Abstract
Deep learning has become a powerful paradigm to analyze the binding sites of regulatory factors including RNA-binding proteins (RBPs), owing to its strength to learn complex features from possibly multiple sources of raw data. However, the interpretability of these models, which is crucial to improve our understanding of RBP binding preferences and functions, has not yet been investigated in significant detail. We have designed a multitask and multimodal deep neural network for characterizing in vivo RBP targets. The model incorporates not only the sequence but also the region type of the binding sites as input, which helps the model to boost the prediction performance. To interpret the model, we quantified the contribution of the input features to the predictive score of each RBP. Learning across multiple RBPs at once, we are able to avoid experimental biases and to identify the RNA sequence motifs and transcript context patterns that are the most important for the predictions of each individual RBP. Our findings are consistent with known motifs and binding behaviors and can provide new insights about the regulatory functions of RBPs.
RNA-binding proteins (RBPs) play important roles in all aspects of post-transcriptional gene regulation including splicing, polyadenylation, transport, translation, and degradation of RNA transcripts (Gerstberger et al. 2014). It is therefore not surprising that misregulation of RBPs as well as mutations in their protein sequence and/or their RNA targets can result in diseases including cancer (Cooper et al. 2009; Siddiqui and Borden 2012). Hence, it is essential to identify RBP binding preferences to understand their function and reveal their disease promoting mechanisms. Although we are reaching a consensus annotation of all human RBPs (Ascano et al. 2012), and recent large-scale efforts have generated data on the targets of many RBPs (Van Nostrand et al. 2016), the binding preferences of comparatively few of these are well determined (Wheeler et al. 2018).
Cross-linking and immunoprecipitation followed by sequencing (CLIP-seq) protocols have made it possible to characterize transcriptome-wide binding sites of RBPs (Hafner et al. 2010; König et al. 2010; Van Nostrand et al. 2016). Despite providing a valuable resource, CLIP data need to be regarded with caution. Compared to alternatives such as RNA-binding and immunoprecipitation (RIP), CLIP results in significantly larger numbers of target sites, indicating possible cross-linking of low-specificity events or that only few mRNA copies of a given gene are actually bound in the same cell (Mukherjee et al. 2011; Plass et al. 2017). On the other hand, CLIP-seq is sensitive to expression levels, meaning that binding events on lowly expressed transcripts may not be detected. Finally, CLIP protocols are variable, and aspects of the protocol can introduce significant biases, most notably owing to the type and concentration of RNase that is used (Kishore et al. 2011). To derive binding sites from CLIP-seq reads, several specialized peak detection methods have been developed to capture high-fidelity RBP binding sites from different CLIP protocols (Corcoran et al. 2011).
Motif finding approaches can extract the dominant shared sequence/structure motifs that characterize the binding sites, ranging from those based on sequence only (Georgiev et al. 2010; Bailey 2011) to more recent ones that also take aspects of RNA structure into account (Kazan et al. 2010; Heller et al. 2017; Munteanu et al. 2018). These approaches aim at deriving short, optimal continuous sequence/structure motifs based on, for example, an information theoretic objective function. Alternatively, binding sites can also be analyzed by classification approaches, for instance, to distinguish between bound and unbound sites. Models with this aim use large numbers of binding sites (and possibly their flanking regions), typically for one RBP in one cell type at a time. The trained model can then be used to reveal missing targets of the RBP in the specific cell type, or to identify putative target sites that are bound in other cell types without available in vivo binding data (Maticzka et al. 2014; Stražar et al. 2016). However, interpreting these classifiers, for example, to derive consensus motifs as in motif finding, is usually not straightforward.
The rise of deep learning has spurred the development of deep neural networks (DNNs) to predict TF or RBP binding sites. Alipanahi et al. 2015 first showed that convolutional neural networks (CNNs) can learn TF/RBP binding sites with high accuracy compared to state-of-the-art methods, using only the DNA/RNA sequences as input. Since then, several convolutional and recurrent neural network models for genomics data have improved prediction accuracy (Quang and Xie 2016; Ben-Bassat et al. 2018). For example, iDeep (Pan and Shen 2017) leverages a multimodal DNN to integrate different sources of data to infer RBP binding sites. A study concurrent to ours additionally included relative distances of binding sites to various positional landmarks such as splice sites, using spline transformations (Avsec et al. 2018).
Although these deep networks show great promise to push the accuracy of predictions, it is generally unclear what the models base these predictions on. Using CNNs with sequence as input makes it possible to inspect the kernels or convolutional filters in the first DNN layers. One can extract the weights of these kernels or aggregate input subsequences that maximally activate the kernels and visualize them as position weight matrices (PWMs) (Alipanahi et al. 2015; Pan and Shen 2017; Avsec et al. 2018). These patterns give general insight about low-level representations that the model has learned, but they do not provide information about the decision itself, especially for DNNs with multiple layers. This challenging problem of explaining predictions has become an active field of study, and several methods have been developed over the last couple of years (Lanchantin et al. 2017; Shrikumar et al. 2017; Sundararajan et al. 2017).
In this work, we propose a multitask and multimodal DNN model, Deep RBP binding preference (DeepRiPe), set up with the aim to characterize RBP binding preferences. DeepRiPe uses a modular structure to learn informative features from DNA sequence and transcript region types, because many RBPs have preferences for binding to specific regions of a transcript. We frame RBP site prediction as multitask learning problem, that is, predicting binding sites for several RBPs simultaneously. This enables the model to use shared information among tasks and helps it to focus on the distinctive features of each RBP. In turn, because several RBPs may possess similar binding patterns, sharing information among their predictors may help the model when training data are limited. We evaluate DeepRiPe on a large compendium of PAR-CLIP and eCLIP data sets and use integrated gradients (IG) to study the impact of different model choices on the interpretation of the model (Sundararajan et al. 2017). Finally, we quantify the potential of DeepRiPe to study the impact of sequence variants on binding events.
Results
DeepRiPe
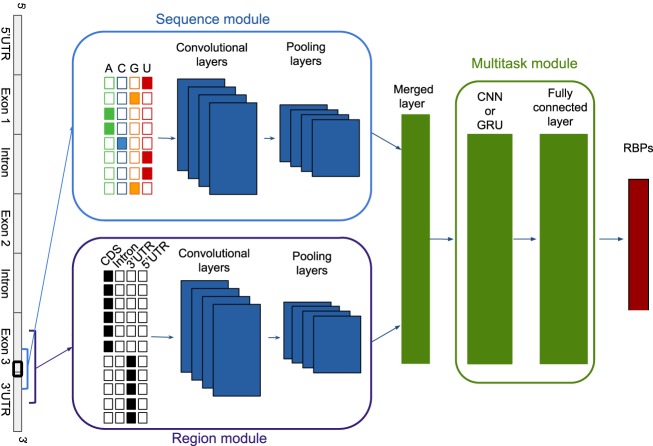
DeepRiPe consists of a sequence module that extracts features from the RNA sequence and a region module that extracts features from transcript locations. The features of these modules are then merged and fed to a multitask module to predict the binding sites of multiple RBPs simultaneously. Figure 1 shows a simplified architecture of the model. The sequence and region modules both consist of convolutional neural networks (CNNs) (Goodfellow et al. 2016). CNNs use a weight-sharing strategy, and they are highly successful to locate motifs, for example, in a sequence, independent of their position within the sequence. The multitask module contains a CNN or recurrent neural network (RNN) (Goodfellow et al. 2016). RNNs have a “memory” that allows information to persist so that they can learn dependencies in sequential data (for more details about the model structure, see Methods).
Figure 1.
A simplified graphic illustration of the model. The model consists of a sequence module that extracts features from the RNA sequence and a region module that extracts features from genomic locations. The features of these modules are then merged and fed to a multitask module to predict the binding sites of multiple RBPs simultaneously.
