09-0402 [DOI] [PubMed] [Google Scholar]
- De Vos M, Schreiber V, Dantzer F. 2012. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol 84: 137–146. 10.1016/j.bcp.2012.03.018 [DOI] [PubMed] [Google Scholar]
- Diestel A, Aktas O, Hackel D, Häke I, Meier S, Raine CS, Nitsch R, Zipp F, Ullrich O. 2003. Activation of microglial poly(ADP-ribose)-polymerase-1 by cholesterol breakdown products during neuroinflammation: a link between demyelination and neuronal damage. J Exp Med 198: 1729–1740. 10.1084/jem.20030975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doles JD, Hogan KA, O'Connor J, Wahner Hendrickson AE, Huston O, Jatoi A. 2018. Does the Poly (ADP-Ribose) polymerase inhibitor veliparib merit further study for cancer-associated weight loss? Observations and conclusions from 60 prospectively treated patients. J Palliat Med 21: 1334–1338. 10.1089/jpm.2018.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Matsumura T, Edelstein D, Rossetti L, Zsengeller Z, Szabo C, Brownlee M. 2003. Inhibition of GAPDH activity by poly(ADP-ribose) polymerase activates three major pathways of hyperglycemic damage in endothelial cells. J Clin Invest 112: 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hamoly T, Hegedus C, Lakatos P, Kovacs K, Bai P, El-Ghazaly MA, El-Denshary ES, Szabo E, Virag L. 2014. Activation of poly(ADP-ribose) polymerase-1 delays wound healing by regulating keratinocyte migration and production of inflammatory mediators. Mol Med 8: 00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Hesse M, Kostadinova R, Hottiger MO. 2012a. Poly(ADP-Ribose)Polymerase-1 (PARP1) controls adipogenic gene expression and adipocyte function. Mol Endocrinol 26: 79–86. 10.1210/me.2011-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Mirsaidi A, Hesse M, Tiaden AN, Ellingsgaard H, Kostadinova R, Donath MY, Richards PJ, Hottiger MO. 2012b. ARTD1 deletion causes increased hepatic lipid accumulation in mice fed a high-fat diet and impairs adipocyte function and differentiation. FASEB J 26: 2631–2638. 10.1096/fj.11-200212 [DOI] [PubMed] [Google Scholar]
- Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, et al. 1997. The organization, promoter analysis, and expression of the human PPARγ gene. J Biol Chem 272: 18779–18789. [DOI] [PubMed] [Google Scholar]
- Fajas L, Fruchart JC, Auwerx J. 1998. Transcriptional control of adipogenesis. Curr Opin Cell Biol 10: 165–173. [DOI] [PubMed] [Google Scholar]
- Faraone-Mennella MR, Masi A, Ferreri C. 2019. Regulatory roles of PARP-1 and lipids in epigenetic mechanisms. In Handbook of Nutrition, Diet, and Epigenetics (ed. Patel VB, Preedy VR), pp. 369–386. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- Farrés J, Martín-Caballero J, Martínez C, Lozano JJ, Llacuna L, Ampurdanés C, Ruiz-Herguido C, Dantzer F, Schreiber V, Villunger A, et al. 2013. PARP-2 is required to maintain hematopoiesis following sublethal γ-irradiation in mice. Blood 122: 44–54. 10.1182/blood-2012-12-472845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrés J, Llacuna L, Martín-Caballero J, Martinez C, Lozano JJ, Ampurdanés C, López-Contreras AJ, Florensa L, Navarro J, Ottina E, et al. 2015. PARP-2 sustains erythropoiesis in mice by limiting replicative stress in erythroid progenitors. Cell Death Differ 22: 1144–1157. 10.1038/cdd.2014.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatokun AA, Dawson VL, Dawson TM. 2014. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol 171: 2000–2016. 10.1111/bph.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A, Singh SA, Kerr CM, Mukai S, Higashi H, Aikawa M. 2020. The impact of PARPs and ADP-ribosylation on inflammation and host–pathogen interactions. Genes Dev (this issue) 10.1101/gad.334425.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijs KL, Forst AH, Verheugd P, Lüscher B. 2013. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat Rev Mol Cell Biol 14: 443–451. 10.1038/nrm3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, Wheeler D, Redpath P, Li J, Romero G, et al. 2014. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD depletion. Cell Rep 10: 00712–00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné JP, Moreel X, Gagné P, Labelle Y, Droit A, Chevalier-Paré M, Bourassa S, McDonald D, Hendzel MJ, Prigent C, et al. 2009. Proteomic investigation of phosphorylation sites in poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase. J Proteome Res 8: 1014–1029. 10.1021/pr800810n [DOI] [PubMed] [Google Scholar]
- Garaulet M, Hernandez-Morante JJ, Lujan J, Tebar FJ, Zamora S. 2006. Relationship between fat cell size and number and fatty acid composition in adipose tissue from different fat depots in overweight/obese humans. Int J Obes (Lond) 30: 899–905. 10.1038/sj.ijo.0803219 [DOI] [PubMed] [Google Scholar]
- Gariani K, Ryu D, Menzies KJ, Yi HS, Stein S, Zhang H, Perino A, Lemos V, Katsyuba E, Jha P, et al. 2017. Inhibiting poly ADP-ribosylation increases fatty acid oxidation and protects against fatty liver disease. J Hepatol 66: 132–141. 10.1016/j.jhep.2016.08.024 [DOI] [PubMed] [Google Scholar]
- Gelman L, Zhou G, Fajas L, Raspé E, Fruchart JC, Auwerx J. 1999. p300 interacts with the N- and C-terminal part of PPARγ2 in a ligand-independent and -dependent manner, respectively. J Biol Chem 274: 7681–7688. 10.1074/jbc.274.12.7681 [DOI] [PubMed] [Google Scholar]
