Abstract
Replication and amplification of the viral genome is a key process for all viruses. For hepatitis C virus (HCV), a positive-strand RNA virus, amplification of the viral genome requires the synthesis of a negative-sense RNA template, which is in turn used for the production of new genomic RNA. This process is governed by numerous proteins, both host and viral, as well as distinct lipids and specific RNA elements within the positive- and negative-strand RNAs. Moreover, this process requires specific changes to host cell ultrastructure to create microenvironments conducive to viral replication. This review will focus on describing the processes and factors involved in facilitating or regulating HCV genome replication.
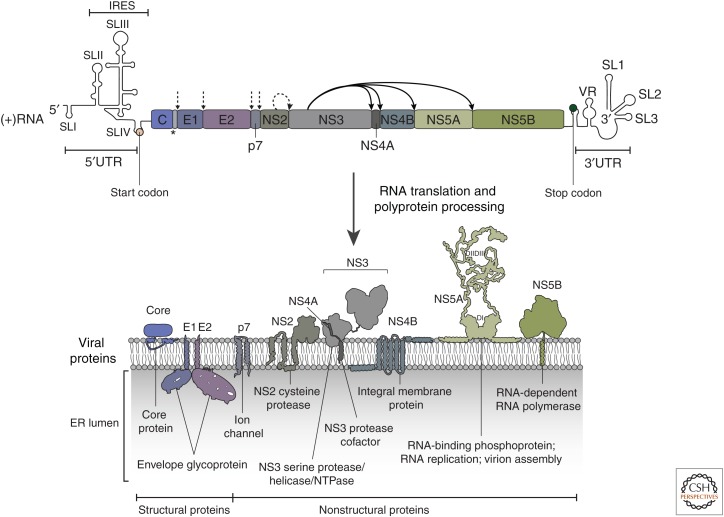
The ∼9600-nucleotide hepatitis C virus (HCV) RNA genome possesses one large open reading frame (ORF) that is flanked by highly structured 5′ and 3′ untranslated regions (UTRs) (Fig. 1). cis-acting RNA elements (CREs) present within the UTRs as well as the protein-coding region contribute to RNA translation and/or genome replication (for review, see Adams et al. 2017). Synthesis of the HCV proteins is mediated by the internal ribosome entry site (IRES) located in the 5′UTR and facilitated by distinct elements in the 3′UTR as well as CREs within the protein-coding region (Fig. 1). The IRES is composed of three stem-loop (SL) domains. Of these, SLII and SLIII reside in the 5′UTR and adopt an extended structure; SLIV overlaps with the 5′ end of the core coding region and forms a short, rather unstable stem that contains the start codon of the HCV ORF (Fig. 1; Pérard et al. 2013; Quade et al. 2015). Mechanistically, SLIII plays an important role in IRES function because it facilitates the IRES–40S ribosome subunit interaction by a conserved base-pairing between the 18S rRNA and a sequence in SLIII (Matsuda and Mauro 2014). In addition, SLII tightly associates with the head of the 40S subunit, whereas SLIII displaces eIF3 to allow the assembly of a translation-competent ribosome (Hashem et al. 2013; Yamamoto et al. 2015). Apart from the SL domains in the 5′UTR, several elements downstream of the IRES impact RNA translation, including SL47 and SL87 (also called SLV and SLVI, respectively) in the core coding region (see below) (McMullan et al. 2007; Vassilaki et al. 2008). RNA translation is also modulated by the liver-specific microRNA (miR)-122 that binds to numerous sites within the viral genome (Jopling et al. 2005).
Figure 1.
Hepatitis C virus (HCV) genome organization and membrane topology of viral proteins. The open reading frame (ORF) encoding the HCV polyprotein and the predicted secondary structures of the flanking 5′ and 3′ untranslated region (UTR) are depicted at the top. Membrane topology of mature viral proteins and their function are shown at the bottom. Co- and posttranslational cleavage of the viral polyprotein are indicated as follows: (dashed vertical arrows) signal peptidase, (star) signal peptide peptidase removing the E1 signal sequence from the carboxyl terminus of core, (dashed curved arrow) NS2-3 protease, (solid arrows) NS3-4A protease. Note that only NS5A is shown as a dimer, but other viral proteins also may form homo- and heterodimers or oligomeric complexes. (D) domain, (ER) endoplasmic reticulum. (Figure adapted from data in Bartenschlager et al. 2013, with permission, from the authors.)
Translation of the viral RNA leads to the production of an ∼3000 amino acid polyprotein from which the individual viral proteins are liberated through the cumulative activity of both host and viral proteases (Fig. 1; Table 1). The structural proteins (i.e., core and the envelope glycoproteins E1 and E2) are the main constituents of HCV particles, whereas the viroporin p7 and nonstructural protein (NS) 2 are involved in virion assembly but are not incorporated into the virus particle (see Shimotohno 2019). The remaining nonstructural proteins (i.e., NS3, NS4A, NS4B, NS5A, and NS5B) have specific roles in viral genome amplification. NS3 has several functions. The amino-terminal domain, together with the cofactor NS4A, is a serine-type protease required for polyprotein cleavage and proteolytic processing of host cell factors (Failla et al. 1994; Bartenschlager et al. 1995; Meylan et al. 2005). The carboxy-terminal NS3 domain is a helicase and possesses an additional NTPase activity that is required for RNA unwinding (see below). Moreover, NS3, via interaction with the NS2 protease domain, is involved in the assembly of HCV particles (Counihan et al. 2011). NS4B is a highly hydrophobic protein involved in inducing membrane alterations that are required for the biogenesis of the viral replication organelle (RO). NS5A is a multifunctional phosphoprotein required for both RNA replication and assembly. NS5B shows RNA-dependent RNA polymerase activity and therefore plays the central role for the amplification of the viral genome. Each of the viral proteins are bound to intracellular membranes by various means, including complex transmembrane domains (e.g., NS2, NS4B), a monotopic α-helix (e.g., NS5A), or a single transmembrane helix (e.g., NS5B) (Fig. 1). Therefore, the HCV replication cycle occurs on membrane surfaces, thus reducing dimensionality.
Table 1.
Viral RNA elements, selected cellular proteins, and lipids involved in hepatitis C virus (HCV) replication covered herein
| Function in virus replication | References | |
|---|---|---|
| Viral RNA element | ||
| SLI | Involved in genome replication | Friebe et al. 2001 |
| SLII | Associates tightly with the head of the 40S subunit to enable RNA translation Involved in genome replication |
Friebe et al. 2001; Hashem et al. 2013; Yamamoto et al. 2015 |
| SLIII | Facilitates the interaction between the IRES and the 40S subunit of the ribosome Displaces eIF3 to allow the assembly of a translation-competent ribosome |
Hashem et al. 2013; Matsuda and Mauro 2014; Yamamoto et al. 2015 |
| SL47 | Impacts RNA translation | McMullan et al. 2007; Vassilaki et al. 2008 |
| SL87 | Impacts RNA translation and replication | McMullan et al. 2007; Vassilaki et al. 2008 |
| SL248 | Interactions with SL87 might serve as a molecular switch between genome replication and packaging | Pirakitikulr et al. 2016 |
| 5BSL3.2 | Forms a kissing loop interaction with the 3′X SL2, which is essential for replication Forms long-range interactions with SLIII in the 5′UTR and has been proposed to regulate switching between replication and translation |
Lee et al. 2004; You et al. 2004; Friebe et al. 2005; Romero-López and Berzal-Herranz 2009, 2012, 2017 |
| 3′X and poly(U/UC) | The 3′X and a minimal poly(U/UC) region are essential for genome replication Initiation of negative-strand RNA synthesis occurs at the terminal uridine that is base-paired to a guanosine in the 3′X SL1 |
Tanaka et al. 1995, 1996; Kolykhalov et al. 1996, 2000; Blight and Rice 1997; Yanagi et al. 1999; Friebe and Bartenschlager 2002; Smith et al. 2002; Yi and Lemon 2003a,b; Friebe et al. 2005; You and Rice 2008 |
| Variable region | Contributes to efficient genome replication | Yi and Lemon 2003a |
| Host protein | ||
| PI4KA | Produces PI4P at HCV protein-containing membranes | Berger et al. 2011; Reiss et al. 2011 |
| VAP-A | Interacts with a specific NS5A phosphoform | Evans et al. 2004 |
| VAP-B | Interacts with NS5A and NS5B | Hamamoto et al. 2005 |
| OSBP | Releases cholesterol in exchange for PI4P into the membrane of DMVs | Wang et al. 2014 |
| NPC1 and NPC2 | Transport unesterified cholesterol to DMVs | Stoeck et al. 2017 |
| FAPP2 | Might release glucosylceramide in exchange for PI4P into the membrane of DMVs | Khan et al. 2014 |
| SREBP | Activates transcription of target lipogenic genes | Waris et al. 2007; Park et al. 2009 |
| DDX3X | Activates a signaling cascade that induces the expression of SREBP | Li et al. 2013a |
| CypA | Catalyzes the isomerization of peptidyl-prolyl bonds in NS5A domain I and domain II stimulating the RNA-binding capacity of NS5A Might be involved in the formation of RO |
Rosnoblet et al. 2012; Madan et al. 2014; Ngure et al. 2016; Badillo et al. 2017 |
| Sec14L2 | Enhances vitamin E–mediated inhibition of lipid peroxidation | Saeed et al. 2015 |
| Lipids | ||
| PI4P | Recruits OSBP and FAPP2 | Khan et al. 2014; Wang et al. 2014 |
| Cholesterol | Required for replicase activity and impacts replication complex architecture | Paul et al. 2013 |
| Sphingolipids | Stimulates replicase activity | Weng et al. 2010; Hirata et al. 2012 |
(IRES) internal ribosome entry site, (UTR) untranslated region, (DMVs) double-membrane vesicles, (RO) replication organelle.
