Abstract
Within the realm of zoological study, the question of how an organism reaches a specific size has been largely unexplored. Recently, studies performed to understand the regulation of organ size have revealed that both cellular signals and external cues contribute toward the determination of total cell mass within each organ. The establishment of final organ size requires the precise coordination of cell growth, proliferation, and survival throughout development and postnatal life. In the mammalian heart, the regulation of size is biphasic. During development, cardiomyocyte proliferation predominantly determines cardiac growth, whereas in the adult heart, total cell mass is governed by signals that regulate cardiac hypertrophy. Here, we review the current state of knowledge regarding the extrinsic factors and intrinsic mechanisms that control heart size during development. We also discuss the metabolic switch that occurs in the heart after birth and precedes homeostatic control of postnatal heart size.
What controls organ size? How is organ growth regulated to achieve exact dimensions? These long-standing, fundamental questions have received a considerable amount of attention from developmental biologists over the years, but thanks to recent efforts, we are now beginning to understand how external and genetic determinants coordinate organ size. Among mammalian species, basic molecular components such as nucleic and amino acids are mostly similar in size. However, impressively, the total organism mass of mammals ranges from grams to tons (Fig. 1), and organ sizes can vary greatly within this animal class. Given that these significant disparities exist, the systemic and autonomous regulation of mammalian organ size and their relationship to overall organism size remain important areas of study. Recent data have indicated that scaling of organ size is coordinated by genetic and environmental determinants. Transplantation and tissue regeneration studies performed during the past century have revealed that organ–body ratios normalize in an organism-specific manner, indicating that intrinsic organ dimension-sensing mechanisms exist. Notable examples include the near-complete regeneration of the mammalian liver after surgical amputation (Michalopoulos and DeFrances 1997) and the regulated growth of limb buds to an organism-specific size when transplanted onto differing host salamander species (Harrison 1924). Moreover, organ growth is subject to a range of ecological constraints such as nutrient availability, survival needs, environmental conditions, and evolutionary adaptation. In addition, a wealth of studies has revealed that organ size is intrinsically governed by signaling pathways in response to mechanical and spatial cues. In this review, we summarize what is known regarding the regulation of heart size in mammals, from embryonic development throughout postnatal life. We discuss how growth factors, transcription factors, and signaling pathways promote hyperplastic growth of the heart, and we detail the signaling cross talk that occurs between pathways during cardiogenesis to specify heart size and patterning from precursor to the determination of final organ size. In addition, we describe how the regenerative neonatal heart undergoes a metabolic switch to promote differentiation and the cell-cycle exit of cardiac myocytes. Finally, we discuss the hypertrophic and homeostatic pathways that govern postnatal heart growth and renewal, respectively.
Figure 1.
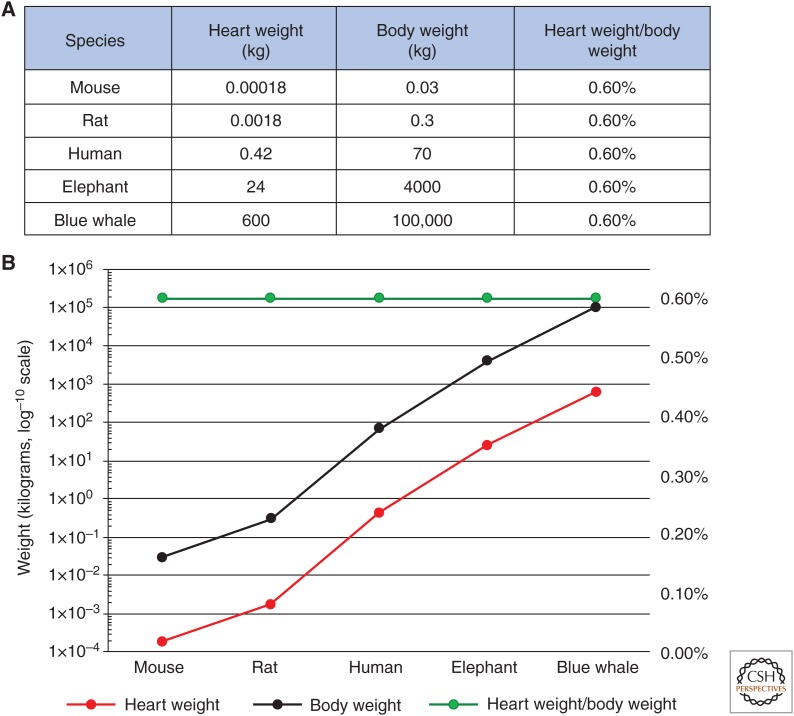
Total organism mass and heart mass of selective mammalian species. (A) Heart and total body weights (kg) and heart weight/total body weight ratios of indicated mammalian species. (B) Graphical representation of A. Secondary y axis plots heart weight/total body weight ratio percentages. (Panels created from data in Dobson 2003.)
REGULATION OF HEART SIZE DURING CARDIOGENESIS
Heart size and function are complementary in that the heart is large enough to generate physiologic output but not too large to cause circulatory obstruction. Mammals have evolved tightly regulated mechanisms to ensure that the heart attains proper dimensions. In the adult mammalian heart, cell-cycle activity and regenerative potential are greatly diminished (Porrello et al. 2011, 2013). Therefore, investigators have focused on defining the cellular mechanism(s) that control cardiomyocyte cell division. Several recent reports have revealed that growth factors, intrinsic signaling pathways, and cell-cycle regulators coordinate cardiomyocyte proliferation during cardiogenesis. The expansion of the two cardiac progenitor fields during early cardiogenesis transpires under the tight control of extrinsic growth regulatory signals, which include the fibroblast growth factor, bone morphogenetic protein, and both canonical and noncanonical Wnt signaling pathways (for a review, see Galdos et al. 2017). For example, data from mice revealed that the genetic deletion of the Wnt effector β-catenin in the second heart field gives rise to hearts bearing stunted right ventricles (Ai et al. 2007), underscoring the importance of this pathway during early stages of cardiac growth.
In recent years, investigators have discovered that heart size is intrinsically controlled during development and that the antigrowth Hippo kinase pathway is at the center of this regulation. At the turn of the 21st century, genetic screens for tumor suppressors in the fly species Drosophila melanogaster revealed that mutation of the hippo, salvador, and warts genes in larval tissues produces flies with dramatically overgrown organs, including the eyes, wings, and legs (for a review, see Pan 2007). In subsequent years, these genes have been established as the major genetic components of the evolutionarily conserved Hippo signaling pathway that restricts tissue growth and organ size (Harvey and Tapon 2007; Pan 2007; Halder and Johnson 2011). The core vertebrate Hippo components are known as MST1/2 (Hippo), SAV1 (Salvador), and LATS1/2 (LATS) (Harvey and Tapon 2007; Halder and Johnson 2011). The Hippo signal is relayed by an intracellular kinase cascade, whereby MST/Hippo is aided by the scaffolding protein SAV1 to phosphorylate LATS (Meng et al. 2016). In turn, LATS phosphorylates the transcriptional coactivator protein Yes-associated protein 1 (YAP). Phosphorylated YAP is sequestered in the cytoplasm, thereby inhibiting its binding to TEA domain (TEAD)/transcription enhancer factor Tef transcription factors in the nucleus to activate the expression of progrowth and prosurvival genes (Meng et al. 2016).