Initial model development and testing made use of extensive PAR-CLIP data sets for 59 RBPs from different publications, which were profiled with the same flag-tagged construct in the HEK293 cell line. These libraries were compiled, quality controlled, and processed with the same pipeline, including PARalyzer (Corcoran et al. 2011) for peak calling and the human GRCh37/hg19 release as reference, in a recent study (Mukherjee et al. 2019). To prepare the input data, we obtained 50-bp nonoverlapping genome bins and assigned a label vector with k entries corresponding to all RBPs of interest (Methods). The input for the sequence module is the one-hot encoded RNA sequence from a 150-bp window; the input for the region module is the vector of one-hot encoded region features from a 250-bp window, both centered on each 50-bp bin. Whereas sequence features denote the nucleotide (A,C,G,U), region features denote each position within mRNA as being in a 3′ UTR, 5′ UTR, CDS, or intron region and otherwise N, meaning no information. The flanking regions can give insight about the context of binding sites, and by using single-nucleotide resolution, we can capture whether binding sites occur at boundaries of region types (e.g., exon/intron junctions, cleavage sites) near cross-linked sites. To account for the drastic differences in the number of called peaks (ranging from approximately 1000 to 1,000,000 sites) (Supplemental Fig. S1A), DeepRiPe consists of three networks with identical architectures (Fig. 1), each of which is trained on a subset of CLIP data sets with comparable binding site numbers, which we refer to model-high, model-mid, and model-low (Methods). We used 20% and 10% of the bins for validation and test of the model, respectively, and the rest of the bins for training the model. All downstream analyses in this study are based on the independent test data.
Performance of DeepRiPe
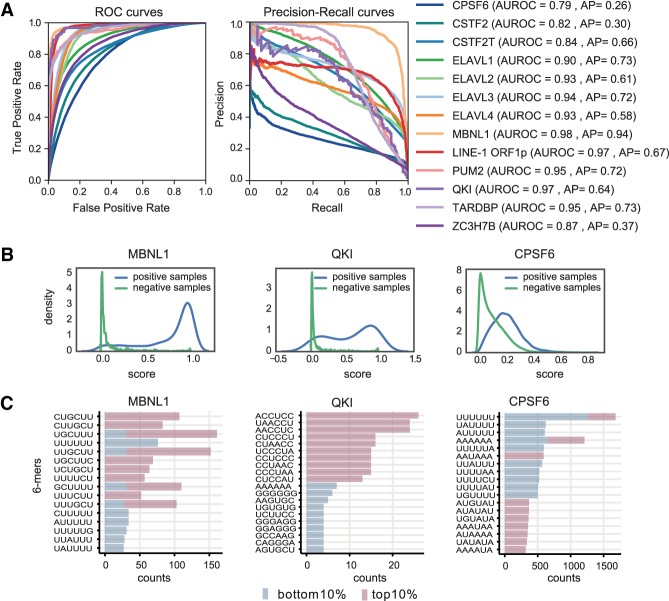
The main goal of our study is to establish interpretable classifiers as a first step toward models that can quantify the impact of sequence variation on post-transcriptional gene regulation. To start, we established the baseline performance of DeepRiPe using receiver-operating characteristics (ROC) and precision-recall (PR) curves. Figure 2A shows the ROC and PR curves, as well as the corresponding area under the ROC (AUROC) and average precision (AP) values, for a subset of 15 RBPs that we investigate in more detail below. The AUROC and AP values for all RBPs are provided in Supplemental Table S1. Although all AUROC values are above 0.7, the AP scores show a wide range. The detailed distribution of prediction scores for three RBPs shows that cases with high AUROC and AP scores (MBNL1 and QKI) show a clear difference between positive and negative samples, whereas there is little discrimination between positives and negatives for RBPs with lower scores (CPSF) (Fig. 2B).
Figure 2.
Performance of DeepRiPe. (A) ROC and precision-recall curves for several RBPs. The corresponding AUROC and AP scores are shown in parentheses. (B) Prediction score distributions for positive and negative samples for MBNL1, QKI, and CPSF6. (C) The 6-mer counts at the top and bottom 10% of the positive samples for MBNL1, QKI, and CPSF6, ranked based on their prediction scores.
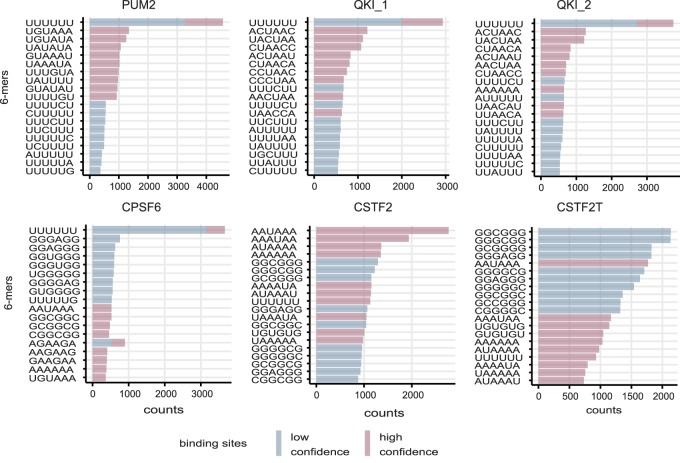
Variation in classification performance can result from the different quality of individual CLIP data sets, which may on the one hand miss genuine binding sites (false negatives), and may on the other hand contain substantial amounts of false positive, low-affinity cross-linked sites. Furthermore, RBPs may belong to complexes in which not all proteins directly bind to RNA in a sequence-specific manner. To investigate this, we ranked candidate binding sites of each RBP (positive CLIP samples of test data) based on the prediction score and extracted the 6-mers from the bottom 10% as well as the top 10% of the sites (Fig. 2C). Although the top 6-mers in the high-ranking sites are in line with the corresponding RBP motif(s), this is not necessarily the case for low ranking sites, especially for RBPs with low scores. As an example, the high-ranking binding sites for CPSF6 (AP of 0.26) contain mostly AAUAAA and UGUA elements, that is, the polyadenylation signal and upstream motif recognized by the CFIm complex that CPSF6 is part of (Martin et al. 2012). Low ranking CPSF6 sites are enriched in U-rich elements that have been previously reported as CLIP artifacts (Krakau et al. 2017). This indicates that the RBP data used in our study vary in terms of the fraction of sequence-specific sites in them, indicating a potentially high rate of false positives in some of the (PAR-)CLIP data sets or, alternatively, specification of sites by features not accounted for in our DNNs. The aim of our study is therefore not to achieve the best performance according to some metric; simply striving for classification performance can be highly misleading if the data are subject to considerable biases.
We also observed that using GRU instead of CNN for the multitask module of DeepRiPe does not significantly improve the performance scores (Supplemental Fig. S2), most likely because of the lack of data for training GRU with more parameters compared to CNN.
