Abstract
Liver organogenesis and development are composed of a series of complex, well-orchestrated events. Identifying key factors and pathways governing liver development will help elucidate the physiological and pathological processes including those of cancer. We conducted multidimensional omics measurements including protein, mRNA, and transcription factor (TF) DNA-binding activity for mouse liver tissues collected from embryonic day 12.5 (E12.5) to postnatal week 8 (W8), encompassing major developmental stages. These data sets reveal dynamic changes of core liver functions and canonical signaling pathways governing development at both mRNA and protein levels. The TF DNA-binding activity data set highlights the importance of TF activity in early embryonic development. A comparison between mouse liver development and human hepatocellular carcinoma (HCC) proteomic profiles reveal that more aggressive tumors are characterized with the activation of early embryonic development pathways, whereas less aggressive ones maintain liver function–related pathways that are elevated in the mature liver. This work offers a panoramic view of mouse liver development and provides a rich resource to explore in-depth functional characterization.
The liver is a metabolic hub that produces, stores, and recycles many life-essential molecules (Rui 2011). Liver cancer is the third most common cause of cancer death worldwide. Previous studies of animal models have shed light on the mechanisms of liver organogenesis and development (Zhao and Duncan 2005; Si-Tayeb et al. 2010; Gordillo et al. 2015; Gruppuso and Sanders 2016). One of the most significant findings is the identification of key signaling events in orchestrating liver development, including Hippo (Yimlamai et al. 2014; Lee et al. 2016; Patel et al. 2017; Lu et al. 2018), Wnt/CTNNB1 (Thompson and Monga 2007; Nejak-Bowen and Monga 2008; Yang et al. 2014; Russell and Monga 2018), transforming growth factor beta (TGFB) (Bissell et al. 2001; Clotman et al. 2005), fibroblast growth factor (FGF) (Jung et al. 1999; Zhang et al. 2004; Calmont et al. 2006; Shin et al. 2007), hepatocyte growth factor (HGF) (Huh et al. 2004; Shin and Monga 2013; Ye et al. 2015), Notch (Zong et al. 2009; Morell and Strazzabosco 2014; Geisler and Strazzabosco 2015), and bone morphogenetic protein (BMP) signaling pathways (Zhang et al. 2004; Shin et al. 2007; Wang et al. 2014; Choi et al. 2017). Additionally, critical transcription factors and epigenetic regulators such as FOXA and GATA families, Homeobox genes, Hepatocyte nuclear factors (HNFs), and NR5A2 (Zhao and Duncan 2005; Si-Tayeb et al. 2010; Sheaffer and Kaestner 2012; Gordillo et al. 2015) have been discovered as key regulators. Deregulation of these pathways is also linked to liver diseases. For example, activation of Wnt/CTNNB1 pathway has been shown to contribute to liver disease (Monga 2015), Hippo signaling can restrain liver overgrowth and tumorigenesis (Wu et al. 2015), defective Notch signaling leads to the impaired ability to repair liver damage, and excessive Notch activation could contribute to liver cancer (Morell et al. 2013).
RNA-seq has been used to monitor dynamic changes in liver transcriptome in development (Gunewardena et al. 2015), to quantify mRNA abundance of transporter genes and their alternative transcript isoforms (Cui et al. 2012), and to determine the expressions of the cytochromes P450 family of genes and their isoforms (Peng et al. 2012). Temporal transcriptomic and proteomic studies have been performed in flies and mice (Casas-Vila et al. 2017; Gao et al. 2017). More recently, single-cell RNA-seq (scRNA-seq) has been used to measure the differentiation and maturation trajectories of stem/progenitor cells in fetal livers (Su et al. 2017) and outline the characteristics of resident cells of the human liver and its immune microenvironment. Studies on expression quantitative trait loci and protein quantitative trait loci show the distinct transcriptional and post-transcriptional regulations in human cells and mouse liver, highlighting the unique value of proteomes in annotating complex trait analysis together with transcriptomics, genomics, and metabolomics (Battle et al. 2015; Chick et al. 2016; Williams et al. 2016). The RNA results still need to be verified at the protein level. Moreover, the information on the activity of the drivers of development transcription factors (TFs) is still lacking.
Here, we conducted an integrative proteomic and transcriptomic characterization of developing mouse liver from embryonic day 12.5 (E12.5) to postnatal week 8 (W8), and the analysis of TF DNA binding activities during the developmental period. Our data present a multidimensional omics overview of the mouse liver covering major stages of liver development.
Results
A dynamic mouse liver proteome during development
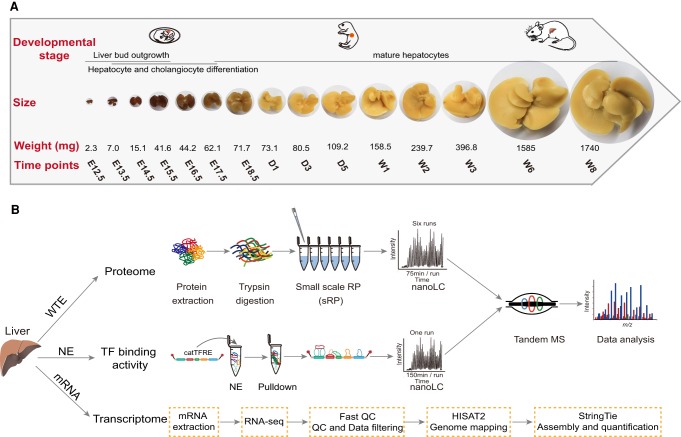
We conducted a temporal, in-depth multi-omics analysis on mouse liver tissues collected at 15 time points from embryonic day 12.5 (E12.5, the earliest date that fetal liver is visible when the resection is possible) to postnatal week 8 (W8) (Fig. 1A). The sampling time window covers the embryonic period (E12.5, E13.5, E14.5, E15.5, E16.5, E17.5, and E18.5) when liver forms, the postnatal days (D1, D3, D5) when liver undergoes early developmental stages, and the postnatal weeks (W1, W2, W3, W6, W8) when liver development is complete and the liver is fully functional (Fig. 1B).
Figure 1.
Temporal, multi-omics profiling of the developing mouse liver. (A) Liver sampling time window and the coverage of developmental stages. (B) Experimental procedure and data analysis workflow of the mouse liver proteome, transcription factor binding activities, and transcriptome.
We performed the whole proteome profiling using a prefractionation strategy with a small-scale reversed phase (sRP) chromatography (Fig. 1B), a workflow that has proven to achieve high sensitivity and reproducibility (r > 0.8) in the quality control runs (Supplemental Fig. S1A). We quantified 7261 ± 392 gene products (GPs) (Mean ± SD) on a single time point and 11,984 GPs in total at 1% peptide-level false discovery rate (FDR) (Supplemental Fig. S1B; Supplemental Table S1). The 2242 GPs that were present in all stages were designated as the core mouse liver proteome (Supplemental Fig. S1C). The main functional categories of the core proteome included cell–cell adhesion, translation, oxidation–reduction process, protein transport, and the metabolic process (Supplemental Fig. S1D). The relative abundance of the GPs (normalized as iFOT) spanned nearly six orders of magnitude (Supplemental Fig. S1E). The abundance of the housekeeping proteins such as TUBB4B and HSP90B1 remained largely unchanged, whereas the hepatic cell markers (e.g., albumin [ALB]; alpha fetoprotein [AFP]) showed highly dynamic behavior during liver development (Supplemental Fig. S1F). For comparison, we also performed RNA sequencing on samples collected during the same developmental period. The transcriptome achieved a deeper coverage with a total of 15,061 expressed genes (FPKM > 1) (Supplemental Table S2).