Since the Hippo signaling pathway was discovered to be an intrinsic cell-growth control pathway, substantial progress has been made toward defining the roles of this pathway in the context of cardiac development, regeneration, cardiomyocyte homeostasis, and cardiovascular disease (Wang et al. 2018). Genetic studies in mice revealed that the cardiac-specific deletion of Sav1 during embryonic development gives rise to severe cardiomegaly at birth as a result of elevated cardiomyocyte proliferation (Fig. 2; Heallen et al. 2011). In the ventricular myocardium of these mice, cardiomyocyte proliferation is up-regulated by fourfold, whereas proliferation of noncardiomyocytes is unaffected. In a similar fashion, the embryonic knockout of Mst1/2 or Lats1/2 kinase genes produces cardiac overgrowth phenotypes, albeit lethal ones (Heallen et al. 2011). Remarkably, tissue patterning is preserved in Hippo mutant hearts, indicating that Hippo restrains organ size and does not regulate morphogenesis during cardiac development (Fig. 2; Heallen et al. 2011). Furthermore, cardiomyocyte cell size is unaffected by Hippo deletion in embryonic hearts, revealing that Hippo restricts hyperplasia and not hypertrophy of cardiomyocytes during prenatal cardiac growth. Inactivation of Yap in embryonic mouse hearts results in lethality and the significant thinning of the myocardial layers because of reduced cardiomyocyte proliferation (Xin et al. 2011; von Gise et al. 2012). In contrast, when Yap is activated in embryonic mouse hearts, the proliferation rate in the ventricular myocardium is increased and heart size is significantly enhanced (Xin et al. 2011; von Gise et al. 2012). These data are consistent with the finding that Hippo loss-of-function hearts display cardiomyocyte hyperproliferation. Disruption of the Yap-binding partner and transcription factor TEAD1 in embryonic mouse hearts results in myocardial thinning and prenatal lethality (Chen et al. 1994). Moreover, the YAP-Tead1 interaction is required for the Yap-dependent activation of cell-cycle genes and promotion of cardiomyocyte proliferation (von Gise et al. 2012). In another study, activation of Yap target gene transcriptional reporters was enhanced by Tead2 (Heallen et al. 2011). These data collectively suggest that Tead proteins are the principle Yap-associated transcription factors that promote cardiac proliferation and organ growth.
Figure 2.
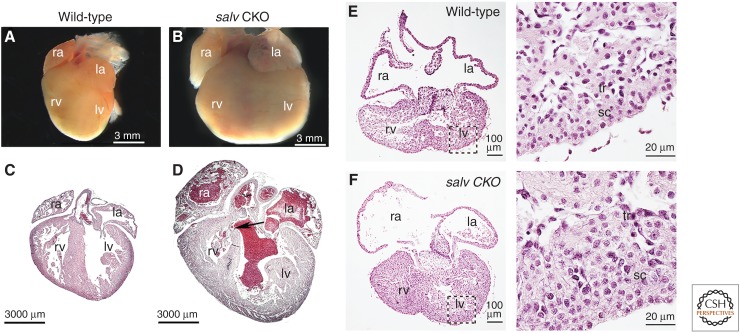
Cardiomegaly of Hippo-deficient mouse hearts. (A–D) Neonate hearts from postnatal day 2 littermate wild-type (A,C), and Sav1 knockout (B,D) (Salv CKO) mice. (C,D) Hearts from A and B were sectioned and stained with hematoxylin and eosin (H&E). The arrow shows a ventricular septum defect. (ra) right atrium, (la) left atrium, (rv) right ventricle, (lv) left ventricle. H&E-stained (E) embryonic wild-type and (F) Salv CKO hearts. (Inset) Images on the right show respective high-magnification views. sc (subcompact) and tr (trabecular) myocardium. (From Heallen et al. 2011; adapted, with permission, from The American Association for the Advancement of Science © 2011.)
Hippo and Other Signaling Pathways
Hippo-YAP signaling cross talks with other signaling pathways during embryogenesis to regulate heart size (Fig. 3). Notably, Wnt/β-catenin signaling is critical for Hippo/Yap-regulated overgrowth of the heart. Transcriptome analyses revealed that the expression of canonical Wnt pathway targets are up-regulated in both Sav1 knockout and Yap gain-of-function embryonic hearts (Heallen et al. 2011; Xin et al. 2011). These studies also revealed that during embryogenesis, nuclear β-catenin levels (a Wnt signal readout) are increased in Sav1 knockout hearts, whereas β-catenin protein levels are stabilized in Yap gain-of-function hearts (Heallen et al. 2011; Xin et al. 2011). Cell culture studies revealed that the Hippo effector TAZ suppresses WNT signaling by binding to the Wnt protein dishevelled, which promotes degradation of β-catenin (Varelas et al. 2010). These observations suggest that Hippo antagonizes canonical Wnt signaling during cardiogenesis on multiple levels. In support of this, Wnt/β-catenin loss-of function rescues the phenotypes of cardiomegaly, myocardial expansion, and increased cardiomyocyte proliferation produced by Hippo deficiency (Heallen et al. 2011). Biochemical examination of the Hippo–Wnt interaction revealed that Yap-Tead and β-catenin-Tcf/Lef co-occupy DNA promoter elements of cardiac growth genes (Heallen et al. 2011), suggesting that these proteins cooperatively activate the transcription of common targets. This raises the intriguing possibility that inhibiting Hippo or activating Yap enhances the Yap-β-catenin interaction to stimulate cardiomyocyte proliferation and cardiac growth. Indeed, integrated genomic analyses of embryonic and postnatal hearts indicate that expression of Yap targets that promote cell-cycle progression and proliferation is up-regulated in hyperplastic Hippo-deficient tissues (Heallen et al. 2011; Lange et al. 2015; Morikawa et al. 2015). Among these are the cytokinesis genes Birc5, Aurora B kinase, and Ect2 and the cell-cycle genes Cdk6 and Ccne2, to name a few. The list of Hippo targets that regulate heart growth is growing and ongoing next-generation sequencing studies should undoubtedly yield mechanistic insights into how growth pathways coordinate heart size.
Figure 3.
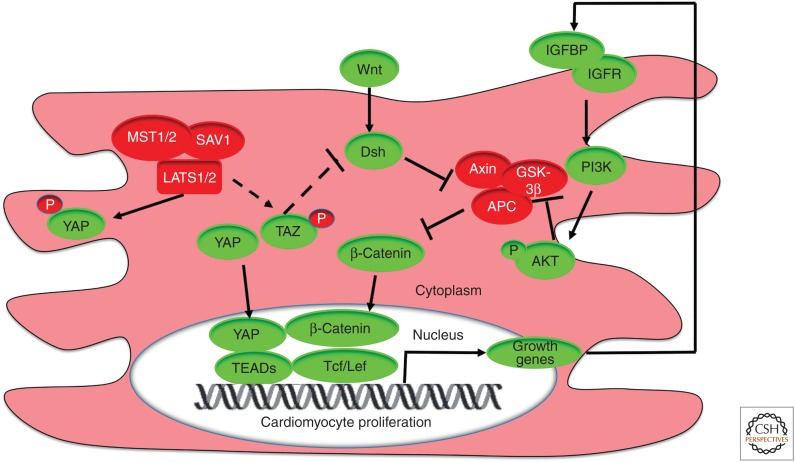
Signaling pathways that developmentally regulate cardiac growth. Hippo, canonical Wnt, and insulin-like growth factor (IGF) pathways coordinate heart size during development via the repression of cardiomyocyte proliferation. Core Hippo pathway components (red) trigger the phosphorylation of YAP (yes-associated protein), and Yap is retained in the cytosol. In the absence of Hippo signaling, YAP can enter the nucleus and form a complex with TEA domain (TEAD) transcription factors and β-catenin/T-cell factor/lymphoid enhancer factor (Tcf/Lef) on DNA promoters to activate expression of cardiac growth genes, which is dependent on active Wnt signaling (green). YAP/β-catenin signaling drives expression of IGF signaling pathway genes. Consequently, increased IGF signaling triggers phosphoinositide 3-kinase (PI3K)-mediated phosphorylation of RACα serine/threonine-protein kinase (AKT) and inhibition of the key β-catenin destruction complex (red) member glycogen synthase kinase 3β (GSK3β). In this context, β-catenin levels are stabilized to stimulate YAP/β-catenin-mediated cardiomyocyte proliferation, myocardial expansion, and cardiomegaly. Additionally, Hippo potentially represses canonical Wnt signaling via the phosphorylation of transcriptional coactivator with PDZ-binding motif (TAZ; dashed line). In turn, TAZ inhibits dishevelled (Dvl; dashed line), a destruction complex inhibitor. However, it has not been directly demonstrated that TAZ regulates cardiac growth. (APC) adenomatous polyposis coli protein, (IGFBP) IGF-binding protein, (IGFR) IGF receptor, (LATS) large tumor suppressor homolog, (MST) mammalian STE20-like protein kinase, (P) phosphate, (SAV1) Salvador homologue 1.