Interpretation of DeepRiPe
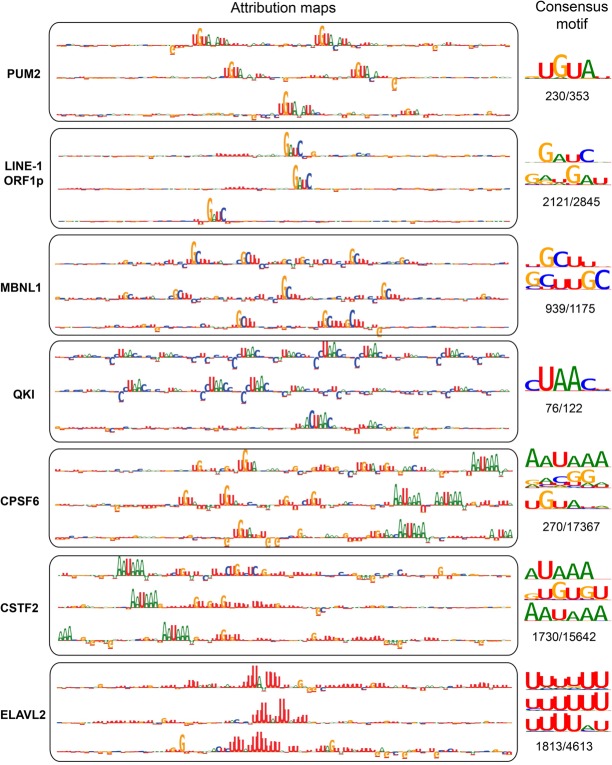
The results so far emphasize the need for an interpretable classifier to better understand what the driving input features are behind a good or poor performance. To this end, we applied methods that provide model interpretability to determine which sequence and region type patterns are informative for predicting RBP binding sites (Methods). For each RBP and any given input sequence, we compute an attribution map that indicates the individual nucleotides that were most important for classification of the input sequence as the target site for this RBP. Attribution maps for several RBPs, for positive samples of the test data with the highest prediction scores, illustrate that the model is able to learn and highlight important sequence motifs (Fig. 3; Supplemental Fig. S3). Despite drastic variability in the size of the data sets and the proportion of high-scoring peaks, these motifs in fact agree with the known motifs. For each RBP, 10 attribution maps corresponding to the inputs with the highest prediction scores (when higher than 0.5) can be found as Supplemental Files and the GitHub repository of the model.
Figure 3.
Interpretation of the model using attribution maps obtained from the IG method. For each RBP, the sequence logos corresponding to the attribution maps of three true binding sites with the highest DeepRiPe prediction scores are shown. Consensus motifs, obtained from attribution maps of all true binding sites of the RBP with prediction scores larger than 0.5, are shown beside the attribution maps. The ratio of the number of binding sites used to obtain the consensus motif to the number of all true binding sites is written below the corresponding consensus motif. The observed patterns in both the attribution maps and the consensus motifs resemble the known motif(s) for the specific RBPs: PUM2 (UGUAHAUA), QKI (ACUAAY), MBNL1 (YGCU/GCUU), CPSF6 (AAUAAA and UGUA), and CSTF2 and CSTF2T (AAUAAA and U/GU-rich).
Looking at specific RBPs in more detail highlights a crucial advantage of DNNs for regulatory sequence interpretation: The models are able to locate both simple and complex patterns in the input, such as one to several occurrences of one motif and composite motifs, without additional prior knowledge. As examples for simple patterns, we observe the well-established UGUAHAUA binding motif in attribution maps corresponding to PUM2. LINE-1 ORF1p is a protein encoded by the transcripts of LINE-1 retrotransposable elements and responsible for its retrotransposition; attribution maps of its target sequences delineate with high precision its GAUC target motif (Mandal et al. 2013).
MBNL1 and QKI are splicing factors with reported YGCU/GCUU (Lambert et al. 2014) and ACUAAY (Hafner et al. 2010) binding motifs, respectively, and their attribution maps reveal several occurrences of the motifs in the mRNAs. ELAVL2, ELAVL3, and ELAVL4 are RBPs that regulate mRNA stability and translation through the 3′ UTR and bind to U-rich elements (Keene 2001). The patterns observed in their attribution maps are consistent with this knowledge. ELAVL1 additionally binds to pre-mRNAs in the nucleus and thus to additional region types, and it also showed similar preference for U- and AU-rich patterns (Keene 2001). Depending on the input sequence, we are also able to identify variable numbers of the core U-rich pentamer.
The model is also able to locate combinations of motifs. For example, we observe RNA polyadenylation/cleavage-related sequence elements—namely, AAUAAA and U/GU-rich elements located in preferred distances to the actual site of cleavage (Darmon and Lutz 2012)—in the attribution maps of cleavage and polyadenylation specificity factors (CPSFs) and cleavage stimulatory factors (CSTFs), respectively. Additionally, the previously reported motif UGUA is observed in attribution maps of CPSF6, which is involved in 3′-end cleavage of RNA transcripts (Brown and Gilmartin 2003; Yang et al. 2011).
Composite motifs may reflect multiple binding modes of one protein, sites of interacting proteins, genomic landmarks such as start codons, or sites that are related to a process but engaged at different times. If the resolution of CLIP experiment is sufficient, our method is able to discriminate among some of these possibilities. As an example, Supplemental Figure S4 shows several attribution maps of CPSF6 targets, in which the position of actual (PAR-CLIP) peaks along the input sequences are marked. We can observe that UGUA motif is always located inside the peak, but this is not the case for the AAUAAA motif. This rules out that the AAUAAA motif is involved in direct interactions. In fact, CPSF1, the largest subunit of CPSF, binds to the AAUAAA polyadenylation signal, whereas UGUA is the target of the CPSF5/6 complex that interacts with UGUA upstream of poly(A) sites (Brown and Gilmartin 2003; Yang et al. 2011).
Altogether, patterns observed in attribution maps were consistent with previously reported motifs, in spite of not optimizing an objective function that directly quantifies the presence of common, strong motifs as in traditional motif finding. It also adds confidence that the model has learned genuine sequence features. Notably, the DNN enables us to see the actual occurrence of the motif in the sequence, and it is intrinsically able to identify complex motif patterns, such as combinations of motifs. This characteristic inherent flexibility of the DNN is a clear advantage over classical regulatory sequence analysis, with its rich literature of highly specific approaches for complex motif configurations.
Consensus motifs
To obtain consensus representations for each RBP, we aggregated the patterns in attribution maps from all positive samples (the whole input sequence) with prediction scores larger than 0.5. We reasoned that high confidence binding sites most probably contain the target motifs, but those with low probability may result from spurious binding. To do so, we first identified the top motif of length 6 in each attribution map and then clustered and aligned the motifs to obtain consensus motifs (Fig. 3; Supplemental Fig. S3; Methods). In line with patterns observed in individual attribution maps, the consensus patterns obtained from high confidence attribution maps are also consistent with previously reported motifs.
Benefits of the multimodal model
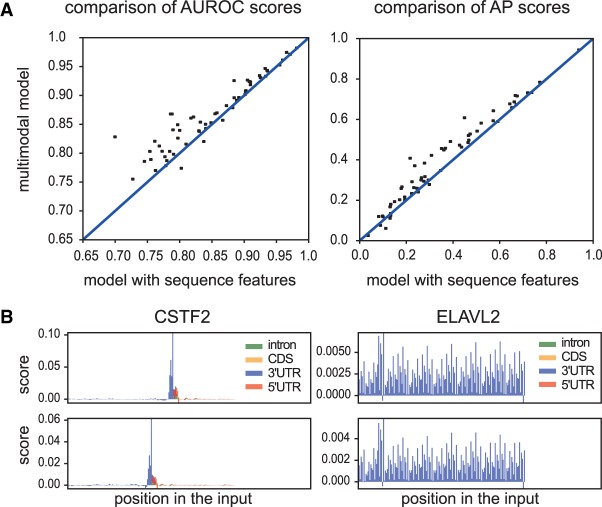
To assess the benefit of the multimodal model that uses both sequence and region type as input, and to evaluate the impact of region type in the performance of the method, we trained the DeepRiPe model without using region type information as input. Both sets of models were trained with similar structure and the same hyperparameters.