HCV-INDUCED MEMBRANE ALTERATIONS
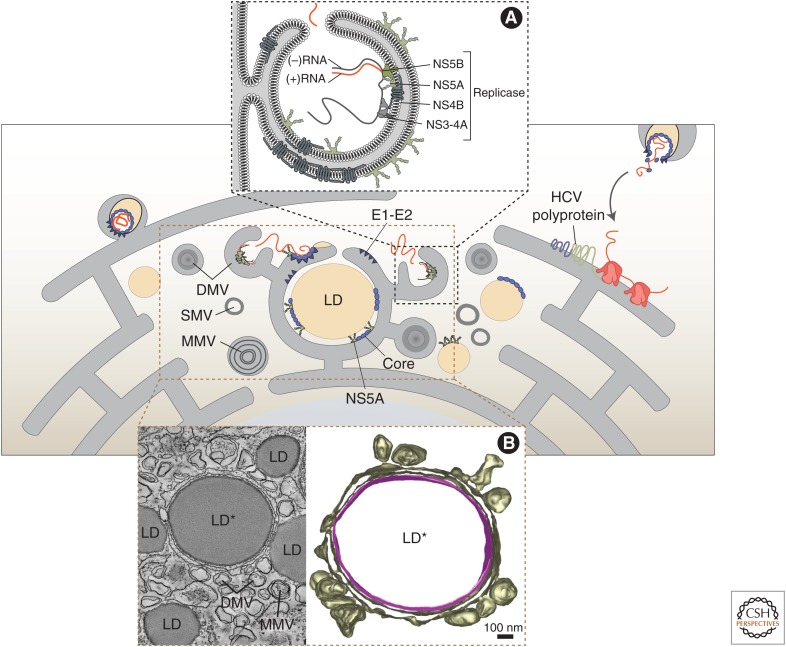
Early electron microscopy (EM) studies of liver tissues from infected patients or chimpanzees indicated that HCV induces membrane alterations in infected hepatocytes (Jackson et al. 1979; Shimizu et al. 1990; Shimizu 1992), which is a hallmark of all positive-strand RNA viruses. With the advent of robust cell culture models for HCV, it became possible to study structure–function relationships of HCV proteins. It was found that the expression of individual viral proteins induced membrane alterations (Egger et al. 2002). Notably, the sole expression of NS4B induces a condensed membrane structure, consisting of single-membrane vesicles in a membranous matrix. This structure was given the designation “membranous web,” and it was also found in cells containing a subgenomic replicon of moderate replication competence (Gosert et al. 2003). However, in hepatoma cells containing a highly replication-competent HCV isolate, designated JFH-1, because it was isolated from a Japanese patient with fulminant hepatitis (Kato et al. 2003), the predominant virus-induced membrane structure is double-membrane vesicles (DMVs) that accumulate in the cytoplasm, often in close proximity of lipid droplets (Fig. 2). Although single-membrane vesicles were detected rather sporadically, multimembrane vesicles were observed primarily at late time points after infection and are thought to reflect a stress response induced by high-level virus replication (Ferraris et al. 2010; Romero-Brey et al. 2012; Paul et al. 2013). DMVs are heterogeneous in size, with an average diameter of ∼200 nm, and are morphologically similar to membrane alterations identified in cells infected with coronaviruses (Gosert et al. 2002; Knoops et al. 2008) or picornaviruses (Belov et al. 2012). In the case of HCV, double-stranded RNA (dsRNA) and nonstructural proteins have been found in association with DMV membranes (Ferraris et al. 2010; Romero-Brey et al. 2012; Paul et al. 2013). This result and the observed correlation between DMV abundance and viral RNA replication argue that DMVs are the sites of viral genome replication (Ferraris et al. 2010; Romero-Brey et al. 2012; Paul et al. 2013). However, it is still unclear whether all DMVs are engaged in HCV replication and whether RNA replication occurs on the interior or exterior membrane surface of the DMV.
Figure 2.
HCV replication organelle. After entering the cell, the HCV genome is released into the cytosol and translated at the rough endoplasmic reticulum (ER). Viral proteins, in cooperation with host factors, induce intracellular membrane alterations consisting of double-membrane vesicles (DMVs), single-membrane vesicles (SMVs), and multimembrane vesicles (MMVs). DMVs, usually found in close association with lipid droplets (LDs), are protrusions of the ER that contain nonstructural proteins required for genome amplification (inset A). These vesicles are open toward the cytosol or are closed (represented as gray shaded vesicles), possibly reflecting different stages of DMV “maturation” (early and late, respectively). Viral RNA amplification may occur inside DMVs, which would allow the exit of newly synthetized viral genomes as long as the DMV is open. RNA molecules might be delivered by NS5A and NS3 to nearby assembly sites enriched in core protein and E1-E2 envelope glycoprotein complexes that are associated with p7 and NS2. Alternatively, replication might occur on the outer surface of DMVs (not represented). Particles are formed by budding into the lumen of the ER. (Inset B) DMVs emanate from ER membranes that are tightly wrapped around LDs as revealed by a combination of live cell imaging and electron tomography. (Left) Single tomographic slice of an HCV-infected cell revealing two classes of LDs. First, an LD (LD*) that is tightly wrapped by the ER and that stains positive for E2 and NS5A as revealed by fluorescence microscopy (not shown) and, second, several LDs that are not wrapped by the ER and that do not stain for E2 and NS5A (LD) suggesting that HCV proteins trigger LD wrapping by ER membranes. (Right) 3D reconstruction of the membranes surrounding LD*. ER membrane and DMVs are shown in yellow-gray; the LD monolayer membrane is shown in violet. Note the DMVs originating from the wrapping ER membrane. In some cases, a stalk-like connection between DMVs and the ER is visible. Assuming that RNA replication occurs in these DMVs, only short-distance trafficking of viral RNA would be required to the ER lumen to allow virus budding (as indicated in the schematic above). (Images in inset B are adapted from images in Lee et al. 2019 under the terms of the Creative Commons Attribution License [CC BY].)
Several studies have reported that viral proteins and RNA associated with the viral ROs are protected from exogenously added proteases or nucleases, indicating that RNA replication occurs in a membranous environment that is segregated from the surrounding cytoplasm (Miyanari et al. 2003; Quinkert et al. 2005; Hsu et al. 2010; Paul et al. 2013). Interestingly, most DMVs appear to be closed structures and only a minority (∼8%) have an opening pore toward the cytosol (Fig. 2; Romero-Brey et al. 2012). This morphology suggests that if replication occurs on the interior surface of DMVs, then a transport mechanism must be present to allow import of metabolites required for replication as well as export of viral RNA for translation or virion assembly. Alternatively, in a model in which replication occurs on the outer surface of DMVs or on open DMVs, a more complex architecture of the RO must exist to explain the protection of viral RNA from attack by exogenously added nucleases. In either case, the protected nature of the viral RNA indicates a transport mechanism to mediate the movement of macromolecules between cellular compartments. This model is supported by observations that HCV hijacks specific cellular components involved in nucleocytoplasmic transport, and that these cellular factors are involved in maintaining a selective barrier between the cytosol and the interior of viral ROs (Neufeldt et al. 2013, 2016). However, the mechanisms underlying this transport process as well as the nature of the barrier between replication compartments and the surrounding cytosol remain unclear.
Viral Factors Involved in HCV Replication Organelle Formation
Although individual expression of the nonstructural proteins induces membrane alterations, NS5A was found to be the only protein capable of inducing DMV formation on its own (Egger et al. 2002; Romero-Brey et al. 2012). However, the efficiency of DMV formation is rather low, but is greatly enhanced when NS5A is expressed as part of an NS3–NS5B polyprotein fragment. Mutation analyses identified important motifs in HCV proteins required for DMV formation. These include the helicase domain in NS3, glycine zipper motifs in NS4B, the amino-terminal NS5A membrane anchor, and domain 1 of NS5A (Romero-Brey et al. 2015; Paul et al. 2018). The important role of NS5A in HCV RO formation is best illustrated by the observation that highly active NS5A inhibitors such as daclatasvir block formation of the HCV RO independent of RNA replication (Berger et al. 2014). Molecular docking studies and the positioning of daclatasvir resistance mutations suggest that the drug binds to the membrane-proximal side of NS5A domain I (Berger et al. 2014; Nettles et al. 2014). Therefore, NS5A inhibitors appear to block formation of the HCV RO through disturbing the positioning, folding, and/or flexibility of the linker segment connecting the amino-terminal α-helix and domain I.
In addition to NS5A, NS4B also plays a central role in HCV RO biogenesis. NS4B has a complex membrane topology that is comprised of four amphipathic α-helices, two at the amino-terminal and two at the carboxy-terminal region, which flank four transmembrane-spanning α-helices (Fig. 1; for review, see Bartenschlager et al. 2013). These terminal protein domains can alter membrane properties and likely undergo posttranslational membrane topology changes (Palomares-Jerez et al. 2012, 2013), presumably in an NS5A-regulated manner (Lundin et al. 2006). Additionally, NS4B forms oligomeric complexes via self-interaction, which is mediated in part by a glycine zipper within transmembrane segments 2 and 3 (Paul et al. 2018). This self-interaction is required for HCV RNA replication and the formation of functional ROs (Gouttenoire et al. 2010, 2014; Paul et al. 2011).
The insertion of the amino-terminal amphipathic α-helix of NS5A, which was shown to alter model membranes in vitro (Palomares-Jerez et al. 2010), into just one membrane layer and the formation of conically shaped protein complexes in the membrane, as might be the case with NS4B, could facilitate membrane curvature required for RO formations (McMahon and Gallop 2005; McMahon and Boucrot 2015). In addition to these direct roles, viral proteins, especially NS5A, might contribute to membrane alterations by recruiting host factors required for RO biogenesis.
Host Factors and Processes Involved in HCV Replication Organelle Formation
In addition to viral proteins and the RNA genome, host cell factors and machineries also contribute to DMV biogenesis. One example is macro-autophagy, the bulk degradation and recycling of cytosolic proteins or organelles. It was shown that HCV induces autophagy, and that the depletion of specific autophagy factors impairs HCV RNA replication and reduces the number of DMVs (for review, see Wang and Ou 2018). Moreover, HCV-induced DMVs have a striking morphological similarity to autophagosomes, which are also cytosolic double-membrane structures. However, DMVs induced by HCV have an average diameter of ∼200 nm and thus are smaller than autophagosomes, which have a diameter of 500–1000 nm. It may be that HCV induces DMV formation by using only specific components of the autophagy pathway, in combination with viral proteins, leading to the formation of vesicular structures that have similar membrane topology but differ in overall size.
