Abstract
Temozolomide (TMZ) is a frequently used chemotherapy for glioma; however, chemoresistance is a major problem limiting its effectiveness. Thus, knowledge of mechanisms underlying this outcome could improve patient prognosis. Here, we report that deletion of a regulatory element in the HOTAIR locus increases glioma cell sensitivity to TMZ and alters transcription of multiple genes. Analysis of a combination of RNA-seq, Capture Hi-C, and patient survival data suggests that CALCOCO1 and ZC3H10 are target genes repressed by the HOTAIR regulatory element and that both function in regulating glioma cell sensitivity to TMZ. Rescue experiments and 3C data confirmed this hypothesis. We propose a new regulatory mechanism governing glioma cell TMZ sensitivity.
Glioma is the most common primary brain tumor, and ∼80% of malignant primary tumors in the central nervous system are glioma (Weller et al. 2015). Glioblastoma (GBM) is the most aggressive glioma and constitutes >50% of all gliomas. Despite treatment advances, prognosis for glioblastoma is still very poor, and median survival time for patients is only 14–17 mo (Reifenberger et al. 2017). Current standard treatment options include surgery, radiation, and chemotherapy, specifically temozolomide (TMZ), which is the most frequently used clinical chemotherapy for GBM (Wick et al. 2012; Sandmann et al. 2015). TMZ efficiently crosses the blood brain barrier and induces glioma cell apoptosis; however, chemoresistance often develops, representing a major therapeutic obstacle. Thus, strategies to increase glioma cell sensitivity to TMZ could serve as useful interventions to improve patient prognosis.
Glioma resistance to TMZ is associated with multiple factors. Some report that high expression of O-6-methylguanine-DNA methyltransferase (MGMT) is related to robust TMZ resistance (Kitange et al. 2009; Wick et al. 2014). In addition, factors such as P4HB (Sun et al. 2013), ALDH1A1 (Schäfer et al. 2012), and poly (ADP-ribose) polymerase (Clarke et al. 2009) have been associated with TMZ resistance in glioma cells. However, clinical tests of strategies designed to attenuate TMZ resistance have not yet produced satisfactory results (Quinn et al. 2009; Warren et al. 2012), requiring further investigations.
Zhang et al. recently identified a regulatory element in the locus of the long noncoding RNA HOTAIR (Zhang et al. 2014). That study suggested that this element functions as an enhancer associated with progression of esophageal squamous cell carcinoma (ESCC) (Zhang et al. 2014). Follow-up studies report that this regulatory element or single-nucleotide polymorphisms (SNPs) within it are associated with risk or progression of uterine cervical (Weng et al. 2018), head and neck (Pan et al. 2017), and lung (Wang et al. 2018) cancers, while few studies report association of this element with glioma.
Based on ChIP data reported by Rheinbay and colleagues (Rheinbay et al. 2013), we found that in glioma cells and in glioblastoma stem cells derived from human tumors, the HOTAIR regulatory element is marked by H3K4me1, H3K4me3, and H3K27me3. Overall, H3K4me1 marks candidate enhancers, and the H3K4me3 mark is an active mark of transcriptional initiation (Zhou et al. 2011; Rheinbay et al. 2013). H3K27me3 is a repressive mark prototypical of the polycomb repressive complex (Boyer et al. 2006; Zhou et al. 2011). Thus, the epigenetic pattern at the HOTAIR regulatory element in glioma cells and glioma stem cells suggests a possible bivalent function.
Development of high-order chromosome conformation capture techniques has facilitated analysis of long-range DNA interactions and enabled analysis of regulatory elements that enhance or repress gene expression through long-range interactions (Wei et al. 2013; Liu et al. 2017; Luo et al. 2017; Qian et al. 2019; Su et al. 2019). We hypothesized that the HOTAIR element may function in glioma progression or chemosensitivity. Here, we investigate the regulatory network governed by the element and discover that the HOTAIR regulatory element modulates glioma cell TMZ sensitivity through long-range regulation of both CALCOCO1 and ZC3H10.
Results
Deletion of the HOTAIR regulatory element increases glioma cell sensitivity to TMZ
The HOTAIR regulatory element is marked by H3K4me1, H3K4me3, and H3K27me3 in glioma cells and glioblastoma stem cells derived from human tumors based on ChIP data reported by Rheinbay and colleagues (Fig. 1A; Rheinbay et al. 2013). To investigate whether the HOTAIR regulatory element functions in glioma cell sensitivity to TMZ, we used the CRISPR-Cas9 system to delete that element in U251 cells, a human GBM line. Two knockout lines (KO-1 and KO-2) were used in all of the following experiments (Supplemental Fig. S1A,B). We then treated both KO lines plus WT U251 cells with high (1 mM) doses of TMZ to assess drug sensitivity, based on lactate dehydrogenase (LDH) release and caspase 3/7 activity. LDH is a stable cytosolic enzyme that is released upon cell lysis which indicates cell death induced by TMZ, while increased caspase 3/7 activity indicates apoptosis. LDH release was evaluated 24 h and 48 h after TMZ treatment and Caspase 3/7 activities at 48 h and 72 h (Fig. 1B). Relative to WT U251 cells treated with TMZ, LDH release in KO-1 and KO-2 glioma cells was significantly increased approximately two- to threefold at 24 h and two- to fourfold at 48 h (Fig. 1C,D). Caspase 3/7 activities also increased markedly in TMZ-treated KO-1 and KO-2 glioma cells relative to WT U251 cells treated with TMZ at 48 h and 72 h (Fig. 1E,F). These data suggest that loss of the HOTAIR regulatory element increases glioma cells’ sensitivity to TMZ. RT-qPCR analysis showed that HOTAIR mRNA is expressed at a low level in U251 cells (Supplemental Fig. S2A), and no significant change of HOTAIR’s expression was detected in HOTAIR RE KO cells (Supplemental Fig. S2B). Moreover, overexpression of the HOTAIR lncRNA did not antagonize the increase in sensitivity to TMZ in either KO line (Fig. 1G,H): LDH release and caspase 3/7 activity were comparable in KO cells transduced with control vector and KO cells overexpressing HOTAIR following TMZ treatment (1 mM). U251 cells were transfected with HOTAIR overexpression plasmids, and overexpression efficiency was verified by RT-qPCR (Fig. 1I). RNA-seq analysis of control and KO U251 cells indicated global changes in mRNA levels in KO relative to WT cells treated with TMZ (1 mM, 72 h) (Fig. 1J). Gene Ontology analysis of significantly changed genes (Q value < 0.01) based on the DAVID knowledgebase (https://david.ncifcrf.gov) showed that the top pathway altered was PI3K-AKT signaling (Fig. 1K). Accordingly, the PI3K-AKT pathway is reportedly associated with TMZ resistance in glioma cells (Shi et al. 2017; Zhong et al. 2018).
Figure 1.