The multimodal model using both sequence and region type outperforms the model that uses only sequence for nearly all of the RBPs (Fig. 4A). This indicates the importance of region type for prediction. The model uses region information and assigns higher scores to the peaks that fall in a specific region. Attribution maps also revealed that the model uses specific region preferences that are consistent with current knowledge (Fig. 4B). The network detects not only the specific region type but also the boundaries of region types near cross-linked sites. For example, functional ELAVL2 binding sites are predominantly located in the 3′ UTRs, and CSTF2 binds to the 3′ end of the gene. Regional features could also provide information about the function of RBPs. For example, RBPs with regional impact of 3′ UTR (ELAVL2) may be involved in RNA stability, whereas RBPs bound to the end of the genes (CSTF2) are likely involved in termination/polyadenylation.
Figure 4.
Assessing the performance of the multimodal model. (A) Scatter plots comparing the AUROC and AP scores of DeepRiPe and the singlemodal model (the model using only sequence features). Each data point represents an RBP and it falls above the diagonal when DeepRiPe outperforms the singlemodal model. (B) Two examples of attribution maps that correspond to region inputs obtained from the multimodal model using IG method for the positives samples of two RBPs, CSTF2 and ELAVL2.
Benefits of multitask learning
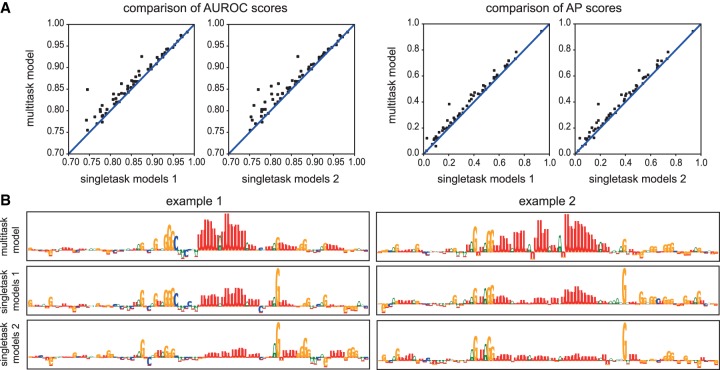
To assess the benefit of multitask learning, we compared the results of the model to those obtained by its singletask counterparts, for which we used the same hyperparameters as for the multitask model. We evaluated multiple strategies to define singletask training data. In the first strategy (single models 1), we oversampled from positive samples of the training and validation data sets for each RBP to ensure an equal number of positive samples as negative samples. In the second strategy (single models 2), we used random negative samples obtained from unbound transcripts for each RBP. We compared the performance (Fig. 5A) and interpretability (Fig. 5B) of two approaches. Finally, we also subsampled from negative samples of the training and validation data sets to ensure an equal number of negative samples as positive samples in these data sets (single models 3) (Supplemental Fig. S5).
Figure 5.
Assessing the performance of the multitask model. (A) Scatter plots comparing the AUROC and AP scores of DeepRiPe and the singletask models. Single models 1 and single models 2 are trained on random negative samples from binding sites of other RBPs and unbound transcript, respectively. Each data point represents an RBP and it falls above the diagonal when DeepRiPe outperforms its singletask counterpart. (B) Comparing the attribution maps obtained from the multitask and singletask models using the IG method for two positives samples of ELAVL2.
The overall results indicate that for some RBPs, the multitask learning indeed boosts the performance. Assessing each of the three DeepRiPe submodels (model-high, model-mid, and model-low) (Supplemental Fig. S5) shows that RBPs with a low number of samples benefit the most, which is in line with the promise of multitask learning.
Although there is consistent but limited performance improvement between single- and multitask models, the interpretability of single- and multitask models differed considerably. Comparison of attribution maps of ELAVL2 (Fig. 5B) revealed that the singletask models showed reduced importance of the known motif and were heavily misled by the PAR-CLIP sequence bias from RNase T1, which cleaves after guanines and is very prominent in especially early PAR-CLIP data sets (Kishore et al. 2011). Although the strategy of using binding sites of other RBPs as negative samples (single models 1) rather than using random negatives from unbound transcript (single models 2) already leads to a slightly better delineation of the target motif, the multitask learning approach can reveal the actual motif clearly: When learning the preferences of multiple RBPs simultaneously, the cleavage bias does not constitute useful information to discriminate between target sites of different RBPs, because many PAR-CLIP peaks will be equally affected by it. Multitask learning thus puts much less weight on protocol biases that are shared between several RBP libraries.
DeepRiPe as a potential tool to study the effects of sequence variants
We developed DeepRiPe as a tool to identify and score sequence variants with potential impact on RBP binding. To assess this aspect specifically, we first used the trained model to compute and compare the attribution maps of wild-type and mutated reporter constructs with known differences in binding efficiency for two RBPs.
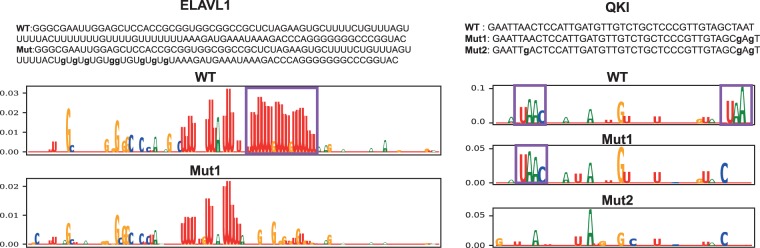
ELAVL1 binds to the 3′ UTR of the ERBB2 oncogene mRNA. In a recent study (Epis et al. 2011), ELAVL1 was shown to oppose the repression effect of microRNA miR-331-3p in ERBB2 by binding to a U-rich element (URE) near the miRNA target region. Mutation of the URE results in an experimentally detected shift of ELAVL1 binding to an upstream site with reduced binding affinity and weakens the repressive effect of ELAVL1 on miR-331-3p. In line with the reported observation, the attribution maps show the loss of ELAVL1 binding site at the mutated site, while upstream sites were not affected (Fig. 6).
Figure 6.
Assessing the impact of sequence variants using attribution maps. Sequences of wild-type and mutant constructs, in which mutations are shown in bold lowercase letters, and their corresponding attribution maps for ELAVL1 and QKI. The potential binding sites are shown in boxes. (WT) wild-type; (Mut) mutant.
As a second example, we examined the effect of mutations in potential QKI binding sites in NUMB pre-mRNA. In a study that investigated the role of QKI in regulating NUMB alternative splicing (Zong et al. 2014), two mutant sequences, Mut1 and Mut2, were generated targeting two potential binding sites of QKI in the regions surrounding the 3′ splice site of intron 12. Although Mut1 contains mutations only in the second binding sites, Mut2 has mutations in both sites (Fig. 6). Compared to wild-type RNA, with binding affinity comparable to that of a control RNA that carries a bipartite QKI consensus sequence, Mut1 RNA showed reduced QKI binding, but Mut2 RNA lost QKI binding completely. The attribution map of the wild-type sequence reveals a strong binding for the second binding site and a weak binding for the first binding site, the attribution map of Mut1 has lost the strong binding but preserves the weak binding, and the attribution map of Mut2 has lost both binding sites.
Identification of potentially disease-causing sequence variants
A major challenge in human genetics is to reveal the role and impact of single-nucleotide variants (SNVs) that are located in noncoding regions, especially in the context of congenital disorders or cancer. For instance, a recent study (Kelley et al. 2018) used DNNs to predict the influence of genomic variants on gene expression, by using thousands of epigenetic and transcriptional regulatory features. In post-transcriptional gene regulation, variants also play roles, for instance by altering RBP binding sites. The naive approach to associate SNVs with alteration of RBP binding sites is to find mutations that have been mapped to RBP targets obtained from CLIP experiments. However, the resolution of peaks is typically not sufficient to conclude that any mutation will alter RBP binding. Furthermore, a CLIP experiment of one cell type or tissue may miss targets in other cell types owing to tissue-specific gene expression. Here, our interpretable model provides an opportunity for the identification of mutations that potentially alter the RBP binding sites.