- Ghabreau L, Roux JP, Frappart PO, Mathevet P, Patricot LM, Mokni M, Korbi S, Wang ZQ, Tong WM, Frappart L. 2004. Poly(ADP-ribose) polymerase-1, a novel partner of progesterone receptors in endometrial cancer and its precursors. Int J Cancer 109: 317–321. 10.1002/ijc.11731 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411–424. 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, Schwede F, Yu Y, Kraus WL. 2016. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353: 45–50. 10.1126/science.aaf7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum MP, Kotas ME, Erion DM, Kursawe R, Chatterjee P, Nead KT, Muise ES, Hsiao JJ, Frederick DW, Yonemitsu S, et al. 2011. SirT1 regulates adipose tissue inflammation. Diabetes 60: 3235–3245. 10.2337/db11-0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, Menissier de Murcia JM, Molinete M, Simonin F, Koken M, Hoeijmakers JH, de Murcia G. 1990. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc Natl Acad Sci 87: 2990–2994. 10.1073/pnas.87.8.2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero G, Brundo MV, Labar S, Bianchi AR, Trocchia S, Rabbito D, Palumbo G, Abdel-Gawad FK, De Maio A. 2018. Frog (Pelophylax bergeri, Gunther 1986) endocrine disruption assessment: characterization and role of skin poly(ADP-ribose) polymerases. Environ Sci Pollut Res Int 25: 18303–18313. 10.1007/s11356-017-0395-2 [DOI] [PubMed] [Google Scholar]
- Gui B, Gui F, Takai T, Feng C, Bai X, Fazli L, Dong X, Liu S, Zhang X, Zhang W, et al. 2019. Selective targeting of PARP-2 inhibits androgen receptor signaling and prostate cancer growth through disruption of FOXA1 function. Proc Natl Acad Sci 116: 14573–14582. 10.1073/pnas.1908547116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans CP, Zerfaoui M, Naura AS, Catling A, Boulares AH. 2008. Differential effects of PARP inhibition on vascular cell survival and ACAT-1 expression favouring atherosclerotic plaque stability. Cardiovasc Res 78: 429–439. 10.1093/cvr/cvn018 [DOI] [PubMed] [Google Scholar]
- Hans CP, Feng Y, Naura AS, Zerfaoui M, Rezk BM, Xia H, Kaye AD, Matrougui K, Lazartigues E, Boulares AH. 2009a. Protective effects of PARP-1 knockout on dyslipidemia-induced autonomic and vascular dysfunction in ApoE mice: effects on eNOS and oxidative stress. PLoS One 4: e7430 10.1371/journal.pone.0007430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans CP, Zerfaoui M, Naura AS, Troxclair D, Strong JP, Matrougui K, Boulares AH. 2009b. Thieno[2,3-c]isoquinolin-5-one, a potent poly(ADP-ribose) polymerase inhibitor, promotes atherosclerotic plaque regression in high-fat diet-fed apolipoprotein E-deficient mice: effects on inflammatory markers and lipid content. J Pharmacol Exp Ther 329: 150–158. 10.1124/jpet.108.145938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. 2012. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15: 848–860. 10.1016/j.cmet.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holechek J, Lease R, Thorsell AG, Karlberg T, McCadden C, Grant R, Keen A, Callahan E, Schüler H, Ferraris D. 2018. Design, synthesis and evaluation of potent and selective inhibitors of mono-(ADP-ribosyl)transferases PARP10 and PARP14. Bioorg Med Chem Lett 28: 2050–2054. 10.1016/j.bmcl.2018.04.056 [DOI] [PubMed] [Google Scholar]
- Hopp AK, Grüter P, Hottiger MO. 2019. Regulation of glucose metabolism by NAD+ and ADP-ribosylation. Cells 8: 890 10.3390/cells8080890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath EM, Benkő R, Gerő D, Kiss L, Szabó C. 2008. Treatment with insulin inhibits poly(ADP-ribose)polymerase activation in a rat model of endotoxemia. Life Sci 82: 205–209. 10.1016/j.lfs.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO. 2015. Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu Rev Biochem 84: 227–263. 10.1146/annurev-biochem-060614-034506 [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. 2010. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35: 208–219. 10.1016/j.tibs.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. 2010. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31: 194–223. 10.1210/er.2009-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wu Z, Hergert P, Henke CA, Bitterman PB, Phan SH. 2013. Regulation of myofibroblast differentiation by poly(ADP-ribose) polymerase 1. Am J Pathol 182: 71–83. 10.1016/j.ajpath.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Yang C, Wang Y, Liao Y, Huang K. 2009. PARP-1 suppresses adiponectin expression through poly(ADP-ribosyl)ation of PPARγ in cardiac fibroblasts. Cardiovasc Res 81: 98–107. 10.1093/cvr/cvn264 [DOI] [PubMed] [Google Scholar]
- Huang K, Du M, Tan X, Yang L, Li X, Jiang Y, Wang C, Zhang F, Zhu F, Cheng M, et al. 2017. PARP1-mediated PPARα poly(ADP-ribosyl)ation suppresses fatty acid oxidation in non-alcoholic fatty liver disease. J Hepatol 66: 962–977. 10.1016/j.jhep.2016.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhang B, Chen Y, Liu H, Liu Y, Li X, Bao Z, Song Z, Wang Z. 2018. Poly(ADP-ribose) polymerase inhibitor PJ34 attenuated hepatic triglyceride accumulation in alcoholic fatty liver disease in mice. J Pharmacol Exp Ther 364: 452–461. 10.1124/jpet.117.243105 [DOI] [PubMed] [Google Scholar]
- Huang W, Su L, Zhang X, Xu X, Li R. 2019. Endocrinological characterization of pancreatic ducts in HFD and HGD fed mice. J Cell Biochem 120: 16153–16159. 10.1002/jcb.28896 [DOI] [PubMed] [Google Scholar]
- Hutin D, Tamblyn L, Gomez A, Grimaldi G, Soedling H, Cho T, Ahmed S, Lucas C, Kanduri C, Grant DM, et al. 2018. Hepatocyte-specific deletion of TIPARP, a negative regulator of the aryl hydrocarbon receptor, is sufficient to increase sensitivity to dioxin-induced wasting syndrome. Toxicol Sci 165: 347–360. 10.1093/toxsci/kfy136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idogawa M, Yamada T, Honda K, Sato S, Imai K, Hirohashi S. 2005. Poly(ADP-ribose) polymerase-1 is a component of the oncogenic T-cell factor-4/β-catenin complex. Gastroenterology 128: 1919–1936. 10.1053/j.gastro.2005.03.007 [DOI] [PubMed] [Google Scholar]
- Javle M, Curtin NJ. 2011. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer 105: 1114–1122. 10.1038/bjc.2011.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAX. 2019. B6(Cg)-Parp1tm1c(EUCOMM)Hmgu/WlkrJ mice. The Jackson Laboratory, https://www.jax.org/strain/032650. [Google Scholar]
- Jog NR, Caricchio R. 2013. Differential regulation of cell death programs in males and females by Poly (ADP-Ribose) Polymerase-1 and 17β estradiol. Cell Death Dis 4: e758 10.1038/cddis.2013.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Mahfooz S, Maurya VK, Kumar V, Basanna CS, Kaur G, Hanif K, Jha RK. 2014. PARP1 during embryo implantation and its upregulation by oestradiol in mice. Reproduction 147: 765–780. 10.1530/REP-13-0588 [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. 2006. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312: 1798–1802. 10.1126/science.1127196 [DOI] [PubMed] [Google Scholar]