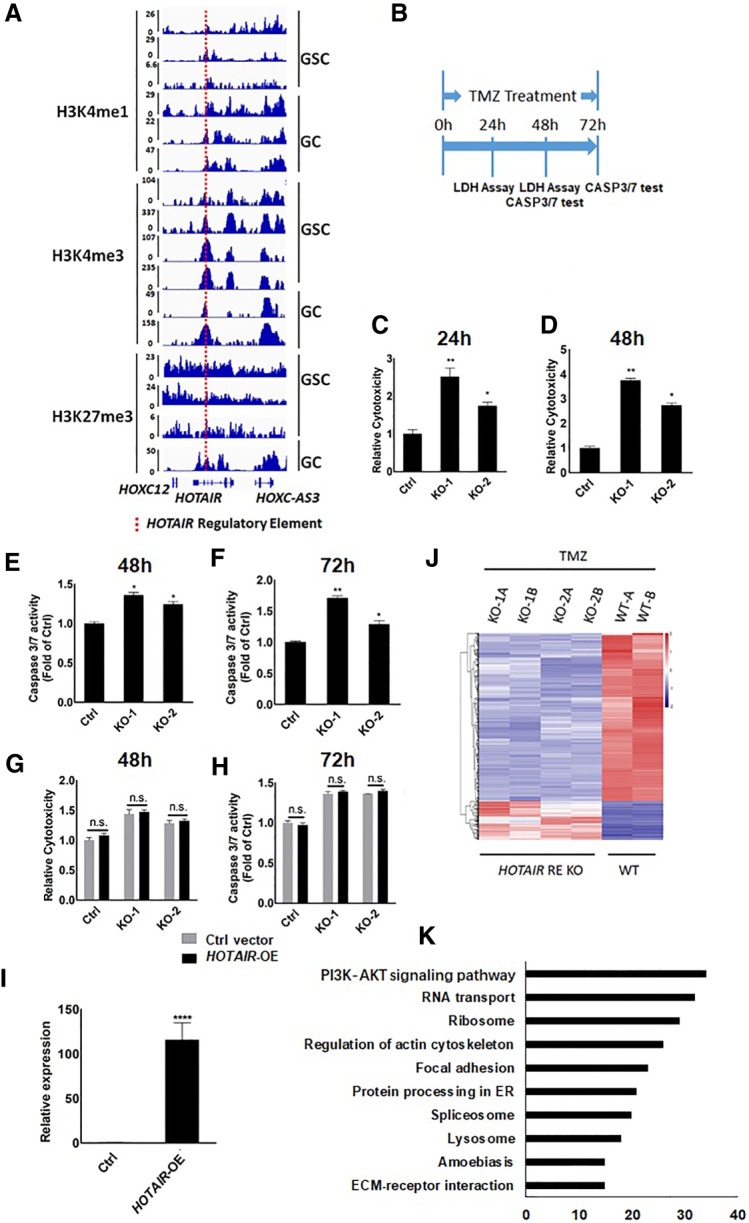
Deletion of the HOTAIR regulatory element increases glioma cells’ sensitivity to TMZ. (A) H3K4me1, H3K4me3, H3K27me3 ChIP of the region containing the HOTAIR regulatory element (red line) in glioma cells (GC) and glioma stem cells (GSC) derived from human tumor. (B) Schematic showing the timing of LDH and Caspase 3/7 assays. U251 glioma cells were treated with TMZ (1 mM) for 24–72 h. (C,D) An LDH assay was used to detect LDH release in wild-type U251 cells (Ctrl) and HOTAIR regulatory element knockout U251 lines (KO-1, KO-2). LDH release was assayed after cells were treated 24 h and 48 h with TMZ (1 mM). Data represent means ± S.E.M. of three independent experiments. (*) P < 0.05, (**) P < 0.01 compared with control. (E,F) Proteolytic activities of caspase 3/7 were tested in wild-type U251 cells (Ctrl) and HOTAIR regulatory element knockout U251 cells (KO-1, KO-2) treated 48 h and 72 h with TMZ. Data represent means ± S.E.M. of three independent experiments. (*) P < 0.05, (**) P < 0.01 compared with control. (G) LDH assay was used to detect LDH release in wild-type U251 cells (Ctrl) and HOTAIR regulatory element knockout U251 lines (KO-1, KO-2) with HOTAIR overexpression (HOTAIR-OE) or transfected with control vector. LDH release was tested after cells were treated 48 h with TMZ. Data represent means ± S.E.M. of three independent experiments. (H) Proteolytic activities of caspase 3/7 were tested in wild-type U251 cells (Ctrl) and HOTAIR regulatory element knockout U251 cells (KO-1, KO-2) with HOTAIR overexpression (HOTAIR-OE) or transfected with control vector. Caspase 3/7 activities were tested after cells were treated 72 h with TMZ. Data represent means ± S.E.M. of three independent experiments. (I) RT-qPCR analysis of HOTAIR in wild-type U251 cells transfected with control plasmid or HOTAIR overexpression plasmid. Data represent means ± S.E.M. of three independent experiments. (****) P < 0.0001 compared with corresponding cells transfected with Ctrl vector. (J) Comparison of gene expression in TMZ-treated (1 mM, 72 h) HOTAIR regulatory knockout (KO) and wild-type (WT) cell lines. Heat map shows clustering of differentially expressed genes in KO and WT glioma cells. KO-1A and B, KO-2A and B, WT-A and B refer to two biological replicates, respectively. (K) Function of genes (Q-value < 0.01) as analyzed by DAVID.
Long-range interactions of the HOTAIR regulatory element with multiple target genes
Given that HOTAIR overexpression did not reverse increases in TMZ sensitivity in KO glioma cells, we surmised that the HOTAIR regulatory element itself has a complex function in regulatory networks in glioma cells. To search for target genes potentially regulated by this element, we performed Capture Hi-C in U251 glioma cells to analyze the interactome. To do so, we selected a 723-bp bait region (Fig. 2A) and performed Capture Hi-C based on a previously reported protocol (Davies et al. 2016; Belaghzal et al. 2017). Target sites from two biological replicates are shown in Circos plots (Krzywinski et al. 2009) in Figure 2, B and C, where red and blue lines, respectively, represent possible interactions in the two replicates. That analysis revealed that 67% and 63.2% are in cis interaction sites, respectively, in the two biological replicates. Mapping of these interaction sites to the human genome led to identification of 89 target genes reproducible in both. These genes represent possible interactions with the HOTAIR regulatory element. Among the 89, 84 were located in cis with the element, and five were in trans.
Figure 2.