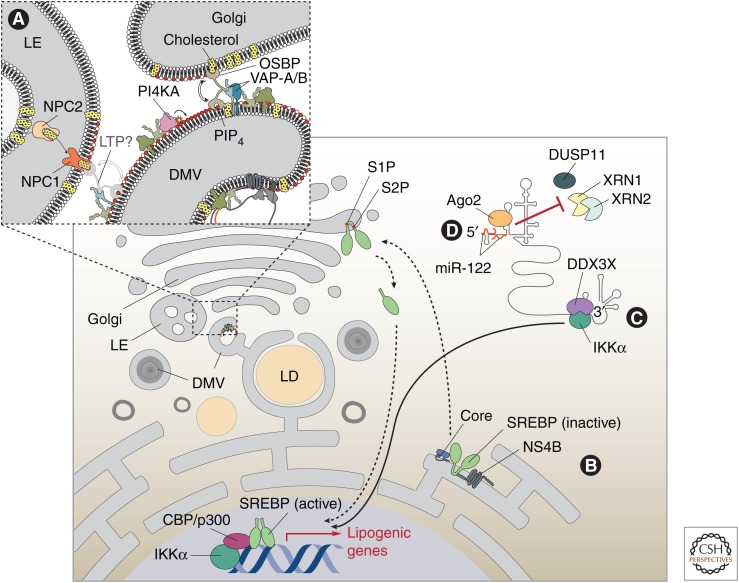
Although HCV-induced DMVs are derived from the ER membrane, their lipid composition is different from the originating membrane, having much higher levels of cholesterol and sphingolipids. NS5A recruits and activates the lipid kinase phosphatidylinositol 4-kinase IIIα (PI4KA) (Berger et al. 2011; Reiss et al. 2011), which produces phosphatidylinositol 4-phosphate (PI4P) at HCV protein-containing membranes (Fig. 3, inset). NS5A also binds to and recruits, likely via VAP-A/B, lipid-transfer proteins, most notably oxysterol-binding protein (Wang et al. 2014). By analogy to results obtained in the yeast system (Mesmin et al. 2013, 2017), we hypothesize that oxysterol-binding protein (OSBP) delivers cholesterol into the DMV membrane in exchange for PI4P (Paul and Bartenschlager 2015). Another lipid-transfer protein, NPC1, possibly together with NPC2 transports unesterified cholesterol to DMVs and contributes to the establishment of a microenvironment conducive for efficient HCV RNA replication (Fig. 3, inset; Stoeck et al. 2017). Additionally, the glucosylceramide transfer protein FAPP2 might be recruited to DMVs to release lipids in exchange for PI4P into the membrane of DMVs (Khan et al. 2014). Overall, these cellular lipid transfer proteins contribute to the enrichment of distinct lipids that could form lipid rafts assumed to be required for high-level HCV replicase activity and possibly the assembly of infectious HCV particles.
Figure 3.
Exploitation of lipid pathways and miR-122 by hepatitis C virus (HCV). (A) HCV alters the lipid composition of rearranged membranes. NS5A and NS5B recruit and activate phosphatidylinositol 4-kinase-α (PI4KA) to produce a local accumulation of phosphatidylinositol 4-phosphate (PI4P). This may determine the directionality of cholesterol transfer by lipid transfer proteins (LTPs), such as oxysterol-binding protein (OSBP), which is recruited by NS5A via VAP-A/B and releases cholesterol in exchange for PIP4 at these membrane contact sites. VAP proteins might serve as anchors for additional host proteins promoting the formation of endoplasmic reticulum (ER)–late endosome (LE) membrane contacts. Here NPC1, possibly in coordination with NPC2, mediates the export of unesterified cholesterol that might be accepted by lipid transfer proteins recruited by HCV (indicated with a question mark). (B,C) HCV infection activates the transcription of lipogenic genes by two distinct pathways. (B) The inactive SREBP precursor traffics from the ER to the Golgi on HCV infection or expression of core or NS4B. There, the transcriptionally active amino-terminal segment is released after two-step proteolytic processing by the site 1 protease (S1P) and S2P. Upon dimerization, the active SREBP enters the nucleus and activates the transcription of lipogenic genes. (C) The HCV 3′UTR interacts with DEAD box polypeptide 3X-linked (DDX3X). This RNA-binding protein activates IKK-α, which stimulates CBP-p300 to promote SREBP-mediated transcription. (D) miR-122, in association with Agonaute-2 (Ago2), binds to the HCV 5′UTR and protects the viral genome from 5′ triphosphate removal and nucleolytic degradation by 5′-3′ exoribonucleases 1 (XRN1) and XRN2.
Although numerous studies have provided important insights into the biogenesis and function of HCV-induced ROs, many questions related to DMV formation remain unanswered. For instance, what is the exact site of HCV RNA replication? Is it SMVs or DMVs? What is the relationship between these two vesicle species? Assuming that DMVs are the site of RNA replication, are they all engaged in replication or only a subpopulation? The observation that only ∼8% of DMVs analyzed at a given time point have a pore-like opening argues that the latter is correct. Alternatively, DMVs might be transient structures that are actively engaged in RNA replication as long as they have an opening to the cytosol, but on closure might become inactive and released out of the cell as extracellular vesicles (Grünvogel et al. 2018). Other questions relate to the molecular mechanisms by which DMVs and MMVs are formed from intracellular (ER-derived) membranes. What is the role of autophagy in this process? Are MMVs a by-product resulting from a cellular stress response induced by HCV? And how is viral cargo transported between HCV-induced subcellular compartments (i.e., sites of RNA translation, RNA replication, and virion assembly)? How is this transport coordinated? These questions illustrate that more studies are required to clearly define the roles of virus-induced membrane alterations in the replication cycle of HCV, and positive-strand RNA viruses in general.
GENOME REPLICATION
RNA Elements Involved in Genome Amplification
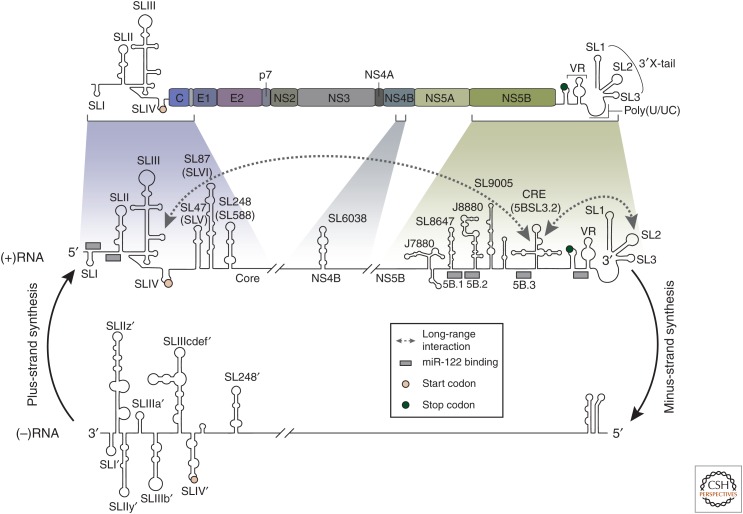
Amplification of the viral genome requires a concerted effort from viral proteins and RNA elements as well as specific host factors. Outside the context of protein production, RNA structural elements found in both the positive-sense viral genome and the negative-strand replication intermediate are essential for viral genome replication. RNA structures found in the 5′UTR of the positive-strand RNA are primarily involved in translation initiation, but several SL structures designated SLI and SLII have been linked to genome replication (Fig. 4; Friebe et al. 2001; Friebe and Bartenschlager 2002). This link is likely associated with the 3′ end of the negative-strand RNA, which forms distinct secondary structures from those found in the positive strand (Fig. 4). In fact, several independent studies have confirmed SL elements in the 3′ terminal region of the negative-strand RNA, including SLI′, SLIIz′, SLIIy′, SLIIIa′, and SLIIIb′; additional elements have been suggested but specific structures and functions remain uncertain (Schuster et al. 2002; Dutkiewicz et al. 2008). Sequence pertaining to the regions of SLI′ and SLIIz′ has been genetically mapped as the minimal requirements for synthesis of the positive-strand RNA from the negative-strand RNA template, and functional assays show that SLIIy′ is also required for RNA synthesis (Friebe et al. 2001; Friebe and Bartenschlager 2009). The importance of the SLI′ element has also been confirmed by in vitro approaches, which also indicate a role for SLIIIb′ in promoting RNA synthesis. However, contrary to the in cellulo analysis, biochemical assays suggested that SLIIz′ inhibits rather than promotes RNA synthesis (Astier-Gin et al. 2005; Masante et al. 2008; Mahias et al. 2010). This discrepancy may indicate the presence of additional control elements in host cells that are required to regulate the structure of the SLIIz′ region or its function in viral genome replication.
Figure 4.
RNA elements within the positive-strand hepatitis C virus (HCV) genome and its negative-strand replication intermediate. The HCV genome organization is represented on the top as in Figure 1. A magnification of three regions within the positive-strand (+) RNA are illustrated below, each representing predicted RNA structures. Long-range RNA–RNA interactions are indicated with dashed arrows, whereas predicted binding sites of miR-122 are shown as gray rectangles. Structures within the negative-strand (−) RNA are shown on the bottom. Alternative nomenclatures of the structures are given in parentheses. (RNA stem-loop (SL) structures are adapted from data in Niepmann et al. 2018 and Bartenschlager et al. 2013.)
Downstream of the positive-strand RNA 5′UTR, several RNA elements have been identified in the core coding region that promote full genome replication, including SL87 (also designated SLVI or SL427) and the SL248 element (also called SL588) (Fig. 4; McMullan et al. 2007; Vassilaki et al. 2008; Pirakitikulr et al. 2016). For SL87, which has previously been described as a translational promoter, this may indicate a dual role in both RNA translation and genome replication. The SL248 element has been linked to replication in the context of full genome virus, but genomes in which this structure has been genetically altered are still replication-competent and produce infectious virus (Vassilaki et al. 2008; Pirakitikulr et al. 2016). Interestingly, mutually exclusive interactions between SL248 and the adjacent SL87 might serve as a molecular switch between efficient replication and infectious virus production (Pirakitikulr et al. 2016). This suggests that RNA structures found throughout the viral genome contribute complex mechanisms involved in regulating different viral processes.
