Abstract
Interactions of nonionic poly(ethylene oxide)-b-poly(propylene oxide) (PEO–PPO) block copolymers, known as Pluronics or poloxamers, with cell membranes have been widely studied for a host of biomedical applications. Herein, we report how cholesterol within phosphatidylcholine (POPC) lipid bilayer liposomes and bilayer curvature affects the binding of several PPO–PEO–PPO triblocks with varying PPO content and a tPPO–PEO diblock, where t refers to a tert-butyl end group. Pulsed-field-gradient NMR was employed to quantify the extent of copolymer associated with liposomes prepared with cholesterol concentrations ranging from 0 to 30 mol % relative to the total content of POPC and cholesterol and vesicle extrusion radii of 25, 50, or 100 nm. The fraction of polymer bound to the liposomes was extracted from NMR data on the basis of the very different mobilities of the bound and free polymers in aqueous solution. Cholesterol concentration was manipulated by varying the molar percentage of this sterol in the POPC bilayer preparation. The membrane curvature was varied by adjusting the liposome size through a conventional pore extrusion technique. Although the PPO content significantly influences the overall amount of block copolymer adsorbed to the liposome, we found that polymer binding decreases with increasing cholesterol concentration in a universal fashion, with the fraction of bound polymer dropping 10-fold between 0 and 30 mol % cholesterol relative to the total content of POPC and cholesterol. Increasing the bilayer curvature (decreasing the radius of the liposome) in the absence of cholesterol increases polymer binding between 2- and 4-fold over the range of liposome sizes studied. These results demonstrate that cholesterol plays a dominant role, and bilayer curvature has a less significant impact as the curvature decreases, on polymer–membrane association.
Graphical abstract
INTRODUCTION
Pluronics, also known as poloxamers, are one of the most widely used classes of commercial nonionic surfactants.1−3 These compounds are symmetric triblock copolymers, composed of a central hydrophobic block of poly(propylene oxide) (PPO) flanked by two hydrophilic blocks of poly(ethylene oxide) (PEO). Their biocompatibility and amphiphilicity provide them with great potential for biomedical applications. Hydrophilic dominant Pluronics, such as F68 (also referred to as poloxamer 188), which contains 80 wt % PEO (27 and 75 repeat units per PPO and PEO block, respectively), have been used as membrane stabilizers to block dystrophic heart failure,4 protect dystrophic skeletal muscle,5 and prevent reperfusion injury after myocardial infarction.6 In contrast, Pluronics with longer hydrophobic PPO blocks, such as F127 (ca. 64 and 108 repeat units per PPO and PEO block, respectively), or with lower PEO content, such as L61 (ca. 10 wt % PEO), have been applied to facilitate drug and gene delivery due to their facile incorporation into the cell membrane.7–11 Recently, diblock analogues of Pluronics have been investigated in cellular assays by Kim et al., which demonstrate the protective effect of the diblock architecture on cell membranes against hypo-osmotic stress in vitro.12 Houang and co-workers also found that modifying the PPO end group chemistry of PPO–PEO diblock polymers can significantly affect the protection efficacy using dystrophic animal models in vivo.13 These, and many other results, indicate that polymer structure plays a critical role in the interactions of Pluronictype surfactants with cell membranes.
Numerous studies have focused on developing a basic understanding of polymer–membrane association mechanisms, using lipid bilayer systems as model membranes.14–24 Cheng et al. suggested that the hydrophobic PPO block inserts into the membrane interior whereas the hydrophilic PEO block weakly adsorbs onto the membrane surface.14 Smallangle X-ray scattering experiments and coarse-grained molecular dynamic simulations indicate that the PPO block length is the key determinant for association with lipid membranes.18,19,21 In our previous work, we systematically investigated how polymer structure dictates the polymer interaction with lipid bilayers by quantifying the binding of both triblocks and diblocks to unilamellar liposomes.25 Larger molecular weight and more hydrophobic PPO−PEO block polymers were found to have stronger interactions with lipid bilayers.