Concurrent studies have shown that the insulin-like growth factor (IGF) signaling pathway relays multitiered downstream regulation of Hippo/Wnt-dependent cardiac growth (Fig. 3). In embryonic mouse hearts overexpressing constitutively active Yap (i.e., Yap-S112A), core IGF pathway genes are activated (Xin et al. 2011). IGF pathway activity is also stimulated in these hearts as a result of up-regulated levels of phosphoinositide 3-kinase (PI3K) and increased phosphorylation of Akt and β-catenin destruction complex protein glycogen synthase kinase-3 (Gsk3β) (Xin et al. 2011). Increased inhibitory phosphorylation of Gsk3β promotes β-catenin stability, thereby enhancing Yap-regulated cardiomyocyte proliferation. In addition, YAP activity and β-catenin levels are significantly reduced when IGF signaling is inhibited in cardiomyocytes (Xin et al. 2011). These findings support a model in which β-catenin activation is dependent on Yap transcriptional activation of the IGF pathway (Fig. 3), revealing an additional regulatory node of Hippo-Wnt–regulated cardiomyocyte proliferation.
Given what is known about intrinsic heart growth mechanisms, a largely unexplored area in mammalian heart size regulation is whether non-cell-autonomous signals prevent the continued growth of the heart within a scaled body cavity. To this end, identification of upstream Hippo regulators in the context of cardiac growth represents an intriguing area of study. Regulation of the core Hippo pathway cascade by membrane proteins has been well documented. These proteins include the tumor suppressor Neurofibromin 2 (NF2, Merlin), cell polarity angiomotin complex members at tight junctions, and intercalated disc proteins (for a review, see Zhou et al. 2015). Whether these proteins function non-cell-autonomously to restrain cardiac growth remains unanswered.
As mentioned previously, mammalian heart growth spans two major phases: cell replication–driven hyperplastic growth during early development and postnatal hypertrophic growth in which postmitotic cells increase in size (Li et al. 1996). Whether Hippo-Yap signaling regulates postnatal cardiac growth remains unclear. However, the data currently available indicate that Hippo has important roles in cardiac repair, survival, and homeostasis throughout life (Heallen et al. 2019), suggesting that the manipulation of Hippo-Yap signaling can be therapeutically harnessed to promote the cell-cycle reentry of postnatal cardiomyocytes. Altogether, these studies indicate that in concert with other growth signaling pathways, Hippo signaling developmentally regulates heart size by suppressing prenatal cardiomyocyte proliferation.
PHYSIOLOGIC CHANGES IN THE HEART AFTER BIRTH
Key Transitions during Fetal to Adult Heart Growth
Shortly after birth, several transitions are triggered in the neonatal heart that initiate complex remodeling from a fetal state into an adult heart. As eluded to above, the early postnatal heart undergoes a growth mode switch in which cardiomyocytes become terminally differentiated, cease proliferating, and undergo hypertrophic growth in which both cell diameter and mass increase. In rat hearts, this hyperplasia-to-hypertrophy switch occurs rapidly between postnatal day (P)3 and 4, and cardiomyocytes largely cease dividing by P4 (Li et al. 1996). In developing mouse hearts, cardiomyocytes proliferate and maintain a relatively constant cellular volume as heart weight increases until they reach P4 (Leu et al. 2001). At this stage, mouse heart growth switches to a state of physiologic cardiomyocyte hypertrophy, with dramatically increased cardiomyocyte volume from P5 until P14 that is concomitant with significantly increased heart weight (Leu et al. 2001). After P14, the growth curve of cardiomyocyte volume levels off, and cardiomyocytes undergo hypertrophic growth to reach their adult volume when mice are ∼3 months of age (Leu et al. 2001). This postnatal cardiac hypertrophic growth is regulated by multiple signals such as the PI3K/AKT/insulin pathway and thyroid hormone tri-iodo-l-thyronine (T3). A more detailed discussion of this hypertrophic growth is provided below. Exciting recent data reveal that extracellular matrix (ECM) composition changes occur shortly after birth and strongly influence Yap localization and cardiomyocyte proliferation. A 2017 study showed that in neonatal cardiomyocytes, the proteoglycan Agrin promotes disassembly of the dystrophin glycoprotein complex (DGC) and Yap from the plasma membrane to induce cardiomyocyte proliferation (Bassat et al. 2017). However, Agrin levels decline sharply after the first week of life, coincident with the loss of cardiac regenerative potential (Porrello et al. 2011). A separate study showed that Yap is bound by the DGC protein dystroglycan 1 to inhibit cardiomyocyte proliferation (Morikawa et al. 2017). These findings are consistent with the data that cardiac ECM stiffens after birth (Bhana et al. 2010; Jacot et al. 2010) and that ECM stiffening promotes cardiomyocyte maturation and cell-cycle exit (Yahalom-Ronen et al. 2015), thereby unveiling an additional mechanism to restrain cardiomyocyte proliferation throughout life.
To meet the metabolic demands that arise from increased heart size and the burgeoning workload that follows the transition from fetal to postnatal circulation, the early postnatal heart undergoes an essential metabolic switch from a predominantly anaerobic and glycolysis-dependent fetal-like state to an adult state that uses oxidative metabolism. After this switch, the early postnatal heart primarily utilizes substrates to catabolize fatty acids, in contrast to the fetal heart that largely relies on carbohydrates as a secondary source of energy (Girard et al. 1992; Lopaschuk et al. 1992). Data from lambs revealed that glucose and lactate are the primary substrates for fetal myocardial oxygen consumption, whereas neonatal hearts primarily use long-chain fatty acids and lactate (Bartelds et al. 2000). Coinciding with the metabolic switch during early postnatal heart development, mitochondrial density doubles in cardiomyocytes, and mitochondrial shape remodels from the small, round, and tubular mitochondria in fetal hearts into large, ovoid, and rectangular mitochondria in adult hearts (Sheldon et al. 1976). This important metabolic switch involves numerous molecular signals such as the PPARγ coactivator-1 (PGC-1)/estrogen-related receptor (ERR) circuit, and failure of this switch to occur produces cardiac defects and shortens the life span of mice to 2 weeks (Fig. 4; Lehman et al. 2000; Michalik et al. 2002; Alaynick et al. 2007; Lai et al. 2008; Kolwicz et al. 2013).
Figure 4.
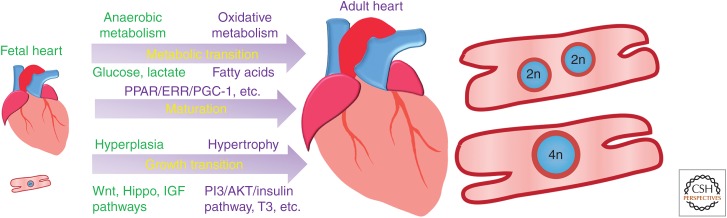
Summary of key transitions during fetal to adult heart growth. Fetal heart growth features hyperplasia and anaerobic metabolism that primarily uses the substrates glucose and lactate, whereas adult heart growth is marked by terminal differentiation, cell-cycle exit, hypertrophic growth, and oxidative metabolism that mainly uses fatty acid substrates. Growth mechanisms and regulatory signals are highlighted in green for the fetal heart and in purple for the adult heart. (PPAR) peroxisome proliferator-activated receptor, (ERR) estrogen-related receptor, (PGC-1) PPARγ coactivator-1, (IGF) insulin-like growth factor.