In addition to the 5′ end of positive-strand RNA, several RNA elements present in the 3′UTR and the NS5B-coding region are essential for genome replication. The 3′UTR is comprised of a variable region, a poly(U/UC) tract of variable length and a highly conserved 3′ region designated the 3′X-tail (Fig. 4; Tanaka et al. 1995; Kolykhalov et al. 1996). Each of these regions contribute to viral replication with the 3′X and a minimal poly(U/UC) region being essential and the variable region contributing to efficient replication (Tanaka et al. 1995, 1996; Kolykhalov et al. 1996, 2000; Blight and Rice 1997; Yanagi et al. 1999; Friebe and Bartenschlager 2002; Smith et al. 2002; Yi and Lemon 2003a,b; Friebe et al. 2005; You and Rice 2008). Initiation of negative-strand RNA synthesis begins at the terminal uridine that is base-paired to a guanosine in the 3′X SL1. In this structure, access of the terminal nucleotide for the NS5B polymerase is limited, suggesting that alternative RNA structures are involved in regulating the initiation of RNA synthesis (Fricke et al. 2015). In addition, in the NS5B-coding region, a cis-acting RNA element (CRE; also called 5BSL3.2) has been identified as a crucifix-like structure and forms a kissing loop interaction with the 3′X SL2, which is essential for replication (You et al. 2004; Friebe et al. 2005). This CRE also forms long-range interactions with SLIII in the 5′UTR (Fig. 4) and has been proposed to regulate switching between replication and translation (Lee et al. 2004; You et al. 2004; Romero-López and Berzal-Herranz 2009, 2012, 2017). Additional long-range interactions between 3′ and 5′ elements, facilitated by trans-acting host factors, have been shown to potentiate viral genome replication (for review, see Niepmann et al. 2018). Three other SL elements within the coding region have also been linked to replication, SL6038, J7880, and J8880 (Fig. 4), but the specific functions of these elements remain to be determined (Mauger et al. 2015; Pirakitikulr et al. 2016).
Virus Protein Contributions to Genome Replication
NS5B Polymerase
A plethora of studies examining NS5B, which harbors RNA-dependent RNA polymerase (RdRp) activity (Behrens et al. 1996; Lohmann et al. 1997), have given us significant insights into its structural and functional properties. NS5B is a tail-anchored protein composed of an amino-terminal catalytic domain that makes up the majority of NS5B, and a carboxy-terminal trans-membrane domain tethering the protein in the membrane (Fig. 1). The trans-membrane domain is essential for RNA replication in cells, most likely to allow proper insertion into the membranous replicase machinery, yet dispensable for enzymatic activity and is therefore deleted in most biochemical or structural assays to increase solubility of the protein. Like all viral RdRps, NS5B has a “right-hand” shape containing palm, thumb, and fingers domains (for review, see Sesmero and Thorpe 2015). Additionally, HCV NS5B contains a β-flap domain that is specific to Flaviviridae RdRps and a linker domain that is common to de novo initiating enzymes. Through well-defined mechanisms, each of these domains contributes to specific steps in viral RNA synthesis (for review, see Sesmero and Thorpe 2015).
RNA synthesis processes governed by NS5B can be divided into four steps: RNA binding, initiation, processive elongation, and termination at the end of the template. Although early reports show that NS5B can initiate RNA synthesis using both primer-based and de novo mechanisms, structural evidence suggests that NS5B uses de novo initiation to replicate the viral genome in cells (Behrens et al. 1996; Lohmann et al. 1997; Luo et al. 2000; Sun et al. 2000; Zhong et al. 2000). It is thought that initiation starts directly at the 3′ end of the viral RNA genome and requires high levels of GTP that bind to an allosteric site in the enzyme to act as a structural support to prime the initiation process (Lohmann et al. 1999b). As discussed above, the 3′ end of the positive-strand RNA is a poor template for de novo initiation, as it is concealed within an SL structure. In contrast, the 3′ end of the negative-strand RNA consists of a stem loop with an overhang (Fig. 4) that serves as a highly efficient initiator of RNA synthesis. This difference likely has a role in regulation of the replication processes and might contribute to the 10-fold excess of positive- over negative-strand RNA. It is highly advantageous for HCV to regulate production of negative-strand RNA, as increased RNA synthesis interferes with translation and excess dsRNA intermediates would stimulate innate immune responses.
NS5B can bind to many RNA target sequences and initiate RNA syntheses both internally within the viral genome and on circular RNA templates, arguing for a lack of specificity for the viral genomic RNA ends (Lohmann et al. 1997; Shim et al. 2002; Ranjith-Kumar and Kao 2006). These properties indicate that the enzyme is not limited to the “closed” conformation that is suggested by most structural analysis. In fact, recent studies identified an NS5B open state that can also accommodate primer and template RNA, which is consistent with early reports that purified NS5B can initiate using both primer-based and de novo mechanisms. These observations argue that, in solution, there is equilibrium between open and closed states that may serve different functions in the replication cycle. Additionally, several reports have shown that NS5B can bind to 3′UTRs of specific host mRNAs (Yuhashi et al. 2014). In combination with low specificity for RNA targets, these observations lend to the hypothesis that NS5B can amplify host RNAs, which could be involved in regulation of host protein production or in activation of innate immune responses (Yu et al. 2012). Although NS5B alone does not seem to have specificity for the viral genome, genetic studies argue that an interaction between the NS3 helicase, NS5A, and NS5B is required for initiation of RNA synthesis (Binder et al. 2007) indicating that template specificity is conferred by a combination of viral factors. In addition, the HCV replicase is tethered in intracellular membranes, which limits access to cellular RNAs.
NS3-4A
HCV NS3 is a bifunctional enzymatic protein containing a carboxy-terminal DExD-box helicase domain (NS3h) and an amino-terminal protease domain that function in conjunction with the cofactor, NS4A (Fig. 1). Whereas the protease domain is indirectly involved in replication through its role in polyprotein processing, the helicase domain has a more direct role in RNA synthesis. Consistent with this assumption, specific NS3h mutations alter HCV replication fitness and a correlation between nucleic acid unwinding activity and replicative ability was observed (Stross et al. 2016; Zhou et al. 2018). Several studies have indicated an allosteric mechanism that governs a switch between protease activity and helicase activity, with the “open or extended form” showing higher DNA unwinding function and representing the biologically relevant state for RNA replication (Ding et al. 2011; Saalau-Bethell et al. 2012). Thus, in the current model, the protease domain also contributes to the activity of the helicase domain (Beran et al. 2007). Although the DNA unwinding mechanism of HCV NS3h is well characterized (Gu and Rice 2010), its precise function within the viral replication cycle remains elusive. Specifically, the helicase activity could be required for the dissociation of highly structured single-stranded RNA elements before NS5B-mediated template-guided RNA synthesis or it could function in dissociating dsRNA following replication. In either case, the NS3 helicase activity is critically required for HCV RNA replication.
NS5A
NS5A is a promiscuous viral protein that has roles at several stages of the viral life cycle (for review, see Ross-Thriepland and Harris 2015). NS5A can bind to RNA and forms homodimers and probably also higher-order structures. It contains three defined domains, linked by low-complexity sequences (LCSs), and an amino-terminal amphipathic α-helix that facilitates membrane association and is essential for genome replication (Fig. 1; Penin et al. 2004). Domain I contains an RNA-binding motif and is linked to virus replication as well as RO biogenesis (Huang et al. 2005; Romero-Brey et al. 2015), whereas domain III functions primarily in virion assembly (Appel et al. 2008). Although the major part of domain II is dispensable for replication, specific residues in the carboxyl terminus are required for genome replication (Appel et al. 2008; Tellinghuisen et al. 2008; Ross-Thriepland and Harris 2014). These different NS5A functions seem to be regulated via differential phosphorylation states. Early studies described two prominent phospho-isoforms for NS5A that could be separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were designated the basally phosphorylated p56 and the hyperphosphorylated p58 (Kaneko et al. 1994). However, this view has been challenged by opposing data sets obtained from experiments performed with genotype 1b versus 2a HCV strains. Additionally, recent reports showed that the NS5A p58 isoform consists of several independently phosphorylated species that perform different functions and argue for an essential role of p58 in both assembly and RNA replication (Masaki et al. 2014; Harak et al. 2016; Schenk et al. 2018). In any case, reports have consistently linked several phospho-acceptor-sites in the LCS1 region to viral genome replication, and showed that limiting phosphorylation of these sites by blocking the casein kinase I isoform α (CKIα) limits replication and virion assembly (Appel et al. 2005; Quintavalle et al. 2006; Pietschmann et al. 2009; Masaki et al. 2014; Harak et al. 2016; Goonawardane et al. 2018). This suggests that phosphorylation in this region may function as a regulatory switch between RNA replication and virion assembly. The combined data on NS5A highlight the multipurpose nature of this protein and the complexity of regulating its different functions through multiple phosphorylation events. This complexity makes experimental characterization of specific NS5A functions exceedingly difficult. At the same time, the multiple functions exerted by NS5A might explain the exceptional antiviral potency of NS5A inhibitors that most likely block several steps of the viral life cycle such as RNA replication and virion assembly (Berger et al. 2014; McGivern et al. 2014). Although the precise mechanisms for how this multifunctionality is achieved are unknown, it is tempting to speculate that various NS5A phosphovariants bind to distinct host cell factors, such as PI4KA, VAPA/B, or apolipoprotein E, that exert the required function.