Despite advances made in this field, most reported work has aimed at understanding the mechanism of polymer–membrane association from the polymer perspective, whereas only a few studies explored how membrane composition influences the interactions between lipid bilayers and amphiphilic polymers.24,26 Composition has a significant impact on multiple membrane properties. Cholesterol, e.g., is one of the most common molecules found in eukaryotic cell membranes and can make up as much as 50 mol % of plasma membranes.27 It is a predominantly hydrophobic molecule, composed of steroid rings and a hydroxyl group. The moderate amphiphilicity due to the presence of the hydroxyl group tends to orient the cholesterol molecule such that the hydroxyl group is located at the interface between the membrane interior and aqueous medium via hydrogen bonding with lipid headgroups and water molecules whereas the steroid rings sit in the membrane interior, parallel to the hydrocarbon chains.28–31 Cholesterol is a vital component for maintaining the membrane integrity, controlling lipid packing, and regulating membrane fluidity.28,31–34 Earlier work suggested that the addition of cholesterol into a lipid bilayer could increase the ordering of the alkyl chains of the lipids,35,36 increase membrane density,37–40 decrease membrane permeability,31 decrease membrane fluidity,41,42 increase mechanical strength of the membrane,34 and induce the formation of liquid domains enriched with particular lipid species.43−45 Additionally, membrane curvature could also play a role in the interactions with polymers. Work by Ahmed et al. using Raman spectroscopy indicated that the curvature of nanoparticlesupported lipid bilayers also has a significant effect on the conformational order and alkyl chain packing.46 By investigating the effect of cholesterol and bilayer curvature on the interactions between PPO–PEO type block copolymers and membranes, we aim to develop a deeper understanding of the mechanism of polymer–membrane association and bring the model membrane systems one step closer to real cell membranes.
In this paper, we quantify the copolymer interactions with model membranes at various cholesterol concentrations and bilayer curvatures, using pulsed-field-gradient NMR (PFGNMR). PPO–PEO block copolymers examined include commercial Pluronics (Scheme 1a) and a lab-synthesized diblock analogue containing a tert-butyl group attached to the end of the PPO block (Scheme 1b). Unilamellar liposomes were used as model membranes due to the ease of tuning composition and curvature. Phosphatidylcholine (POPC, Scheme 1c), one of the most common lipids found in plasma membranes, has been widely used as a major component of model systems.15–17 Herein, the lipid bilayer composition was manipulated by mixing cholesterol (Scheme 1d) at various concentrations with POPC. Pure POPC bilayers were prepared as controls. Bilayer curvature was manipulated by varying the liposome size. Polymer binding to liposomes was quantified by PFG-NMR. This technique has been applied to determine self-diffusion coefficients of small molecules,47 to assess drug partitioning to polymer,48,49 and to quantify the molecular exchange rate through membranes of polymersomes.50–52 In the case of polymers mixed with liposomes, the diffusivity of polymers bound to liposomes can be distinguished from that of free polymers by PFG-NMR, from which the polymer binding can be quantified. Our results demonstrate that the cholesterol concentration has a significant effect on polymer−membrane association and bilayer curvature less so with increasing liposome size.
Scheme 1.
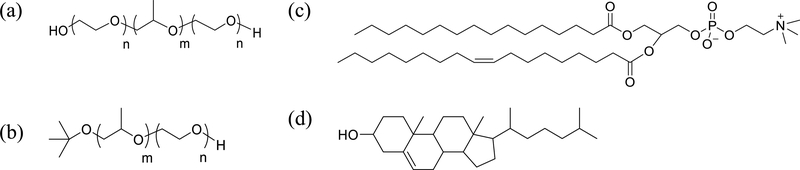
Chemical Structures of (a) Pluronic Triblock Copolymer, (b) a PPO−PEO Diblock Copolymer with a tert-Butyl End Group on the PPO Block, (c) Phosphatidylcholine (POPC), and (d) Cholesterol
MATERIALS AND METHODS
Materials.
Pluronics F68, F127, and P103 were provided by BASF. The materials used for anionic polymerization, including ethylene oxide (EO, ≥99.5%), propylene oxide (PO, ≥99%), potassium tert-butoxide, n-butyllithium, naphthalene, potassium, and 18-crown-6 ether, were purchased from Sigma-Aldrich and used as received. 1Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1,2dioleoyl-sn-glycero-3-phosphocholine (DOPC) in chloroform were purchased from Avanti Polar Lipids (Alabaster, AL) and used as received. Cholesterol (≥99%) and the chemicals used for matrix-assisted laser desorption ionization (MALDI) characterization, including silver trifluoroacetate (AgTFA) and α-cyano-4-hydroxycinnamic acid (CHCA, ≥98%), were also purchased from Sigma-Aldrich. Deuterium oxide (D2O, 99.9 atom % D) and chloroform-d (CDCl3, 99.8 atom % D + 0.05% v/v tetramethylsilane) were purchased from Cambridge Isotope Laboratories, Inc.
Polymer Synthesis.