Metabolic Changes during Heart Growth
Changes in cardiac metabolism have important effects on cardiomyocyte proliferation, differentiation, and postnatal maturation (Lopaschuk and Jaswal 2010). The metabolic switch from anaerobic to oxidative metabolism in cardiomyocytes after birth is one of the most significant transitions during heart growth. The fetal heart adapts to an environment with low oxygen and low levels of circulating fatty acids and is therefore primarily dependent on glycolysis as the energy source to produce ATP, whereas mitochondrial oxidative metabolism is still poorly developed (Chung et al. 2007, 2010). In addition, the fetal heart produces ATP via lactate oxidation because lactate is relatively abundant. To meet the high metabolic demands for continuous beating, mature cardiomyocytes need to consume an exorbitant amount of ATP to continuously fuel cells and therefore switch to predominately relying on oxidative metabolism (Lopaschuk and Jaswal 2010). Glycolysis accounts for more than half of cardiac ATP production immediately after birth. However, by P7, this amount decreases quickly to <10% of overall ATP production, when fatty acids become the primary fuel source (Lopaschuk and Jaswal 2010). Throughout adulthood, oxidative metabolism remains the principle metabolic state, primarily utilizing fatty acids as an energy source. However, in certain pathologic conditions such as cardiac hypertrophy and heart failure, fetal carbohydrate utilization is reactivated.
Although the switch to oxidative phosphorylation is critical to supply the large amount of ATP necessary to fuel cardiomyocyte contractile function and postnatal hypertrophic growth, it also has consequences for cellular proliferation. It has been demonstrated that postnatal cardiomyocytes have a greater amount of reactive oxygen species (ROS), which are in part generated through oxidative phosphorylation. ROS can cause oxidative DNA damage, which has been shown to trigger cell-cycle arrest. However, reducing ROS via genetic overexpression of a mitochondrial-targeted catalase enzyme, administration of the antioxidant N-acetylcysteine, or exposing mice to chronic hypoxia has been shown to increase cardiomyocyte cell-cycle activity in the contexts of normal postnatal development and ischemic injury (Puente et al. 2014; Nakada et al. 2017). These experiments demonstrate a clear trade-off between a glycolytic cellular environment more permissive to cell division with an oxidative phosphorylation dependent environment that allows for greater ATP production required for cellular contraction and growth.
Regulation of the Metabolic Switch during Heart Growth
The metabolic switch from anaerobic to oxidative metabolism is a key component of cardiomyocyte maturation during heart growth and is a hallmark of the transition from the fetal stage to the adult stage. The nuclear receptor superfamily plays a pivotal role in the transcriptional regulation of metabolic function. Several nuclear receptors and their coactivators have been identified in the perinatal heart (Sucov et al. 1994; Mangelsdorf et al. 1995; Barak et al. 1999; Huss et al. 2002; Huss and Kelly 2004; Bookout et al. 2006; Chung et al. 2007, 2010; Lopaschuk and Jaswal 2010; Ellen Kreipke et al. 2016). Transcriptional changes in nuclear receptor pathways that control cardiac metabolism such as the hypoxia-inducible factor 1a (HIF-1a) pathway, the PGC-1/peroxisome proliferator-activated receptor (PPAR)a pathway, and the PGC-1/PPARd pathway can alter the level of enzymes regulating both glycolysis and mitochondrial oxidative metabolism. For example, the ERRγ gene Esrrg is highly expressed in the mouse heart from embryonic day 8.5 through development and in the postnatal heart. ERRγ regulates a network of nuclear-encoded mitochondrial genes that control oxidative metabolism and the postnatal metabolic transition (Alaynick et al. 2007). Disruption of ERRγ blocks the transition to oxidative metabolism and results in lactatemia, electrocardiographic abnormalities, and postnatal death within the first week after birth (Alaynick et al. 2007). In the adult mouse heart, ERRγ functions together with ERRα to regulate genes involved in energy substrate uptake, ATP production and transport, intracellular fuel sensing, and calcium handling (Dufour et al. 2007). Recently, additional factors have been identified that regulate the metabolic maturation of cardiomyocytes such as microRNAs, which are small noncoding RNAs (Ellen Kreipke et al. 2016). In vitro studies of human embryonic stem-cell-derived cardiomyocytes (hESC-CMs) have revealed that the let-7 family microRNAs are sufficient and required for hESC-CM maturation. Let-7 family microRNAs accelerate cardiomyocyte maturation most likely by increasing fatty acid metabolism and repressing genes of the PI3K/AKT/insulin pathway (Kuppusamy et al. 2015). Although numerous factors have been identified as regulators of the cardiac metabolic switch (Sucov et al. 1994; Mangelsdorf et al. 1995; Barak et al. 1999; Huss et al. 2002; Huss and Kelly 2004; Bookout et al. 2006; Chung et al. 2007, 2010; Lopaschuk and Jaswal 2010; Ellen Kreipke et al. 2016), the molecular mechanisms that direct this process remain to be elucidated.
POSTNATAL REGULATION OF HEART SIZE
As mentioned above, the formation of the heart's structure is heavily reliant on cell proliferation during development. In contrast, postnatal growth of the mammalian heart relies almost exclusively on cellular hypertrophy. In particular, the cardiomyocytes—the contractile cells that compose most of the heart by mass—are largely responsible for the cellular hypertrophy required for normal heart growth after birth. The cellular mechanisms that govern the cell-cycle exit and the subsequent hypertrophy of cardiomyocytes are of great interest because they have the potential to be exploited for developing therapeutic methods of stimulating cardiac regeneration. Below, we summarize what is currently known about cell-cycle exit and the postnatal hypertrophic growth of cardiomyocytes in the mammalian heart.
Cardiomyocyte Cell-Cycle Exit
It has been well established that mammalian cardiomyocytes lose their proliferative potential shortly after birth. Although the rate of decline of cardiomyocyte division after birth has previously been debated, it is now generally accepted that cardiomyocyte division occurs only rarely after postnatal day 11 in the mouse heart (Alkass et al. 2015). This topic has been similarly debated for the human heart, but a recent study provided strong evidence from the 14C dating of carbon integrated into cardiomyocyte DNA after atmospheric nuclear-bomb testing showing that the final cardiomyocyte number in the human heart is reached within the first month after birth (Bergmann et al. 2015). Once this total number of cardiomyocytes has been reached, the turnover of existing cardiomyocytes is limited. At birth, ∼5% of cardiomyocytes are estimated to turnover each year, and this turnover rate decreases throughout adolescence, resulting in a <1% rate by adulthood (Mollova et al. 2013).