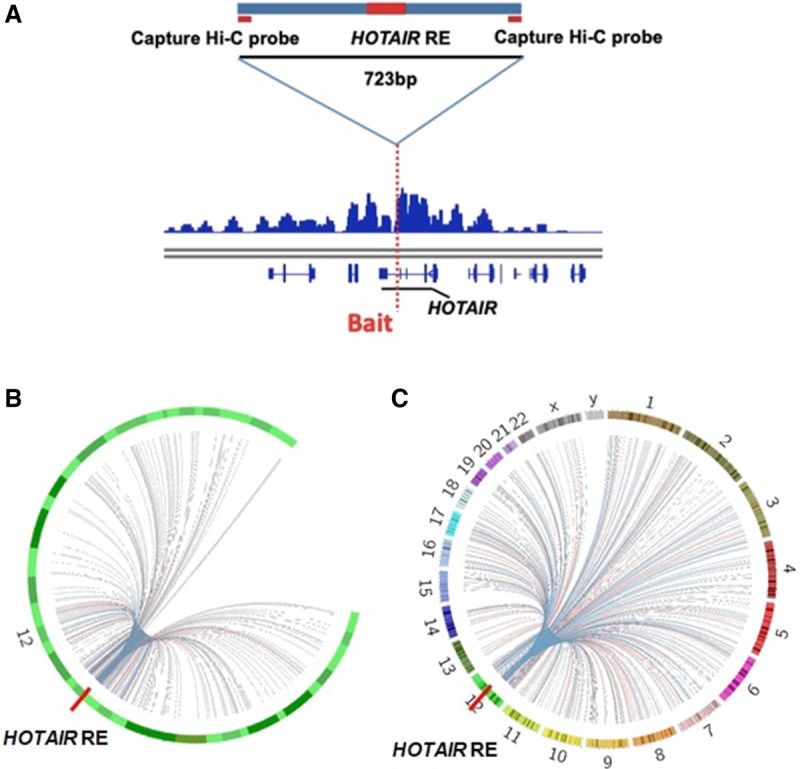
Intra- and inter-chromosomal interactions with the HOTAIR regulatory element. (A) The 723-bp Capture Hi-C bait region and Capture Hi-C signal near the bait region. (B) Circos plot showing genome-wide intra-interactions indicated by curves extending from the bait region; the two biological replicates are in red and blue, respectively. HOTAIR RE: HOTAIR regulatory element. (C) Circos plot showing genome-wide intra- and inter-interactions indicated by curves extending from the bait region; the two biological replicates are in red and blue, respectively. HOTAIR RE: HOTAIR regulatory element.
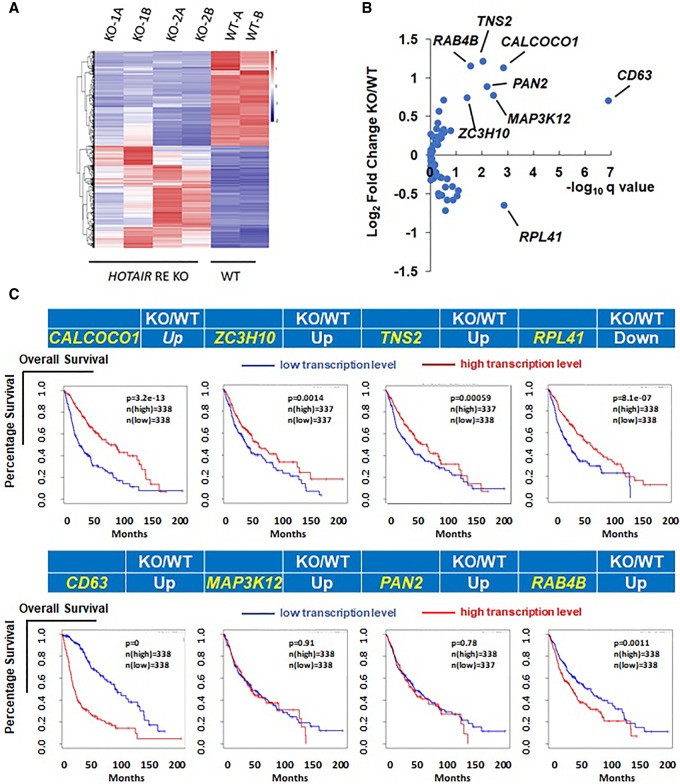
To identify genes potentially regulated by the HOTAIR regulatory element, we performed RNA-seq on both wild-type and HOTAIR KO U251 cell lines (Fig. 3A). These genes may be direct or indirect targets of the regulatory fragment. When combined with Capture Hi-C, it may reveal genes that are directly regulated by the regulatory fragment. Positive candidate genes identified in both RNA-seq and Capture Hi-C are shown in Fig. 3B. Genes with significant changes in mRNA levels in enhancer KO relative to WT U251 cells (fold-change >1.5 or <−1.5; Q value < 0.05) are denoted by gene names (Fig. 3B). Overall, eight genes may be directly regulated by the HOTAIR regulatory element (Fig. 3B), including seven up-regulated in KO cells and one down-regulated.
Figure 3.
Potential target genes regulated by the HOTAIR regulatory element. (A) Comparison of gene expression in HOTAIR regulatory knockout (KO) and wild-type (WT) glioma lines. Heat map shows clustering of differentially expressed genes in KO and WT glioma cells. KO-1A and B, KO-2A and B, WT-A and B refer to two biological replicates, respectively. (B) Comparison of fold-changes in gene expression based on RNA-seq data and corresponding Capture Hi-C signal. Eighty-nine potential target genes reproducible in two biological replicates are shown on the plot. Among them, eight with significant expression change between KO and WT (adjusted P-value < 0.05, and fold-change >1.5 or <−1.5) are marked with the gene name. (C) Shown are the eight target genes and associated survival data from glioma patients.
Given that deletion of the HOTAIR regulatory element increased glioma cell sensitivity to TMZ, we asked which target genes function in this outcome. To do so, we first analyzed patient survival data associated with the eight target genes (Fig. 3C), which were available through The Cancer Genome Atlas (TCGA) database. Our analysis of survival curves indicated that three candidates (CALCOCO1, ZC3H10, and TNS2) showed a pattern consistent with our in vitro data: All three were up-regulated in HOTAIR regulatory element KO cells, and accordingly, high levels of all three corresponded to longer survival time in glioma patients. These observations suggest overall that the HOTAIR regulatory element may function as a silencer of these candidate genes in glioma and that this activity is associated with TMZ sensitivity.