A PPO–PEO diblock polymer with a tertbutyl end group, tPPO14–PEO46, was synthesized by anionic polymerization. The subscripts in the designation denote the number-average degrees of polymerization, and “t” represents the tert-butyl end group on the PPO block. The synthesis followed procedures described elsewhere and was reported previously.25,53–55 The main steps are summarized as follows. First, PO monomers were initiated using potassium tert-butoxide in tetrahydrofuran (THF) and reacted for 48 h under an air- and water-free environment at room temperature. Excess 18-crown-6 ether was present to reduce side reactions.53 The reaction was terminated by excess acidic methanol. Next, the PEO block was added to the PPO block by reinitiating the hydroxyl end group on the PPO block with freshly made potassium naphthalenide at 45 °C and reacting for 24 h before termination. Finally, the monohydroxyl terminated PPO–PEO block polymer was obtained by removing naphthalene and potassium salts through dialysis, followed by freeze-drying in benzene. The resulting fine white polymer powder was then characterized.
Polymer Characterization.
The number-average molecular weight (Mn) and dispersity (Đ). of each polymer were characterized by both matrix-assisted laser desorption ionization (MALDI) mass spectroscopy (AB SCIEX TOF/TOF 5800) and size exclusion chromatography (SEC) (Phenogel columns, Phenomenex) with a multiangle laser light scattering detector (Wyatt DAWN). Polymer compositions (wPEO) were determined on the basis of the mole fraction obtained by 1H NMR spectroscopy (Bruker AV-500). MALDI samples were prepared by mixing a polymer solution (1 mg/mL in THF, 0.1 mL) with a matrix solution of CHCA (20 mg/mL in THF, 0.2 mL) and a salt solution of AgTFA (1 mg/mL in THF, 5 μL). Sample solutions of 10 mg/mL polymers in THF were prepared for SEC measurements, using THF as the mobile phase at a flow rate of 1 mL/min. The refractive index increment (dn/dc) for each polymer was estimated from the weight average of PPO (dn/dc = 0.087 mL/g) and PEO (dn/dc = 0.068 mL/g).56,57 1H NMR samples were prepared as 10 mg/mL polymers in CDCl3. Table 1 summarizes the number-average molecular weight Mn, dispersity Đ, weight fraction of PEO wPEO, and the corresponding average number of repeat units of PO (NPO) and EO (NEO). These data were also reported in our previous study.25
Table 1.
Polymer Characterization
| Mn,1a (kg/mol) | Mn,2b (kg/mol) | Mn,3c (kg/mol) | Đb | Đc | wPEO,1a | wPEO,2d | NPOc | NEOe | |
|---|---|---|---|---|---|---|---|---|---|
| F68 | 8.4 | 8.2 | 7.9 | 1.01 | 1.04 | 0.80 | 0.81 | 27 | 75 |
| F127 | 12.6 | 13.2 | 10.8 | 1.01 | 1.07 | 0.70 | 0.72 | 64 | 108 |
| P103 | 4.95 | 5.2 | 4.6 | 1.02 | 1.14 | 0.30 | 0.33 | 60 | 19 |
| tPPO14–PEO46 | 2.9 | 3.0 | 1.03 | 1.07 | 0.70 | 14 | 46 |
Specified by supplier.
Determined by MALDI mass spectroscopy.
Determined by SEC. Weight average dn/dc was used with dn/dc = 0.068 for PEO and 0.087 for PPO.
Determined from molar ratios by 1H NMR spectroscopy.
Calculated from number-average molecular weight (Mn,2) and weight fraction of PEO (wPEO,2). For triblock Pluronics, NEO represents the number of repeat units of EO in one PEO block.
Liposome Preparation.