- Jukarainen S, Heinonen S, Rämö JT, Rinnankoski-Tuikka R, Rappou E, Tummers M, Muniandy M, Hakkarainen A, Lundbom J, Lundbom N, et al. 2016. Obesity is associated with low NAD+/SIRT pathway expression in adipose tissue of BMI-discordant monozygotic twins. J Clin Endocrinol Metab 101: 275–283. 10.1210/jc.2015-3095 [DOI] [PubMed] [Google Scholar]
- Kajimura S. 2015. Promoting brown and beige adipocyte biogenesis through the PRDM16 pathway. Int J Obes Suppl 5: S11–S14. 10.1038/ijosup.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg T, Langelier MF, Pascal JM, Schüler H. 2013. Structural biology of the writers, readers, and erasers in mono- and poly(ADP-ribose) mediated signaling. Mol Aspects Med 34: 1088–1108. 10.1016/j.mam.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaichi M, Oka J, Zhang J, Ueda K, Hayaishi O. 1983. Properties of poly(ADP-ribose) synthetase and ADP-ribosyl histone splitting enzyme. Princess Takamatsu Symp 13: 121–128. [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Lu GZ, Jedrychowski MP, Bare CJ, Mina AI, Kumari M, Zhang S, Vuckovic I, Laznik-Bogoslavski D, et al. 2017. Genetic depletion of adipocyte creatine metabolism inhibits diet-induced thermogenesis and drives obesity. Cell Metab 26: 660–671.e3. 10.1016/j.cmet.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner NM, Mayo SA, Hua J, Lee C, Moore DD, Fu L. 2015. Circadian dysfunction induces leptin resistance in mice. Cell Metab 22: 448–459. 10.1016/j.cmet.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanh VC, Zulkifli AF, Tokunaga C, Yamashita T, Hiramatsu Y, Ohneda O. 2018. Aging impairs beige adipocyte differentiation of mesenchymal stem cells via the reduced expression of Sirtuin 1. Biochem Biophys Res Commun 500: 682–690. 10.1016/j.bbrc.2018.04.136 [DOI] [PubMed] [Google Scholar]
- Kim KH, Lee MS. 2014. Autophagy as a crosstalk mediator of metabolic organs in regulation of energy metabolism. Rev Endocr Metab Disord 15: 11–20. 10.1007/s11154-013-9272-6 [DOI] [PubMed] [Google Scholar]
- Kiss L, Chen M, Gero D, Modis K, Lacza Z, Szabo C. 2006. Effects of 7-ketocholesterol on the activity of endothelial poly(ADP-ribose) polymerase and on endothelium-dependent relaxant function. Int J Mol Med 18: 1113–1117. [PubMed] [Google Scholar]
- Kiss B, Szántó M, Szklenár M, Brunyánszki A, Marosvölgyi T, Sárosi E, Remenyik E, Gergely P, Virág L, Decsi T, et al. 2015. Poly(ADP) ribose polymerase-1 ablation alters eicosanoid and docosanoid signaling and metabolism in a murine model of contact hypersensitivity. Mol Med Rep 11: 2861–2867. 10.3892/mmr.2014.3044 [DOI] [PubMed] [Google Scholar]
- Kiss B, Szántó M, Hegedűs C, Antal D, Szödényi A, Márton J, Méhes G, Virág L, Szegedi A, Bai P. 2019. Poly(ADP-ribose) polymerase-1 depletion enhances the severity of inflammation in an imiquimod-induced model of psoriasis. Exp Dermatol 10.1111/exd.14061. [DOI] [PubMed] [Google Scholar]
- Kleine H, Herrmann A, Lamark T, Forst AH, Verheugd P, Lüscher-Firzlaff J, Lippok B, Feijs KL, Herzog N, Kremmer E, et al. 2012. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun Signal 10: 28 10.1186/1478-811X-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc A, Aydin Sayitoglu M, Karakurt F, Batar B, Niyazoglu M, Celik O, Onaran I, Tasan E, Sultuybek GK. 2014. Association of three SNPs in the PARP-1 gene with Hashimoto's thyroiditis. Hum Genome Var 1: 14016 10.1038/hgv.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komjati K, Mabley JG, Virag L, Southan GJ, Salzman AL, Szabo C. 2004. Poly(ADP-ribose) polymerase inhibition protect neurons and the white matter and regulates the translocation of apoptosis-inducing factor in stroke. Int J Mol Med 13: 373–382. [PubMed] [Google Scholar]
- Kraus WL, Hottiger MO. 2013. PARP-1 and gene regulation: progress and puzzles. Mol Aspects Med 34: 1109–1123. 10.1016/j.mam.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319: 819–821. 10.1126/science.1149250 [DOI] [PubMed] [Google Scholar]
- Kristóf E, Doan-Xuan QM, Sárvari AK, Klusóczki A, Fischer-Posovszky P, Wabitsch M, Bacso Z, Bai P, Balajthy Z, Fésus L. 2016. Clozapine modifies the differentiation program of human adipocytes inducing browning. Transl Psychiatry 6: e963 10.1038/tp.2016.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutuzov MM, Khodyreva SN, Amé JC, Ilina ES, Sukhanova MV, Schreiber V, Lavrik OI. 2013. Interaction of PARP-2 with DNA structures mimicking DNA repair intermediates and consequences on activity of base excision repair proteins. Biochimie 95: 1208–1215. 10.1016/j.biochi.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Kutuzov MM, Khodyreva SN, Ilina ES, Sukhanova MV, Amé JC, Lavrik OI. 2015. Interaction of PARP-2 with AP site containing DNA. Biochimie 112: 10–19. 10.1016/j.biochi.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Larmonier CB, Shehab KW, Laubitz D, Jamwal DR, Ghishan FK, Kiela PR. 2016. Transcriptional reprogramming and resistance to colonic mucosal injury in poly(ADP-ribose) polymerase 1 (PARP1)-deficient mice. J Biol Chem 291: 8918–8930. 10.1074/jbc.M116.714386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léger K, Bär D, Savić N, Santoro R, Hottiger MO. 2014. ARTD2 activity is stimulated by RNA. Nucleic Acids Res 42: 5072–5082. 10.1093/nar/gku131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Pirinen E, Mirsaidi A, Kunze FA, Richards PJ, Auwerx J, Hottiger MO. 2015. ARTD1-induced poly-ADP-ribose formation enhances PPARγ ligand binding and co-factor exchange. Nucleic Acids Res 43: 129–142. 10.1093/nar/gku1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie Pedrioli DM, Leutert M, Bilan V, Nowak K, Gunasekera K, Ferrari E, Imhof R, Malmstrom L, Hottiger MO. 2018. Comprehensive ADP-ribosylome analysis identifies tyrosine as an ADP-ribose acceptor site. EMBO Rep 19: e45310 10.15252/embr.201745310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li L, Li M, Lam SM, Wang G, Wu Y, Zhang H, Niu C, Zhang X, Liu X, et al. 2019. Microbiota depletion impairs thermogenesis of brown adipose tissue and browning of white adipose tissue. Cell Rep 26: 2720–2737.e5. 10.1016/j.celrep.2019.02.015 [DOI] [PubMed] [Google Scholar]