Long-range interactions between HOTAIR regulatory element and target genes are associated with TMZ sensitivity
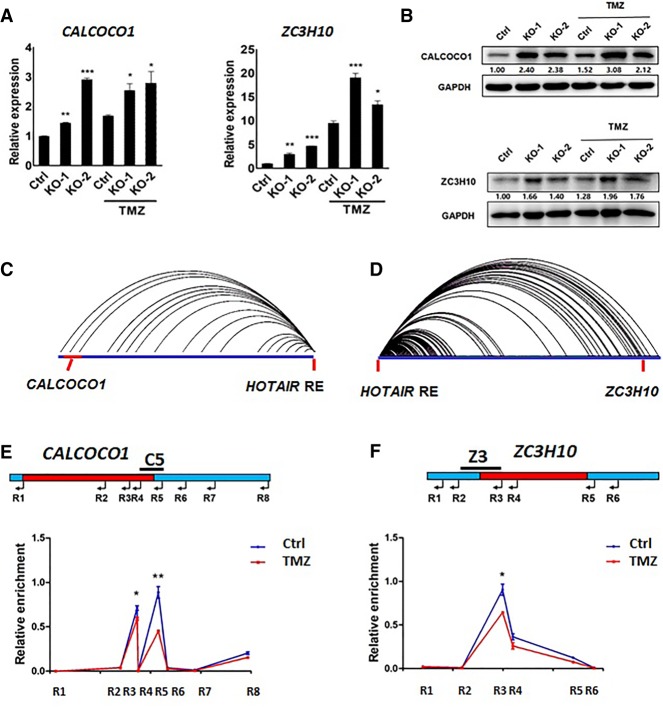
We next assessed CALCOCO1, ZC3H10, and TNS2 expression in WT U251 glioma cells and the two KO lines. RT-qPCR analysis showed increased CALCOCO1 and ZC3H10 transcript levels in KO lines relative to WT U251 cells with or without treatment of TMZ (1 mM, 72 h), while TNS2 mRNA levels were comparable in both (Supplemental Fig. S3), suggesting that TNS2 is a false positive. In addition, TMZ treatment (1 mM, 72 h) significantly up-regulated CALCOCO1 and ZC3H10 in both WT U251 cells and KO lines as shown by RT-qPCR (Fig. 4A) and western blot (Fig. 4B). Thus, we confined our analysis in the rest of the study to CALCOCO1 and ZC3H10.
Figure 4.
CALCOCO1 and ZC3H10 are potentially regulated by the HOTAIR regulatory element. (A) RT-qPCR analysis of mRNA (independent mRNA library) levels of potential target genes CALCOCO1 and ZC3H10. Data represent means ± S.E.M. of three independent experiments. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 compared with corresponding Ctrl cells. (B) Western blot analysis of CALCOCO1 and ZC3H10 in wild-type U251 cells (Ctrl) and HOTAIR regulatory element knockout U251 lines (KO-1, KO-2) that were treated with or without TMZ (1 mM, 72 h). Relative protein levels were quantified by using ImageJ. (C) Capture Hi-C loop view shows interactions between HOTAIR RE and CALCOCO1. (D) Capture Hi-C loop view shows interactions between HOTAIR RE and ZC3H10. (E) CALCOCO1-HOTAIR RE interaction analyzed by 3C assay in U251 cells treated with or without TMZ (1 mM, 72 h). Interaction frequency was calculated as described in the Methods. Data represent means ± S.E.M. of three independent experiments. (*) P < 0.05, (**) P < 0.01, compared with corresponding relative enrichment in U251 cells treated with TMZ. (F) ZC3H10-HOTAIR RE interaction analyzed by 3C assay in U251 cells treated with or without TMZ (1 mM, 72 h). Interaction frequency was calculated as described in the Methods. Data represent means ± S.E.M. of three independent experiments. (*) P < 0.05, compared with corresponding relative enrichment in U251 cells treated with TMZ.
The CALCOCO1 and ZC3H10 loci are ∼0.25 Mb and ∼2.2 Mb distant from the HOTAIR regulatory element region, respectively. The interactions between the HOTAIR regulatory element and CALCOCO1 or ZC3H10 genes, respectively, are shown by Capture Hi-C (Fig. 4C,D). A Chromatin Conformation Capture (3C) assay was then performed to further define these interactions (Fig. 4E,F). A C5 fragment at the CALCOCO1 promoter and Z3 fragment at the ZC3H10 promoter showed significantly higher interaction frequency with the HOTAIR regulatory element region compared to the neighboring regions. The interaction frequencies between the HOTAIR regulatory element and C5 and Z3 fragments were significantly decreased in U251 cells treated with TMZ (Fig. 4E,F). In addition, we performed Hi-C based on a previously reported protocol (Belaghzal et al. 2017) to measure changes of interactions between the HOTAIR element and CALCOCO1 or ZC3H10 in U251 cells treated with TMZ (1 mM, 72 h). Analysis of Z-scores showed widespread changes in long-range interactions near the HOTAIR regulatory element in TMZ-treated U251 cells compared with U251 cells not treated with the drug and decreased interactions between the HOTAIR element and CALCOCO1 or ZC3H10 in U251 cells treated with TMZ (Supplemental Fig. S4). These data suggest that interactions between the HOTAIR regulatory element and CALCOCO1 and ZC3H10 are associated with TMZ-induced cell death.
CALCOCO1 and ZC3H10 down-regulation decreases TMZ sensitivity in glioma cells lacking the HOTAIR regulatory element
We next asked whether CALCOCO1 or ZC3H10 regulation by the HOTAIR regulatory element modulates glioma cell sensitivity to TMZ. To do so, we undertook CALCOCO1 or ZC3H10 knockdown using siRNAs. RT-qPCR analysis of U251 cells and U251 cells transfected with siRNAs verified that CALCOCO1 or ZC3H10 siRNA transfection down-regulated CALCOCO1 or ZC3H10 ∼0.3-fold or 0.4-fold relative to the control, respectively (Fig. 5A–D). Transfection of cells with both CALCOCO1 and ZC3H10 siRNAs down-regulated CALCOCO1 and ZC3H10 ∼0.3-fold or less relative to the control (Fig. 5E,F). When we knocked down CALCOCO1 or ZC3H10 separately in WT and KO lines, we observed no significant change in LDH release (Fig. 5G,H) or in caspase 3/7 activities (Fig. 5I,J) in si-control TMZ-treated (1 mM) KO relative to TMZ-treated (1 mM) KO cells transfected with CALCOCO1 or ZC3H10 siRNAs. However, combined knockdown of CALCOCO1 and ZC3H10 markedly decreased TMZ sensitivity in HOTAIR regulatory element KO glioma lines based on decreased LDH release and caspase 3/7 activities (Fig. 5K,L), suggesting that CALCOCO1 and ZC3H10 function synergistically to modulate TMZ sensitivity.
Figure 5.