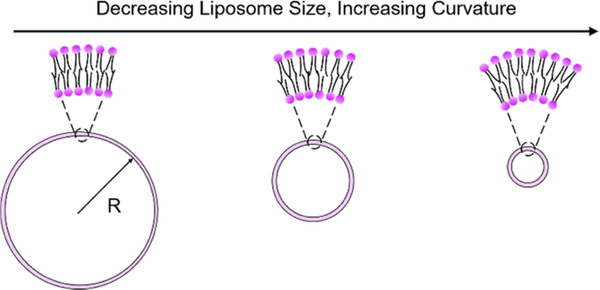
The procedures for liposome preparation were modified from the established method described elsewhere.15,25,58–60 First, the organic solvent present in the commercial phospholipid solution (e.g., 10 mg/mL POPC in chloroform) was evaporated under a nitrogen atmosphere to yield a thin lipid film on the wall of a glass vial. The thin film was further dried under vacuum for 30 min to remove the organic solvent completely. Next, the dried lipid film was hydrated with D2O, which gave a final lipid concentration of 10 mM. During the hydration process, the sample vial was kept in a water bath at 37 °C for 1 h, with vortex applied every 5 min to the lipid suspension to facilitate the formation of large multilamellar lipid vesicles. Last, the lipid suspension was extruded through a polycarbonate membrane back and forth 27 times to obtain the final liposome solution, using an Avanti Mini-Extruder assembly. The curvature κ of the lipid bilayer was manipulated by varying the liposome size (Scheme 2), as κ = 1/R, where R is the liposome radius. This was accomplished by using polycarbonate membranes with 25, 50, and 100 nm radius pores. The hydrodynamic radius Rh of the liposomes obtained from dynamic light scattering (DLS) was used to calculate κ. In the case of bicomponent liposome preparation (i.e., POPC/cholesterol), POPC lipids were mixed with cholesterol in chloroform at a specific ratio calculated from the desired composition, followed by the same procedures described above.
Scheme 2.
Schematic of Manipulating Bilayer Curvature by Varying the Liposome Size
Dynamic Light Scattering (DLS).
Multiangle DLS measurements were performed on a Brookhaven BI-200SM system with a laser wavelength of 637 nm at 27 °C. Each sample was measured at five scattering angles ranging from 60 to 120° in 15° increments in a temperature-controlled decalin bath that matches the refractive index of the sample cell. The resulting intensity autocorrelation functions were fit to a second-order cumulant expansion to acquire the mean decay rate Γ and dispersity, represented by the reduced second cumulant μ2/Γ2, at each angle. The mutual diffusion coefficient Dm was obtained from the linear fit of Γ versus q2, where q is the scattering wavevector (eq 1). The average hydrodynamic radius Rh was further calculated from Dm through the Stokes−Einstein relationship (eq 2). Liposome samples were diluted with D2O to 100 μM from the original 10 mM stock solution for DLS measurements. All samples were filtered through 0.2 μm filters before measurements.
| (1) |
| (2) |
Cryogenic Transmission Electron Microscopy (Cryo-TEM).
Solution samples were prepared for imaging using a FEI Vitrobot Mark IV vitrification system with the climate control chamber set to 27 °C and 100% humidity. Five microliters of the sample were pipetted onto a lacy carbon/Formvar grid (Ted Pella, 200 mesh) that was previously cleaned and hydrophilized using a PELCO easiGlow glow discharge cleaning system. The sample was annealed in the chamber for 1 s before the grid was blotted for 7 s with the instrument-defined blot force set to −2, equilibrated for 3 s, and plunged into liquid ethane cooled by liquid nitrogen. The grids were then transferred into liquid nitrogen for storage prior to imaging. The vitrified sample grids were transferred to a Gatan-626 single tilt cryoholder and imaged using an FEI Tecnai G2 Spirit BioTWIN microscope operated at 120 kV coupled with a FEI Eagle chargecoupled device camera (2048 × 2048 pixels).
NMR Spectroscopy.
A Bruker Avance III 500 MHz NMR instrument with a 5 mm broadband fluorine observe probe was employed for PFG-NMR measurements, using the “ledbpgp2s” stimulated pulse sequence (longitudinal eddy current delay experiment using bipolar gradients acquired in two dimensions).61 The echo-attenuated intensity I was recorded from each one-dimensional 1H spectrum corresponding to increasing gradient strengths G (from 2 to 95% of the maximum gradient strength). The translational diffusivity (D) of the samples was extracted from the linear fit on the basis of the following relationship
| (3) |
where I0 is the intensity with respect to zero gradient strength and γ is the gyromagnetic ratio of 1H (42.6 MHz/T). The length of the gradient pulse δ was set to 5 ms, and the diffusion time Δ was set to 300, 500, and 700 ms for polymer–liposome mixtures and 700 ms for pure polymer solutions. Polymer diffusivity was acquired from the intensity attenuation of the PEO peak due to its strong signal at G = 0.