The reduction cardiomyocyte proliferation that occurs after birth is followed by short bursts of alternative cell-cycle variations known as polyploidization and binucleation. The typical cell cycle consists of cellular growth, DNA synthesis, mitosis, and cytokinesis, resulting in two daughter cells with a single nucleus and a normal amount of chromosomal material (1 × 2n). The polyploidization cell-cycle variation occurs when the cell progresses normally through the cell cycle until exiting before the G2/M phase transition, resulting in a cell with double the normal amount of chromosomal material in a single nucleus (1 × 4n). In the binucleation cell-cycle variation, the cell enters M phase but exits before cytokinesis occurs, resulting in a cell with two nuclei that each have a normal amount of chromosomal material (2 × 2n). Both of these variants occur in mouse and human cardiomyocytes after birth, but at relatively different rates. In mice, binucleation has been established as the predominant cell-cycle variant in cardiomyocytes, peaking around postnatal day 7, after which a small round of polyploidization peaks at postnatal day 14 (Alkass et al. 2015). In humans, polyploidization is the predominant variant, occurring mostly between the ages of 10–20 years (Bergmann et al. 2015). Practical application of this suggests that the adult mouse heart is primarily composed of binucleated cardiomyocytes (2 × 2n), whereas the adult human heart is primarily composed of polyploid cardiomyocytes with a single nucleus (1 × 4n). However, the molecular underpinnings responsible for the relative differences in cardiomyocyte cell-cycle differences between species remain unknown
The functional consequence(s) of cardiomyocyte polyploidy are yet to be investigated. No clear explanation for the presence of polyploidy in cardiomyocytes has been established; however, one could surmise that a major advantage of polyploidy is that increased gene copy numbers allows for the greater transcript production required for hypertrophic growth. Other benefits of polyploidy include increased resistance to somatic mutations and a greater ability to adapt to stress and injury without undergoing mitosis (Øvrebø and Edgar 2018). Polyploidy also presents drawbacks, including an apparent reduction in proliferative potential. In the mouse, there is growing evidence that diploid cardiomyocytes are more likely to undergo proliferation following cardiac injury. Furthermore, mouse strains with a greater number of mononucleated cardiomyocytes have exhibited improved cardiac function following myocardial infarction (Patterson et al. 2017). These findings are encouraging and indicate that mononuclear diploid cardiomyocytes may have increased responsiveness to regenerative therapies, but much greater investigation into these emerging functional differences is necessary, as much of this evidence is only correlative.
Although injury is sufficient to stimulate proliferation in many other mammalian cell types, this is not the case for differentiated cardiomyocytes. Cardiomyocyte cell-cycle reentry has been shown to occur after cardiac injury such as myocardial infarction, but this cell-cycle activity results largely in polyploidization and not in proliferation (Herget et al. 1997; Meckert et al. 2005). Thus, the mammalian heart is incapable of meaningful endogenous repair after injury through cardiomyocyte proliferation.
Postnatal Hypertrophic Cardiac Growth
The human postnatal heart grows an estimated 15 times larger in volume during adolescence—an amazing increase in size. Stereological measurements from human hearts of varying ages have shown that the increase in cardiac size during the first 25 years of life can be explained by the increase in cardiomyocyte size that occurs through cellular hypertrophy (Bergmann et al. 2015). Studies of the molecular regulation of postnatal hypertrophic growth are ongoing; however, multiple signaling molecules have been shown to be crucial for this process, including IGF1, thyroid hormone, and insulin.
IGF1 has long been known to be a critical stimulus for postnatal cardiac hypertrophy (Baker et al. 1993; Lupu et al. 2001). The majority of IGF1 is produced systemically by the liver, with the heart itself also producing small amounts of IGF1 that can act in an autocrine manner. IGF1 can directly bind to both the IGF1 receptor (IGFR1) and insulin receptor (IR) on the extracellular membrane of cardiomyocytes. These are both transmembrane receptor tyrosine kinases that, when stimulated through IGF1, activate the PI3K/AKT pathway. Ultimately, this signaling event results in AKT phosphorylation, which leads to the stimulation of mTOR and inhibition of GSK-3, resulting in a net product of increased protein synthesis in cardiomyocytes (Dorn 2007). Insulin is another ligand capable of stimulating postnatal hypertrophy in cardiomyocytes. By directly binding IR, it also leads to PI3K/AKT signaling and stimulates protein synthesis.
The thyroid hormone T3 is another important molecule for stimulating postnatal hypertrophy in cardiomyocytes. Unlike the signaling cascades activated by IGF1 and insulin, T3 affects transcription directly. After birth, a short spike occurs in circulating thyroid hormones. At an increased systemic concentration, T3 can directly enter the nucleus of the cardiomyocytes, where it directly binds one of two thyroid hormone receptors, TRα or TRβ. Upon binding T3, either thyroid hormone receptor can form a dimer with the retinoic acid receptor (RAR) and function as a transcription factor complex to up-regulate critical sarcomere components such as α myosin heavy chain (αMHC) and cardiac troponin I cTnI (Kenessey and Ojamaa 2006). In addition to in vitro studies of these pathways in isolated rat ventricular cardiomyocytes, in vivo evidence has been generated with various transgenic mouse models, substantiating their specific roles in postnatal hypertrophy (Table 1).
Table 1.
In vivo evidence generated with various transgenic mouse models, substantiating their specific roles in postnatal hypertrophy
| Gene | Genetic mouse model | Cardiac growth/size phenotype | References |
|---|---|---|---|
| Igf1 | Germline knockout (Igf1−/−) | Reduced heart size at birth and throughout adulthood | Powell-Braxton et al. 1993; Wang et al. 1999 |
| Cardiomyocyte-specific overexpression (αMHC-Igf1 Tg) | Cardiac hypertrophy with intact systolic function; speculation of cardiac hypertrophy due to increased cardiomyocyte number | Reiss et al. 1996 | |
| Cardiomyocyte-specific overexpression (α-actin-Igf1 Tg) | Cardiac hypertrophy due to cardiomyocyte hypertrophy with enhanced systolic function at 10 weeks but depressed function by 1 year | Delaughter et al. 1999 | |
| Igf1r | Cardiomyocyte-specific knockout (αMHC-cre) | No change in heart size | Kim et al. 2008 |
| Cardiomyocyte-specific overexpression (αMHC-Igf1r Tg) | Cardiac hypertrophy due to cardiomyocyte hypertrophy through 16 months of age | McMullen et al. 2004 | |
| Insr | Cardiomyocyte-specific knockout (αMHC-cre) | Reduced heart size with reduced cardiac output | Belke et al. 2002 |
| Pik3ca | Cardiomyocyte-specific overexpression of constitutively active Pik3ca (αMHC-caPik3ca Tg) | Cardiac hypertrophy with intact systolic function at 1 year | Shioi et al. 2000 |
| Cardiomyocyte-specific overexpression of dominant-negative Pik3ca (αMHC-dnPik3ca Tg) | Reduced heart size with intact systolic function at 1 year | Shioi et al. 2000 | |
| Conditional cardiomyocyte-specific knockout (αMHC-iCre) | Deletion induced at 8 weeks resulted in reduced heart size | Lu et al. 2009 | |
| Pten | Cardiomyocyte-specific knockout (MCK-cre) | Cardiac hypertrophy due to cardiomyocyte hypertrophy with reduced cardiac contractility | Crackower et al. 2002 |
| Pdk1 | Cardiomyocyte-specific knockout (MCK-cre) | Reduced heart size due to reduced cardiomyocyte size with reduced systolic function | Mora et al. 2003 |
| Akt1 | Cardiomyocyte-specific overexpression of constitutively active Akt1 (αMHC-caAkt1 Tg) | Cardiac hypertrophy due to cardiomyocyte hypertrophy | Condorelli et al. 2002; Matsui et al. 2002; Shioi et al. 2002 |
| Inducible cardiomyocyte-specific overexpression of membrane-targeted Akt1 (αMHC-myrAkt1 Tg) | 2-Week induction resulted in cardiac hypertrophy due to cardiomyocyte hypertrophy; 6 weeks resulted in heart failure | Shiojima et al. 2005 | |
| Gsk3b | Cardiomyocyte-specific overexpression (αMHC-Gsk3b Tg) | Reduced heart size due to reduced cardiomyocyte size | Michael et al. 2004 |
| Foxo3 | Adenovirus overexpression in cardiomyocytes (Ad-Foxo3) | Reduced cardiomyocyte size | Skurk et al. 2005 |
| Map2k1 | Cardiomyocyte-specific overexpression of active Map2k1 (αMHC-aPik3ca Tg) | Cardiac hypertrophy due to cardiomyocyte hypertrophy | Bueno et al. 2000 |
| Tsc1 | Cardiomyocyte-specific knockout (SM22-cre) | Cardiac hypertrophy due to cardiomyocyte hypertrophy | Malhowski et al. 2011 |
| Cebpb | Germline heterozygote null (Cebpb+/−) | Cardiomyocyte hypertrophy | Boström et al. 2010 |
| Vegfb | Germline knockout (Vegfb−/−) | Reduced heart size | Bellomo et al. 2000 |
| Cardiomyocyte-specific overexpression (αMHC-Vegfb Tg) | Concentric cardiac hypertrophy with intact cardiac function due to cardiomyocyte hypertrophy | Karpanen et al. 2008 | |
| Stk11 | Cardiomyocyte-specific knockout (MCK-cre) | Reduced heart size, no measurement of cardiomyocyte size | Sakamoto et al. 2006 |
| Cardiomyocyte-specific knockout (αMHC-cre) | Atrial enlargement due to atrial cardiomyocyte hypertrophy at 4 weeks, cardiac hypertrophy due to both atrial and ventricular cardiomyocyte hypertrophy at 12 weeks | Ikeda et al. 2009 | |
| Mapk8/ Mapk9 | Germline heterozygous Mapk8 knockout and homozygous Mapk9 knockout (Mapk8+/−; Mapk9−/−) | Cardiac hypertrophy due to cardiomyocyte hypertrophy at 7 months (but not at 3 months) | Liang et al. 2003 |
| Cardiomyocyte-specific overexpression of dominant-negative Mapk8 and Mapk9 (αMHC-dnMapk8; αMHC-dnMapk9 Tg) | Cardiac hypertrophy due to cardiomyocyte hypertrophy at 7 months (but not at 3 months) | Liang et al. 2003 |
(MHC) myosin heavy chain.