We therefore analyzed the potential mutational effects of known pathogenic SNVs obtained from COSMIC (v89) (Forbes et al. 2015) of three RBPs with known motifs and high DeepRiPe performance (MBNL1, QKI, and PUM2).
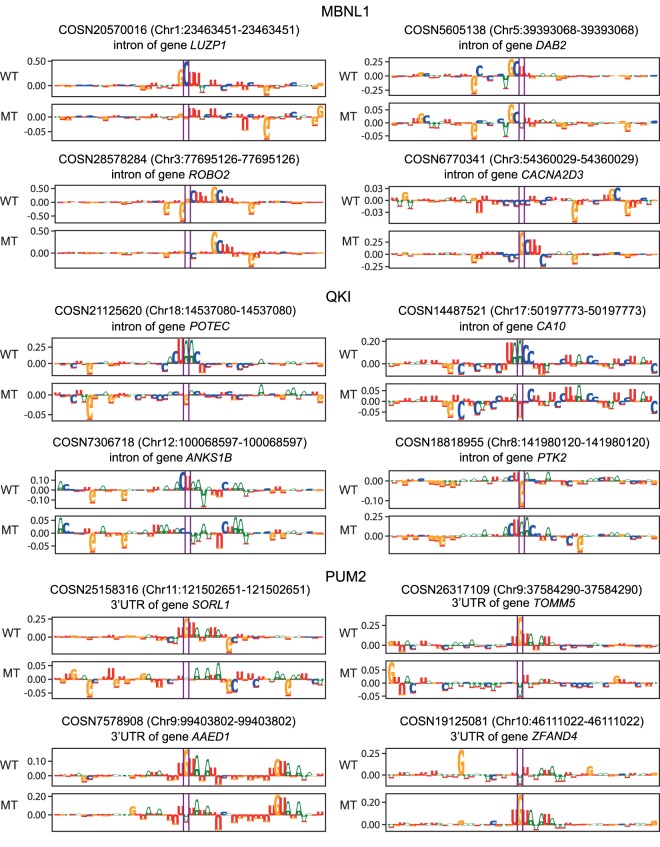
For MBNL1 and QKI, we scored the variants that are residing in the intronic location, and for PUM2, those that are located in the 3′ UTRs. For each variant, we obtained the 150-bp DNA sequence centered on the annotated site and fed both wild-type and variant sequences to the network to compare their prediction scores. When we observed a sufficient difference in scores (> 0.1), we assessed the effect of the mutation by comparing their corresponding attribution maps. Figure 7 shows that variants predicted to alter the binding sites for the specific RBP can do so in different ways: Variants can disrupt the binding sites, create new potential targets, or increase/decrease RBP binding in cases in which there are multiple potential target sites close to each other. Among the variants with score differences higher than two, 180, 36, and 48 variants are located within CLIP peaks of MBNL1, QKI, and PUM2, respectively.
Figure 7.
Examples of effects of noncoding SNVs on the binding sites of MBNL1, QKI, and PUM2. The attribution maps corresponding to the wild-type (WT) and mutant (MT) sequences for different noncoding SNVs obtained from the COSMIC database. The COSMIC ID as well as the position of the SNV is provided for each example.
Generalization power of DeepRiPe
DeepRiPe as a classification method should be able to distinguish between bound and unbound sites for a specific RBP regardless of experimental conditions and therefore to identify putative binding sites in other cell types for which there are no PAR-CLIP data. To assert this ability to generalize, we used six data sets of RBPs that were profiled by both eCLIP and PAR-CLIP in different cell lines, namely CPSF6, CSTF2T, CSTF2, PUM2, and QKI (two additional cell lines) (Van Nostrand et al. 2016). For each RBP we used processed binding sites (intersection between two replicates) provided by the ENCODE Project (https://www.encodeproject.org) and predicted binding for them using DeepRiPe trained on PAR-CLIP. To define comparable input vectors for DeepRiPe, we extended the middle of each eCLIP peak with 75 bp and 125 bp both upstream and downstream for sequence and region modules, respectively.
We ran the PAR-CLIP trained models on eCLIP targets, ranked eCLIP peaks for each RBP based on their DeepRiPe prediction score, and counted all possible 6-mers in the top 2000 (high confidence) and bottom 2000 (low confidence) binding sites. Figure 8 shows the top 10 6-mers in each set. Although the top 6-mers in high confidence binding sites resemble the motif(s) for the specific RBP, this is not the case for low confidence binding sites. As we observed on PAR-CLIP data, low-scoring eCLIP peaks are therefore likely to represent weak affinity or spurious binding sites.
Figure 8.
Performance of DeepRiPe on eCLIP data conducted in different cell types. The top 6-mers from both the set of high- and low-confidence binding sites, based on the prediction scores obtained from DeepRiPe. The top 6-mers from the high confidence binding sites resemble the known motif(s) for the specific RBPs: PUM2 (UGUAHAUA), QKI (ACUAAY), MBNL1 (YGCU/GCUU), CPSF6 (AAUAAA and UGUA), and CSTF2 and CSTF2T (AAUAAA and U/GU-rich).
Performance and interpretation of DeepRiPe on eCLIP data
DeepRiPe is not limited to PAR-CLIP data sets; although it generalizes well, it will typically be advantageous to be retrained on data obtained from other CLIP protocols and cell lines. For example, we applied our method on eCLIP data generated by the ENCODE Project (https://www.encodeproject.org) to find relevant sequence patterns. The eCLIP data consist of target data sets for approximately 150 RBPs profiled across two cell lines, K562 and HepG2. As we had done for PAR-CLIP data, we again trained several different models (here five) with the same parameters for each cell line to account for differences in the number of peaks (Methods). The performance of the models in terms of AUROC and AP are provided in Supplemental Table S2. For each RBP, 10 attribution maps corresponding to the inputs with the highest prediction scores (when higher than 0.5) can be found at Supplemental Files and the GitHub repository of the model.
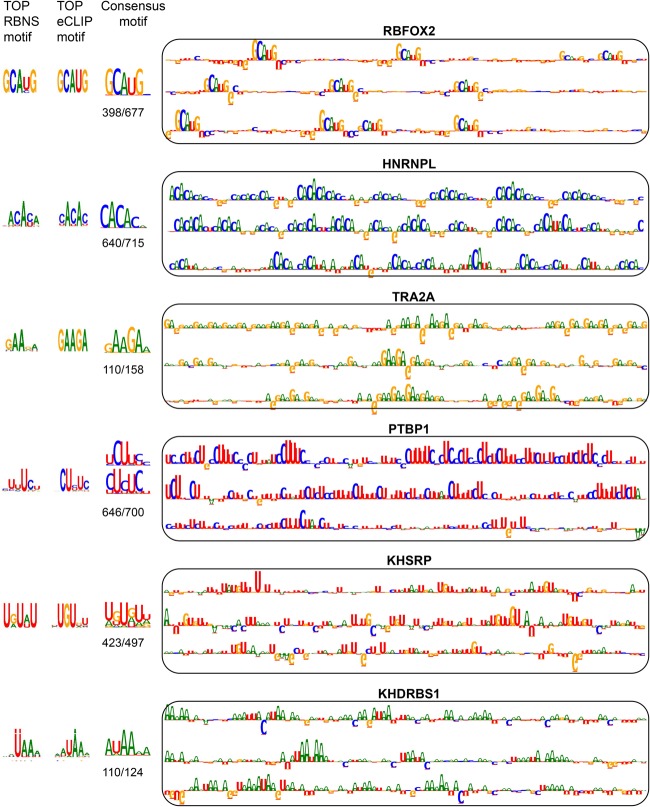
Complementing in vivo CLIP data, the ENCODE Project applied RNA Bind-n-Seq (RBNS), an in vitro method to characterize RBP binding preferences. Dominguez and colleagues compared the top k-mers in RBNS and eCLIP data sets for RBPs profiled in both assays (24 RBPs) (Dominguez et al. 2018) and found agreement between eCLIP peaks and corresponding RBNS motifs for most cases (17 RBPs). For RBPs with significant agreement between in vitro and in vivo motifs, we compared the patterns in attribution maps to the in vivo and in vitro motifs (Fig. 9). In all those cases, the networks detect the relevant motifs. We next examined the attribution maps corresponding to two RBPs (IGF2BP2 and RBP15) with no agreement between ENCODE in vivo and in vitro motifs (Supplemental Fig. S6). Although the reported eCLIP motif is CG-rich for both RBPs, the network detects different motifs that are similar to the RBNS motif.