Key phases of the liver development and functions
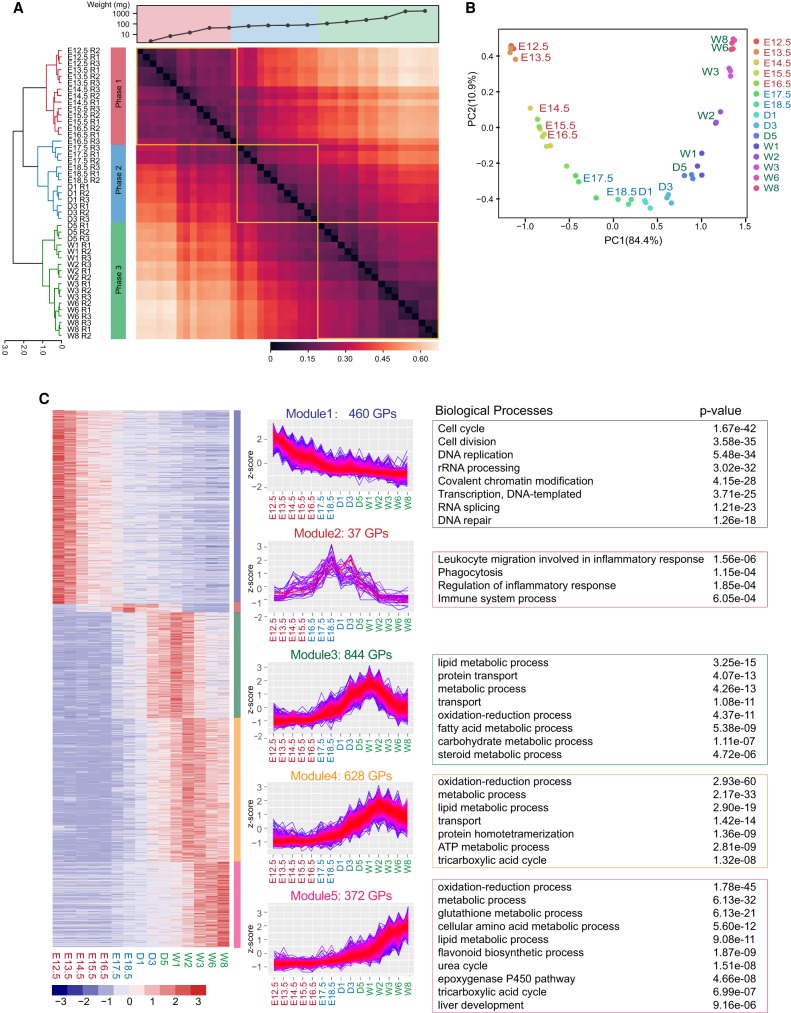
To achieve higher confidence, 6458 GPs identified in at least two of the three replicates in at least one time point were used for further statistical analyses (Supplemental Fig. S1G; Supplemental Table S1). Hierarchical clustering (Fig. 2A) partitioned samples into three clusters: Phase 1 (E12.5–E16.5), the early embryo phase, when the liver rapidly gained weight; Phase 2 (E17.5–W2), the perinatal phase, when the weight gain was slower but constant; and Phase 3 (W3–W8), the matured phase, when the weight gain again became pronounced. Principal component analysis (PCA) showed the partition of data points in a time-dependent manner (Fig. 2B). One-way ANOVA analysis identified 3100 GPs that were differentially expressed (FDR < 0.01), and 2341 proteins whose expressions were at least twofold higher in one phase than in any other phase were defined as phase-specific proteins (Supplemental Table S1).
Figure 2.
Three major developmental phases and five temporal expression modules revealed by the proteomics analysis. (A) Hierarchical clustering analyses of temporal proteomics data separated the liver development into three phases. (B) Principal component analysis of the temporal proteomics data. Each dot represents an independent biological replicate. (C) Five temporal expression modules revealed by k-means clustering and the major biological processes they govern.
To gain insights into the temporal behavior of the differentially expressed proteins, we performed an unsupervised k-means clustering analysis (Supplemental Methods), resulting in five modules enriched with distinct Gene Ontology (GO) terms (Fig. 2C; Supplemental Fig. S2A,B; Supplemental Table S1). Proteins in Module 1 are mainly involved in cell-cycle and RNA transcription, including DNA replication and repair, cell division, rRNA processing, ribosome biogenesis, and RNA transport. Module 2 proteins participate in inflammatory response, phagocytosis, and immune response. Although proteins in Modules 3–5 differ in timing for peak intensity, they are enriched in similar biological processes including oxidation–reduction, metabolism, and transport, which are all essential for adult liver function. It appears that cell proliferation is the predominant pathway during early development, whereas the metabolism-related functions emerge when the liver becomes mature.
Hierarchical clustering and PCA were also used to separate the RNA-seq data into three phases with minor difference from the proteome-based classification (Supplemental Fig. S3A,B). Likewise, k-means clustering of the differentially expressed RNA transcripts revealed five gene modules with similar expression profiles and functional annotations as those obtained from the proteomic data (Supplemental Fig. S3C; Supplemental Table S2).
Dynamic changes in core liver functions and hepatic cell types
Fetal liver is a major organ for hematopoiesis, whereas the adult liver plays a key role in metabolism (Gebhardt 1992; Morrison et al. 1995). These functions are carried out by specialized hepatic cell types either individually or cooperatively. A question is when the specialization takes place.
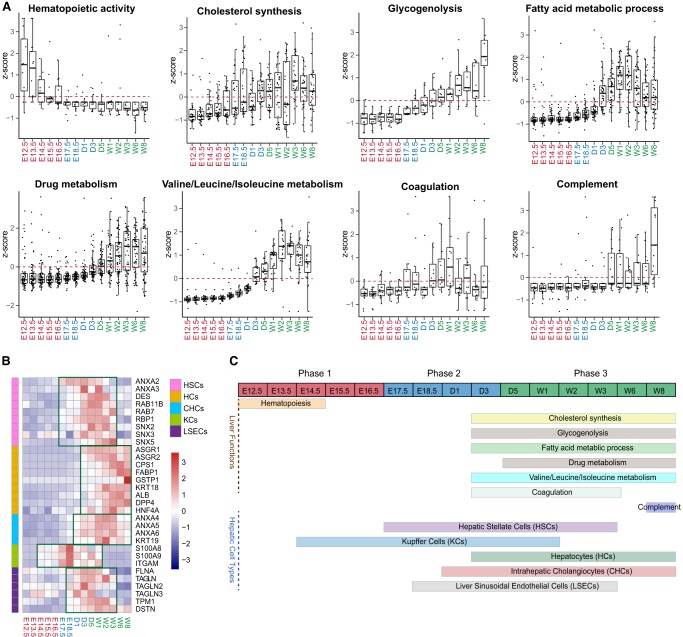
We retrieved key cell-type markers from a previous publication (Azimifar et al. 2014) and proteins involved in the eight core liver functions from the common knowledge databases (KEGG, Reactome, and Gene Ontology) and mapped their temporal patterns along the developmental time scale (Fig. 3A–C; Supplemental Table S3). As expected, hematopoiesis occurs exclusively in Phase 1 with the highest activity observed during E12.5 to E14.5; cholesterol synthesis, glycogenolysis, drug metabolism, Valine/Leucine/Isoleucine metabolism, fatty acid metabolic process, coagulation and complement are predominantly enriched in Phase 3 when the liver growth plateaus (Fig. 3A,C).