Of note, several other forms of hypertrophy can occur in the adult heart. These include physiologic hypertrophy induced by pregnancy and exercise training, as well as pathologic hypertrophy that is characteristic of cardiomyopathies (Maillet et al. 2013). Although the signaling mechanisms that regulate these other forms of hypertrophy overlap somewhat with those that regulate postnatal hypertrophy, such as the IGF1 and PI3K/AKT signaling pathway, many mechanisms function independently. For example, mechanosensors such as the transient receptor potential (TRP) channels have a well-documented role in physiologic hypertrophy induced through exercise training, but their role in postnatal hypertrophy needs clarification (Seth et al. 2009). Furthermore, calcineurin-NFAT signaling has been shown to be critical in multiple models of pathologic hypertrophy but less critical for physiologic hypertrophy, which may be leveraged for therapeutic benefit (Wilkins et al. 2004).
CONCLUDING REMARKS
As the mechanisms that underlie cardiac growth come into focus, future studies should carefully address several key unanswered questions. For example, what are the precise roles for the Hippo signaling pathway in the context of hypertrophy, homeostasis, and cardiac protection? How can cardiomyocytes be reprogrammed to dedifferentiate and reenter the cell cycle for regenerative therapies? Specifically, concerted effort should be placed on leveraging the known signaling pathways that regulate cardiac growth to create therapies that benefit patients with heart failure. For many of these patients, especially those with ischemic heart disease, simple hypertrophy of cardiomyocytes will most likely be insufficient because the number of functional cardiomyocytes has been reduced. Therefore, cardiomyocyte cell-cycle reentry and proliferation must be stimulated to replenish the myocardium and achieve functional recovery. As discussed here, endogenous pathways that regulate cardiomyocyte hyperplasia, such as the Hippo pathway, have shown exciting potential for stimulating cardiomyogenesis (Leach et al. 2017). After cardiomyogenesis, a means to drive cardiomyocyte maturation and hypertrophy is needed to properly incorporate new cardiomyocytes into functional myocardium. Continued research that focuses on the determinants of cardiac growth has the potential to lead to novel therapies for heart failure that directly address the loss of functional myocardium.
ACKNOWLEDGMENTS
The authors apologize to investigators whose work is not cited here due to space constraints. We offer our gratitude to the House and Senate of the Texas Legislature for financially supporting Texas Heart's Regenerative Medicine Program, to Mr. Don McGill for his support of our research, and to the National Institutes of Health, Department of Defense, and the American Heart Association for continued funding support. Nicole Stancel, PhD, ELS(D), of the Texas Heart Institute, provided editorial support.
Footnotes
Editors: Benoit G. Bruneau and Paul R. Riley
Additional Perspectives on Heart Development and Disease available at www.cshperspectives.org
REFERENCES
- Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini A, Klein WH, Martin JF. 2007. Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc Natl Acad Sci 104: 9319–9324. 10.1073/pnas.0701212104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, et al. 2007. ERRγ directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab 6: 13–24. 10.1016/j.cmet.2007.06.007 [DOI] [PubMed] [Google Scholar]
- Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL, Bergmann O. 2015. No evidence for cardiomyocyte number expansion in preadolescent mice. Cell 163: 1026–1036. 10.1016/j.cell.2015.10.035 [DOI] [PubMed] [Google Scholar]
- Baker J, Liu JP, Robertson EJ, Efstratiadis A. 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75: 73–82. 10.1016/S0092-8674(05)80085-6 [DOI] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell 4: 585–595. 10.1016/S1097-2765(00)80209-9 [DOI] [PubMed] [Google Scholar]
- Bartelds B, Knoester H, Smid GB, Takens J, Visser GH, Penninga L, van der Leij FR, Beaufort-Krol GC, Zijlstra WG, Heymans HS, et al. 2000. Perinatal changes in myocardial metabolism in lambs. Circulation 102: 926–931. 10.1161/01.CIR.102.8.926 [DOI] [PubMed] [Google Scholar]
- Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, Kain D, Rajchman D, Leach J, Riabov Bassat D, et al. 2017. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547: 179–184. 10.1038/nature22978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belke DD, Betuing S, Tuttle MJ, Graveleau C, Young ME, Pham M, Zhang D, Cooksey RC, McClain DA, Litwin SE, et al. 2002. Insulin signaling coordinately regulates cardiac size, metabolism, and contractile protein isoform expression. J Clin Invest 109: 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellomo D, Headrick JP, Silins GU, Paterson CA, Thomas PS, Gartside M, Mould A, Cahill MM, Tonks ID, Grimmond SM, et al. 2000. Mice lacking the vascular endothelial growth factor-B gene (Vegfb) have smaller hearts, dysfunctional coronary vasculature, and impaired recovery from cardiac ischemia. Circulation Res 86: E29–E35. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, dos Remedios C, et al. 2015. Dynamics of cell generation and turnover in the human heart. Cell 161: 1566–1575. 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- Bhana B, Iyer RK, Chen WL, Zhao R, Sider KL, Likhitpanichkul M, Simmons CA, Radisic M. 2010. Influence of substrate stiffness on the phenotype of heart cells. Biotechnol Bioeng 105: 1148–1160. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126: 789–799. 10.1016/j.cell.2006.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström P, Mann N, Wu J, Quintero PA, Plovie ER, Panáková D, Gupta RK, Xiao C, MacRae CA, Rosenzweig A, et al. 2010. C/EBPβ controls exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell 143: 1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno OF, Windt LJD, Tymitz KM, Witt SA, Kimball TR, Klevitsky R, Hewett TE, Jones SP, Lefer DJ, Peng CF, et al. 2000. The MEK1–ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J 19: 6341–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Friedrich GA, Soriano P. 1994. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev 8: 2293–2301. 10.1101/gad.8.19.2293 [DOI] [PubMed] [Google Scholar]
- Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, Terzic A. 2007. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med 4(Suppl 1): S60–S67. 10.1038/ncpcardio0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP. 2010. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J Mol Cell Cardiol 48: 725–734. 10.1016/j.yjmcc.2009.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli G, Drusco A, Stassi G, Bellacosa A, Roncarati R, Iaccarino G, Russo MA, Gu Y, Dalton N, Chung C, et al. 2002. Akt induces enhanced myocardial contractility and cell size in vivo in transgenic mice. Proc Natl Acad Sci 99: 12333–12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower MA, Oudit GY, Kozieradzki I, Sarao R, Sun H, Sasaki T, Hirsch E, Suzuki A, Shioi T, Irie-Sasaki J, et al. 2002. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell 110: 737–749. [DOI] [PubMed] [Google Scholar]
- Delaughter MC, Taffet GE, Fiorotto ML, Entman ML, Schwartz RJ. 1999. Local insulin-like growth factor I expression induces physiologic, then pathologic, cardiac hypertrophy in transgenic mice. FASEB J 13: 1923–1929. [DOI] [PubMed] [Google Scholar]