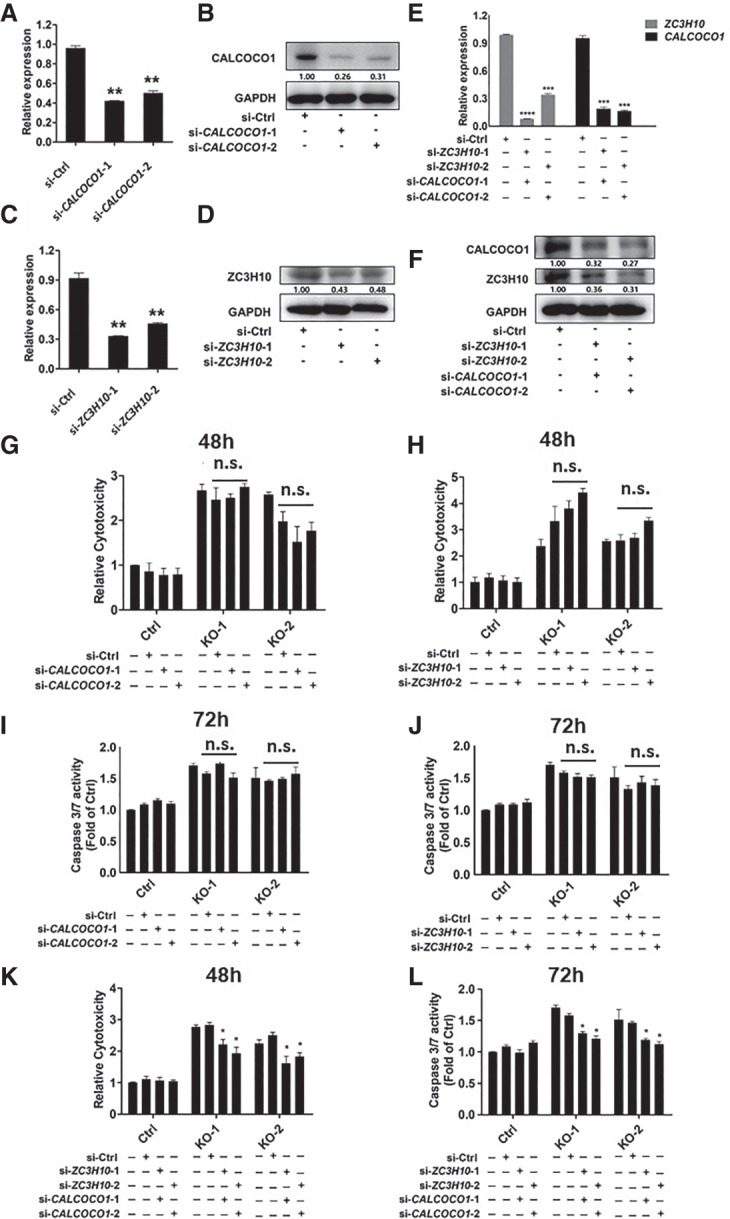
Knockdown of both CALCOCO1 and ZC3H10 rescues increased TMZ sensitivity in HOTAIR regulatory element KO glioma cells. (A) RT-qPCR verification of CALCOCO1 siRNA knockdown efficiency. Data represent means ± S.E.M. of three independent experiments. (**) P < 0.01 compared with Ctrl. (B) Western blot verification of CALCOCO1 siRNA knockdown efficiency. Relative protein levels were quantified by using ImageJ. (C) RT-qPCR verification of ZC3H10 siRNA knockdown efficiency. Data represent means ± S.E.M. of three independent experiments. (**) P < 0.01 compared with Ctrl. (D) Western blot verification of ZC3H10 siRNA knockdown efficiency. Relative protein levels were quantified by using ImageJ. (E) RT-qPCR verification of CALCOCO1 siRNA and ZC3H10 siRNA knockdown efficiency. Data represent means ± S.E.M. of three independent experiments. (***) P < 0.001, (****) P < 0.0001, compared with Ctrl. (F) Western blot verification of CALCOCO1 siRNA and ZC3H10 siRNA knockdown efficiency. Relative protein levels were quantified by using ImageJ. (G,H) LDH release was tested in control U251 glioma cells and HOTAIR regulatory element KO glioma cells transfected with control siRNAs and CALCOCO1 or ZC3H10 siRNAs. LDH release was tested after 48 h of TMZ (1 mM) treatment. Data represent means ± S.E.M. of three independent experiments. (I,J) Caspase 3/7 activities were tested in control U251 glioma cells and HOTAIR regulatory element KO glioma cells transfected with control siRNAs or CALCOCO1 or ZC3H10 siRNAs. Caspase 3/7 activities were tested after cells were treated 72 h with TMZ (1 mM). Data represent means ± S.E.M. of three independent experiments. (K) LDH release was tested in control U251 glioma cells and HOTAIR regulatory element KO glioma cells transfected with control siRNAs or CALCOCO1 and ZC3H10 siRNAs. LDH release was tested after cells were treated 48 h with TMZ (1 mM). Data represent means ± S.E.M. of three independent experiments. (*) P < 0.05 compared with corresponding Ctrl. (L) Caspase 3/7 activities were tested in control U251 glioma cells and HOTAIR regulatory element KO glioma cells transfected with control siRNAs or CALCOCO1 and ZC3H10 siRNAs. Caspase 3/7 activities were tested after cells were treated 72 h with TMZ (1 mM). Data represent means ± S.E.M. of three independent experiments. (*) P < 0.05 compared with corresponding Ctrl.
Discussion
Development of chromosome conformation capture technologies has facilitated study of long-range gene regulation by regulatory elements (Dekker et al. 2002; Dostie et al. 2006; Simonis et al. 2006; Lieberman-Aiden et al. 2009; Davies et al. 2016). Using those technologies, here, we show that deletion of a recently identified HOTAIR regulatory element in U251 glioma cells increased sensitivity of those cells to TMZ, and HOTAIR overexpression could not reverse this change in TMZ sensitivity. Thus, we suggest that the element modulates glioma cell TMZ sensitivity through long-range regulation of other target genes, based in part on our review of the pattern of epigenetic marks at this locus. Analysis of Capture Hi-C, RNA-seq, and patient survival data revealed that CALCOCO1 and ZC3H10 may be key target genes regulated long-range by the HOTAIR element and that these genes regulate glioma cell sensitivity to TMZ.
Bivalent chromatin structure has been previously reported (Azuara et al. 2006; Bernstein et al. 2006; Ku et al. 2008; Vastenhouw and Schier 2012). Bernstein et al. proposed that bivalent chromatin domains marked by H3K4 methylation and H3K27 methylation silence some developmental genes in embryonic stem cells but keep these genes poised for activation (Bernstein et al. 2006). A similar function for bivalent chromatin has also been reported in cancer cells (Zaidi et al. 2017). Moreover, Zhang et al. identified the HOTAIR regulatory element as an enhancer in ESCC (Zhang et al. 2014), while our data suggest that this regulatory element silences CALCOCO1 and ZC3H10 in glioma and also functions as an enhancer for other genes, like RPL41 (Supplemental Fig. S5). Our combining of Capture Hi-C with RNA-seq data revealed eight target genes potentially regulated by the HOTAIR element, seven up-regulated in cells lacking the element and one down-regulated, again suggesting a bivalent function in glioma cells. More investigations are needed to confirm this function. We focused on regulation of two of them, CALCOCO1 and ZC3H10, as their expression patterns are consistent with patient survival data. Moreover, experiments in which we knocked out the regulatory element in glioma cells supported the idea that these two genes function together to modulate glioma cells’ TMZ sensitivity. CALCOCO1 and ZC3H10 reside on the same chromosome as the HOTAIR regulatory element in humans and are 0.25 Mb and 2.2 Mb, respectively, distant from it. Thus, they may be regulated by the element through long-range interactions. A previous study reports such repressive long-range regulations: Luo and colleagues identified a prostate cancer risk element that regulates HOXA13 through a repressive chromatin loop (Luo et al. 2017). Our study reveals the repressive function of the HOTAIR regulatory element.