Figure 9.
Comparison of motifs obtained from in vitro (RBNS) and in vivo (eCLIP) experiments with patterns observed in attribution maps. For each RBP, the motifs obtained from RBNS, eCLIP, and the attribution maps, along with attribution maps for the top three inputs with the highest prediction scores, are shown. The consensus motifs obtained from the attribution maps correspond to all true binding sites with prediction scores larger than 0.5. For the attribution map motifs, the ratio of the number of binding sites used to obtain consensus motif to the number of all true binding sites is written below the corresponding consensus motif.
On investigating the attribution maps of other eCLIP-profiled RBPs, we found additional cases in which the model can detect complex sequence patterns (Supplemental Fig. S7). Particularly, the model highlighted 5′ or 3′ splice sites (GGUAG, CAG) in the attribution maps of several splicing factors. Although these motifs are not involved in direct interactions of the RBPs, they can provide information for the annotation and function of RBPs.
Studying the impact of sequence variants using allele-specific binding events of RBPs
Allele-specific binding (ASB) of RBPs provides a natural source of data to assess the ability of DeepRiPe to predict the impact of variants. Having a full compendium of models trained on eCLIP data allowed us to make use of the results of recent methods that have been developed specifically to identify ASB events (Bahrami-Samani and Xing 2019; Yang et al. 2019).
Specifically, BEAPR predicts ASB events using the allele-specific mRNA expression as null hypothesis, as quantified by eCLIP input (Yang et al. 2019). For each reported significant BEAPR SNV, we computed three scores: motif score, model score, and IG-score. Motif score is defined as the maximum log-odds scores of 10-bp windows flanking the ASB SNV (both alleles) against the reported RBP motif (position weight matrix), obtained from pentamers identified by an RBNS assay of corresponding RBP (if available) or from the literature. Model score is calculated as the difference between DeepRiPe prediction scores of RNA sequences centered on minor and major alleles. IG-score is obtained as the difference between the sum of the attribution scores in 6-bp windows flanking ASB SNV alleles. To account for the potential of multiple binding sites in the input window of 200 bp, we here used just the 30-bp sequence centered at the ASB SNV (the remaining positions are filled with N, meaning equal probability of being A, C, G, or U).
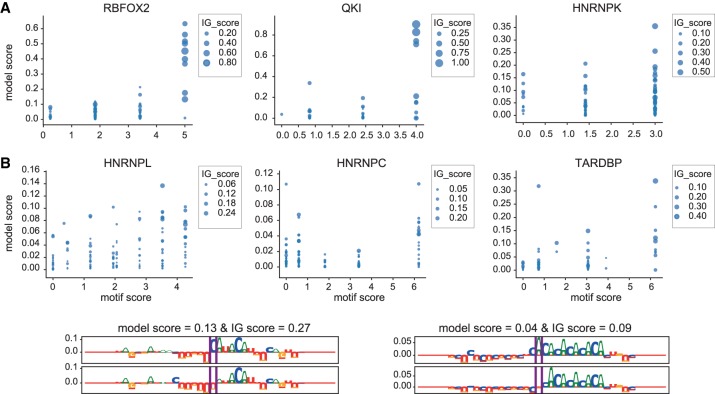
For RBPs with well-defined distinct, short motifs like RBFOX2 or QKI, ASB events with high motif scores also have high model scores and IG-scores, indicating that ASB SNPs that impact the core motif may be causal for the observed ASB (Fig. 10A). For RBPs like HNRNPL that bind to longer, mono- or dinucleutide repeats, the mutation can lead to a weaker or stronger binding effect depending on its position as well as the length of the repeat. Here, the DeepRiPe results suggest that it is more likely that the variant leads to an altered binding affinity when the repeat sequence is short (i.e., with a lower motif score) compared to when the repeat sequence is long (i.e., with the highest motif score), and this effect can again be visualized using attribution maps (as is the case for HNRNPL with AC-rich motif) (Fig. 10B).
Figure 10.
Assessing the ability of DeepRiPe to predict the impact of variants using ASB events. (A) Relationships between different scores of ASB events for different RBPs. As motif score increases for ASB events, we observe a larger difference between the model prediction scores (model score) and attribution values (IG-score) corresponding to sequences with minor and major alleles. (B) Examples of attribution maps corresponding to ASB events of HNRNPL with a different length of AC-rich motif in their flanking regions.
Discussion
We have developed a multimodal and multitask deep learning approach to model genuine, specific RBP binding events, and to extract informative features about RBP binding characteristics from dozens of high-throughput, noisy CLIP-seq data sets. The model recovers known sequence motifs and provides insight about RBP binding preferences. It can also locate the sequence motifs along the input sample and identify co-occurrences of motifs in a flexible manner. Comparing our approach, which determines the influence of input features on the output, with interpreting the convolutional layers as in previous studies (Supplemental Figs. S8–S13), we noticed that the filters in DeepRiPe's first layer typically represented only parts of the motifs. The network as a whole can detect complete motifs or take nonlinear dependencies into account by assembling multiple filters in the downstream layers, which complicates direct interpretation of these filters. Additionally, some of these filters may represent motifs in the negative set or the bias in the data. Therefore, if there is no prior knowledge about the true motif, it is very hard to decipher complete, biologically relevant motifs from filters. This issue gets confounded even further in the case of multitask learning, because individual filters may now not be specific to one RBP.
We observed considerable variability of success across different RBPs, and we were able to relate this to the absence of known motifs in low-scoring peaks; CLIP-seq experiments can result in tens of thousands of peaks, and it is highly unlikely that all of these represent targets with defined functional consequences of binding. Rather, large numbers of peaks may reflect poor antibody quality, sequencing artifacts, or interaction patterns of RBPs beyond specific sequence/structure target site definitions, such as helicases. As many peak callers do, our model assumes site-specific binding, and for libraries for which this assumption holds true, we are moving closer to a scenario in which we can now use the model to judge the quality of experiments, rather than to take noisy data as “ground truth.”
Singletask and multitask models solve different classification problems. Although the overall performance of multitask and singletask reported here appear superficially similar, the multitask formulation of learning allows the model to focus on the features that are shared across the tasks. In this way, it is able to ignore possible protocol-inherent biases, as these will be present in data sets across different RBPs. We illustrate that this leads to notable differences in the features that a model uses for its predictions, with the multitask models relying more strongly on the presence of known motifs compared to the singletask methods. Choosing negative samples for each RBP from binding sites of other RBPs makes the prediction task harder, but at the same time it guides the model to learn specific motifs. Most previous RBP target classification approaches have been set up as singletask problems, which means that we cannot directly benchmark against them. In turn, many singletask models have been evaluated on cross-validated, held-out data from the same experiment. For some of these, the reported results will likely be overly optimistic—the models will not generalize well, as we have recently observed anecdotally (Munteanu et al. 2018).