HOST CELL FACTORS OF RELEVANCE TO HCV GENOME REPLICATION
We can assume that for each individual step of the HCV life cycle, host cell factors are required. These can be proteins, lipids, and/or nucleic acids. During the last years, numerous factors of this kind have been identified and characterized, but only a few key examples can be mentioned here because of space limitations. The reader interested in more in-depth discussion of this aspect is referred to more recent reviews (Ross-Thriepland and Harris 2015; Sarnow and Sagan 2016; Wang and Tai 2016).
miRNAs are a class of small noncoding RNAs with an approximate length of 22 nucleotides, which are involved in the posttranscriptional regulation of gene expression. Certain miRNAs are expressed ubiquitously, whereas miR-122 is specifically expressed in the liver. Interestingly, binding sites for miR-122 are present in the 5′UTR, 3′UTR, and the NS5B-coding regions of the HCV positive-strand RNA genome (Fig. 4) and are linked to various viral processes (for reviews, see Sarnow and Sagan 2016; Bernier and Sagan 2018). Original studies showed that HCV RNA mutated in miR-122-targeting 3′UTR sequence was replicated as well as wild-type, whereas HCV RNA mutated in 5′UTR target sequence affected RNA accumulation and translation (Jopling et al. 2005), suggesting that HCV replication is dependent on miR-122 binding. On one hand, in typical interactions of miRNAs with mRNA, miRNAs promote translational repression and/or degradation of the target RNA. On the other hand, in the case of HCV, miR-122 has a positive effect by binding to the viral RNA genome in association with Agonaute-2 (Ago2) and protecting the viral genome from nucleolytic degradation by host 5′-3′ exoribonucleases 1 (XRN1) and XRN2 (Fig. 3; Shimakami et al. 2012; Li et al. 2013b, 2015). XRN activity is usually specific to 5′ monophosphate transcripts and not the 5′ triphosphate product of viral polymerase. In this case, the 5′ RNA triphosphatase, DUSP11, functions together with XRNs to restrict HCV, a process that is prevented by miR-122 binding (Fig. 3; Amador-Cañizares et al. 2018a; Kincaid et al. 2018). It is also reported that miR-122 binding contributes to the folding of a functional IRES by suppressing energetically favorable alternative secondary structures (Amador-Cañizares et al. 2018b; Schult et al. 2018; Chahal et al. 2019). Additionally, a recent study of the miR-122-binding sites in the NS5B-coding region correlated miR-122 binding with genome replication (Gerresheim et al. 2017). Specifically, mutations in the miR-122-binding site denoted 5B.2 (Fig. 4) caused a significant decrease in HCV RNA accumulation. However, compensatory mutations in this region that restore miR-122 binding failed to rescue viral RNA accumulation indicating that this RNA region has a role beyond that of direct miR-122 binding (Bernier and Sagan 2019). Together, these studies show that miR-122 is intricately involved in regulating different stages of the HCV infection cycle. It is an essential HCV host dependency factor and its depletion renders cells nonpermissive for this virus under physiological conditions.
Another host factor required for HCV replication is the prolyl-peptidyl cis-trans isomerase cyclophilin A (CypA), which alters the conformation of proteins by interconverting the cis and trans isomers of peptide bonds with the amino acid proline. CypA is ubiquitously expressed in tissues and inhibited by cyclosporine A (CsA) or nonimmunosuppressive compounds binding tightly to CypA. Initial studies showed that CsA treatment efficiently suppressed viral replication (Watashi et al. 2003). Subsequent studies showed that CypA interacts with NS5A, and that binding appears to catalyze the isomerization of peptidyl-prolyl bonds in NS5A domain I and domain II, resulting in conformational changes in NS5A (Badillo et al. 2017, and references cited therein). These alterations may stimulate the RNA-binding capacity of NS5A and enhance viral replication. Interestingly, CypA and NS5B competitively bind to a similar region of NS5A suggesting a regulatory role for CypA in the formation of virus protein interactions and genome replication (Rosnoblet et al. 2012; Ngure et al. 2016). CypA activity also seems to be required for the formation of HCV Ros, because CypA antagonists such as cyclosporine D block de novo formation of ROs (Madan et al. 2014).
The development of tools such as subgenomic replicons (Lohmann et al. 1999a), the infectious HCV cell culture system that was based on the unique viral isolate JFH-1 (Wakita et al. 2005) and highly permissive hepatoma cell lines (Blight et al. 2002; Friebe et al. 2005; Zhong et al. 2005), enabled us to study different aspects of the HCV life cycle. However, replication of primary isolates contained in patient serum has been notoriously difficult. Recently, two discoveries have been made that provide explanations for this difficulty. The first discovery was reported by Saeed and colleagues who found that overexpression of SEC14L2 in Huh7.5 cells allowed some replication of nonadapted HCV isolates (Saeed et al. 2015). SEC14L2 is a phosphatidylinositol transfer protein involved in the regulation of the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway, cholesterol synthesis, and vitamin E metabolism (Kempná et al. 2003; Mokashi and Porter 2005; Mokashi et al. 2005; Ni et al. 2005; Neuzil et al. 2006). Although the precise mode of action of SEC14L2 in the HCV replication cycle is unknown, part of the mechanism appears to be an enhanced vitamin E–mediated inhibition of lipid peroxidation (Saeed et al. 2015). Consistently, replication of HCV isolates resistant to lipid peroxidation, such as JFH-1, is not stimulated by SEC14L2 expression. The second discovery was made by Harak and coworkers, who studied the mechanism by which RNA replication–enhancing mutations that accumulate in natural HCV sequences on passage in cell culture stimulate HCV replication in hepatoma cell lines. The authors found that these genetic changes are loss-of-function mutations that attenuate the interaction between HCV NS5A and PI4KA (Harak et al. 2016). As alluded to above (see Host Factors and Processes Involved in HCV Replication Organelle Formation), this lipid kinase is required to render HCV-remodeled membranes conducive to RNA replication (Fig. 3, inset). Of note, the majority of hepatoma cell lines express high levels of PI4KA, which appears to be deleterious to robust HCV replication. This defect can be compensated either by mutations that impair the interaction with this lipid kinase, which is the case with HCV replication–enhancing mutations, or by pharmacological inhibition of PI4KA (Harak et al. 2016). Thus, enhanced HCV replication in cell culture is mediated, at least in part, by loss-of-function mutations.
LIPID-MODULATING HOST CELL FACTORS OF RELEVANCE TO HCV GENOME REPLICATION
The extensive remodeling of intracellular membranes induced by HCV and the assembly of highly lipidated HCV particles require the production and accumulation of distinct lipids. Consistently, lipidomic profiling of HCV-infected cells revealed distinct alterations of host cell lipid composition (Diamond et al. 2010). In addition to the importance of cholesterol in HCV replication described above, fatty acid biosynthetic pathways are also required for efficient HCV replication. Indeed, inhibition of fatty acid synthesis by blocking acetyl-CoA carboxylase decreases viral replication (Kapadia and Chisari 2005). Furthermore, increases in saturated and monounsaturated fatty acids enhance HCV replication, whereas increases in polyunsaturated fatty acids suppress replication (Kapadia and Chisari 2005). These results suggest that specific lipids and membrane fluidity are important for the function of the membranous HCV RO. Moreover, distinct lipids might form lipid rafts required for assembly and activity of the viral replicase. Consistent with this assumption, extraction of cholesterol from the replicase complex impairs replicase activity (Paul et al. 2013). In addition, sphingolipids were shown to stimulate HCV replicase activity (Weng et al. 2010; Hirata et al. 2012).
Lipid metabolism is regulated by a family of sterol regulatory element–binding proteins (SREBPs). SREBPs are transcription factors that activate the expression of more than 30 genes involved in biosynthesis or uptake of cholesterol, fatty acids, triglycerides, and phospholipids. In the case of HCV, virus-induced ER stress or viral proteins such as NS4B (Waris et al. 2007; Park et al. 2009) trigger the normally ER-localized inactive SREBP precursor to traffic to the Golgi, where it is proteolytically cleaved by site 1 protease (S1P) and S2P (Fig. 3). The released amino-terminal fragment is transported into the nucleus and activates transcription of target genes such as fatty acid synthase and 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMG-CoA), which is the rate-limiting enzyme of the cholesterol biosynthetic mevalonate pathway. Moreover, SREBPs are also activated by the cellular RNA helicase DDX3X. It was shown that the 3′UTR in the HCV RNA genome binds to DDX3X, which acts as an intracellular sensor to activate a signaling cascade that induces, via CBP-p300, the expression of SREBP, which in turns activates the expression of lipogenic genes (Li et al. 2013a). Finally, NS5B was reported to bind to fatty acid synthase, which activates the viral polymerase; however, this activation appears to be mediated by direct protein–protein interaction rather than by lipids produced by the synthase (Huang et al. 2013). Although these results illustrate the profound effect of HCV on host cell lipid metabolism, further studies are needed to clarify how distinct lipids contribute to or regulate viral RNA replication.
CONCLUDING REMARKS
The plethora of studies focused on uncovering the mechanisms of HCV RNA replication have had a significant impact on both our basic understanding of these biological processes and in the production of HCV-specific direct-acting antiviral drugs. However, there remain many unanswered questions in regard to these viral processes, and addressing them will improve our understanding of the basic principles of HCV replication. Some of the key unresolved topics in HCV replication are the mechanisms driving RO biogenesis, the precise subcellular location of viral RNA replication, the 3D architecture and functionality of the viral replicase machinery, the mechanisms responsible for governing the fate of newly synthesized viral RNA, including the trafficking of this RNA between different viral compartments, the host cell factors involved in HCV RNA replication, the mechanism whereby NS5A exerts its multiplicity of functions, and how HCV replication intermediates are concealed from the immune response. A greater mechanistic understanding of these processes will not only give insight into HCV replication strategies but also will generate knowledge that might be applicable to a large range of human pathogens. For instance, all positive-strand RNA viruses induce the reorganization of cellular membranes, several of which, including coronaviruses and picornaviruses, induce membrane structures similar to HCV (Paul and Bartenschlager 2013; Neufeldt et al. 2018). The similarity of these structures invites speculation that similar mechanisms are operating with these viruses. Therefore, insights gained from the study of HCV replication and membrane manipulation have the potential for application to related viruses and could lead to the identification of novel strategies for development of broad-spectrum antivirals by targeting host cell pathways and factors that are commonly used by these viruses. Moreover, the study of how HCV interacts with its host to create an environment conducive to replication has also contributed to our understanding of basic cellular processes. Examples of this include important insights into polymerase or helicase structure and activity, cellular lipid metabolism and the role of distinct lipids for robust viral replication, ER membrane dynamics, exploitation of miRNAs to protect the viral genome from degradation and to alter RNA structure, and sensing of viral RNA and viral countermeasures to suppress innate immunity. Thus, the combined research on HCV replication has had an enormous impact on various fields, far beyond HCV itself. Taking into account the excellent toolbox that has been generated to study this virus, we can expect that continued research of HCV–host cell interaction will continue to make important discoveries with far-reaching implications.