CALCOCO1, also known as CoCoA, is a nuclear receptor coactivator that binds to the MYBBP1A N terminus to enhance transcriptional activation (Kim et al. 2003). Other studies report that CALCOCO1 and CCAR1, a different coactivator, synergize to coactivate the tumor suppressor TP53 (Kim et al. 2008). One possibility is that increased TMZ sensitivity seen following enhancer KO is partially due to changes in TP53 activity, although further investigations are needed. Our second target gene ZC3H10 reportedly functions as a tumor suppressor and is down-regulated in highly malignant breast cancer cells, and its expression significantly inhibits colony formation by breast cancer cells in vitro (van ’t Veer et al. 2002; Guardiola-Serrano et al. 2008). Overexpression of CALCOCO1 or ZC3H10 also induced an increase of caspase 3/7 activity in U251 cells treated with TMZ (Supplemental Fig. S6). The reported functions of both CALCOCO1 and ZC3H10 are consistent with our findings, as up-regulation of both following deletion of the HOTAIR regulatory element increases TMZ sensitivity of glioma cells. Moreover, TMZ treatment decreased the interactions between the HOTAIR RE and CALCOCO1 or ZC3H10 and up-regulated expression of CALCOCO1 and ZC3H10. We also showed that individual knockdown of CALCOCO1 and ZC3H10 cannot rescue the increase in TMZ sensitivity, suggesting that the HOTAIR regulatory element governs a network of genes functioning in drug sensitivity.
Development of methods to enhance TMZ as chemotherapy is important to improve the prognosis of glioma patients. Several studies of the mechanism of TMZ sensitivity/resistance have focused on downstream factors that regulate DNA methylation, as TMZ is an alkylating chemotherapeutic agent (Wick et al. 2014). However, ours is one of the few studies to investigate this mechanism in view of long-range gene regulation networks. Our study reveals a new mechanism underlying TMZ sensitivity related to high-order chromatin structures in which the HOTAIR element modulates TMZ sensitivity through long-range regulation networks. Previous studies have reported methods to modify high-order chromatin structure as a means to change target gene expression (Deng et al. 2012, 2014; Morgan et al. 2017). Future analysis should focus on whether such approaches could represent an alternative method to improve prognosis of glioma patients.
In summary, our study shows that a recently identified HOTAIR regulatory element functions as a silencer to modulate glioma cell sensitivity to TMZ through long-range regulation of the targets CALCOCO1 and ZC3H10.
Methods
Cell culture
The GBM cell line U251 was obtained from the National Infrastructure of Cell Line Resource (China). U251 cells were incubated at 37°C with 5% CO2 and cultured in Dulbecco's Modified Eagle Medium (GIBCO), supplemented with 10% fetal bovine serum (FBS) (BI) and 1% penicillin-streptomycin (GIBCO). All cells were tested for mycoplasma contamination each month. All cells used in this paper were mycoplasma-free.
CRISPR-Cas9-mediated deletions
U251 glioma cells were transfected with Cas9 and guide RNA plasmids that target the HOTAIR regulatory element region (hg19_Chr 12: 54,360,188–54,360,822). Guide RNAs were designed using the MIT CRISPR Design website (https://zlab.bio/guide-design-resources). To minimize potential off-target effects of guide RNA, only high-score guide RNAs (score > 85) were used. Guide RNA sequences are listed in Supplemental Table S1. Cell clones were genotyped by PCR, and two clones with homozygous deletion of the HOTAIR regulatory element were used for experiments. Genotyping PCR primers are listed in Supplemental Table S1. GRCh37 (hg19) was used in this study. The differences between GRCh37 and GRCh38 did not affect the deletion region of the regulatory element and the relative position of the regulatory element and its targets; therefore, using GRCh38 would not significantly affect the conclusions in this study.
Lactate dehydrogenase assay
Lactate dehydrogenase in the culture medium was evaluated using the LDH cytotoxicity kit (Promega). In brief, cells were cultured in 96-well plates. After cells were treated with TMZ or transfected with siRNA or overexpression plasmids, the culture medium was collected and LDH activity was assayed in accordance with the manufacturer's instructions. The OD value at 490 nm was measured using a microplate reader. Levels of released LDH of each group were calculated as a percentage of the total amount (positive control as described in the kit protocol).
Caspase 3/7 activity measurements
A Caspase-Glo 3/7 kit (Promega) was used to evaluate caspase 3/7 activities in U251 cells based on the manufacturer's instruction. In brief, cells were harvested and incubated with 100 μL Caspase-Glo 3/7 reagent for 1 h on a rotary shaker in the dark. Luminescence of each sample was measured at 485/530 nm using a microplate reader. Relative caspase 3/7 activity was calculated to evaluate fold-changes of samples treated with TMZ or transfected with siRNA or overexpression plasmids relative to controls.
HOTAIR overexpression
HOTAIR was amplified by PCR with NEBNext High-Fidelity 2× PCR Master Mix (New England BioLabs) and subsequently cloned into the pCDH-CMV-MCS-EF1-Puro overexpression plasmid (System Biosciences) using restriction enzymes NheI (New England BioLabs) and NotI (New England BioLabs). Plasmids were amplified in One Shot Stbl3 chemically competent E. coli (Thermo Fisher Scientific) with ampicillin (Bio Basic Canada, Inc.) selection. Plasmids were transiently transfected using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's instructions.
RNA-seq
Total RNA was extracted from U251 glioma cells or HOTAIR regulatory element KO U251 cells treated with or without TMZ (1 mM, 72 h). Barcoded RNA-seq libraries were sequenced as 150-bp paired-end reads using the Illumina HiSeq 4000 platform. Reads were mapped to the human reference genome (hg19) using HISAT (Kim et al. 2015) with a GENCODE GTF file supplied as gene model annotations. HTSeq (Anders et al. 2015) was used to quantitate transcript abundance for each gene. DESeq2 (Love et al. 2014) was used to perform normalization and regularized log transformations on read counts.
Survival curve
Clinical glioma patients’ survival data were obtained from TCGA. Patient samples were divided into two groups based on the expression level of specific genes indicated in the main text. Samples with gene expression levels higher or lower than the median were marked as “high expression group” or “low expression group,” respectively. P-value < 0.0001 indicates a significant survival difference between the two groups.