- Liu FQ, Zhang XL, Gong L, Wang XP, Wang J, Hou XG, Sun Y, Qin WD, Wei SJ, Zhang Y, et al. 2011. Glucagon-like peptide 1 protects microvascular endothelial cells by inactivating the PARP-1/iNOS/NO pathway. Mol Cell Endocrinol 339: 25–33. 10.1016/j.mce.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Lönn P, van der Heide LP, Dahl M, Hellman U, Heldin CH, Moustakas A. 2010. PARP-1 attenuates Smad-mediated transcription. Mol Cell 40: 521–532. 10.1016/j.molcel.2010.10.029 [DOI] [PubMed] [Google Scholar]
- Luche E, Sengenès C, Arnaud E, Laharrague P, Casteilla L, Cousin B. 2015. Differential hematopoietic activity in white adipose tissue depending on its localization. J Cell Physiol 230: 3076–3083. 10.1002/jcp.25045 [DOI] [PubMed] [Google Scholar]
- Luo X, Ryu KW, Kim DS, Nandu T, Medina CJ, Gupte R, Gibson BA, Soccio RE, Yu Y, Gupta RK, et al. 2017. PARP-1 controls the adipogenic transcriptional program by PARylating C/EBPβ and modulating its transcriptional activity. Mol Cell 65: 260–271. 10.1016/j.molcel.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabley JG, Horváth EM, Murthy KG, Zsengellér Z, Vaslin A, Benkő R, Kollai M, Szabó C. 2005. Gender differences in the endotoxin-induced inflammatory and vascular responses: potential role of poly(ADP-ribose) polymerase activation. J Pharmacol Exp Ther 315: 812–820. 10.1124/jpet.105.090480 [DOI] [PubMed] [Google Scholar]
- Mangerich A, Herbach N, Hanf B, Fischbach A, Popp O, Moreno-Villanueva M, Bruns OT, Bürkle A. 2010. Inflammatory and age-related pathologies in mice with ectopic expression of human PARP-1. Mech Ageing Dev 131: 389–404. 10.1016/j.mad.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Manunza A, Casellas J, Quintanilla R, González-Prendes R, Pena RN, Tibau J, Mercade A, Castelló A, Aznárez N, Hernández-Sánchez J, et al. 2014. A genome-wide association analysis for porcine serum lipid traits reveals the existence of age-specific genetic determinants. BMC Genomics 15: 758 10.1186/1471-2164-15-758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti L, Pollock K, Guettler S. 2017. Regulation of Wnt/β-catenin signalling by tankyrase-dependent poly(ADP-ribosyl)ation and scaffolding. Br J Pharmacol 174: 4611–4636. 10.1111/bph.14038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques M, Jangal M, Wang LC, Kazanets A, da Silva SD, Zhao T, Lovato A, Yu H, Jie S, Del Rincon S, et al. 2019. Oncogenic activity of poly (ADP-ribose) glycohydrolase. Oncogene 38: 2177–2191. 10.1038/s41388-018-0568-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet W, Knaapen MW, De Meyer GR, Herman AG, Kockx MM. 2002. Elevated levels of oxidative DNA damage and DNA repair enzymes in human atherosclerotic plaques. Circulation 106: 927–932. 10.1161/01.CIR.0000026393.47805.21 [DOI] [PubMed] [Google Scholar]
- Márton J, Fodor T, Nagy L, Vida A, Kis G, Brunyánszki A, Antal M, Lüscher B, Bai P. 2018a. PARP10 (ARTD10) modulates mitochondrial function. PLoS One 13: e0187789 10.1371/journal.pone.0187789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton J, Peter M, Balogh G, Bodi B, Vida A, Szanto M, Bojcsuk D, Janko L, Bhattoa HP, Gombos I, et al. 2018b. Poly(ADP-ribose) polymerase-2 is a lipid-modulated modulator of muscular lipid homeostasis. Biochim Biophys Acta 2: 30187–30182. [DOI] [PubMed] [Google Scholar]
- Masszi G, Horvath EM, Tarszabo R, Benko R, Novak A, Buday A, Tokes AM, Nadasy GL, Hamar P, Benyó Z, et al. 2013. Reduced estradiol-induced vasodilation and poly-(ADP-ribose) polymerase (PARP) activity in the aortas of rats with experimental polycystic ovary syndrome (PCOS). PLoS One 8: e55589 10.1371/journal.pone.0055589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier-de Murcia J, Molinete M, Gradwohl G, Simonin F, de Murcia G. 1989. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J Mol Biol 210: 229–233. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Kakizawa T, Hashizume K. 1999. Inhibition of nuclear receptor signalling by poly(ADP-ribose) polymerase. Mol Cell Biol 19: 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocchegiani E, Muzzioli M, Giacconi R, Cipriano C, Gasparini N, Franceschi C, Gaetti R, Cavalieri E, Suzuki H. 2003. Metallothioneins/PARP-1/IL-6 interplay on natural killer cell activity in elderly: parallelism with nonagenarians and old infected humans. Effect of zinc supply. Mech Ageing Dev 124: 459–468. 10.1016/S0047-6374(03)00023-X [DOI] [PubMed] [Google Scholar]
- Mocchegiani E, Giacconi R, Cipriano C, Gasparini N, Bernardini G, Malavolta M, Menegazzi M, Cavalieri E, Muzzioli M, Ciampa AR, et al. 2004. The variations during the circadian cycle of liver CD1d-unrestricted NK1.1+ TCRγ/δ+ cells lead to successful ageing. Role of metallothionein/IL-6/gp130/PARP-1 interplay in very old mice. Exp Gerontol 39: 775–788. 10.1016/j.exger.2004.01.014 [DOI] [PubMed] [Google Scholar]
- Módis K, Gerő D, Erdélyi K, Szoleczky P, Dewitt D, Szabo C. 2012. Cellular bioenergetics is regulated by PARP1 under resting conditions and during oxidative stress. Biochem Pharmacol 83: 633–643. 10.1016/j.bcp.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed JS, Hajira A, Pardo PS, Boriek AM. 2014. MicroRNA-149 inhibits PARP-2 and promotes mitochondrial biogenesis via SIRT-1/PGC-1α network in skeletal muscle. Diabetes 63: 1546–1559. 10.2337/db13-1364 [DOI] [PubMed] [Google Scholar]