Figure 3.
The dynamic changes of core liver functions and hepatic cell types. (A) Proteins involved in the eight core liver functions were retrieved from the common knowledge databases (KEGG, Reactome, and Gene Ontology) and their temporal expression patterns were mapped along the developmental time scale. (B) Relative expression levels of five hepatic cell types represented by their selective markers or cell-type-specific proteins. (C) A summary of the eight liver functions and the expressions of the five hepatic cell types on the development time scale.
Temporal expression patterns of the cell-type-specific protein markers indicate that the five cell types are formed in a regulated manner during liver development (Fig. 3B). Markers for Kupffer Cells (KCs) appeared the earliest and spanned all three phases. For instance, S100A8 and S100A9, two calcium-binding proteins known to promote inflammation and autoimmune response via toll-like receptor 4 signaling, show high levels in Phase 2 (Loser et al. 2010). Hepatic stellate cells (HSCs), which are activated in response to liver damage, are highly enriched with intracellular trafficking proteins such as ANXA2/3, SNX2/3/5, and RAB7/11, and are expressed at high levels during Phase 2 and early Phase 3. Markers for HSCs and Liver sinusoidal endothelial cells (LSECs) are more abundantly expressed in Phase 1 and Phase 2. Several LSEC-specific markers, including ACTA1, TAGLN, DSTN, FLNA, TPM1, and MYO are the components of the cytoskeleton and actomyosin machinery. The high expression of these proteins in Phases 2 and 3 reinforces the notion that LSECs function in lining the blood vessels. Hepatocytes (HCs) are the predominant cell type that constitutes nearly 80% of the cells in the adult liver. HC markers start to express at high levels in late Phase 2 and Phase 3. Liver-specific fatty acid binding proteins (FABPs, GSTP1, and CPS1) are the three most abundant proteins in HC. These proteins gradually increased starting from Phase 1 and peaked in Phases 2 and 3 (Fig. 3B). This temporal pattern coincides with the enrichment of GO terms for metabolic process, lipid metabolic process, and urea cycle in the mature liver (Fig. 3C). Intrahepatic cholangiocytes (CHCs) cell markers ANXA4, ANXA5, ANXA6, and KRT19 appeared in late Phase 2 and Phase 3, during the time when the bile duct formed (Gordillo et al. 2015).
The transcriptome revealed similar temporal expression patterns of cell type and core liver functions (Supplemental Fig. S4A,B). In general, the fetal liver is enriched in genes that function in hematopoiesis, but the mature liver is enriched in genes for metabolic functions, with the exception of glycogenolysis. Although the mRNA transcripts for glycogenolysis were mostly seen in the early phases of the development, the proteins were not observed until a later phase, demonstrating a greater discordance between mRNA and proteins (Fig. 3A; Supplemental Fig. S4A). Although the time frames that outlined the formation of HC, CHC, and KC showed nearly identical expression patterns in mRNA and protein, they appeared to be different for HSCs and LSECs (Fig. 3B; Supplemental Fig. S4B).
Global profiling of transcription factor DNA-binding activities
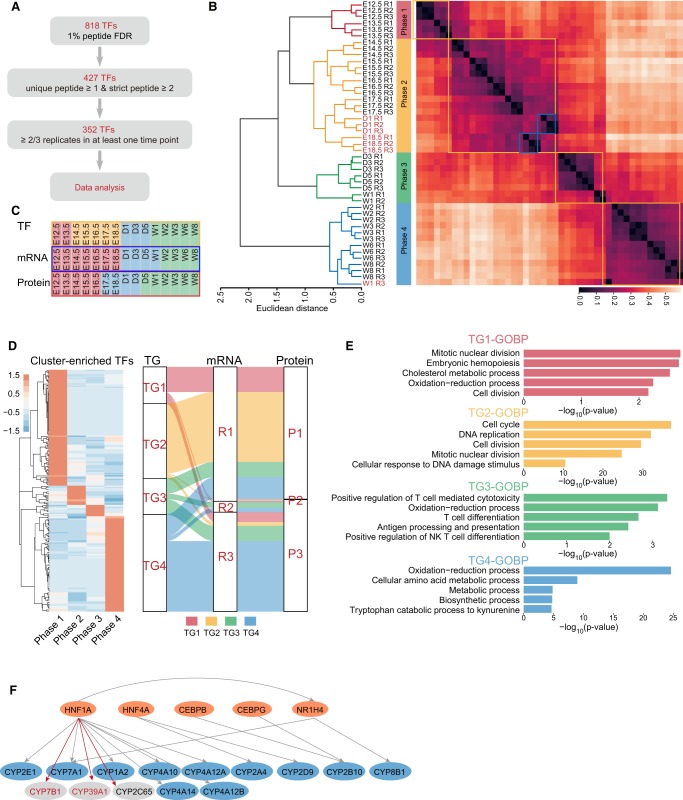
Transcription factors (TFs) control their target gene expressions upon development. Because the transcriptional activity is often regulated by post-translational modifications and protein–protein interactions, the abundance of a TF does not always reflect its activity. Here, we used a concatenated tandem array of the consensus TF response elements (catTFRE) (Ding et al. 2013a) to pull down TFs from mouse liver nuclear extract, which could better reflect TF activities. The TFRE pulldowns captured 818 transcription factors that covered the majority of the TF families (Fig. 4A; Supplemental Table S4) with Zf-C2H2, Homeobox, and bHLH as the top three transcription factor families (Supplemental Fig. S5A; Hu et al. 2019). Based on the expression patterns of the 352 high-confidence TFs, hierarchical clustering (Fig. 4B) and PCA (Supplemental Fig. S5B) separated the developing liver into four phases: TF Phase 1 (E12.5 and E13.5), TF Phase 2 (E14.5 to D1), TF Phase 3 (D1 to D5), and TF Phase 4 (W2–W8). Thus, although the four phases generally corresponded with those derived from the whole proteome and transcriptome profiling, the TF-based classification provided a higher resolution of early development (Fig. 4C). Next, we identified phase-specific TFs, the TFs whose mean values in one phase are at least twofold higher than the means in the other three phases, resulting in 102, 17, 10, and 84 TFs for TF Phases 1–4, respectively (Fig. 4D; Supplemental Table S4).
Figure 4.
Analysis of transcription factors in developing mouse liver. (A) Flowchart for TFRE pulldown data quality control. (B) Hierarchical clustering analysis of the temporal expression of transcription factors separated the liver development into four phases. Each data point represents an independent replicate. (C) A comparison of developmental phases obtained by transcription factor binding activities, RNA expression, and protein expression. (D, left) Cluster-enriched phase-specific TFs; (right) a comparison of predicted downstream transcriptional targets (TG1–TG4) corresponding to TF Phases 1–4 and the distribution of these TGs detected in the RNA- or protein-based clustering. (E) Enrichment of the biological processes by the four TG groups. (F) Prediction of HNF1A as an upstream TF for CYP7B1 (protein and mRNA), CYP39A1 (protein and mRNA), and CYP2C65 (protein) by their coexpression with CYP family members known to be HNF1A TF targets (for heatmaps, see Supplemental Fig. S5D). (Orange) TFs; (blue) TGs known to be regulated by the TFs; (gray) TGs potentially to be regulated by the TF.