Extending our current deep network appears promising in several directions. The method already provides functionality to locate binding sites, score variants, and derive motifs from attribution maps. Owing to the (1) multitask learning process, in which we combine data sources of varying quality and numbers of targets, (2) the occurrence of sometimes multiple sites per CLIP peak, and (3) our strategy to derive motifs from well-scoring (>0.5) inputs regions only, they are rather serving the purpose of illustrating, summarizing, and comparing results. However, we anticipate that changes to the training approach, including solutions to the issue of imbalanced data, can allow for a fully fledged motif finder, in which motifs represent in vivo binding affinities similar to in vitro-derived motifs. This holds promise to alleviating some shortcomings of current approaches for RBP motif discovery that struggle because of the shortness of the binding motif and the potentially large number of false positives in the input data. In this context, interpreting DNNs may provide competitive flexibility, because there is no need to specify parameter like motif length or configuration. For both classification and prediction, future work should address how to adequately consider RNA structure within the framework of deep neural networks to advance the interpretation of noncoding sequence variants.
Methods
Input data
We collected PAR-CLIP data sets for 59 RBPs from different publications, which were profiled with the same flag-tagged construct in the HEK293 cell line. These libraries were quality controlled and processed with the same pipeline, including PARalyzer (Corcoran et al. 2011) for peak calling and the human GRCh37/hg19 release as reference, in a recent study (Mukherjee et al. 2019). We based our models on this consistently processed CLIP data, and we chose not to lift over annotations or completely reanalyze this large compendium on GRCh38 to maintain consistency with previous results. Slight sequence/assembly variation for some individual peaks will not affect the overall results, because our models are based on thousands of CLIP peaks and not on detailed investigations of a small number of individual loci.
We chose RBPs that have between 1000 and 106 peaks and divided them into three categories: RBPs with >105 peaks, RBPs that have between 15,000 and 105 peaks, and RBPs with <15,000 peaks. We used RBPs in each category for training and evaluating a separate DNN, which we refer to as model-high, model-mid, and model-low, respectively. Supplemental Figure S1 shows the number of peaks for each RBP (Supplemental Fig. S1A) as well as the number of shared binding sites for each pair of RBPs (Supplemental Fig. S1B).
To prepare the data for input to the DNN, we first split the genome into 50-bp nonoverlapping bins and kept only bins that overlap with the transcriptome. For each bin, we assigned a label vector with k entries corresponding to all RBPs of interest to define the labeled data for the multitask model. For each bin, the label of an RBP is 1 if more than half of its peak region falls within a 50-bp bin, and 0 otherwise. We kept only bins with at least one binding event. In this way, the negative samples of one RBP may serve as positive samples of other RBPs. We used 20% and 10% of the bins for validation and testing of the model, respectively, and the rest of the bins for training the model.
From eCLIP experiments of human RBPs (hg19), we collected the merged peaks between two replicates for each RBP provided by the ENCODE Project (https://www.encodeproject.org) and kept RBPs with more than 1000 reported peaks. We divided RBPs into five categories for each cell line: RBPs with >104 peaks, RBPs that have between 7000 and 104 peaks, between 4000 and 7000 peaks, between 2000 and 4000 peaks, and between 1000 and 2000 peaks. To prepare the input data for the model, we used a bin size of 100 bp to account for the eCLIP peaks resolutions. Other steps are similar to PAR-CLIP.
Model design and training
In this work, we used two types of DNN architectures, convolutional neural networks (CNNs) and recurrent neural networks (RNNs) (Goodfellow et al. 2016). More specifically, we used a bidirectional gated recurrent network (GRU) (Chung et al. 2014) to account for possible long-range dependencies of the features.
The model consists of a sequence module that extracts features from the RNA sequence and a region module that extracts features from genomic locations. The features of these modules are then merged and fed to a multitask module to predict the binding sites of multiple RBPs simultaneously. Figure 1 shows a simplified architecture of the model.
The sequence and region modules both have a convolution layer followed by a rectified linear unit (Relu), a max pool layer, and a drop out layer with probability of 0.25. We used 90 filters with length 7 for both convolution layers. The multitask module takes as input the concatenated features from sequence and region modules and consists of one CNN (with 100 filters of length 5) or one bidirectional GRU (with 60 units) and one fully connected layer with 250 hidden units and Relu activations. The output layer contains k sigmoid neurons to predict the probability of binding, one for each RBP.
To assess the contribution of different aspects to the success of the DNNs, we also explored variations of the architecture and training of the model; in singletask models, in which the model predicts the binding sites of one RBP, the output layer has only one neuron. In all applications, we used CNNs in the multitask module unless stated otherwise. The training was performed with an Adam optimizer (Kingma and Ba 2014) using a minibatch size of 128 for 20 epochs to minimize the mean multitask binary cross entropy loss function on the training set. To account for imbalanced data, we used a weighted loss function that gives higher penalties for misclassifying samples related to the classes with less samples. The best model was chosen based on the validation loss computed at the end of each epoch. We used early stopping to prevent the possibility of overfitting during the training.
Evaluation scores
We evaluated the DeepRiPe model, which was trained using training and validation sets, on independent test data. Classification performance was assessed by both the receiver-operating characteristic (ROC) and precision-recall (PR) curves, as well as the area under the ROC curves (denoted as AUROC). Average precision (AP) summarizes PR curves and is defined as the precision averaged across all values of recall. AP is more conservative compared to the area under the PR curve, because the latter uses linear interpolation and can be too optimistic. AP is more appropriate than AUROC in the case of imbalanced data with more negative samples, because it does not take into account the number of true negatives.
Interpretation
Although obtaining accurate predictions of RBP/TF binding sites is important, it is at least equally important to understand why the model makes these predictions and which parts of the input contribute the most to the output. The gradient (partial derivatives) of an output neuron with respect to its input indicates how the output value changes with respect to a small change in inputs. This is the basic concept used in gradient-based attribution methods that assign an attribution value to each input feature of the network, indicating how much that feature contributes to the output. Here, the target neuron of interest is the output neuron associated with the corresponding RBP class for a given sample, and an attribution method can specify which nucleotides of the sample input sequence and/or which region part were responsible for the output of the RBP. In this study, we used an attribution method called integrated gradients (IG) (Sundararajan et al. 2017). IG computes the average gradients of the output as the input varies along a linear path from a baseline or reference to the input, to avoid the saturation problem that occurs when computing gradients only at the input. The baseline is defined based on the application and often chosen to be zero. We used zero and 0.25 for the baselines of sequence and region inputs, respectively.
Calculating all the attribution values corresponding to all positions of one input sample leads to an “attribution map” of the sample. By visualizing the attribution map as sequence logos (for sequence) or barplots (for region), we can observe the influence of each position on the prediction. The height of sequence logos or bar plots indicates the importance of that position in the prediction. Positions with large positive attribution values can be interpreted as features that were informative for the prediction of the RBP. Visualization of the attribution maps of each input sample for a specific RBP not only reveals the potential target motif or motifs of the RBP, but it can also be used to locate the potential binding sites of the RBP on a new sequence or to assess the effect of genetic variants on RBP binding site.
To assess the effect of sequence variants, the wild-type and mutant sequences are used as the input for the sequence module. For the region module, we used N for each position in the input, meaning equal probability of being in any region. Then we compared the attribution maps corresponding to the wild type and the mutant.
Consensus motifs
To obtain the consensus motifs for each RBP, we aggregated the results of all the attribution maps corresponding to all the binding sites with prediction scores larger than 0.5. First, we searched for the top k motifs in each attribution map to obtain a list of all potential motifs for each RBP. To find the top motifs for each attribution map, we averaged the scores in sliding windows of the desired length, picked the window with the highest score, removed its neighborhood, and searched again for the next motif. We converted all the negative attribution scores of the obtained windows to zero and normalized them. Subsequently, we used UMAP (McInnes et al. 2018) to embed the top motifs obtained from the attribution maps and clustered the embedded motifs using DBSCAN clustering. Next, we aggregated motifs in each cluster by averaging over corresponding nonembedded motifs and aligned them to find the consensus motifs.