Capture Hi-C and Hi-C
Capture Hi-C and Hi-C were performed as previously described (Davies et al. 2016; Belaghzal et al. 2017). The Hi-C library was generated with DpnII (New England BioLabs) digestion. Two biological replicates were assessed with 1 × 107 U251 cells. The Hi-C library was then sheared to 200- to 300-bp fragments by sonication, and the library or precapture library was prepared using the NEBNext DNA library kit according to the manufacturer's instructions (New England BioLabs). Biotinylated probes and streptavidin beads (Invitrogen) were used to enrich the “bait” region and its linked chromatin fragments by two rounds of hybridization-capture approach. Capture Hi-C libraries were sequenced as 150-bp paired-end reads using the Illumina HiSeq X Ten platform. Capture Hi-C and Hi-C data were analyzed based on a pipeline proposed in previous studies (Supplemental Table S2; Servant et al. 2015; Davies et al. 2016; Belaghzal et al. 2017).
Two biotinylated DNA oligonucleotides were designed for both ends of the region containing the HOTAIR regulatory element, with the following sequence (5′–3′):
Biotin-
GATCGAAAGATAAAAAGAGGCGGGAGGGAGGCAGAAGCAAGGGTCGGCTGTTGCTCTCCCTCGCCCGGGCGCCTGCAGCAGTCTGGGTGGGCGGGGGCGCCATGACAAAGTGAAGGTCAA
Biotin-
GTCTGAATGTTACGGTTTCCTTCAGAAAACAAGGCGGTACACTCGGTAAGGAGAAGTGAGGCAGCAGGGGTGGGGAGGGCTGGTGCGGTAGGAGGCTTGGTTTTATTTCTTTCTCAGATC
Quantitative real-time PCR
Total RNA was extracted using TRIzol Reagent (Invitrogen) following the manufacturer's protocol. Five micrograms total RNA was reverse-transcribed into cDNA using a PrimeScript RT reagent kit (Takara). PCR analysis was performed using an SYBR Premix Ex Taq kit (Takara). Primer sequences used in the study are listed in Supplemental Table S3. GAPDH served as an internal reference, and the relative expression of messenger RNAs (mRNAs) was quantified using the 2−ΔΔCt method.
Chomatin Conformation Capture (3C) assay
The 3C assay was performed as previously described (Hagège et al. 2007). In brief, U251 cells were cross-linked with 1% formaldehyde and quenched with 125 mM glycine. The 3C library was generated with HindIII (New England BioLabs) digestion and followed by ligation with T4 ligase (New England BioLabs). The ligated products were quantified by RT-qPCR. Primers used in the 3C assay are listed in Supplemental Table S4. Control 3C template was generated by using bacterial artificial chromosomes (BACs) RP11-657H18 and RP11-166L22 and DNA fragments that contain the HOTAIR regulatory element region, CALCOCO1 loci, and ZC3H10 loci, respectively. BACs and DNA fragments were digested with HindIII and ligated. The ligated product from the control was used for normalization. The relative interaction frequency was calculated as 2Ct(Control)−Ct(3C).
Western blots
Proteins were collected from glioma cells using RIPA lysis buffer with 1× protease inhibitor cocktail (Roche). Sample loading was based on the results of a Bicinchoninic Acid (BCA) assay. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to polyvinylidene difluoride (PVDF) 0.45-μm membranes (Millipore). The membranes were blocked and incubated with primary antibody overnight at 4°C, followed by secondary antibodies for 1 h at room temperature. Then, the blot bands were detected by Image Quant LAS 4000 with an Enhanced Chemiluminescence kit (Thermo Fisher Scientific). Relative protein levels were quantified by using ImageJ. Antibodies used in western blots were as follows: CALCOCO1 (Bioworld BS71035), ZC3H10 (Abcam ab127693), GAPDH (Santa Cruz Biotechnology sc-365062), goat anti-rabbit IgG-HRP (Sungene Biotech LK2001), and rabbit anti-mouse IgG-HRP (Abcam ab6728).
siRNA transfection
Glioma cells were transfected with siRNAs (Supplemental Table S5) using Lipofectamine 3000 reagent (Thermo Fisher Scientific). siRNA knockdown efficiency was verified by RT-qPCR 48–72 h later.
Statistical analysis
Data represent means ± S.E.M.; statistical analysis was performed using Student's t-test. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
Data access
All sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE125629 and GSE125243.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31530027, 31701129, 81772687), National Key R&D Program of China (Grant No. 2017YFA0102600), and the Natural Science Foundation of Tianjin City of China (Grant No. 18JCQNJC10100). We thank Dr. Lingyi Chen at the College of Life Sciences of Nankai University for providing the Cas9 plasmids. We thank Dr. Mulin Jun Li at the School of Basic Medical Sciences of Tianjin Medical University for his suggestions.
Author contributions: L.Z., A.H., B.C., J.B., J.C., D.G., Y.Q., W.W., T.S., Z.Z., J.S., W.A., and F.A. conducted the experiments; L.Z. and W.L. designed the experiments; and L.Z. and W.L. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.251058.119.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538. 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- Belaghzal H, Dekker J, Gibcus JH. 2017. Hi-C 2.0: an optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123: 56–65. 10.1016/j.ymeth.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Clarke MJ, Mulligan EA, Grogan PT, Mladek AC, Carlson BL, Schroeder MA, Curtin NJ, Lou Z, Decker PA, Wu W, et al. 2009. Effective sensitization of temozolomide by ABT-888 is lost with development of temozolomide resistance in glioblastoma xenograft lines. Mol Cancer Ther 8: 407–414. 10.1158/1535-7163.MCT-08-0854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JO, Telenius JM, McGowan SJ, Roberts NA, Taylor S, Higgs DR, Hughes JR. 2016. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods 13: 74–80. 10.1038/nmeth.3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244. 10.1016/j.cell.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. 2014. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158: 849–860. 10.1016/j.cell.2014.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. 2006. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16: 1299–1309. 10.1101/gr.5571506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardiola-Serrano F, Haendeler J, Lukosz M, Sturm K, Melchner H, Altschmied J. 2008. Gene trapping identifies a putative tumor suppressor and a new inducer of cell migration. Biochem Biophys Res Commun 376: 748–752. 10.1016/j.bbrc.2008.09.070 [DOI] [PubMed] [Google Scholar]
- Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722–1733. 10.1038/nprot.2007.243 [DOI] [PubMed] [Google Scholar]
- Kim JH, Li H, Stallcup MR. 2003. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell 12: 1537–1549. 10.1016/S1097-2765(03)00450-7 [DOI] [PubMed] [Google Scholar]
- Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR. 2008. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell 31: 510–519. 10.1016/j.molcel.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitange GJ, Carlson BL, Schroeder MA, Grogan PT, Lamont JD, Decker PA, Wu W, James CD, Sarkaria JN. 2009. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro-Oncology 11: 281–291. 10.1215/15228517-2008-090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NQ, Ter Huurne M, Nguyen LN, Peng T, Wang SY, Studd JB, Joshi O, Ongen H, Bramsen JB, Yan J, et al. 2017. The non-coding variant rs1800734 enhances DCLK3 expression through long-range interaction and promotes colorectal cancer progression. Nat Commun 8: 14418 10.1038/ncomms14418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Rhie SK, Lay FD, Farnham PJ. 2017. A prostate cancer risk element functions as a repressive loop that regulates HOXA13. Cell Rep 21: 1411–1417. 10.1016/j.celrep.2017.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan SL, Mariano NC, Bermudez A, Arruda NL, Wu F, Luo Y, Shankar G, Jia L, Chen H, Hu JF, et al. 2017. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun 8: 15993 10.1038/ncomms15993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Wu C, Su Z, Duan Z, Li L, Mi F, Li C. 2017. Genetic polymorphisms of non-coding RNAs associated with increased head and neck cancer susceptibility: a systematic review and meta-analysis. Oncotarget 8: 62508–62523. 10.18632/oncotarget.20096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Zhang L, Cai M, Li H, Xu H, Yang H, Zhao Z, Rhie SK, Farnham PJ, Shi J, et al. 2019. The prostate cancer risk variant rs55958994 regulates multiple gene expression through extreme long-range chromatin interaction to control tumor progression. Sci Adv 5: eaaw6710 10.1126/sciadv.aaw6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Friedman AH, Sampson JH, McLendon RE, Herndon JE II, Friedman HS. 2009. Phase II trial of temozolomide (TMZ) plus irinotecan (CPT-11) in adults with newly diagnosed glioblastoma multiforme before radiotherapy. J Neurooncol 95: 393–400. 10.1007/s11060-009-9937-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. 2017. Advances in the molecular genetics of gliomas—implications for classification and therapy. Nat Rev Clin Oncol 14: 434–452. 10.1038/nrclinonc.2016.204 [DOI] [PubMed] [Google Scholar]
- Rheinbay E, Suvà ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, et al. 2013. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep 3: 1567–1579. 10.1016/j.celrep.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Bourgon R, Garcia J, Li C, Cloughesy T, Chinot OL, Wick W, Nishikawa R, Mason W, Henriksson R, et al. 2015. Patients with proneural glioblastoma may derive overall survival benefit from the addition of bevacizumab to first-line radiotherapy and temozolomide: retrospective analysis of the AVAglio trial. J Clin Oncol 33: 2735–2744. 10.1200/JCO.2015.61.5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A, Teufel J, Ringel F, Bettstetter M, Hoepner I, Rasper M, Gempt J, Koeritzer J, Schmidt-Graf F, Meyer B, et al. 2012. Aldehyde dehydrogenase 1A1—a new mediator of resistance to temozolomide in glioblastoma. Neuro-Oncology 14: 1452–1464. 10.1093/neuonc/nos270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E. 2015. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16: 259 10.1186/s13059-015-0831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Guo H, Zhang R, Liu H, Wu L, Wu Q, Liu J, Liu T, Zhang Q. 2017. The PI3K inhibitor GDC-0941 enhances radiosensitization and reduces chemoresistance to temozolomide in GBM cell lines. Neuroscience 346: 298–308. 10.1016/j.neuroscience.2017.01.032 [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354. 10.1038/ng1896 [DOI] [PubMed] [Google Scholar]
- Su G, Guo D, Chen J, Liu M, Zheng J, Wang W, Zhao X, Yin Q, Zhang L, Zhao Z, et al. 2019. A distal enhancer maintaining Hoxa1 expression orchestrates retinoic acid-induced early ESCs differentiation. Nucleic Acids Res 47: 6737–6752. 10.1093/nar/gkz482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Lee D, Ho AS, Pu JK, Zhang XQ, Lee NP, Day PJ, Lui WM, Fung CF, Leung GK. 2013. Inhibition of prolyl 4-hydroxylase, β polypeptide (P4HB) attenuates temozolomide resistance in malignant glioma via the endoplasmic reticulum stress response (ERSR) pathways. Neuro-Oncology 15: 562–577. 10.1093/neuonc/not005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536. 10.1038/415530a [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. 2012. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol 24: 374–386. 10.1016/j.ceb.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Y, Li YW, Zhang HB, Gong H, Yuan Y, Li WT, Liu HY, Chen J. 2018. HOTAIR lncRNA SNPs rs920778 and rs1899663 are associated with smoking, male gender, and squamous cell carcinoma in a Chinese lung cancer population. Acta Pharmacol Sin 39: 1797–1803. 10.1038/s41401-018-0083-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren KE, Gururangan S, Geyer JR, McLendon RE, Poussaint TY, Wallace D, Balis FM, Berg SL, Packer RJ, Goldman S, et al. 2012. A phase II study of O6-benzylguanine and temozolomide in pediatric patients with recurrent or progressive high-grade gliomas and brainstem gliomas: a Pediatric Brain Tumor Consortium study. J Neurooncol 106: 643–649. 10.1007/s11060-011-0709-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. 2013. Klf4 organizes long-range chromosomal interactions with the Oct4 locus in reprogramming and pluripotency. Cell Stem Cell 13: 36–47. 10.1016/j.stem.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R, et al. 2015. Glioma. Nature Rev Dis Prim 1: 15017 10.1038/nrdp.2015.17 [DOI] [PubMed] [Google Scholar]
- Weng SL, Wu WJ, Hsiao YH, Yang SF, Hsu CF, Wang PH. 2018. Significant association of long non-coding RNAs HOTAIR genetic polymorphisms with cancer recurrence and patient survival in patients with uterine cervical cancer. Int J Med Sci 15: 1312–1319. 10.7150/ijms.27505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M, et al. 2012. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol 13: 707–715. 10.1016/S1470-2045(12)70164-X [DOI] [PubMed] [Google Scholar]
- Wick W, Weller M, van den Bent M, Sanson M, Weiler M, von Deimling A, Plass C, Hegi M, Platten M, Reifenberger G. 2014. MGMT testing—the challenges for biomarker-based glioma treatment. Nat Rev Neurol 10: 372–385. 10.1038/nrneurol.2014.100 [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Frietze SE, Gordon JA, Heath JL, Messier T, Hong D, Boyd JR, Kang M, Imbalzano AN, Lian JB, et al. 2017. Bivalent epigenetic control of oncofetal gene expression in cancer. Mol Cell Biol 37 10.1128/MCB.00352-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, Lu C, Zhou C, Yuan Q, Yang M. 2014. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis 35: 2062–2067. 10.1093/carcin/bgu103 [DOI] [PubMed] [Google Scholar]
- Zhong C, Chen Y, Tao B, Peng L, Peng T, Yang X, Xia X, Chen L. 2018. LIM and SH3 protein 1 regulates cell growth and chemosensitivity of human glioblastoma via the PI3K/AKT pathway. BMC Cancer 18: 722 10.1186/s12885-018-4649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18. 10.1038/nrg2905 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.