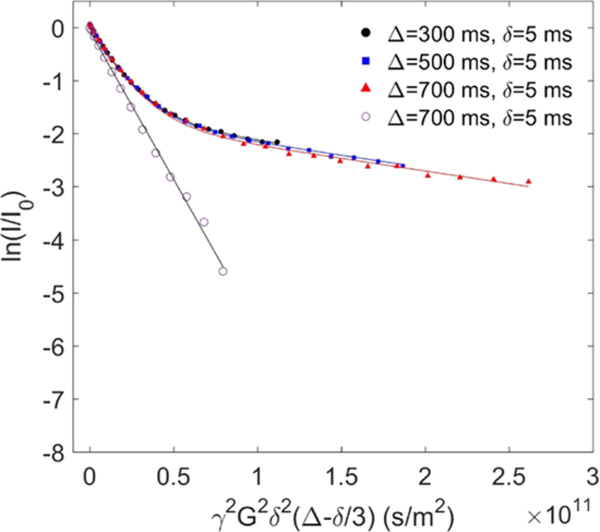
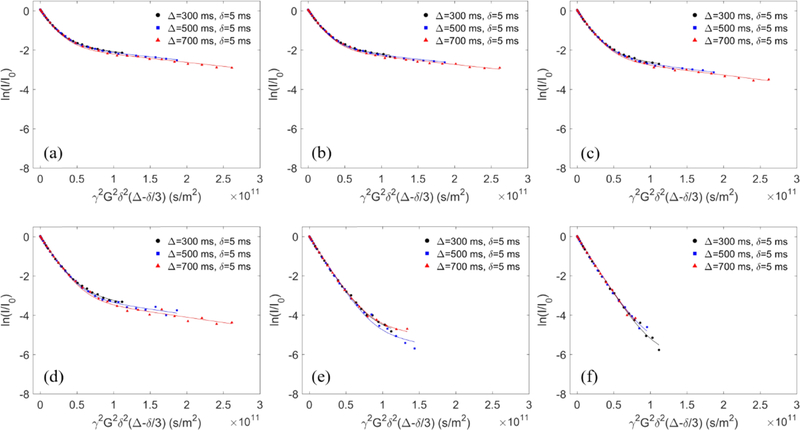
Polymer–liposome mixtures were prepared by combining 0.25 mL of 10 mM liposome stock solution with 0.25 mL of polymer stock solution (0.4 mg/mL for P103 and F127 and 2 mg/mL for F68 and tPPO14−PEO46 in D2O). Note that the polymers P103 and F127 were dissolved at a lower concentration due to their relatively low critical micelle concentrations.62,63 Pure polymer and pure liposome samples were prepared as controls by adding 0.25 mL of D2O to 0.25 mL of the polymer and liposome stock solutions, respectively. Pure polymer solutions measured by PFG-NMR followed single-exponential decays according to eq 3, as depicted by the linear relationship (open symbols) in Figure 2. On the other hand, in the polymer–liposome mixtures, a fraction of polymers bound to the liposome surface results in a contribution to the decay profile that reflects much slower diffusion than that of the remaining free polymers. We associate this slow diffusion with polymers bound to the liposomes, which have diffusivity that is 1 order of magnitude slower than that of free polymers. We further assume that the liposome diffusivity changes little before and after polymer binding. The coexistence of the bound and free states of polymers gave rise to biexponential decays, as modeled in eq 4
| (4) |
where Dfree and Dbound stand for the diffusivity of free and bound polymers, corresponding to the initial and final slope of the decay curves (filled symbols) in Figure 2, respectively. The binding percentage of polymers to liposomes, fbound, was quantified by fitting this biexponential model to the measured decay curves. Note that the molecular exchange rate between free and bound polymers can be neglected as the three decay curves shown in Figure 2 measured at different Δ collapsed onto each other. All samples were measured at 27 °C with a temperature fluctuation less than ±0.3 °C (indicated by the temperature monitor inside the NMR equipment). The temperature of 27 °C was used to ensure that all polymers are free coils and that there is no micelle formation.25 We speculate that at higher temperature, polymer binding would increase as the drive for polymers to insert into hydrophobic bilayer interiors, or to form hydrophobic micellar cores, is stronger. Our previous work found that the incubation time of polymers with liposomes prior to measurements has little influence on polymer binding.25 Therefore, all freshly made samples were measured with no further incubation.
Figure 2.
Experimental and fitted echo decay curves of the protons from PEO of 0.2 mg/mL F127 in the presence of 5 mM POPC liposome in D2O at 27 °C with Δ = 300, 500, and 700 ms (filled circles, filled squares, and filled triangles, respectively) and with fixed δ = 5 ms. The data were fit to eq 4. The open circles denote the proton signal from PEO of 0.2 mg/mL F127 in the absence of liposomes; these data were fit to eq 3.
RESULTS AND DISCUSSION
Liposome Size Characterization.