Software availability
The code for DeepRiPe is available in the Supplemental Code and from https://github.com/ohlerlab/DeepRiPe.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Neelanjan Mukherjee for providing the processed PAR-CLIP data, and Xinshu Xiao and Giovanni Quinones-Valdez for providing the allele-specific binding data. This work has been supported by Bundesministerium für Bildung und Forschung under e:Bio grant CaRNAtion (031L0075A) and the Berlin Center for Machine Learning (Berliner Zentrum für maschinelles Lernen [BZML]) (01IS18037A).
Author contributions: M.G. and U.O. conceived the project; M.G. developed the methodology with contributions by U.O. and implemented the method and performed the analysis. M.G. and U.O. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.247494.118.
References
- Alipanahi B, Delong A, Weirauch MT, Frey BJ. 2015. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 33: 831–838. 10.1038/nbt.3300 [DOI] [PubMed] [Google Scholar]
- Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. 2012. Identification of RNA–protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA 3: 159–177. 10.1002/wrna.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avsec Z, Barekatain M, Cheng J, Gagneur J. 2018. Modeling positional effects of regulatory sequences with spline transformations increases prediction accuracy of deep neural networks. Bioinformatics 34: 1261–1269. 10.1093/bioinformatics/btx727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami-Samani E, Xing Y. 2019. Discovery of allele-specific protein-RNA interactions in human transcriptomes. Am J Hum Genet 104: 492–502. 10.1016/j.ajhg.2019.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. 10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Bassat I, Chor B, Orenstein Y. 2018. A deep neural network approach for learning intrinsic protein-RNA binding preferences. Bioinformatics 34: i638–i646. 10.1093/bioinformatics/bty600 [DOI] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM. 2003. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell 12: 1467–1476. 10.1016/S1097-2765(03)00453-2 [DOI] [PubMed] [Google Scholar]
- Chung J, Gulcehre C, Cho K, Bengio Y. 2014. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv:1412.3555 [cs.NE].
- Cooper TA, Wan L, Dreyfuss G. 2009. RNA and disease. Cell 136: 777–793. 10.1016/j.cell.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. 2011. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 12: R79 10.1186/gb-2011-12-8-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon SK, Lutz CS. 2012. mRNA 3′ end processing factors: a phylogenetic comparison. Comp Funct Genomics 2012: 876893 10.1155/2012/876893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, et al. 2018. Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell 70: 854–867.e9. 10.1016/j.molcel.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epis MR, Barker A, Giles KM, Beveridge DJ, Leedman PJ. 2011. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem 286: 41442–41454. 10.1074/jbc.M111.301481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. 2015. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 43: D805–D811. 10.1093/nar/gku1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev S, Boyle AP, Jayasurya K, Ding X, Mukherjee S, Ohler U. 2010. Evidence-ranked motif identification. Genome Biol 11: R19 10.1186/gb-2010-11-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Bengio Y, Courville A. 2016. Deep learning. MIT Press, Cambridge, MA. [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller D, Krestel R, Ohler U, Vingron M, Marsico A. 2017. ssHMM: extracting intuitive sequence-structure motifs from high-throughput RNA-binding protein data. Nucleic Acids Res 45: 11004–11018. 10.1093/nar/gkx756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. 2010. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comput Biol 6: e1000832 10.1371/journal.pcbi.1000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. 2001. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci 98: 7018–7024. 10.1073/pnas.111145598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Reshef YA, Bileschi M, Belanger D, McLean CY, Snoek J. 2018. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res 28: 739–750. 10.1101/gr.227819.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma DP, Ba J. 2014. Adam: a method for stochastic optimization. arXiv:1412.6980 [cs.LG].
- Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. 2011. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods 8: 559–564. 10.1038/nmeth.1608 [DOI] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakau S, Richard H, Marsico A. 2017. PureCLIP: capturing target-specific protein–RNA interaction footprints from single-nucleotide CLIP-seq data. Genome Biol 18: 240 10.1186/s13059-017-1364-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. 2014. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell 54: 887–900. 10.1016/j.molcel.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanchantin J, Singh R, Wang B, Qi Y. 2017. Deep motif dashboard: visualizing and understanding genomic sequences using deep neural networks. Pac Symp Biocomput 22: 254–265. 10.1142/9789813207813_0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Ewing AD, Hancks DC, Kazazian HH. 2013. Enrichment of processed pseudogene transcripts in L1-ribonucleoprotein particles. Hum Mol Genet 22: 3730–3748. 10.1093/hmg/ddt225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′UTR length. Cell Rep 1: 753–763. 10.1016/j.celrep.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Maticzka D, Lange SJ, Costa F, Backofen R. 2014. GraphProt: modeling binding preferences of RNA-binding proteins. Genome Biol 15: R17 10.1186/gb-2014-15-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Saul N, Großberger L. 2018. UMAP: Uniform Manifold Approximation and Projection. J Open Source Softw 3: 861 10.21105/joss.00861 [DOI] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Tuschl T, Ohler U, Keene JD. 2011. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43: 327–339. 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Wessels HH, Lebedeva S, Sajek M, Ghanbari M, Garzia A, Munteanu A, Yusuf D, Farazi T, Hoell JI, et al. 2019. Deciphering human ribonucleoprotein regulatory networks. Nucleic Acids Res 47: 570–581. 10.1093/nar/gky1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munteanu A, Mukherjee N, Ohler U. 2018. SSMART: sequence-structure motif identification for RNA-binding proteins. Bioinformatics 34: 3990–3998. 10.1093/bioinformatics/bty404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Shen HB. 2017. RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC Bioinformatics 18: 136 10.1186/s12859-017-1561-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass M, Rasmussen SH, Krogh A. 2017. Highly accessible AU-rich regions in 3′ untranslated regions are hotspots for binding of regulatory factors. PLoS Comput Biol 13: e1005460 10.1371/journal.pcbi.1005460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang D, Xie X. 2016. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res 44: e107 10.1093/nar/gkw226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrikumar A, Greenside P, Kundaje A. 2017. Learning important features through propagating activation differences. In Proceedings of the 34th international conference on machine learning, Proceedings of Machine Learning Research (ed. Precup D, Teh YW), Vol. 70, pp. 3145–3153.
- Siddiqui N, Borden KL. 2012. mRNA export and cancer. Wiley Interdiscip Rev RNA 3: 13–25. 10.1002/wrna.101 [DOI] [PubMed] [Google Scholar]
- Stražar M, Žitnik M, Zupan B, Ule J, Curk T. 2016. Orthogonal matrix factorization enables integrative analysis of multiple RNA binding proteins. Bioinformatics 32: 1527–1535. 10.1093/bioinformatics/btw003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan M, Taly A, Yan Q. 2017. Axiomatic attribution for deep networks. In Proceedings of the 34th international conference on machine learning, Proceedings of Machine Learning Research (ed. Precup D, Teh YW), Vol. 70, pp. 3319–3328. [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Van Nostrand EL, Yeo GW. 2018. Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip Rev RNA 9: e1436 10.1002/wrna.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Coseno M, Gilmartin GM, Doublié S. 2011. Crystal structure of a human cleavage factor CFIm25/CFIm68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 19: 368–377. 10.1016/j.str.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EW, Bahn JH, Hsiao EY, Tan BX, Sun Y, Fu T, Zhou B, Van Nostrand EL, Pratt GA, Freese P, et al. 2019. Allele-specific binding of RNA-binding proteins reveals functional genetic variants in the RNA. Nat Commun 10: 1338 10.1038/s41467-019-09292-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong F, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H, et al. 2014. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet 10: e1004289 10.1371/journal.pgen.1004289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.