- Dobson GP. 2003. On being the right size: heart design, mitochondrial efficiency and lifespan potential. Clin Exp Pharmacol Physiol 30: 590–597. 10.1046/j.1440-1681.2003.03876.x [DOI] [PubMed] [Google Scholar]
- Dorn GW II. 2007. The fuzzy logic of physiological cardiac hypertrophy. Hypertension 49: 962–970. 10.1161/HYPERTENSIONAHA.106.079426 [DOI] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguère V. 2007. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metab 5: 345–356. 10.1016/j.cmet.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Ellen Kreipke R, Wang Y, Miklas JW, Mathieu J, Ruohola-Baker H. 2016. Metabolic remodeling in early development and cardiomyocyte maturation. Semin Cell Dev Biol 52: 84–92. 10.1016/j.semcdb.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdos FX, Guo Y, Paige SL, VanDusen NJ, Wu SM, Pu WT. 2017. Cardiac regeneration: Lessons from development. Circ Res 120: 941–959. 10.1161/CIRCRESAHA.116.309040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J, Ferre P, Pegorier JP, Duee PH. 1992. Adaptations of glucose and fatty acid metabolism during perinatal period and suckling-weaning transition. Physiol Rev 72: 507–562. 10.1152/physrev.1992.72.2.507 [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. 2011. Hippo signaling: growth control and beyond. Development 138: 9–22. 10.1242/dev.045500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG. 1924. Some unexpected results of the heteroplastic transplantation of limbs. Proc Natl Acad Sci 10: 69–74. 10.1073/pnas.10.2.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K, Tapon N. 2007. The salvador–warts–hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer 7: 182–191. 10.1038/nrc2070 [DOI] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. 2011. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332: 458–461. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen TR, Kadow ZA, Kim JH, Wang J, Martin JF. 2019. Stimulating cardiogenesis as a treatment for heart failure. Circ Res 124: 1647–1657. 10.1161/CIRCRESAHA.118.313573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herget GW, Neuburger M, Plagwitz R, Adler CP. 1997. DNA content, ploidy level and number of nuclei in the human heart after myocardial infarction. Cardiovasc Res 36: 45–51. 10.1016/S0008-6363(97)00140-5 [DOI] [PubMed] [Google Scholar]
- Huss JM, Kelly DP. 2004. Nuclear receptor signaling and cardiac energetics. Circ Res 95: 568–578. 10.1161/01.RES.0000141774.29937.e3 [DOI] [PubMed] [Google Scholar]
- Huss JM, Kopp RP, Kelly DP. 2002. Peroxisome proliferator-activated receptor coactivator-1α (PGC-1α) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. Identification of novel leucine-rich interaction motif within PGC-1α. J Biol Chem 277: 40265–40274. 10.1074/jbc.M206324200 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Sato K, Pimentel DR, Sam F, Shaw RJ, Dyck JRB, Walsh K. 2009. Cardiac-specific deletion of LKB1 leads to hypertrophy and dysfunction. J Biol Chem 284: 35839–35849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot JG, Martin JC, Hunt DL. 2010. Mechanobiology of cardiomyocyte development. J Biomech 43: 93–98. 10.1016/j.jbiomech.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpanen T, Bry M, Ollila HM, Seppänen-Laakso T, Liimatta E, Leskinen H, Kivelä R, Helkamaa T, Merentie M, Jeltsch M, et al. 2008. Overexpression of vascular endothelial growth factor-B in mouse heart alters cardiac lipid metabolism and induces myocardial hypertrophy. Circulation Res 103: 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenessey A, Ojamaa K. 2006. Thyroid hormone stimulates protein synthesis in the cardiomyocyte by activating the Akt-mTOR and p70S6K pathways. J Biol Chem 281: 20666–20672. 10.1074/jbc.M512671200 [DOI] [PubMed] [Google Scholar]
- Kim J, Wende AR, Sena S, Theobald HA, Soto J, Sloan C, Wayment BE, Litwin SE, Holzenberger M, LeRoith D, et al. 2008. Insulin-like growth factor I receptor signaling is required for exercise-induced cardiac hypertrophy. Molec Endocrinol 22: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolwicz SC Jr, Purohit S, Tian R. 2013. Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res 113: 603–616. 10.1161/CIRCRESAHA.113.302095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu WZ, Tulloch NL, Yang X, et al. 2015. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci 112: E2785–E2794. 10.1073/pnas.1424042112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. 2008. Transcriptional coactivators PGC-1α and PGC-lβ control overlapping programs required for perinatal maturation of the heart. Genes Dev 22: 1948–1961. 10.1101/gad.1661708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange AW, Sridharan A, Xu Y, Stripp BR, Perl AK, Whitsett JA. 2015. Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. J Mol Cell Biol 7: 35–47. 10.1093/jmcb/mju046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach JP, Heallen T, Zhang M, Rahmani M, Morikawa Y, Hill MC, Segura A, Willerson JT, Martin JF. 2017. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550: 260–264. 10.1038/nature24045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. 2000. Peroxisome proliferator–activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest 106: 847–856. 10.1172/JCI10268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu M, Ehler E, Perriard JC. 2001. Characterisation of postnatal growth of the murine heart. Anat Embryol (Berl) 204: 217–224. 10.1007/s004290100206 [DOI] [PubMed] [Google Scholar]
- Li F, Wang X, Capasso JM, Gerdes AM. 1996. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol 28: 1737–1746. 10.1006/jmcc.1996.0163 [DOI] [PubMed] [Google Scholar]
- Liang Q, Bueno OF, Wilkins BJ, Kuan CY, Xia Y, Molkentin JD. 2003. c-Jun N-terminal kinases (JNK) antagonize cardiac growth through cross-talk with calcineurin–NFAT signaling. EMBO J 22: 5079–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopaschuk GD, Jaswal JS. 2010. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol 56: 130–140. 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- Lopaschuk GD, Collins-Nakai RL, Itoi T. 1992. Developmental changes in energy substrate use by the heart. Cardiovasc Res 26: 1172–1180. 10.1093/cvr/26.12.1172 [DOI] [PubMed] [Google Scholar]
- Lu Z, Jiang Y-P, Wang W, Xu X-H, Mathias RT, Entcheva E, Ballou LM, Cohen IS, Lin RZ. 2009. Loss of cardiac phosphoinositide 3-kinase pp110α results in contractile dysfunction. Circulation 120: 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Terwilliger JD, Lee K, Segre GV, Efstratiadis A. 2001. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev Biol 229: 141–162. 10.1006/dbio.2000.9975 [DOI] [PubMed] [Google Scholar]
- Malhowski AJ, Hira H, Bashiruddin S, Warburton R, Goto J, Robert B, Kwiatkowski DJ, Finlay GA. 2011. Smooth muscle protein-22-mediated deletion of Tsc1 results in cardiac hypertrophy that is mTORC1-mediated and reversed by rapamycin. Human Molec Genet 20: 1290–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet M, van Berlo JH, Molkentin JD. 2013. Molecular basis of physiological heart growth: fundamental concepts and new players. Nat Rev Mol Cell Biol 14: 38–48. 10.1038/nrm3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. 1995. The nuclear receptor superfamily: The second decade. Cell 83: 835–839. 10.1016/0092-8674(95)90199-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Li L, Wu JC, Cook SA, Nagoshi T, Picard MH, Liao R, Rosenzweig A. 2002. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J Biol Chem 277: 22896–22901. [DOI] [PubMed] [Google Scholar]