Table 2 summarizes the mean Rh and dispersity (μ2/Γ2) values for the liposomes prepared using the three pore sizes, as characterized by DLS. When extruded through a polycarbonate membrane with a 25 nm pore radius, the Rh of POPC/cholesterol liposomes increases from 42 to 57 nm as the molar percentage of cholesterol in the bilayer increases from 0 to 30 mol %. On the other hand, the Rh of the liposomes at intermediate size (i.e., extruded through a polycarbonate membrane with 50 nm pore radius) fluctuates slightly between 65 and 70 nm with cholesterol composition. This variation in Rh is within experimental uncertainty. These results indicate that the effect of cholesterol on liposome Rh is more significant for smaller liposomes. Previous work found a remarkable increase of liposome Rh when the liposomes extruded with 50 nm pore radius contain more than 20 mol % cholesterol, which could be attributed to increasing membrane stiffness.64 Such an increase in liposome Rh, however, was only observed here in the liposomes extruded with a 25 nm pore radius. The dispersity of liposomes was not affected by the presence of cholesterol or the extrusion pore radius, as the values of μ2/Γ2 are all less than 0.1. Note that the variations in liposome size do not affect the quantification of polymer binding, since the liposome diffusion is still 1 order of magnitude slower than that for the free polymer molecules, and accordingly, the diffusion coefficients of bound and free polymers can still be readily distinguished by PFG-NMR.
Table 2.
Liposome Size Characterization in D2O by DLS
| cholesterol molar percentage (mol %) | extrusion pore radius (nm) | Rha (nm) | μ2/Γ2 at 90° |
|---|---|---|---|
| 0 | 25 | 42 | 0.06 |
| 1 | 25 | 48 | 0.06 |
| 2 | 25 | 47 | 0.07 |
| 5 | 25 | 48 | 0.07 |
| 10 | 25 | 51 | 0.06 |
| 20 | 25 | 55 | 0.06 |
| 30 | 25 | 57 | 0.05 |
| 0 | 50 | 70 | 0.04 |
| 1 | 50 | 70 | 0.03 |
| 2 | 50 | 70 | 0.04 |
| 5 | 50 | 65 | 0.06 |
| 10 | 50 | 67 | 0.04 |
| 20 | 50 | 68 | 0.03 |
| 30 | 50 | 69 | 0.05 |
| 0 | 100 | 109 | 0.07 |
Rh measured by DLS has approximately 5% uncertainty.
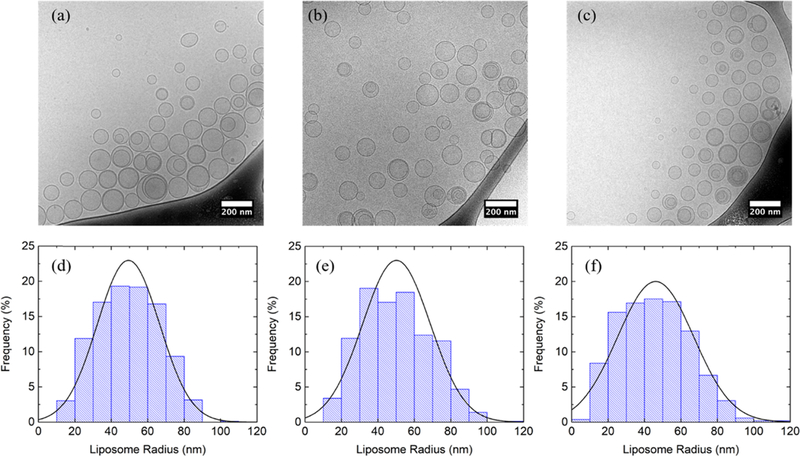
Additionally, cryo-TEM was employed to characterize the size and morphology of liposomes with selected compositions. Liposomes with 0, 10, and 30 mol % cholesterol extruded through a polycarbonate membrane with 50 nm pore radius were selected as representative for cryo-TEM imaging, as shown in Figure 1a–c, respectively. Liposome morphology shows little change as the cholesterol molar percentage increases. A small number of multilamellar liposomes are present at all three cholesterol concentrations. The estimated percentages of liposome surface area within multilamellar liposomes are summarized in Table S1; at all three cholesterol concentrations, a similar small percentage of surface area is sequestered in the multilamellar liposomes (i.e., less than 20%). The sequestered surface area of liposomes could be potentially reduced by increasing the times of extrusion.58 Histograms of liposome radius measured from cryo-TEM are plotted in Figure 1d–f. The mean and standard deviation of the liposome radius calculated from the histograms are summarized in Table S1 (i.e., 49 ± 17, 50 ± 19, and 46 ± 21 nm at 0, 10, and 30 mol % cholesterol, respectively). The mean radius of the liposomes at all three cholesterol concentrations is close to the extrusion pore radius (i.e., 50 nm). The mean radius is apparently constant, as the standard deviation is relatively large. This suggests that increasing cholesterol molar percentage in the lipid bilayer results in little change of liposome size in the case of a 50 nm extrusion pore radius, which agrees with the conclusion obtained from DLS. The histograms are reasonably well represented by a normal distribution based on the mean and the standard deviation obtained from the histograms, as shown in Figure 1. The liposome radii measured by cryo-TEM were found to be smaller than the Rh measured by DLS. We attribute this difference to the following reasons: (1) Rh includes the thickness of the hydration layer of the lipid headgroups, and (2) DLS is skewed toward large size particles, since it measures intensity-averaged size and the scattered intensity of vesicles is proportional to R2, whereas cryo-TEM measures the numberaveraged size.65
Figure 1.