ACKNOWLEDGMENTS
We are grateful to Eliana G. Acosta for excellent editorial assistance and preparation of the figures as well as Ji Young Lee for providing electron microscopy images in Figure 2. Work in the authors’ laboratory was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), Projektnummer 240245660-SFB 1129, Project Number 112927078-TRR 83, and Projektnummer 272983813-TRR 179, all to R.B. C.J.N. was supported by a European Molecular Biology Organization (EMBO) Long-Term Fellowship (ALTF 466-2016).
Footnotes
Editors: Arash Grakoui, Jean-Michel Pawlotsky, and Glenn Randall
Additional Perspectives on Hepatitis C Viruses: The Story of a Scientific and Therapeutic Revolution available atwww.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Adams RL, Pirakitikulr N, Pyle AM. 2017. Functional RNA structures throughout the hepatitis C virus genome. Curr Opin Virol 24: 79–86. 10.1016/j.coviro.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Cañizares Y, Bernier A, Wilson JA, Sagan SM. 2018a. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5′ end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res 46: 5139–5158. 10.1093/nar/gky273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Cañizares Y, Panigrahi M, Huys A, Kunden RD, Adams HM, Schinold MJ, Wilson JA. 2018b. miR-122, small RNA annealing and sequence mutations alter the predicted structure of the hepatitis C virus 5′ UTR RNA to stabilize and promote viral RNA accumulation. Nucleic Acids Res 46: 9776–9792. 10.1093/nar/gky662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N, Pietschmann T, Bartenschlager R. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol 79: 3187–3194. 10.1128/JVI.79.5.3187-3194.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog 4: e1000035 10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier-Gin T, Bellecave P, Litvak S, Ventura M. 2005. Template requirements and binding of hepatitis C virus NS5B polymerase during in vitro RNA synthesis from the 3′-end of virus minus-strand RNA. FEBS J 272: 3872–3886. 10.1111/j.1742-4658.2005.04804.x [DOI] [PubMed] [Google Scholar]
- Badillo A, Receveur-Brechot V, Sarrazin S, Cantrelle FX, Delolme F, Fogeron ML, Molle J, Montserret R, Bockmann A, Bartenschlager R, et al. 2017. Overall structural model of NS5A protein from hepatitis C virus and modulation by mutations conferring resistance of virus replication to cyclosporin A. Biochemistry 56: 3029–3048. 10.1021/acs.biochem.7b00212 [DOI] [PubMed] [Google Scholar]
- Bartenschlager R, Lohmann V, Wilkinson T, Koch JO. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol 69: 7519–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol 11: 482–496. 10.1038/nrmicro3046 [DOI] [PubMed] [Google Scholar]
- Behrens SE, Tomei L, De Francesco R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J 15: 12–22. 10.1002/j.1460-2075.1996.tb00329.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. 2012. Complex dynamic development of poliovirus membranous replication complexes. J Virol 86: 302–312. 10.1128/JVI.05937-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beran RK, Serebrov V, Pyle AM. 2007. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J Biol Chem 282: 34913–34920. 10.1074/jbc.M707165200 [DOI] [PubMed] [Google Scholar]
- Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. 2011. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III α-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol 85: 8870–8883. 10.1128/JVI.00059-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Romero-Brey I, Radujkovic D, Terreux R, Zayas M, Paul D, Harak C, Hoppe S, Gao M, Penin F, et al. 2014. Daclatasvir-like inhibitors of NS5A block early biogenesis of hepatitis C virus-induced membranous replication factories, independent of RNA replication. Gastroenterology 147: 1094–1105.e25. 10.1053/j.gastro.2014.07.019 [DOI] [PubMed] [Google Scholar]
- Bernier A, Sagan SM. 2018. The diverse roles of microRNAs at the host–virus interface. Viruses 10: E440 10.3390/v10080440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier A, Sagan SM. 2019. Beyond sites 1 and 2, miR-122 target sites in the HCV genome have negligible contributions to HCV RNA accumulation in cell culture. J Gen Virol 100: 217–226. 10.1099/jgv.0.001217 [DOI] [PubMed] [Google Scholar]
- Binder M, Quinkert D, Bochkarova O, Klein R, Kezmic N, Bartenschlager R, Lohmann V. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J Virol 81: 5270–5283. 10.1128/JVI.00032-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, Rice CM. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol 71: 7345–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76: 13001–13014. 10.1128/JVI.76.24.13001-13014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahal J, Gebert LFR, Gan HH, Camacho E, Gunsalus KC, MacRae IJ, Sagan SM. 2019. miR-122 and Ago interactions with the HCV genome alter the structure of the viral 5′ terminus. Nucleic Acids Res 47: 5307–5324. 10.1093/nar/gkz194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counihan NA, Rawlinson SM, Lindenbach BD. 2011. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog 7: e1002302 10.1371/journal.ppat.1002302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, et al. 2010. Temporal proteome and lipidome profiles reveal hepatitis C virus–associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog 6: e1000719 10.1371/journal.ppat.1000719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SC, Kohlway AS, Pyle AM. 2011. Unmasking the active helicase conformation of nonstructural protein 3 from hepatitis C virus. J Virol 85: 4343–4353. 10.1128/JVI.02130-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz M, Świa¸tkowska A, Figlerowicz M, Ciesiołka J. 2008. Structural domains of the 3′-terminal sequence of the hepatitis C virus replicative strand. Biochemistry 47: 12197–12207. 10.1021/bi800348g [DOI] [PubMed] [Google Scholar]
- Egger D, Wölk B, Gosert R, Bianchi L, Blum H, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 76: 5974–5984. 10.1128/JVI.76.12.5974-5984.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Rice CM, Goff SP. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci 101: 13038–13043. 10.1073/pnas.0405152101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failla C, Tomei L, De Francesco R. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol 68: 3753–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris P, Blanchard E, Roingeard P. 2010. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol 91: 2230–2237. 10.1099/vir.0.022186-0 [DOI] [PubMed] [Google Scholar]
- Fricke M, Dünnes N, Zayas M, Bartenschlager R, Niepmann M, Marz M. 2015. Conserved RNA secondary structures and long-range interactions in hepatitis C viruses. RNA 21: 1219–1232. 10.1261/rna.049338.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Bartenschlager R. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol 76: 5326–5338. 10.1128/JVI.76.11.5326-5338.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Bartenschlager R. 2009. Role of RNA structures in genome terminal sequences of the hepatitis C virus for replication and assembly. J Virol 83: 11989–11995. 10.1128/JVI.01508-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Lohmann V, Krieger N, Bartenschlager R. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J Virol 75: 12047–12057. 10.1128/JVI.75.24.12047-12057.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol 79: 380–392. 10.1128/JVI.79.1.380-392.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerresheim GK, Dünnes N, Nieder-Röhrmann A, Shalamova LA, Fricke M, Hofacker I, Höner zu Siederdissen C, Marz M, Niepmann M. 2017. microRNA-122 target sites in the hepatitis C virus RNA NS5B coding region and 3′ untranslated region: Function in replication and influence of RNA secondary structure. Cell Mol Life Sci 74: 747–760. 10.1007/s00018-016-2377-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonawardane N, Ross-Thriepland D, Harris M. 2018. Regulation of hepatitis C virus replication via threonine phosphorylation of the NS5A protein. J Gen Virol 99: 62–72. 10.1099/jgv.0.000975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol 76: 3697–3708. 10.1128/JVI.76.8.3697-3708.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum H, Bienz K, Moradpour D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 77: 5487–5492. 10.1128/JVI.77.9.5487-5492.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouttenoire J, Roingeard P, Penin F, Moradpour D. 2010. Amphipathic α-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J Virol 84: 12529–12537. 10.1128/JVI.01798-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouttenoire J, Montserret R, Paul D, Castillo R, Meister S, Bartenschlager R, Penin F, Moradpour D. 2014. Aminoterminal amphipathic α-helix AH1 of hepatitis C virus nonstructural protein 4B possesses a dual role in RNA replication and virus production. PLoS Pathog 10: e1004501 10.1371/journal.ppat.1004501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünvogel O, Colasanti O, Lee JY, Klöss V, Belouzard S, Reustle A, Esser-Nobis K, Hesebeck-Brinckmann J, Mutz P, Hoffmann K, et al. 2018. Secretion of hepatitis C virus replication intermediates reduces activation of Toll-like receptor 3 in hepatocytes. Gastroenterology 154: 2237–2251.e16. 10.1053/j.gastro.2018.03.020 [DOI] [PubMed] [Google Scholar]
- Gu M, Rice CM. 2010. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci 107: 521–528. 10.1073/pnas.0913380107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto I, Nishimura Y, Okamoto T, Aizaki H, Liu M, Mori Y, Abe T, Suzuki T, Lai MMC, Miyamura T, et al. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J Virol 79: 13473–13482. 10.1128/JVI.79.21.13473-13482.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harak C, Meyrath M, Romero-Brey I, Schenk C, Gondeau C, Schult P, Esser-Nobis K, Saeed M, Neddermann P, Schnitzler P, et al. 2016. Tuning a cellular lipid kinase activity adapts hepatitis C virus to replication in cell culture. Nat Microbiol 2: 16247 10.1038/nmicrobiol.2016.247 [DOI] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. 2013. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503: 539–543. 10.1038/nature12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Ohta M, Tobita Y, Okano K, Ozeki K, et al. 2012. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog 8: e1002860 10.1371/journal.ppat.1002860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, et al. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141: 799–811. 10.1016/j.cell.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem 280: 36417–36428. 10.1074/jbc.M508175200 [DOI] [PubMed] [Google Scholar]
- Huang JT, Tseng CP, Liao MH, Lu SC, Yeh WZ, Sakamoto N, Chen CM, Cheng JC. 2013. Hepatitis C virus replication is modulated by the interaction of nonstructural protein NS5B and fatty acid synthase. J Virol 87: 4994–5004. 10.1128/JVI.02526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Tabor E, Gerety RJ. 1979. Acute non-A, non-B hepatitis: specific ultrastructural alterations in endoplasmic reticulum of infected hepatocytes. Lancet 313: 1249–1250. 10.1016/S0140-6736(79)91938-X [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309: 1577–1581. 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun 205: 320–326. 10.1006/bbrc.1994.2667 [DOI] [PubMed] [Google Scholar]