- McMullen JR, Shioi T, Huang W-Y, Zhang L, Tarnavski O, Bisping E, Schinke M, Kong S, Sherwood MC, Brown J, et al. 2004. The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110α) pathway. J Biol Chem 279: 4782–4793. [DOI] [PubMed] [Google Scholar]
- Meckert PC, Rivello HG, Vigliano C, Gonzalez P, Favaloro R, Laguens R. 2005. Endomitosis and polyploidization of myocardial cells in the periphery of human acute myocardial infarction. Cardiovasc Res 67: 116–123. 10.1016/j.cardiores.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan KL. 2016. Mechanisms of Hippo pathway regulation. Genes Dev 30: 1–17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik L, Desvergne B, Dreyer C, Gavillet M, Laurini RN, Wahli W. 2002. PPAR expression and function during vertebrate development. Int J Dev Biol 46: 105–114. [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. 1997. Liver regeneration. Science 276: 60–66. 10.1126/science.276.5309.60 [DOI] [PubMed] [Google Scholar]
- Michael A, Haq S, Chen X, Hsich E, Cui L, Walters B, Shao Z, Bhattacharya K, Kilter H, Huggins G, et al. 2004. Glycogen synthase kinase-3β regulates growth, calcium homeostasis, and diastolic function in the heart. J Biol Chem 279: 21383–21393. [DOI] [PubMed] [Google Scholar]
- Mora A, Davies AM, Bertrand L, Sharif I, Budas GR, Jovanović S, Mouton V, Kahn CR, Lucocq JM, Gray GA, et al. 2003. Deficiency of PDK1 in cardiac muscle results in heart failure and increased sensitivity to hypoxia. EMBO J 22: 4666–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollova M, Bersell K, Walsh S, Savla J, Das L, Park S-Y, Silberstein LE, dos Remedios CG, Graham D, Colan S, et al. 2013. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci 110: 1446–1451. 10.1073/pnas.1214608110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Zhang M, Heallen T, Leach J, Tao G, Xiao Y, Bai Y, Li W, Willerson JT, Martin JF. 2015. Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci Signal 8: ra41 10.1126/scisignal.2005781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Heallen T, Leach J, Xiao Y, Martin JF. 2017. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature 547: 227–231. 10.1038/nature22979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada Y, Canseco DC, Thet S, Abdisalaam S, Asaithamby A, Santos CX, Shah AM, Zhang H, Faber JE, Kinter MT, et al. 2017. Hypoxia induces heart regeneration in adult mice. Nature 541: 222–227. 10.1038/nature20173 [DOI] [PubMed] [Google Scholar]
- Øvrebø JI, Edgar BA. 2018. Polyploidy in tissue homeostasis and regeneration. Development 145: dev.156034 10.1242/dev.156034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. 2007. Hippo signaling in organ size control. Genes Dev 21: 886–897. 10.1101/gad.1536007 [DOI] [PubMed] [Google Scholar]
- Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, Parikh S, Denholtz M, Huang Y, Yamaguchi Y, et al. 2017. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet 49: 1346–1353. 10.1038/ng.3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. 2011. Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. 2013. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci 110: 187–192. 10.1073/pnas.1208863110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell-Braxton L, Hollingshead P, Warburton C, Dowd M, Pitts-Meek S, Dalton D, Gillett N, Stewart TA. 1993. IGF-I is required for normal embryonic growth in mice. Genes Dev 7: 2609–2617. [DOI] [PubMed] [Google Scholar]
- Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, Grinsfelder D, Rothermel BA, Chen R, Garcia JA, et al. 2014. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 157: 565–579. 10.1016/j.cell.2014.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K, Cheng W, Ferber A, Kajstura J, Li P, Li B, Olivetti G, Homcy CJ, Baserga R, Anversa P. 1996. Overexpression of insulin-like growth factor-1 in the heart is coupled with myocyte proliferation in transgenic mice. Proc Natl Acad Sci 93: 8630–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Zarrinpashneh E, Budas GR, Pouleur A-C, Dutta A, Prescott AR, Vanoverschelde J-L, Ashworth A, Jovanović A, Alessi DR, et al. 2006. Deficiency of LKB1 in heart prevents ischemia-mediated activation of AMPKα2 but not AMPKα1. Am J Physiol-Endocrinol Metab 290: E780–E788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi T, Kang PM, Douglas PS, Hampe J, Yballe CM, Lawitts J, Cantley LC, Izumo S. 2000. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J 19: 2537–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi T, McMullen JR, Kang PM, Douglas PS, Obata T, Franke TF, Cantley LC, Izumo S. 2002. Akt/protein kinase B promotes organ growth in transgenic mice. Molec Cell Biol 22: 2799–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I, Sato K, Izumiya Y, Schiekofer S, Ito M, Liao R, Colucci WS, Walsh K. 2005. Disruption of coordinated cardiac hypertrophy and angiogenesis contributes to the transition to heart failure. J Clin Invest 115: 2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurk C, Izumiya Y, Maatz H, Razeghi P, Shiojima I, Sandri M, Sato K, Zeng L, Schiekofer S, Pimentel D, et al. 2005. The FOXO3a transcription factor regulates cardiac myocyte size downstream of AKT signaling. J Biol Chem 280: 20814–20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Zhang ZS, Mao L, Graham V, Burch J, Stiber J, Tsiokas L, Winn M, Abramowitz J, Rockman HA, et al. 2009. TRPC1 channels are critical for hypertrophic signaling in the heart. Circ Res 105: 1023–1030. 10.1161/CIRCRESAHA.109.206581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CA, Friedman WF, Sybers HD. 1976. Scanning electron microscopy of fetal and neonatal lamb cardiac cells. J Mol Cell Cardiol 8: 853–862. 10.1016/0022-2828(76)90068-7 [DOI] [PubMed] [Google Scholar]
- Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. 1994. RXRα mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev 8: 1007–1018. 10.1101/gad.8.9.1007 [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, et al. 2010. The Hippo pathway regulates Wnt/β-catenin signaling. Dev Cell 18: 579–591. 10.1016/j.devcel.2010.03.007 [DOI] [PubMed] [Google Scholar]
- von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, et al. 2012. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci 109: 2394–2399. 10.1073/pnas.1116136109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou J, Powell-Braxton L, Bondy C. 1999. Effects of Igf1 gene deletion on postnatal growth patterns. Endocrinology 140: 3391–3394. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu S, Heallen T, Martin JF. 2018. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol 15: 672–684. 10.1038/s41569-018-0063-3 [DOI] [PubMed] [Google Scholar]
- Wilkins BJ, Dai YS, Bueno OF, Parsons SA, Xu J, Plank DM, Jones F, Kimball TR, Molkentin JD. 2004. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ Res 94: 110–118. 10.1161/01.RES.0000109415.17511.18 [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. 2011. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal 4: ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom-Ronen Y, Rajchman D, Sarig R, Geiger B, Tzahor E. 2015. Reduced matrix rigidity promotes neonatal cardiomyocyte dedifferentiation, proliferation and clonal expansion. eLife 4: e07455 10.7554/eLife.07455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li L, Zhao B, Guan KL. 2015. The hippo pathway in heart development, regeneration, and diseases. Circ Res 116: 1431–1447. 10.1161/CIRCRESAHA.116.303311 [DOI] [PMC free article] [PubMed] [Google Scholar]