Cryo-TEM images of 10 mM liposomes in D2O composed of (a) POPC, (b) 90 mol % POPC + 10 mol % cholesterol, and (c) 70 mol % POPC + 30 mol % cholesterol extruded through a polycarbonate membrane with a 50 nm pore radius. (d)–(f) are corresponding histograms of liposome radius measured from cryo-TEM compared with normal distribution based on the mean and the standard deviation of the population. Nonspherical liposomes were excluded. The radius of a multilamellar liposome was measured at the outermost vesicle.
Effect of Cholesterol.
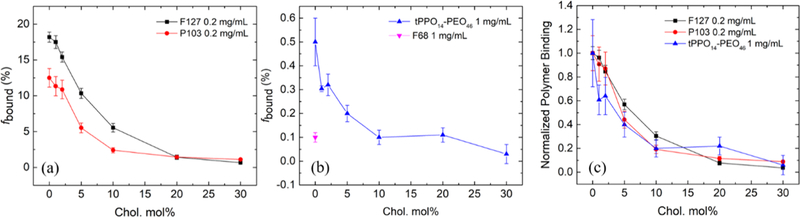
The role of cholesterol was investigated by quantifying the binding of four representative PPO–PEO block copolymers (F127, P103, tPPO14–PEO46, and F68) to the POPC liposomes containing various molar ratios of cholesterol using the intermediate size liposomes (i.e., extruded through a polycarbonate membrane with 50 nm pore radius). F127 and P103 have relatively strong interactions with lipid bilayers compared to F68 and tPPO14–PEO46. More than 10 mol % of F127 and P103 bind to neat POPC liposomes, whereas less than 1 mol % of tPPO14−PEO46 and F68 associated with the same bilayer particles (Table S2). Echo decay curves of F127 in the presence of liposomes with increasing cholesterol concentration are shown in Figure 3 as an example. In the control case of F127 interacting with neat POPC liposomes (Figure 2), the strong signal intensity of the echo decay curves retained at the final slope illustrates that a substantial fraction of polymers is bound to the liposomes. In contrast, the polymer signal at the final slope becomes weaker upon increasing the cholesterol concentration in the lipid bilayer, indicating diminished polymer binding (Figure 3). When the cholesterol content reaches 10 mol %, the final slope of the polymer decay curves becomes noisy and less reliable, due to a lack of bound polymer. The decay curves barely display a final slope when the lipid bilayer contains more than 20 mol % cholesterol; the decay curve closely resembles a single-exponential decay of free polymers, thereby indicating little interaction between the polymers and lipid bilayers. The binding percentages of all four polymers (F127, P103, tPPO14–PEO46, and F68) extracted from the NMR data are plotted as a function of cholesterol molar percentage in the POPC/cholesterol lipid bilayers in Figure 4a,b. The trends found for F127, P103, and tPPO14–PEO46 are similar, where polymer binding decreases as cholesterol concentration increases, despite much weaker interactions between tPPO14–PEO46 and the liposomes. Binding of F68 was only measured with neat POPC liposomes; due to the low extent of association there was inadequate signal to noise to quantify the extent of binding in the presence of cholesterol indicative of little interaction. Note that the tert-butyl ended diblock was selected instead of one with a hydroxyl end because the latter has very weak interaction with pure POPC bilayers and therefore does not show a clear binding trend upon varying cholesterol concentration.25 Also, the tert-butyl ended diblock showed greater potential in cell membrane stabilization, as previous in vitro and in vivo studies found that the tert-butyl end group on the PPO block can significantly enhance the polymer stabilization efficacy for damaged cell membranes.12,13
Figure 3.