- Kapadia SB, Chisari FV. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci 102: 2561–2566. 10.1073/pnas.0409834102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125: 1808–1817. 10.1053/j.gastro.2003.09.023 [DOI] [PubMed] [Google Scholar]
- Kempná P, Zingg JM, Ricciarelli R, Hierl M, Saxena S, Azzi A. 2003. Cloning of novel human SEC14p-like proteins: ligand binding and functional properties. Free Radic Biol Med 34: 1458–1472. 10.1016/S0891-5849(03)00173-4 [DOI] [PubMed] [Google Scholar]
- Khan I, Katikaneni DS, Han Q, Sanchez-Felipe L, Hanada K, Ambrose RL, Mackenzie JM, Konan KV. 2014. Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2. J Virol 88: 12276–12295. 10.1128/JVI.00970-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid RP, Lam VL, Chirayil RP, Randall G, Sullivan CS. 2018. RNA triphosphatase DUSP11 enables exonuclease XRN-mediated restriction of hepatitis C virus. Proc Natl Acad Sci 115: 8197–8202. 10.1073/pnas.1802326115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6: e226 10.1371/journal.pbio.0060226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov AA, Feinstone SM, Rice CM. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol 70: 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. 2000. Hepatitis C virus–encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol 74: 2046–2051. 10.1128/JVI.74.4.2046-2051.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Shin H, Wimmer E, Paul AV. 2004. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J Virol 78: 10865–10877. 10.1128/JVI.78.20.10865-10877.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Cortese M, Haselmann U, Tabata K, Romero-Brey I, Funaya C, Schieber NL, Qiang Y, Bartenschlager M, Kallis S, et al. 2019. Spatiotemporal coupling of the hepatitis C virus replication cycle by creating a lipid droplet-proximal membranous replication compartment. Cell Rep 27: 3602–3617.e5. 10.1016/j.celrep.2019.05.063 [DOI] [PubMed] [Google Scholar]
- Li Q, Pène V, Krishnamurthy S, Cha H, Liang TJ. 2013a. Hepatitis C virus infection activates an innate pathway involving IKK-α in lipogenesis and viral assembly. Nat Med 19: 722–729. 10.1038/nm.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. 2013b. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci 110: 1881–1886. 10.1073/pnas.1213515110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yamane D, Lemon SM. 2015. Dissecting the roles of the 5′ exoribonucleases Xrn1 and Xrn2 in restricting hepatitis C virus replication. J Virol 89: 4857–4865. 10.1128/JVI.03692-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Herian U, Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol 71: 8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999a. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285: 110–113. 10.1126/science.285.5424.110 [DOI] [PubMed] [Google Scholar]
- Lohmann V, Overton H, Bartenschlager R. 1999b. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J Biol Chem 274: 10807–10815. 10.1074/jbc.274.16.10807 [DOI] [PubMed] [Google Scholar]
- Lundin M, Lindstrom H, Gronwall C, Persson MA. 2006. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J Gen Virol 87: 3263–3272. 10.1099/vir.0.82211-0 [DOI] [PubMed] [Google Scholar]
- Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol 74: 851–863. 10.1128/JVI.74.2.851-863.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V, Paul D, Lohmann V, Bartenschlager R. 2014. Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology 146: 1361–1372.e9, e1361–e1369 10.1053/j.gastro.2014.01.055 [DOI] [PubMed] [Google Scholar]
- Mahias K, Ahmed-El-Sayed N, Masante C, Bitard J, Staedel C, Darfeuille F, Ventura M, Astier-Gin T. 2010. Identification of a structural element of the hepatitis C virus minus strand RNA involved in the initiation of RNA synthesis. Nucleic Acids Res 38: 4079–4091. 10.1093/nar/gkq109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Matsunaga S, Takahashi H, Nakashima K, Kimura Y, Ito M, Matsuda M, Murayama A, Kato T, Hirano H, et al. 2014. Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-α in infectious virus production. J Virol 88: 7541–7555. 10.1128/JVI.03170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masante C, Mahias K, Lourenco S, Dumas E, Cahour A, Trimoulet P, Fleury H, Astier-Gin T, Ventura M. 2008. Seven nucleotide changes characteristic of the hepatitis C virus genotype 3 5′ untranslated region: Correlation with reduced in vitro replication. J Gen Virol 89: 212–221. 10.1099/vir.0.83067-0 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Mauro VP. 2014. Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo. Proc Natl Acad Sci 111: 15385–15389. 10.1073/pnas.1413472111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger DM, Golden M, Yamane D, Williford S, Lemon SM, Martin DP, Weeks KM. 2015. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci 112: 3692–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivern DR, Masaki T, Williford S, Ingravallo P, Feng Z, Lahser F, Asante-Appiah E, Neddermann P, De Francesco R, Howe AY, et al. 2014. Kinetic analyses reveal potent and early blockade of hepatitis C virus assembly by NS5A inhibitors. Gastroenterology 147: 453–462.e7. 10.1053/j.gastro.2014.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. 2015. Membrane curvature at a glance. J Cell Sci 128: 1065–1070. 10.1242/jcs.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438: 590–596. 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci 104: 2879–2884. 10.1073/pnas.0611267104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER–Golgi tether OSBP. Cell 155: 830–843. 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Polidori J, Jamecna D, Lacas-Gervais S, Antonny B. 2017. Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J 36: 3156–3174. 10.15252/embj.201796687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172. 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Hijikata M, Yamaji M, Hosaka M, Takahashi H, Shimotohno K. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J Biol Chem 278: 50301–50308. 10.1074/jbc.M305684200 [DOI] [PubMed] [Google Scholar]
- Mokashi V, Porter TD. 2005. Supernatant protein factor requires phosphorylation and interaction with Golgi to stimulate cholesterol synthesis in hepatoma cells. Arch Biochem Biophys 435: 175–181. 10.1016/j.abb.2004.11.030 [DOI] [PubMed] [Google Scholar]
- Mokashi V, Singh DK, Porter TD. 2005. Supernatant protein factor stimulates HMG-CoA reductase in cell culture and in vitro. Arch Biochem Biophys 433: 474–480. 10.1016/j.abb.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Nettles JH, Stanton RA, Broyde J, Amblard F, Zhang H, Zhou L, Shi J, McBrayer TR, Whitaker T, Coats SJ, et al. 2014. Asymmetric binding to NS5A by daclatasvir (BMS-790052) and analogs suggests two novel modes of HCV inhibition. J Med Chem 57: 10031–10043. 10.1021/jm501291c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt CJ, Joyce MA, Levin A, Steenbergen RH, Pang D, Shields J, Tyrrell DL, Wozniak RW. 2013. Hepatitis C virus–induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog 9: e1003744 10.1371/journal.ppat.1003744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt CJ, Joyce MA, Van Burren N, Levin A, Kirkegaard K, Gale MJ, Tyrrell DL, Wozniak RW. 2016. The hepatitis C virus–induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog 12: e1005428 10.1371/journal.ppat.1005428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. 2018. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16: 125–142. 10.1038/nrmicro.2017.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuzil J, Dong LF, Wang XF, Zingg JM. 2006. Tocopherol-associated protein-1 accelerates apoptosis induced by α-tocopheryl succinate in mesothelioma cells. Biochem Biophys Res Commun 343: 1113–1117. 10.1016/j.bbrc.2006.03.052 [DOI] [PubMed] [Google Scholar]
- Ngure M, Issur M, Shkriabai N, Liu HW, Cosa G, Kvaratskhelia M, Götte M. 2016. Interactions of the disordered domain II of hepatitis C virus NS5A with cyclophilin A, NS5B, and viral RNA show extensive overlap. ACS Infect Dis 2: 839–851. 10.1021/acsinfecdis.6b00143 [DOI] [PubMed] [Google Scholar]
- Ni J, Wen X, Yao J, Chang HC, Yin Y, Zhang M, Xie S, Chen M, Simons B, Chang P, et al. 2005. Tocopherol-associated protein suppresses prostate cancer cell growth by inhibition of the phosphoinositide 3-kinase pathway. Cancer Res 65: 9807–9816. 10.1158/0008-5472.CAN-05-1334 [DOI] [PubMed] [Google Scholar]
- Niepmann M, Shalamova LA, Gerresheim GK, Rossbach O. 2018. Signals involved in regulation of hepatitis C virus RNA genome translation and replication. Front Microbiol 9: 395 10.3389/fmicb.2018.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares-Jerez MF, Guillén J, Villalaín J. 2010. Interaction of the N-terminal segment of HCV protein NS5A with model membranes. Biochim Biophys Acta 1798: 1212–1224. 10.1016/j.bbamem.2010.02.007 [DOI] [PubMed] [Google Scholar]
- Palomares-Jerez MF, Nemesio H, Villalaín J. 2012. Interaction with membranes of the full C-terminal domain of protein NS4B from hepatitis C virus. Biochim Biophys Acta 1818: 2536–2549. 10.1016/j.bbamem.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Palomares-Jerez MF, Nemesio H, Franquelim HG, Castanho MA, Villalaín J. 2013. N-terminal AH2 segment of protein NS4B from hepatitis C virus. Binding to and interaction with model biomembranes. Biochim Biophys Acta 1828: 1938–1952. 10.1016/j.bbamem.2013.04.020 [DOI] [PubMed] [Google Scholar]
- Park CY, Jun HJ, Wakita T, Cheong JH, Hwang SB. 2009. Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J Biol Chem 284: 9237–9246. 10.1074/jbc.M808773200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2: 32–48. 10.5501/wjv.v2.i2.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Bartenschlager R. 2015. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu Rev Virol 2: 289–310. 10.1146/annurev-virology-100114-055007 [DOI] [PubMed] [Google Scholar]
- Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. 2011. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J Virol 85: 6963–6976. 10.1128/JVI.00502-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol 87: 10612–10627. 10.1128/JVI.01370-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Madan V, Ramirez O, Bencun M, Stoeck IK, Jirasko V, Bartenschlager R. 2018. Glycine zipper motifs in hepatitis C virus nonstructural protein 4B are required for the establishment of viral replication organelles. J Virol 92: e01890-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penin F, Brass V, Appel N, Ramboarina S, Montserret R, Ficheux D, Blum HE, Bartenschlager R, Moradpour D. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem 279: 40835–40843. 10.1074/jbc.M404761200 [DOI] [PubMed] [Google Scholar]