To find correlations between TF activities and the expression of their downstream transcriptional targets (TGs), we used CellNet (Cahan et al. 2014), a database of gene regulatory networks, to predict the groups of transcripts that are potentially regulated by the phase-specific TFs. This resulted in four TG groups (TG1–TG4) that correspond to protein (TG-Protein1-4) and mRNA (TG-RNA1-4) (Supplemental Table S5). Temporal expression patterns of the majority of TG1s and TG2s as measured by mRNA and protein profiling show that they are expressed at higher levels mainly in R1 and P1 (Fig. 4D; Supplemental Fig. S5C), with GO terms enriched in mitotic nuclear division, embryonic hemopoiesis, and cell division (Fig. 4E). Similarly, the expression of the predicted TG3 and TG4 transcripts as well as proteins is at higher levels mainly during R3 and P3, respectively, with GO terms enriched in metabolic and immune system processes (cf. Fig. 4E with profiling Fig. 2C and transcriptome GO figures Supplemental Fig. S3C). These results suggest that TF activities can correctly predict target gene expressions and functions during early and late development periods. However, few predicted TGs were found to be enriched in R2 and P2. The reason for the discrepancy is not clear.
Cytochrome P450s, a superfamily of proteins for drug metabolism, is highly expressed during Phase 3. Previous studies (Jover et al. 2001; Akiyama and Gonzalez 2003) show that the expression of the hundreds of CYP isoforms is regulated by a number of TFs, including HNF1A, HNF4A, CEBPB, CEBPG, and NR1H4 (Fig. 4F). Consistently, the expressions of several HNF1A-regulated CYPs are highly correlated (Supplemental Fig. S5D). This cluster also includes CYP7B1, CYP39A1, and CYP2C65, whose upstream TFs are not known. The close correlation between these CYPs and the HNF1A target genes raised the possibility that they may also be HNF1A target genes.
Integrative analysis of the transcriptome, proteome, and DNA-bound TFs
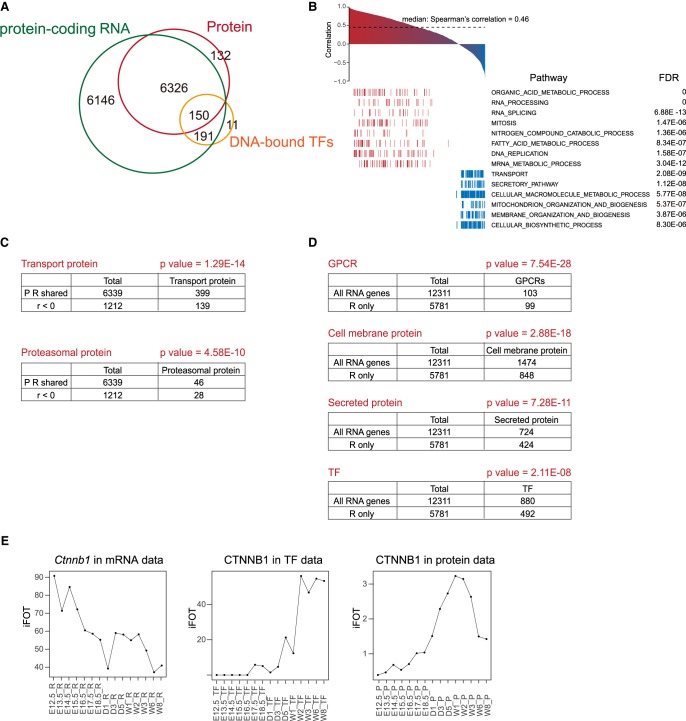
The temporal expression patterns obtained by RNA sequencing and proteomic profiling revealed not only resemblances but also distinct features. In total, 6326 GPs were identified in both RNA and protein data sets with a median Spearman's correlation coefficient of 0.46, similar to previous studies (Fig. 5A; Schwänhausser et al. 2011). Genes with positive correlation are significantly enriched in organic acid metabolism, RNA processing, and RNA splicing; genes with negative correlation are enriched in transport, secretion, and cellular macromolecule metabolism (Fig. 5B,C).
Figure 5.
Integrative analysis of the transcriptome, proteome, and DNA-bound TFs. (A) Venn diagram of total number of transcripts, proteins, and the number of overlapping TFs detected in the TFRE pulldown data set. (B) Protein-mRNA correlation calculated by Spearman's correlation coefficient. Pathways in red are positively correlated (r > 0.3); pathways in blue are negatively correlated (r < 0). (C) Protein-mRNA levels of the transport proteins and proteasomal proteins are negatively correlated (r < 0). P-values were calculated by hypergeometric distribution. (D) P-values of GPCRs, cell membrane protein, secreted protein, and TFs in the RNA-only data set calculated by the hypergeometric distribution. (E) Temporal expression levels of CTNNB1 by the mRNA, transcription factor, and protein measurements.
There are 8544 genes that were detected by the RNA-seq only; among them, 6146 (71.9%) are protein-coding RNAs as annotated in the Ensembl and NCBI database (Fig. 5A; Supplemental Fig. S6A; Supplemental Table S6). Pseudogenes, lncRNAs, snoRNAs, and miRNAs constitute 92.3% of all noncoding genes (Supplemental Fig. S6B). For the 6146 protein-coding genes, RNA transcripts of secreted proteins including chemokines/cytokines, growth factors, and membrane proteins that may be difficult to solubilize (transmembrane, ion channel, and GPCRs) are significantly enriched in this data set, suggesting a possibility that these proteins may have expressed but escaped detection (Fig. 5D). Transcription factors and transcription coregulators are significantly overrepresented in the RNA-only data set, constituting ∼55% of all 893 TFs detected in this study (Fig. 5D).
Protein stability could account for the inability to detect proteins in RNA-only transcripts (Supplemental Table S6). For example, Trp53, which encodes the tumor suppressor TRP53 that is degraded by MDM2 in nonstress conditions, was detected with a maximal FPKM of 97 at early embryonic days. Cdt1, which encodes a component of the prereplication complex, was detected with a maximal FPKM of 101 in E13.5. CDT1 is regulated in part through ubiquitin-dependent proteolysis at the onset of the S-phase to ensure that replication occurs only once per cell cycle.
The AP-1 family of transcription factors Atf4 and Atf5 are among the highest expressing transcripts in the RNA-only data set (Supplemental Table S6). The AP-1 transcription factor family regulates a variety of cellular processes; their activities are regulated at multiple levels, including protein translation. In fact, ATF4/5 is mainly under translational regulation (Vesely et al. 2009).
The TFRE pulldown data set provides an added value in investigating the correlation between protein expression and transcription activities. Overall, the correlation of TFRE-mRNA (median = 0.25) is higher than that of TFRE-Protein (median = 0.19) and both are markedly lower than that of Protein-RNA (Supplemental Fig. S6C,D). For the 149 TFs found in all three data sets, the correlation is the highest between mRNA-Protein (median 0.54), slightly higher than that of all gene products (0.46), and the lowest between mRNA-TFRE (median 0.07) (Supplemental Table S7).
TFs whose activities are positively correlated with their protein abundance include PURA, PURB, and CTF domain TFs NFIA, B, and C (Supplemental Table S6). Protein abundance of CTNNB1 (also known as beta-catenin), a key transcription coregulator of the Wnt pathway, is positively correlated with its DNA-binding activity, but is negatively correlated with its RNA abundance (Fig. 5E). Since CTNNB1 is phosphorylated and degraded through proteolysis in the absence of WNT ligands, the negative RNA-Protein correlation implies that protein stability plays a major regulatory role during early embryonic days. Conversely, the strong positive correlation between DNA binding and protein abundance suggests that the stabilized CTNNB1 is mostly active upon WNT activation, as the timing of the first CTNNB1 activation peak (E16.5–D1) coincides with that of TCF7 and LEF1, key partner TFs of CTNNB1 in transcription, and of Wnt4 RNA.