- Morrow DA, Brickman CM, Murphy SA, Baran K, Krakover R, Dauerman H, Kumar S, Slomowitz N, Grip L, McCabe CH, et al. 2009. A randomized, placebo-controlled trial to evaluate the tolerability, safety, pharmacokinetics, and pharmacodynamics of a potent inhibitor of poly(ADP-ribose) polymerase (INO-1001) in patients with ST-elevation myocardial infarction undergoing primary percutaneous coronary intervention: results of the TIMI 37 trial. J Thromb Thrombolysis 27: 359–364. 10.1007/s11239-008-0230-1 [DOI] [PubMed] [Google Scholar]
- Mota de Sa P, Richard AJ, Hang H, Stephens JM. 2017. Transcriptional regulation of adipogenesis. Compr Physiol 7: 635–674. [DOI] [PubMed] [Google Scholar]
- Muiras ML, Müller M, Schächter F, Bürkle A. 1998. Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians. J Mol Med (Berl) 76: 346–354. 10.1007/s001090050226 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P, Horváth B, Rajesh M, Varga ZV, Gariani K, Ryu D, Cao Z, Holovac E, Park O, Zhou Z, et al. 2017. PARP inhibition protects against alcoholic and non-alcoholic steatohepatitis. J Hepatol 66: 589–600. 10.1016/j.jhep.2016.10.023 [DOI] [PubMed] [Google Scholar]
- Muñoz-Gámez JA, Rodríguez-Vargas JM, Quiles-Perez R, Aguilar-Quesada R, Martin-Oliva D, de Murcia G, de Murcia Murcia J, Almendros A, Ruiz de Almodóvar M, Oliver FJ. 2009. PARP-1 is involved in autophagy induced by DNA damage. Autophagy 5: 61–74. 10.4161/auto.5.1.7272 [DOI] [PubMed] [Google Scholar]
- Muthumani K, Choo AY, Zong WX, Madesh M, Hwang DS, Premkumar A, Thieu KP, Emmanuel J, Kumar S, Thompson CB, et al. 2006. The HIV-1 Vpr and glucocorticoid receptor complex is a gain-of-function interaction that prevents the nuclear localization of PARP-1. Nat Cell Biol 8: 170–179. 10.1038/ncb1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. 1998. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell 93: 229–240. 10.1016/S0092-8674(00)81574-3 [DOI] [PubMed] [Google Scholar]
- Nagy L, Rauch B, Balla N, Ujlaki G, Kis G, Abdul-Rahman O, Kristof E, Sipos A, Antal M, Toth A, et al. 2019. Olaparib induces browning of in vitro cultures of human primary white adipocytes. Biochem Pharmacol 25: 30250–30253. [DOI] [PubMed] [Google Scholar]
- Nomura F, Yaguchi M, Itoga And S, Noda M. 2001. Effects of chronic alcohol consumption on hepatic poly-ADP-ribosylation in the rat. Alcohol Clin Exp Res 25: 35S–38S. 10.1111/j.1530-0277.2001.tb02415.x [DOI] [PubMed] [Google Scholar]
- Nozaki T, Fujihara H, Watanabe M, Tsutsumi M, Nakamoto K, Kusuoka O, Kamada N, Suzuki H, Nakagama H, Sugimura T, et al. 2003. Parp-1 deficiency implicated in colon and liver tumorigenesis induced by azoxymethane. Cancer Sci 94: 497–500. 10.1111/j.1349-7006.2003.tb01472.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T, Fujimori H, Wang J, Suzuki H, Imai H, Watanabe M, Ohura K, Masutani M. 2013. Parp-1 deficiency in ES cells promotes invasive and metastatic lesions accompanying induction of trophoblast giant cells during tumorigenesis in uterine environment. Pathol Int 63: 408–414. 10.1111/pin.12086 [DOI] [PubMed] [Google Scholar]
- Ohkura N, Nagamura Y, Tsukada T. 2008. Differential transactivation by orphan nuclear receptor NOR1 and its fusion gene product EWS/NOR1: possible involvement of poly(ADP-ribose) polymerase I, PARP-1. J Cell Biochem 105: 785–800. 10.1002/jcb.21876 [DOI] [PubMed] [Google Scholar]
- Oka S, Kato J, Moss J. 2006. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem 281: 705–713. 10.1074/jbc.M510290200 [DOI] [PubMed] [Google Scholar]
- Oliver FJ, Ménissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de La RG, Stoclet JC, de Murcia G. 1999. Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J 18: 4446–4454. 10.1093/emboj/18.16.4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan J, Tedim Ferreira M, Gagné JP, Sharma AK, Hendzel MJ, Masson JY, Poirier GG. 2019. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat Commun 10: 1182 10.1038/s41467-019-08859-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oumouna M, Datta R, Oumouna-Benachour K, Suzuki Y, Hans C, Matthews K, Fallon K, Boulares H. 2006. Poly(ADP-ribose) polymerase-1 inhibition prevents eosinophil recruitment by modulating Th2 cytokines in a murine model of allergic airway inflammation: a potential specific effect on IL-5. J Immunol 177: 6489–6496. 10.4049/jimmunol.177.9.6489 [DOI] [PubMed] [Google Scholar]
- Oumouna-Benachour K, Hans CP, Suzuki Y, Naura A, Datta R, Belmadani S, Fallon K, Woods C, Boulares AH. 2007. Poly(ADP-ribose) polymerase inhibition reduces atherosclerotic plaque size and promotes factors of plaque stability in apolipoprotein E-deficient mice: effects on macrophage recruitment, nuclear factor-κB nuclear translocation, and foam cell death. Circulation 115: 2442–2450. 10.1161/CIRCULATIONAHA.106.668756 [DOI] [PubMed] [Google Scholar]
- Pacher P, Szabo C. 2005. Role of poly(ADP-ribose) polymerase-1 activation in the pathogenesis of diabetic complications: endothelial dysfunction, as a common underlying theme. Antioxid Redox Signal 7: 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Szabo C. 2006. Role of peroxynitrite in the pathogenesis of cardiovascular complications of diabetes. Curr Opin Pharmacol 6: 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, Ahel I. 2018. Serine is the major residue for ADP-ribosylation upon DNA damage. Elife 7 10.7554/eLife.34334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. 2005. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell 18: 83–96. [DOI] [PubMed] [Google Scholar]
- Peng H, Zhu QS, Zhong S, Levy D. 2015. Transcription of the human microsomal epoxide hydrolase gene (EPHX1) Is regulated by PARP-1 and Histone H1.2. Association with sodium-dependent bile acid transport. PLoS One 10: e0125318 10.1371/journal.pone.0125318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirinen E, Cantó E, Jo SK, Morato L, Zhang H, Menzies KJ, Williams EG, Mouchiroud L, Moullan N, Hagberg C, et al. 2014. Pharmacological inhibition of Poly(ADP-Ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab 19: 1034–1041. 10.1016/j.cmet.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, et al. 2012. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell 150: 620–632. 10.1016/j.cell.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack JGM, Palazzo L, Ahel I. 2020. (ADP-ribosyl)hydrolases: structure, function, and biology. Genes Dev (this issue) 10.1101/gad.334631.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappou E, Jukarainen S, Rinnankoski-Tuikka R, Kaye S, Heinonen S, Hakkarainen A, Lundbom J, Lundbom N, Saunavaara V, Rissanen A, et al. 2016. Weight loss is associated with increased NAD+/SIRT1 expression but reduced PARP activity in white adipose tissue. J Clin Endocrinol Metab 101: 1263–1273. 10.1210/jc.2015-3054 [DOI] [PubMed] [Google Scholar]