- Pérard J, Leyrat C, Baudin F, Drouet E, Jamin M. 2013. Structure of the full-length HCV IRES in solution. Nat Commun 4: 1612 10.1038/ncomms2611 [DOI] [PubMed] [Google Scholar]
- Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. 2009. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog 5: e1000475 10.1371/journal.ppat.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirakitikulr N, Kohlway A, Lindenbach BD, Pyle AM. 2016. The coding region of the HCV genome contains a network of regulatory RNA structures. Mol Cell 62: 111–120. 10.1016/j.molcel.2016.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N. 2015. Cryo-EM structure of hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat Commun 6: 7646 10.1038/ncomms8646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinkert D, Bartenschlager R, Lohmann V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J Virol 79: 13594–13605. 10.1128/JVI.79.21.13594-13605.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintavalle M, Sambucini S, Di Pietro C, De Francesco R, Neddermann P. 2006. The α isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J Virol 80: 11305–11312. 10.1128/JVI.01465-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar CT, Kao CC. 2006. Recombinant viral RdRps can initiate RNA synthesis from circular templates. RNA 12: 303–312. 10.1261/rna.2163106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, et al. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9: 32–45. 10.1016/j.chom.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, et al. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8: e1003056 10.1371/journal.ppat.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I, Berger C, Kallis S, Kolovou A, Paul D, Lohmann V, Bartenschlager R. 2015. NS5A domain 1 and polyprotein cleavage kinetics are critical for induction of double-membrane vesicles associated with hepatitis C virus replication. MBio 6: e00759 10.1128/mBio.00759-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López C, Berzal-Herranz A. 2009. A long-range RNA–RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 15: 1740–1752. 10.1261/rna.1680809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López C, Berzal-Herranz A. 2012. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell Mol Life Sci 69: 103–113. 10.1007/s00018-011-0729-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López C, Berzal-Herranz A. 2017. The 5BSL3.2 functional RNA domain connects distant regions in the hepatitis C virus genome. Front Microbiol 8: 2093 10.3389/fmicb.2017.02093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnoblet C, Fritzinger B, Legrand D, Launay H, Wieruszeski JM, Lippens G, Hanoulle X. 2012. Hepatitis C virus NS5B and host cyclophilin A share a common binding site on NS5A. J Biol Chem 287: 44249–44260. 10.1074/jbc.M112.392209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Thriepland D, Harris M. 2014. Insights into the complexity and functionality of hepatitis C virus NS5A phosphorylation. J Virol 88: 1421–1432. 10.1128/JVI.03017-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Thriepland D, Harris M. 2015. Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J Gen Virol 96: 727–738. 10.1099/jgv.0.000009 [DOI] [PubMed] [Google Scholar]
- Saalau-Bethell SM, Woodhead AJ, Chessari G, Carr MG, Coyle J, Graham B, Hiscock SD, Murray CW, Pathuri P, Rich SJ, et al. 2012. Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function. Nat Chem Biol 8: 920–925. 10.1038/nchembio.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M, Andreo U, Chung HY, Espiritu C, Branch AD, Silva JM, Rice CM. 2015. SEC14L2 enables pan-genotype HCV replication in cell culture. Nature 524: 471–475. 10.1038/nature14899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P, Sagan SM. 2016. Unraveling the mysterious interactions between hepatitis C virus RNA and liver-specific microRNA-122. Annu Rev Virol 3: 309–332. 10.1146/annurev-virology-110615-042409 [DOI] [PubMed] [Google Scholar]
- Schenk C, Meyrath M, Warnken U, Schnölzer M, Mier W, Harak C, Lohmann V. 2018. Characterization of a threonine-rich cluster in hepatitis C virus nonstructural protein 5A and its contribution to hyperphosphorylation. J Virol 92: e00737-18 10.1128/JVI.00737-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult P, Roth H, Adams RL, Mas C, Imbert L, Orlik C, Ruggieri A, Pyle AM, Lohmann V. 2018. MicroRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat Commun 9: 2613 10.1038/s41467-018-05053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C, Isel C, Imbert I, Ehresmann C, Marquet R, Kieny MP. 2002. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J Virol 76: 8058–8068. 10.1128/JVI.76.16.8058-8068.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesmero E, Thorpe IF. 2015. Using the hepatitis C virus RNA-dependent RNA polymerase as a model to understand viral polymerase structure, function and dynamics. Viruses 7: 3974–3994. 10.3390/v7072808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JH, Larson G, Wu JZ, Hong Z. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J Virol 76: 7030–7039. 10.1128/JVI.76.14.7030-7039.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. 2012. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci 109: 941–946. 10.1073/pnas.1112263109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu YK. 1992. Ultrastructural alterations and expression of cytoplasmic antigen 48-1 in hepatocytes in association with hepatitis C virus infection. Microbiol Immunol 36: 911–922. 10.1111/j.1348-0421.1992.tb02095.x [DOI] [PubMed] [Google Scholar]
- Shimizu YK, Weiner AJ, Rosenblatt J, Wong DC, Shapiro M, Popkin T, Houghton M, Alter HJ, Purcell RH. 1990. Early events in hepatitis C virus infection of chimpanzees. Proc Natl Acad Sci 87: 6441–6444. 10.1073/pnas.87.16.6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Shimotohno K. 2019. Hepatitis C virus (HCV) assembly and egress via modifications in host lipid metabolic systems. Cold Spring Harb Perspect Med 10.1101/cshperspect.a036814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Walton CM, Wu CH, Wu GY. 2002. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J Virol 76: 9563–9574. 10.1128/JVI.76.19.9563-9574.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck IK, Lee JY, Tabata K, Romero-Brey I, Paul D, Schult P, Lohmann V, Kaderali L, Bartenschlager R. 2017. Hepatitis C virus replication depends on endosomal cholesterol homeostasis. J Virol 92: e01196-17 10.1128/JVI.01196-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stross C, Shimakami T, Haselow K, Ahmad MQ, Zeuzem S, Lange CM, Welsch C. 2016. Natural HCV variants with increased replicative fitness due to NS3 helicase mutations in the C-terminal helix α18. Sci Rep 6: 19526 10.1038/srep19526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XL, Johnson RB, Hockman MA, Wang QM. 2000. De novo RNA synthesis catalyzed by HCV RNA-dependent RNA polymerase. Biochem Biophys Res Commun 268: 798–803. 10.1006/bbrc.2000.2120 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kato N, Cho MJ, Shimotohno K. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun 215: 744–749. 10.1006/bbrc.1995.2526 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kato N, Cho MJ, Sugiyama K, Shimotohno K. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol 70: 3307–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol 82: 1073–1083. 10.1128/JVI.00328-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccala G, Leroux-Roels G, Mavromara P, Bartenschlager R. 2008. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol 82: 11503–11515. 10.1128/JVI.01640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11: 791–796. 10.1038/nm1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ou JJ. 2018. Regulation of autophagy by hepatitis C virus for its replication. DNA Cell Biol 37: 287–290. 10.1089/dna.2017.4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tai AW. 2016. Mechanisms of cellular membrane reorganization to support hepatitis C virus replication. Viruses 8: E142 10.3390/v8050142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. 2014. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology 146: 1373–1385.e11. 10.1053/j.gastro.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waris G, Felmlee DJ, Negro F, Siddiqui A. 2007. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol 81: 8122–8130. 10.1128/JVI.00125-07 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Watashi K, Hijikata M, Hosaka M, Yamaji M, Shimotohno K. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38: 1282–1288. 10.1053/jhep.2003.50449 [DOI] [PubMed] [Google Scholar]
- Weng L, Hirata Y, Arai M, Kohara M, Wakita T, Watashi K, Shimotohno K, He Y, Zhong J, Toyoda T. 2010. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype-specific manner. J Virol 84: 11761–11770. 10.1128/JVI.00638-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Bürger J, et al. 2015. Molecular architecture of the ribosome-bound hepatitis C virus internal ribosomal entry site RNA. EMBO J 34: 3042–3058. 10.15252/embj.201592469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi M, St Claire M, Emerson SU, Purcell RH, Bukh J. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci 96: 2291–2295. 10.1073/pnas.96.5.2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Lemon SM. 2003a. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol 77: 3557–3568. 10.1128/JVI.77.6.3557-3568.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Lemon SM. 2003b. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9: 331–345. 10.1261/rna.2144203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S, Rice CM. 2008. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J Virol 82: 184–195. 10.1128/JVI.01796-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S, Stump DD, Branch AD, Rice CM. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol 78: 1352–1366. 10.1128/JVI.78.3.1352-1366.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GY, He G, Li CY, Tang M, Grivennikov S, Tsai WT, Wu MS, Hsu CW, Tsai Y, Wang LH, et al. 2012. Hepatic expression of HCV RNA-dependent RNA polymerase triggers innate immune signaling and cytokine production. Mol Cell 48: 313–321. 10.1016/j.molcel.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhashi K, Ohnishi S, Kodama T, Koike K, Kanamori H. 2014. In vitro selection of the 3′-untranslated regions of the human liver mRNA that bind to the HCV nonstructural protein 5B. Virology 450-451: 13–23. 10.1016/j.virol.2013.11.036 [DOI] [PubMed] [Google Scholar]
- Zhong W, Uss AS, Ferrari E, Lau JY, Hong Z. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol 74: 2017–2022. 10.1128/JVI.74.4.2017-2022.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci 102: 9294–9299. 10.1073/pnas.0503596102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Ren X, Adams RL, Pyle AM. 2018. NS3 from hepatitis C virus strain JFH-1 is an unusually robust helicase that is primed to bind and unwind viral RNA. J Virol 92: e01253-17 10.1128/JVI.01253-17 [DOI] [PMC free article] [PubMed] [Google Scholar]