Estrogen-related receptor gamma (ESRRG) was detected only by the TFRE pulldown. ESRRG is an orphan nuclear receptor and regulates cell growth and tumorigenesis in various cancers, but its function in liver homeostasis is poorly understood. It has been reported that O-GlcNAcylation of ESRRG promotes hepatic gluconeogenesis (Misra et al. 2016). The increased DNA binding of ESRRG in TF Phase 4 is consistent with its role in regulating glucose metabolism in adult liver.
Essential pathways for liver development and their connections to HCC
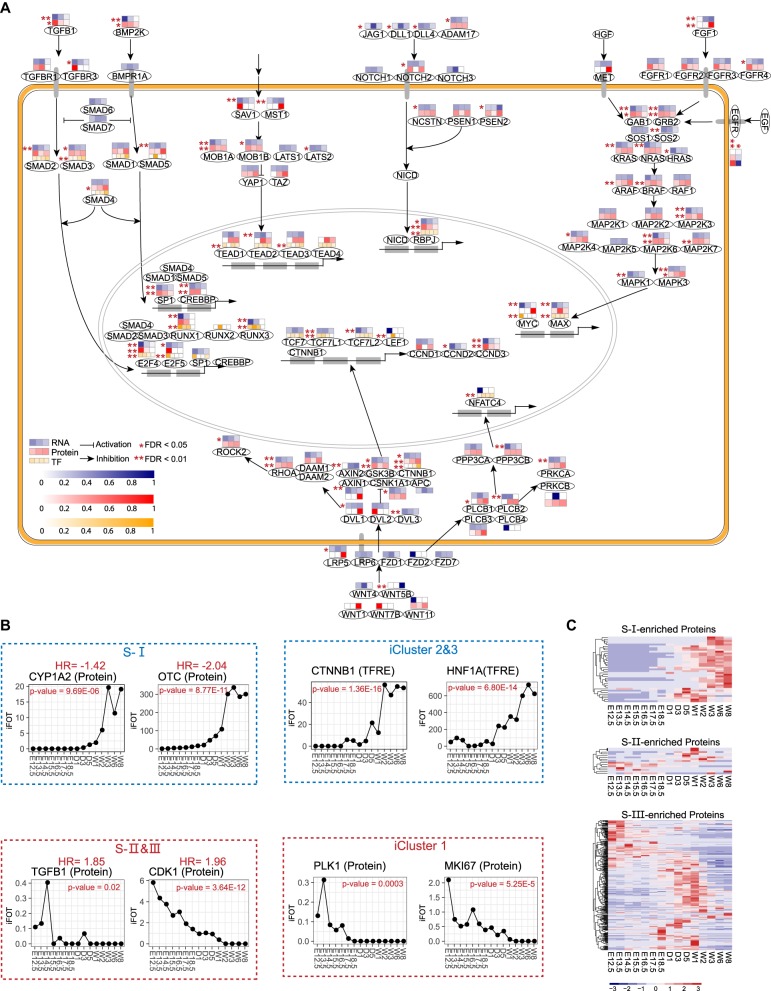
It has been observed that embryogenesis and tumorigenesis share common and distinct characteristics; although both are governed by few key signaling pathways, the former responds to developmental cues and is spatially and temporally regulated, whereas the latter is misregulated, which could result in the reactivation of some embryonic developmental processes that need to be repressed in adult tissues (Aiello and Stanger 2016). We surveyed the three data sets for key factors involved in the signaling pathways implicated in liver development (Fig. 6A). Of the 115 proteins shown, ∼61% (69 proteins) showed significant alterations (ANOVA, Q < 0.01) during the developmental period examined and the majority of these factors are expressed at higher levels in Phase 1 or Phase 2. Changes in mRNA are largely reflected in the changes of their protein counterparts (Fig. 6A).
Figure 6.
Analyses of key signaling pathways related to liver development. (A) Changes in effectors of key signaling pathways based on KEGG and other studies (Yimlamai et al. 2014) (TFs, proteins, or RNAs). An ANOVA test was performed to identify statistically significantly changed proteins or genes. (B) Temporal expression levels of selected prognosis signature proteins across 15 time points during liver development. Proteins in blue boxes are correlated with better prognosis; proteins in red boxes are correlated with poor prognosis. iCluster 1–3 subtypes were obtained from the TCGA paper (The Cancer Genome Atlas Research Network 2017); S-I, -II, -III subtypes were obtained from the HCC paper (Jiang et al. 2019). (C) Heatmaps showing temporal expression patterns of S-I-, S-II-, or S-III-enriched proteins across 15 time points during liver development.
SMAD proteins are key players in the TGFB- and BMP-mediated pathways in cell growth, differentiation, and development (Wu and Hill 2009). Seven SMAD proteins were detected at earlier phases by MS and RNA-seq; highest expressions of the SMADs’ downstream effectors were also found in earlier phases. Three NOTCH proteins (NOTCH1-3) and their downstream transcription factor RBPJ showed higher expression levels in Phase 1 and Phase 2, implying that the biliary differentiation is an early event. RAS, RAF, MAPK, and MAP2K are common effectors in several RTK-mediated pathways, including HGF, EGF, and FGFs. The expression patterns of these proteins in Phase 1 is in line with the time frame of hepatic specification (Gordillo et al. 2015). A subset of WNT target genes are highly expression in Phase 1, whereas other WNT target genes that are required for liver function are highly expressed in Phase 3.
Recent transcriptomic and proteomic analyses of HCC provide new insights into the association of molecular subtypes and clinical relevance. An integrative molecular HCC subtyping incorporating five data sets by The Cancer Genome Atlas (TCGA) identifies three molecular subtypes (iCluster 1–3) (The Cancer Genome Atlas Research Network 2017). iCluster 1 is characterized with overexpression of proliferation marker genes that is associated with poor prognosis and higher tumor grade. In contrast, iCluster 2 and 3, which are associated with low-grade tumors, showed a high frequency in mutations of liver-function genes including CTNNB1 and HNF1A. Most recently, a study reported the subtyping of early-stage hepatocellular carcinoma into three subtypes (named as S-I, S-II, and S-III) that are associated with different clinical outcomes (Jiang et al. 2019). The S-III subtype, which has a significantly lower overall rate of survival, is characterized with the activation of tumor-promoting pathways and the silencing of tumor-suppressor pathways; in contrast, the less aggressive S-I subtype is characterized with the up-regulation of liver-function-related proteins and pathways, suggesting hepatocyte-like characteristics in these tumors.
We examined the expression patterns of the 19 prognosis signature proteins listed in Jiang et al. (2019) during liver development (Supplemental Fig. S7A). As shown in Figure 6B, CYP1A2 and OTC, two proteins that are enriched in S-I with negative Hazard Ratios (HRs) are highly expressed in Phase 3. Similar trends are also observed for iCluster 2 and 3, (two subtypes with better prognosis in the TCGA paper) (The Cancer Genome Atlas Research Network 2017) enriched proteins CTNNB1 and HNF1A (Supplemental Fig. S7B). In contrast, the six proteins (CDK1, HDAC2, PCNA, PRPF4, SLC3A2, and TGFB1) (Supplemental Fig. S7A) with positive HRs that are enriched in S-II or S-III display the highest expressions on E12.5 or E14.5. This expression pattern is also evident for proliferation markers such as PLK1, MKI67, and MYBL2 found in iCluster 1 (Supplemental Fig. S7B), the subtype that has the lowest survival rate.