- Regdon Z, Robaszkiewicz A, Kovács K, Rygielska Z, Hegedűs C, Bodoor K, Szabó E, Virág L. 2019. LPS protects macrophages from AIF-independent parthanatos by downregulation of PARP1 expression, induction of SOD2 expression, and a metabolic shift to aerobic glycolysis. Free Radic Biol Med 131: 184–196. 10.1016/j.freeradbiomed.2018.11.034 [DOI] [PubMed] [Google Scholar]
- Reilly SM, Saltiel AR. 2017. Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol 13: 633–643. 10.1038/nrendo.2017.90 [DOI] [PubMed] [Google Scholar]
- Robaszkiewicz A, Valkó Z, Kovács K, Hegedűs C, Bakondi E, Bai P, Virág L. 2014. The role of p38 signaling and poly(ADP-ribosyl)ation-induced metabolic collapse in the osteogenic differentiation-coupled cell death pathway. Free Radic Biol Med 76C: 69–79. 10.1016/j.freeradbiomed.2014.07.027 [DOI] [PubMed] [Google Scholar]
- Roca-Rivada A, Alonso J, Al-Massadi O, Castelao C, Peinado JR, Seoane LM, Casanueva FF, Pardo M. 2011. Secretome analysis of rat adipose tissues shows location-specific roles for each depot type. J Proteomics 74: 1068–1079. 10.1016/j.jprot.2011.03.010 [DOI] [PubMed] [Google Scholar]
- Roper SJ, Chrysanthou S, Senner CE, Sienerth A, Gnan S, Murray A, Masutani M, Latos P, Hemberger M. 2014. ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells. Nucleic Acids Res 42: 8914–8927. 10.1093/nar/gku591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. 2014. What we talk about when we talk about fat. Cell 156: 20–44. 10.1016/j.cell.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ojeda FJ, Rupérez AI, Gomez-Llorente C, Gil A, Aguilera CM. 2016. Cell models and their application for studying adipogenic differentiation in relation to obesity: a review. Int J Mol Sci 17: 1040 10.3390/ijms17071040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, Kraus WL. 2018. Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360: eaan5780 10.1126/science.aan5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks HS, Fain JN, Bahouth SW, Ojha S, Frontini A, Budge H, Cinti S, Symonds ME. 2013. Adult epicardial fat exhibits beige features. J Clin Endocrinol Metab 98: E1448–E1455. 10.1210/jc.2013-1265 [DOI] [PubMed] [Google Scholar]
- Salomone F, Barbagallo I, Godos J, Lembo V, Currenti W, Cinà D, Avola R, D'Orazio N, Morisco F, Galvano F, et al. 2017. Silibinin restores NAD+ levels and induces the SIRT1/AMPK pathway in non-alcoholic fatty liver. Nutrients 9: 1086 10.3390/nu9101086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Guertin DA. 2014a. Adipocyte lineages: tracing back the origins of fat. Biochim Biophys Acta 1842: 340–351. 10.1016/j.bbadis.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Guertin DA. 2014b. Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat Commun 5: 4099 10.1038/ncomms5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheja L, Heeren J. 2019. The endocrine function of adipose tissues in health and cardiometabolic disease. Nat Rev Endocrinol 15: 507–524. 10.1038/s41574-019-0230-6 [DOI] [PubMed] [Google Scholar]
- Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, Liu F, Planck JL, Ravindranathan P, Chinnaiyan AM, et al. 2012. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov 2012: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Amé JC, Dollé P, Schultz I, Rinaldi B, Fraulob V, Ménissier-de Murcia J, de Murcia G. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277: 23028–23036. 10.1074/jbc.M202390200 [DOI] [PubMed] [Google Scholar]
- Shan T, Liang X, Bi P, Kuang S. 2013. Myostatin knockout drives browning of white adipose tissue through activating the AMPK–PGC1α–Fndc5 pathway in muscle. FASEB J 27: 1981–1989. 10.1096/fj.12-225755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Wang W, Wang L, Houde C, Wu W, Tudor M, Thompson JR, Sisk CM, Hubbard B, Li J. 2012. Identification of genes affecting apolipoprotein B secretion following siRNA-mediated gene knockdown in primary human hepatocytes. Atherosclerosis 222: 154–157. 10.1016/j.atherosclerosis.2012.02.012 [DOI] [PubMed] [Google Scholar]
- Shi L, Ko S, Kim S, Echchgadda I, Oh TS, Song CS, Chatterjee B. 2008. Loss of androgen receptor in aging and oxidative stress through Myb protooncoprotein-regulated reciprocal chromatin dynamics of p53 and poly(ADP-ribose) polymerase PARP-1. J Biol Chem 283: 36474–36485. 10.1074/jbc.M805980200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Macey TA, Quillinan N, Klawitter J, Perraud AL, Traystman RJ, Herson PS. 2013. Androgen and PARP-1 regulation of TRPM2 channels after ischemic injury. J Cereb Blood Flow Metab 33: 1549–1555. 10.1038/jcbfm.2013.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha E, Hussein MA, Savas JN, Ouimet M, Barrett TJ, Leone S, Yates JR III, Moore KJ, Fisher EA, Garabedian MJ. 2016. Poly(ADP-ribose) polymerase 1 represses liver X receptor-mediated ABCA1 expression and cholesterol efflux in macrophages. J Biol Chem 291: 11172–11184. 10.1074/jbc.M116.726729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D. 2020. PARP and PARG inhibitors in cancer treatment. Genes Dev (this issue) 10.1101/gad.334516.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulson ME, Kang VH, Ntambi JM, Rosenthal DS, Ding R, Simbulan CM. 1995. Requirement for the expression of poly(ADP-ribose) polymerase during the early stages of differentiation of 3T3-L1 preadipocytes, as studied by antisense RNA induction. J Biol Chem 270: 119–127. [DOI] [PubMed] [Google Scholar]