To obtain additional evidence of association between liver development and the proteomic HCC subtypes, we identified proteins that are differentially expressed in each HCC subtype. For the 1269 proteins that show the most variations among HCC subtypes, 1033 are differentially expressed in each subtype (One-way ANOVA, FDR < 0.01) (Supplemental Table S8), and 472 proteins whose mean expressions in the highest expressing subtype are two times higher than those in the second highest subtype are defined as subtype signature proteins (Supplemental Table S8). Of the 472 signature proteins, 345 mouse counterparts are identified in the liver development data set (Fig. 6C; Supplemental Table S8). The 37 S-I signature proteins are differentially expressed almost exclusively in late Phase 2 or Phase 3, particularly from W3–W8. This feature is consistent with the hepatic characteristics of the S-I tumors and indicates that tumors that retain their hepatic function have a good prognosis. In contrast, the majority of the S-III signature proteins are differentially expressed in either early embryo days (E12.5–E13.5) or during D1–W2 with functional enrichment in organ development, immune system process, defense response, and signal transduction. The S-II has the least number of signature proteins. It appears that many proteins that are highly expressed in S-II are also highly expressed in either S-I or S-III. Together these analyses suggest that tumors with aberrant expression of proliferation proteins or activated in pathways that need to be silenced in adult liver are correlated with poorer clinical outcomes, whereas tumors that overexpress liver-function-related proteins are associated with better clinical outcomes.
Discussion
The transcriptomic, proteomic, and DNA-binding activity profiling during liver development in this study revealed a multidimensional, high-resolution atlas that can be further characterized in detail. The streamlined proteomic workflow allowed the accurate measurement of protein abundance of 11,984 GPs across the 15 sampling time points. The TFRE data set contains 818 TFs compared with 370 TFs from protein profiling, allowing not only for more accurate quantification of low abundant TFs, but inferring their regulatory roles during development. Moreover, the TFRE-based clustering separated the earliest development phase obtained by RNA and protein profiling further into two phases, highlighting the high TF activity during E12.5 and E13.5.
Although the RNA-seq-based transcriptome and the proteome have moderate correlation, they do share many common features, including the six functionally similar gene modules, similar temporal expression profiles of genes that govern core liver functions, and features of the five hepatic cell types. An exception is the discordance between RNA and protein markers of HSCs and LSECs (Fig. 3B; Supplemental Fig. S4B). This discordance is not likely caused by technical issues because both the data quality and the abundance are high. Because the peaks of mRNA in both cells precede those of protein, it could be rationalized that protein synthesis takes time. One possible explanation is that the HSC and LSEC markers are inaccurate, which is exacerbated by the low abundance of HSC and LSEC cells.
Our integrative analysis of mRNA transcripts, proteins, and DNA-bound TFs links transcription factor activities to mouse liver development. Although the low correlation between mRNA and TF DNA-binding activity may be understandable as post-transcriptional and translational regulation are in play, the much lower correlation between protein abundance and DNA-bound TF activity suggests that additional regulatory mechanisms, such as post-translational modification and nuclear translocation, play an important part for TF activation. The combined analyses of the three data sets will facilitate the investigation of underlying regulatory mechanism and select candidate TFs that are post-transcriptionally and/or post-translationally regulated. For example, our data showed that the protein abundance of CTNNB1 is negatively correlated with its mRNA, but positively correlated with its DNA-binding activity. Because CTNNB1 is regulated through proteolysis, these data imply that the majority of CTNNB1 is post-transcriptionally regulated, but the stabilized CTNNB1 is transcriptionally active during early embryonic days.
Connecting TFs with their target genes is one of the most challenging issues in the field. Several databases are available but the overlap among them is low (Keenan et al. 2019). Additionally, few TGs are regulated by a single TF, making the one-to-one correlation even more difficult. In our TF-TG analysis, we predicted development cluster–specific TFs with their TGs using a database from CellNet and showed that the RNAs and proteins that were actually detected were enriched in the same GO terms. This avoided the one-to-one match but examined TGs belonging to the same functional categories that are likely regulated by a group of TFs with either physical or functional interactions. Although the majority of TGs predicted by TF Phases 1 and 4 were indeed detected and enriched in these time frames by the RNA and protein measurement, predicted TF-TG relationship in the other two phases did not agree with the measurements. The reason for such a discordance is not clear. We speculate that because the R2/P2 phase spans the period during which the embryos are matured and the pups are born, the drastic changes in their living environment require complex signaling cross-talks with multiple TFs. Moreover, other factors, including post-translational modifications, protein–protein interaction between TFs and their coregulators, also contribute to the transcriptional regulation.
Our data sets also reveal an intriguing relationship between liver development and HCC subtypes that are associated with clinical outcomes. More aggressive tumors are associated with aberrant overexpression of proliferation markers or activation of early embryonic development pathways, whereas less aggressive tumors are associated with overexpression of liver function–related proteins that are also elevated in mature livers. This correlation raises the possibility that different HCC subtypes may originate from different oncogenic events.
In summary, the multi-omics data in the present study offer a valuable resource to study the mechanisms of liver organogenesis, development, and tools to understand liver tumorigenesis, from the origins of cancer cells to metastasis.
Methods
Animals and tissue collection
Eight-week-old C57BL/6 mice were mated at night, and pregnancy was determined by the appearance of a viscous vaginal plug in the female mice the following morning, which was designated as gestational day 0.5. Livers were collected at 15 time points spanning the major developmental stages as indicated. Three livers were collected from three mice at each time point. Multiple livers were combined for each sample before E16.5.
Sample preparation and LC-MS/MS analysis
Liver tissues were subjected to protein extraction in lysis buffer (8 M Urea, 100 mM Tris-HCl at pH 8.0) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific). Lysates were centrifuged at 16,000g for 10 min at 4°C. The supernatant was digested with sequencing-grade trypsin and the resulting peptides were fractionated with a small-scale C18 reversed phase (sRP) chromatography (Ding et al. 2013b; Jung et al. 2017) to nine fractions at pH = 10.
Nuclear extracts were incubated with biotinylated DNA preimmobilized on Dynabeads (M-280 streptavidin) for 2 h at 4°C. Proteins bound to Dynabeads were washed with NETN (100 mM NaCl, 20 mM Tris-HCl, 0.5 mM EDTA, and 0.5% [vol/vol] NP-40) buffer twice and water once; they were then digested with trypsin in 50 mM ammonium bicarbonate for 6 h at 37°C.
The digested peptides were vacuum dried and then redissolved in 0.1% formic acid and resolved on an UltiMate 3000 RSLCnano System (Thermo Fisher Scientific) operating on a 75-min linear gradient (5%–35% acetonitrile in 0.1% formic acid) at a flow rate of 600 nL/min. Tandem mass spectra were acquired on a QExactive HF mass spectrometer (Thermo Fisher Scientific) in the data-dependent mode.
Protein identification and quantification
MS data were processed on the Firmiana platform (Feng et al. 2017). Proteins were identified against the RefSeq mouse protein database (version 04/07/2013, derived from the protein-coding GENCODE.vM2) using the MASCOT search engine (Matrix Science, version 2.3.01). Mass tolerances were 20 ppm for precursor ions and 0.05 Da for product ions, respectively. Up to one missed cleavage was allowed for trypsin digestion. Cysteine carbamidomethylation was considered as a fixed modification, N-terminal acetylation and methionine oxidation were considered as dynamic modifications. One percent FDR on both the peptide and protein levels estimated by searching a decoy database were allowed. Gene symbols were updated to GENCODE.vM23 (09/19/2019). The list of converted genes is provided in Supplemental Table S1. Proteins were quantified by a label-free, intensity-based absolute quantification (iBAQ) approach (Schwänhausser et al. 2011) and further normalized into iFOT (intensity-based the fraction of total multiplied by 105).
High-throughput RNA sequencing and data analysis
RNA sequencing (RNA-seq) was performed on Illumina HiSeq 4000. The quality control and data filtering were processed by the FastQC software (Version 0.11.5). The processed reads were mapped onto the mouse reference genome (GRCm38.p2. genome, 12/10/2013) using the HISAT2 software (Version 2.1.0) (Kim et al. 2015). Transcripts were assembled and quantified using the StringTie software (Version 1.3.1) with a reference genome annotation file (Pertea et al. 2015). Relative quantification of transcripts was measured as fragments per kilobase of transcript per million mapped reads (FPKM).
Bioinformatics and statistical analysis
Principal component analysis (PCA) and unsupervised hierarchical clustering analysis were carried out. Gene Ontology (GO) term enrichment analysis was performed by using DAVID Bioinformatics Resources 6.8, an online bioinformatics tool for gene functional annotation (Huang da et al. 2009). One-way ANOVA test was performed to identify differentially expressed proteins.
Genes linked to specific liver functions were selected by KEGG, Gene Ontology, and Reactome. Hepatic cell-type markers were selected from the studies (Kinoshita et al. 2010; Azimifar et al. 2014; Gordillo et al. 2015). Genes with similar functions were grouped by k-means clustering analysis of differentially expressed proteins. Proteins or genes that were highly correlated in the same cluster (r > 0.8) were used for GO-term enrichment analysis. Potential transcriptional targets were identified from CellNet (Cahan et al. 2014).
Data access
All RNA-seq data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE132034. The MS raw data generated in this study have been submitted to ProteomeXchange database (www.proteomexchange.org) via the iProX partner repository (Ma et al. 2019) under accession number PXD014144.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFC0908404, 2018YFA0507503, 2017YFA0505102), the Natural Science Foundation of China (81874237), and Beijing Municipal Training Program for the Talents (Grant No. 2015000021223TD04). We thank Aileen W. Qin for editing.
Author contributions: T.G., C.Z., C.D., J.Q., and Y.W. conceived the project; T.G., X.L., M.L., Q.Z., and L.S. performed the experiments; T.G., C.Z., X.N., J.L., X.X., and D.Z. analyzed the data; and T.G., C.Z., J.Q., and Y.W. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.253328.119.
References
- Aiello NM, Stanger BZ. 2016. Echoes of the embryo: using the developmental biology toolkit to study cancer. Dis Model Mech 9: 105–114. 10.1242/dmm.023184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama TE, Gonzalez FJ. 2003. Regulation of P450 genes by liver-enriched transcription factors and nuclear receptors. Biochim Biophys Acta 1619: 223–234. 10.1016/S0304-4165(02)00480-4 [DOI] [PubMed] [Google Scholar]
- Azimifar SB, Nagaraj N, Cox J, Mann M. 2014. Cell-type-resolved quantitative proteomics of murine liver. Cell Metab 20: 1076–1087. 10.1016/j.cmet.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, Gilad Y. 2015. Genomic variation. Impact of regulatory variation from RNA to protein. Science 347: 664–667. 10.1126/science.1260793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell DM, Roulot D, George J. 2001. Transforming growth factor β and the liver. Hepatology 34: 859–867. 10.1053/jhep.2001.28457 [DOI] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. 2014. CellNet: network biology applied to stem cell engineering. Cell 158: 903–915. 10.1016/j.cell.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmont A, Wandzioch E, Tremblay KD, Minowada G, Kaestner KH, Martin GR, Zaret KS. 2006. An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev Cell 11: 339–348. 10.1016/j.devcel.2006.06.015 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2017. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169: 1327–1341.e23. 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Vila N, Bluhm A, Sayols S, Dinges N, Dejung M, Altenhein T, Kappei D, Altenhein B, Roignant JY, Butter F. 2017. The developmental proteome of Drosophila melanogaster. Genome Res 27: 1273–1285. 10.1101/gr.213694.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP. 2016. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534: 500–505. 10.1038/nature18270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi TY, Khaliq M, Tsurusaki S, Ninov N, Stainier DYR, Tanaka M, Shin D. 2017. Bone morphogenetic protein signaling governs biliary-driven liver regeneration in zebrafish through Tbx2b and Id2a. Hepatology 66: 1616–1630. 10.1002/hep.29309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. 2005. Control of liver cell fate decision by a gradient of TGFβ signaling modulated by Onecut transcription factors. Genes Dev 19: 1849–1854. 10.1101/gad.340305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Gunewardena SS, Yoo B, Liu J, Renaud HJ, Lu H, Zhong XB, Klaassen CD. 2012. RNA-Seq reveals different mRNA abundance of transporters and their alternative transcript isoforms during liver development. Toxicol Sci 127: 592–608. 10.1093/toxsci/kfs107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Chan DW, Liu W, Liu M, Li D, Song L, Li C, Jin J, Malovannaya A, Jung SY, et al. 2013a. Proteome-wide profiling of activated transcription factors with a concatenated tandem array of transcription factor response elements. Proc Natl Acad Sci 110: 6771–6776. 10.1073/pnas.1217657110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Jiang J, Wei J, Liu W, Zhang W, Liu M, Fu T, Lu T, Song L, Ying W, et al. 2013b. A fast workflow for identification and quantification of proteomes. Mol Cell Proteomics 12: 2370–2380. 10.1074/mcp.O112.025023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Ding C, Qiu N, Ni X, Zhan D, Liu W, Xia X, Li P, Lu B, Zhao Q, et al. 2017. Firmiana: towards a one-stop proteomic cloud platform for data processing and analysis. Nat Biotechnol 35: 409–412. 10.1038/nbt.3825 [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu X, Tang B, Li C, Kou Z, Li L, Liu W, Wu Y, Kou X, Li J, et al. 2017. Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep 21: 3957–3969. 10.1016/j.celrep.2017.11.111 [DOI] [PubMed] [Google Scholar]
- Gebhardt R. 1992. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol Ther 53: 275–354. 10.1016/0163-7258(92)90055-5 [DOI] [PubMed] [Google Scholar]
- Geisler F, Strazzabosco M. 2015. Emerging roles of notch signaling in liver disease. Hepatology 61: 382–392. 10.1002/hep.27268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo M, Evans T, Gouon-Evans V. 2015. Orchestrating liver development. Development 142: 2094–2108. 10.1242/dev.114215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruppuso PA, Sanders JA. 2016. Regulation of liver development: implications for liver biology across the lifespan. J Mol Endocrinol 56: R115–R125. 10.1530/JME-15-0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunewardena SS, Yoo B, Peng L, Lu H, Zhong X, Klaassen CD, Cui JY. 2015. Deciphering the developmental dynamics of the mouse liver transcriptome. PLoS One 10: e0141220 10.1371/journal.pone.0141220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY. 2019. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res 47: D33–D38. 10.1093/nar/gky822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huh CG, Factor VM, Sánchez A, Uchida K, Conner EA, Thorgeirsson SS. 2004. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci 101: 4477–4482. 10.1073/pnas.0306068101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, Xing B, Sun W, Ren L, Hu B, et al. 2019. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 567: 257–261. 10.1038/s41586-019-0987-8 [DOI] [PubMed] [Google Scholar]