- Soriano FG, Pacher P, Mabley J, Liaudet L, Szabo C. 2001. Rapid reversal of the diabetic endothelial dysfunction by pharmacological inhibition of poly(ADP-ribose) polymerase. Circ Res 89: 684–691. [DOI] [PubMed] [Google Scholar]
- Suárez-Zamorano N, Fabbiano S, Chevalier C, Stojanović O, Colin DJ, Stevanović A, Veyrat-Durebex C, Tarallo V, Rigo D, Germain S, et al. 2015. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat Med 21: 1497–1501. 10.1038/nm.3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh SW, Aoyama K, Matsumori Y, Liu J, Swanson RA. 2005. Pyruvate administered after severe hypoglycemia reduces neuronal death and cognitive impairment. Diabetes 54: 1452–1458. 10.2337/diabetes.54.5.1452 [DOI] [PubMed] [Google Scholar]
- Sun K, Gao Z, Kolonin MG. 2018. Transient inflammatory signaling promotes beige adipogenesis. Sci Signal 11: eaat3192 10.1126/scisignal.aat3192 [DOI] [PubMed] [Google Scholar]
- Sun Q, Gatie M II, Kelly GM. 2019. Serum-dependent and independent regulation of PARP2. Biochem Cell Biol 97: 600–611. 10.1139/bcb-2018-0345 [DOI] [PubMed] [Google Scholar]
- Sunderland PT, Woon EC, Dhami A, Bergin AB, Mahon MF, Wood PJ, Jones LA, Tully SR, Lloyd MD, Thompson AS, et al. 2011. 5-Benzamidoisoquinolin-1-ones and 5-(ω-carboxyalkyl)isoquinolin-1-ones as isoform-selective inhibitors of poly(ADP-ribose) polymerase 2 (PARP-2). J Med Chem 54: 2049–2059. 10.1021/jm1010918 [DOI] [PubMed] [Google Scholar]
- Szabo C. 2005. Roles of poly(ADP-ribose) polymerase activation in the pathogenesis of diabetes mellitus and its complications. Pharmacol Res 52: 60–71. [DOI] [PubMed] [Google Scholar]
- Szabo C, Biser A, Benko R, Bottinger E, Susztak K. 2006. Poly(ADP-ribose) polymerase inhibitors ameliorate nephropathy of type 2 diabetic Leprdb/db mice. Diabetes 55: 3004–3012. 10.2337/db06-0147 [DOI] [PubMed] [Google Scholar]
- Szanto M, Rutkai I, Hegedűs C, Czikora A, Rózsahegyi M, Kiss B, Virág L, Gergely P, Tóth A, Bai P. 2011. Poly(ADP-ribose) polymerase-2 depletion reduces doxorubicin-induced damage through SIRT1 induction. Cardiovasc Res 92: 430–438. 10.1093/cvr/cvr246 [DOI] [PubMed] [Google Scholar]
- Szántó M, Brunyánszki A, Kiss B, Nagy L, Gergely P, Virág L, Bai P. 2012. Poly(ADP-ribose) polymerase-2: emerging transcriptional roles of a DNA repair protein. Cell Mol Life Sci 69: 4079–4092. 10.1007/s00018-012-1003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szántó M, Brunyánszki A, Márton J, Vámosi G, Nagy L, Fodor T, Kiss B, Virag L, Gergely P, Bai P. 2014. Deletion of PARP-2 induces hepatic cholesterol accumulation and decrease in HDL levels. Biochem Biophys Acta 1842: 594–602. 10.1016/j.bbadis.2013.12.006 [DOI] [PubMed] [Google Scholar]
- Upton K, Meyers M, Thorsell AG, Karlberg T, Holechek J, Lease R, Schey G, Wolf E, Lucente A, Schüler H, et al. 2017. Design and synthesis of potent inhibitors of the mono(ADP-ribosyl)transferase, PARP14. Bioorg Med Chem Lett 27: 2907–2911. 10.1016/j.bmcl.2017.04.089 [DOI] [PubMed] [Google Scholar]
- Venkannagari H, Verheugd P, Koivunen J, Haikarainen T, Obaji E, Ashok Y, Narwal M, Pihlajaniemi T, Lüscher B, Lehtiö L. 2016. Small-molecule chemical probe rescues cells from Mono-ADP-ribosyltransferase ARTD10/PARP10-induced apoptosis and sensitizes cancer cells to DNA damage. Cell Chem Biol 23: 1251–1260. 10.1016/j.chembiol.2016.08.012 [DOI] [PubMed] [Google Scholar]
- Vida A, Abdul-Rahman O, Mikó E, Brunyanszki A, Bai P. 2016. Poly(ADP-Ribose) polymerases in aging—friend or foe? Curr Protein Pept Sci 17: 705–712. 10.2174/1389203717666160419144959 [DOI] [PubMed] [Google Scholar]
- Vida A, Kardos G, Kovacs T, Bodrogi BL, Bai P. 2018. Deletion of poly(ADPribose) polymerase-1 changes the composition of the microbiome in the gut. Mol Med Rep 18: 4335–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag L, Salzman AL, Szabo C. 1998a. Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death. J Immunol 161: 3753–3759. [PubMed] [Google Scholar]
- Virág L, Scott GS, Cuzzocrea S, Marmer D, Salzman AL, Szabó C. 1998b. Peroxynitrite-induced thymocyte apoptosis: the role of caspases and poly (ADP-ribose) synthetase (PARS) activation. Immunology 94: 345–355. 10.1046/j.1365-2567.1998.00534.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishvanath L, Gupta RK. 2019. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J Clin Invest 129: 4022–4031. 10.1172/JCI129191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivelo CA, Wat R, Agrawal C, Tee HY, Leung AKL. 2017. ADPriboDB: the database of ADP-ribosylated proteins. Nucleic Acids Res 45: 6254 10.1093/nar/gkw706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lukowicz T, Hassa PO, Lohmann C, Borén J, Braunersreuther V, Mach F, Odermatt B, Gersbach M, Camici GG, Stähli BE, et al. 2008. PARP1 is required for adhesion molecule expression in atherogenesis. Cardiovasc Res 78: 158–166. 10.1093/cvr/cvm110 [DOI] [PubMed] [Google Scholar]
- Vyas DR, McCarthy JJ, Tsika GL, Tsika RW. 2001. Multiprotein complex formation at the β myosin heavy chain distal muscle CAT element correlates with slow muscle expression but not mechanical overload responsiveness. J Biol Chem 276: 1173–1184. 10.1074/jbc.M007750200 [DOI] [PubMed] [Google Scholar]
- Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. 2007. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol 27: 7475–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Öncu D, et al. 2012. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol 30: 283–288. 10.1038/nbt.2121 [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev 9: 509–520. 10.1101/gad.9.5.509 [DOI] [PubMed] [Google Scholar]