- Jover R, Bort R, Gomez-Lechon MJ, Castell JV. 2001. Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology 33: 668–675. 10.1053/jhep.2001.22176 [DOI] [PubMed] [Google Scholar]
- Jung J, Zheng M, Goldfarb M, Zaret KS. 1999. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science 284: 1998–2003. 10.1126/science.284.5422.1998 [DOI] [PubMed] [Google Scholar]
- Jung SY, Choi JM, Rousseaux MW, Malovannaya A, Kim JJ, Kutzera J, Wang Y, Huang Y, Zhu W, Maity S, et al. 2017. An anatomically resolved mouse brain proteome reveals Parkinson disease-relevant pathways. Mol Cell Proteomics 16: 581–593. 10.1074/mcp.M116.061440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, Jagodnik KM, Kropiwnicki E, Wang Z, Ma'ayan A. 2019. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res 47: W212–W224. 10.1093/nar/gkz446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Uchida T, Sato A, Nakashima M, Nakashima H, Shono S, Habu Y, Miyazaki H, Hiroi S, Seki S. 2010. Characterization of two F4/80-positive Kupffer cell subsets by their function and phenotype in mice. J Hepatol 53: 903–910. 10.1016/j.jhep.2010.04.037 [DOI] [PubMed] [Google Scholar]
- Lee DH, Park JO, Kim TS, Kim SK, Kim TH, Kim MC, Park GS, Kim JH, Kuninaka S, Olson EN, et al. 2016. LATS-YAP/TAZ controls lineage specification by regulating TGFβ signaling and Hnf4α expression during liver development. Nat Commun 7: 11961 10.1038/ncomms11961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loser K, Vogl T, Voskort M, Lueken A, Kupas V, Nacken W, Klenner L, Kuhn A, Foell D, Sorokin L, et al. 2010. The Toll-like receptor 4 ligands Mrp8 and Mrp14 are crucial in the development of autoreactive CD8+ T cells. Nat Med 16: 713–717. 10.1038/nm.2150 [DOI] [PubMed] [Google Scholar]
- Lu L, Finegold MJ, Johnson RL. 2018. Hippo pathway coactivators Yap and Taz are required to coordinate mammalian liver regeneration. Exp Mol Med 50: e423 10.1038/emm.2017.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Chen T, Wu S, Yang C, Bai M, Shu K, Li K, Zhang G, Jin Z, He F, et al. 2019. iProX: an integrated proteome resource. Nucleic Acids Res 47: D1211–D1217. 10.1093/nar/gky869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J, Kim DK, Jung YS, Kim HB, Kim YH, Yoo EK, Kim BG, Kim S, Lee IK, Harris RA, et al. 2016. O-GlcNAcylation of orphan nuclear receptor estrogen-related receptor γ promotes hepatic gluconeogenesis. Diabetes 65: 2835–2848. 10.2337/db15-1523 [DOI] [PubMed] [Google Scholar]
- Monga SP. 2015. β-catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology 148: 1294–1310. 10.1053/j.gastro.2015.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell CM, Strazzabosco M. 2014. Notch signaling and new therapeutic options in liver disease. J Hepatol 60: 885–890. 10.1016/j.jhep.2013.11.028 [DOI] [PubMed] [Google Scholar]
- Morell CM, Fiorotto R, Fabris L, Strazzabosco M. 2013. Notch signalling beyond liver development: emerging concepts in liver repair and oncogenesis. Clin Res Hepatol Gastroenterol 37: 447–454. 10.1016/j.clinre.2013.05.008 [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. 1995. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci 92: 10302–10306. 10.1073/pnas.92.22.10302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejak-Bowen K, Monga SP. 2008. Wnt/β-catenin signaling in hepatic organogenesis. Organogenesis 4: 92–99. 10.4161/org.4.2.5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SH, Camargo FD, Yimlamai D. 2017. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology 152: 533–545. 10.1053/j.gastro.2016.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong XB. 2012. RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes P450 and their alternative transcripts during mouse liver development. Drug Metab Dispos 40: 1198–1209. 10.1124/dmd.112.045088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L. 2011. Energy metabolism in the liver. Compr Physiol 4: 177–197. 10.1002/cphy.c130024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JO, Monga SS. 2018. Wnt/β-catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol 13: 351–378. 10.1146/annurev-pathol-020117-044010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwänhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Sheaffer KL, Kaestner KH. 2012. Transcriptional networks in liver and intestinal development. Cold Spring Harb Perspect Biol 4: a008284 10.1101/cshperspect.a008284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Monga SP. 2013. Cellular and molecular basis of liver development. Compr Physiol 3: 799–815. 10.1002/cphy.c120022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. 2007. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development 134: 2041–2050. 10.1242/dev.000281 [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre FP, Duncan SA. 2010. Organogenesis and development of the liver. Dev Cell 18: 175–189. 10.1016/j.devcel.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Su X, Shi Y, Zou X, Lu ZN, Xie G, Yang JYH, Wu CC, Cui XF, He KY, Luo Q, et al. 2017. Single-cell RNA-Seq analysis reveals dynamic trajectories during mouse liver development. BMC Genomics 18: 946 10.1186/s12864-017-4342-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MD, Monga SP. 2007. WNT/β-catenin signaling in liver health and disease. Hepatology 45: 1298–1305. 10.1002/hep.21651 [DOI] [PubMed] [Google Scholar]
- Vesely PW, Staber PB, Hoefler G, Kenner L. 2009. Translational regulation mechanisms of AP-1 proteins. Mutat Res 682: 7–12. 10.1016/j.mrrev.2009.01.001 [DOI] [PubMed] [Google Scholar]
- Wang RN, Green J, Wang Z, Deng Y, Qiao M, Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S, et al. 2014. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis 1: 87–105. 10.1016/j.gendis.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, Houten SM, Amariuta T, Wolski W, Zamboni N, et al. 2016. Systems proteomics of liver mitochondria function. Science 352: aad0189 10.1126/science.aad0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Hill CS. 2009. TGF-β superfamily signaling in embryonic development and homeostasis. Dev Cell 16: 329–343. 10.1016/j.devcel.2009.02.012 [DOI] [PubMed] [Google Scholar]
- Wu H, Wei L, Fan F, Ji S, Zhang S, Geng J, Hong L, Fan X, Chen Q, Tian J, et al. 2015. Integration of Hippo signalling and the unfolded protein response to restrain liver overgrowth and tumorigenesis. Nat Commun 6: 6239 10.1038/ncomms7239 [DOI] [PubMed] [Google Scholar]
- Yang J, Mowry LE, Nejak-Bowen KN, Okabe H, Diegel CR, Lang RA, Williams BO, Monga SP. 2014. β-catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation! Hepatology 60: 964–976. 10.1002/hep.27082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JS, Su XS, Stoltz JF, de Isla N, Zhang L. 2015. Signalling pathways involved in the process of mesenchymal stem cells differentiating into hepatocytes. Cell Prolif 48: 157–165. 10.1111/cpr.12165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ, Camargo FD. 2014. Hippo pathway activity influences liver cell fate. Cell 157: 1324–1338. 10.1016/j.cell.2014.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yatskievych TA, Baker RK, Antin PB. 2004. Regulation of Hex gene expression and initial stages of avian hepatogenesis by Bmp and Fgf signaling. Dev Biol 268: 312–326. 10.1016/j.ydbio.2004.01.019 [DOI] [PubMed] [Google Scholar]
- Zhao R, Duncan SA. 2005. Embryonic development of the liver. Hepatology 41: 956–967. 10.1002/hep.20691 [DOI] [PubMed] [Google Scholar]
- Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. 2009. Notch signaling controls liver development by regulating biliary differentiation. Development 136: 1727–1739. 10.1242/dev.029140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.