- Wang S, Liang X, Yang Q, Fu X, Rogers CJ, Zhu M, Rodgers BD, Jiang Q, Dodson MV, Du M. 2015. Resveratrol induces brown-like adipocyte formation in white fat through activation of AMP-activated protein kinase (AMPK) α1. Int J Obes (Lond) 39: 967–976. 10.1038/ijo.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Cui NH, Zhang S, Guo SR, Liu ZJ, Ming L. 2017. PARP-1 Variant Rs1136410 confers protection against coronary artery disease in a Chinese Han population: a two-stage case-control study involving 5643 subjects. Front Physiol 8: 916 10.3389/fphys.2017.00916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Xing JH, Wang BL, Xue L, Wang JL, Li R, Qin WD, Wang J, Wang XP, Zhang MX, et al. 2013. Poly(ADP-ribose) polymerase inhibition prevents reactive oxygen species induced inhibition of aldehyde dehydrogenase2 activity. Biochim Biophys Acta 1833: 479–486. 10.1016/j.bbamcr.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. 2012. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150: 366–376. 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Dong Z, Wang CJ, Barlow LJ, Fako V, Serrano MA, Zou Y, Liu JY, Zhang JT. 2016. FASN regulates cellular response to genotoxic treatments by increasing PARP-1 expression and DNA repair activity via NF-κB and SP1. Proc Natl Acad Sci 113: E6965–E6973. 10.1073/pnas.1609934113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Bai P, Little PJ, Liu P. 2014. Poly(ADP-ribose) polymerase 1 (PARP1) in atherosclerosis: from molecular mechanisms to therapeutic implications. Med Res Rev 34: 644–675. 10.1002/med.21300 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Franczyk MP, Chondronikola M, Qi N, Gunawardana SC, Stromsdorfer KL, Porter LC, Wozniak DF, Sasaki Y, Rensing N, et al. 2019. Adipose tissue NAD+ biosynthesis is required for regulating adaptive thermogenesis and whole-body energy homeostasis in mice. Proc Natl Acad Sci 116: 23822–23828. 10.1073/pnas.1909917116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa T, Funasaka T, Tsutsumi S, Hu H, Watanabe H, Raz A. 2007. Regulation of phosphoglucose isomerase/autocrine motility factor activities by the poly(ADP-ribose) polymerase family-14. Cancer Res 67: 8682–8689. 10.1158/0008-5472.CAN-07-1586 [DOI] [PubMed] [Google Scholar]
- Ye DZ, Tai MH, Linning KD, Szabo C, Olson LK. 2006. MafA expression and insulin promoter activity are induced by nicotinamide and related compounds in INS-1 pancreatic β-cells. Diabetes 55: 742–750. 10.2337/diabetes.55.03.06.db05-0653 [DOI] [PubMed] [Google Scholar]
- Yeh TY, Sbodio JI, Tsun ZY, Luo B, Chi NW. 2007. Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem J 402: 279–290. 10.1042/BJ20060793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TY, Beiswenger KK, Li P, Bolin KE, Lee RM, Tsao TS, Murphy AN, Hevener AL, Chi NW. 2009. Hypermetabolism, hyperphagia, and reduced adiposity in tankyrase-deficient mice. Diabetes 11: 2476–2485. 10.2337/db08-1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yélamos J, Monreal Y, Saenz L, Aguado E, Schreiber V, Mota R, Fuente T, Minguela A, Parrilla P, de Murcia G, et al. 2006. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J 25: 4350–4360. 10.1038/sj.emboj.7601301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Chen Y, Alano CC, Swanson RA. 2002. Tricarboxylic acid cycle substrates prevent PARP-mediated death of neurons and astrocytes. J Cereb Blood Flow Metab 22: 774–779. 10.1097/00004647-200207000-00002 [DOI] [PubMed] [Google Scholar]
- Ying W, Garnier P, Swanson RA. 2003. NAD+ repletion prevents PARP-1-induced glycolytic blockade and cell death in cultured mouse astrocytes. Biochem Biophys Res Commun 308: 809–813. 10.1016/S0006-291X(03)01483-9 [DOI] [PubMed] [Google Scholar]
- Zaremba T, Thomas HD, Cole M, Coulthard SA, Plummer ER, Curtin NJ. 2011. Poly(ADP-ribose) polymerase-1 (PARP-1) pharmacogenetics, activity and expression analysis in cancer patients and healthy volunteers. Biochem J 436: 671–679. 10.1042/BJ20101723 [DOI] [PubMed] [Google Scholar]
- Zarrinpar A, Chaix A, Panda S. 2016. Daily eating patterns and their impact on health and disease. Trends Endocrinol Metab 27: 69–83. 10.1016/j.tem.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Yang GY, Ying W, Kelly M, Hirai K, James TL, Swanson RA, Litt L. 2007. Pyruvate improves recovery after PARP-1-associated energy failure induced by oxidative stress in neonatal rat cerebrocortical slices. J Cereb Blood Flow Metab 27: 304–315. 10.1038/sj.jcbfm.9600335 [DOI] [PubMed] [Google Scholar]
- Zerfaoui M, Suzuki Y, Naura AS, Hans CP, Nichols C, Boulares AH. 2008. Nuclear translocation of p65 NF-κB is sufficient for VCAM-1, but not ICAM-1, expression in TNF-stimulated smooth muscle cells: differential requirement for PARP-1 expression and interaction. Cell Signal 20: 186–194. 10.1016/j.cellsig.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Yao J, DuMond ME, Krishnakumar R, Ruhl DD, Ryu KW, Gamble MJ, Kraus WL. 2012. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J Biol Chem 287: 12405–12416. 10.1074/jbc.M111.304469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang Y, Wang L, Luo X, Huang K, Wang C, Du M, Liu F, Luo T, Huang D, et al. 2013. Poly(ADP-ribose) polymerase 1 is a key regulator of estrogen receptor α-dependent gene transcription. J Biol Chem 288: 11348–11357. 10.1074/jbc.M112.429134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zou J, Chai E, Qi Y, Zhang Y. 2014. α-Lipoic acid attenuates cardiac hypertrophy via downregulation of PARP-2 and subsequent activation of SIRT-1. Eur J Pharmacol 744: 203–210. 10.1016/j.ejphar.2014.09.037 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang C, Tian Y, Zhang F, Xu W, Li X, Shu Z, Wang Y, Huang K, Huang D. 2016. Inhibition of Poly(ADP-Ribose) polymerase-1 protects chronic alcoholic liver injury. Am J Pathol 186: 3117–3130. 10.1016/j.ajpath.2016.08.016 [DOI] [PubMed] [Google Scholar]
- Zhao H, Sifakis EG, Sumida N, Millán-Arino L, Scholz BA, Svensson JP, Chen X, Ronnegren AL, de Lima CD M, Varnoosfaderani FS, et al. 2015. PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol Cell 59: 984–997. 10.1016/j.molcel.2015.07.019 [DOI] [PubMed] [Google Scholar]
- Zhou X, Patel D, Sen S, Shanmugam V, Sidawy A, Mishra L, Nguyen BN. 2017. Poly-ADP-ribose polymerase inhibition enhances ischemic and diabetic wound healing by promoting angiogenesis. J Vasc Surg 65: 1161–1169. 10.1016/j.jvs.2016.03.407 [DOI] [PubMed] [Google Scholar]