Experimental and fitted echo decay curves of the protons from PEO of 0.2 mg/mL F127 in the presence of 5 mM POPC/cholesterol liposome solution in D2O at 27 °C with (a) 1 mol %, (b) 2 mol %, (c) 5 mol %, (d) 10 mol %, (e) 20 mol %, and (f) 30 mol % cholesterol.
Figure 4.
Polymer binding percentage of (a) two strongly bound polymer species, and (b) two weakly bound polymer species as a function of cholesterol molar percentage in POPC/cholesterol lipid bilayers. (c) Binding trend of three polymer species as a function of cholesterol molar percentage in the lipid bilayer, normalized to their binding with pure POPC lipid bilayers.
These results are broadly consistent with earlier studies, which suggested that the addition of cholesterol decreases the polymer association with liposomes.24,26 Johnsson et al. studied the morphology of liposomes prepared with premixed Pluronics and lipids, with and without cholesterol.24 They found that inclusion of cholesterol in the liposomal preparations reduces the incorporation of copolymers in the lipid bilayer, and thus reduces the liposomal morphological changes due to the presence of copolymers observed in the absence of cholesterol. According to their study, we speculate that the binding trend would be similar if the polymer were added at the time of liposome formation (i.e., fewer polymers can incorporate into lipid bilayers as cholesterol increases). In order to compare the cholesterol effect on F127, P103, and tPPO14–PEO46, the polymer binding at various cholesterol concentrations was normalized to the binding with neat POPC liposomes (i.e., no cholesterol addition) so that the binding trend of the three polymer species can be plotted on the same scale, as shown in Figure 4c. It can be seen that all three polymers exhibit a universal trend, with a remarkable ten-fold decrease in binding as the cholesterol concentration increases to 30 mol %. This cholesterol weakening effect on polymer binding has little relative dependence on polymer species across a wide range of relative hydrophobicities, an observation that will be rationalized subsequently. The diffusion coefficients of free and bound polymers used for fitting and the resulting binding percentage of polymers to POPC liposomes at increasing cholesterol concentration are summarized in Table S2. Data for polymer binding with neat POPC liposomes were reported previously.25
Significantly reduced polymer binding to liposomes could result from the change of membrane properties due to the presence of cholesterol. Membrane stiffness is a likely candidate, since the addition of cholesterol can induce ordering in lipid hydrocarbon chains, leading to membrane structural condensation and thereby increasing the bending modulus of the membrane.64,66 Previous work of Arriaga and co-workers demonstrated that the bending modulus of POPC lipid bilayers significantly increases with added cholesterol.64 Pan et al. also investigated the effect of cholesterol on the membrane bending modulus, and reported that the stiffening effect of cholesterol on lipid bilayers is largely reduced when the phospholipids contain two mono-unsaturated hydrocarbon chains, compared to one or none.66 In order to test if the weakening effect of cholesterol on polymer binding depends on the degree of unsaturation of lipids, we compared the polymer binding to POPC liposomes with that to DOPC liposomes at increasing cholesterol molar percentage in the membrane. Since DOPC contains two mono-unsaturated hydrocarbon chains (Figure S1a), while POPC only contains one, DOPC lipid bilayers remain more flexible (i.e., lower bending modulus) than POPC bilayers when cholesterol is present, based on the study of Pan and co-workers. As shown in Figure S1b, the polymer binding to DOPC liposomes is always greater than with POPC, despite the same overall trend that the binding decreases as the cholesterol molar percentage increases. After normalizing to the binding in the absence of cholesterol (Figure S1c), it can be seen that the polymer binding to DOPC liposomes decays more slowly than for POPC with increasing cholesterol. This indicates that cholesterol is less effective in reducing polymer binding to the lipid bilayers containing more mono-unsaturated hydrocarbon chains, which indirectly supports the hypothesis that the lipid bilayers with lower stiffness (i.e., lower bending modulus) favor more polymer binding. Additionally, the hydrophobicity of the membrane alkyl chain region may also play an important role, as the hydrophobic interaction between the lipid bilayer interior and the PPO block of the polymers is a key determinant of the amount of polymer binding.25 The addition of cholesterol may disturb such interactions since the steroid rings of cholesterol sitting in the bilayer interior are much more hydrophobic than lipid alkyl chains or the PPO blocks.67,68 We suspect that it is less favorable for the PPO blocks to associate with a much more hydrophobic component in the bilayer interior. It is also possible that the copolymer interacts less favorably with a more ordered lipid, or that increased membrane organization retards the initial stages of polymer association.
Bilayer Curvature Effect.
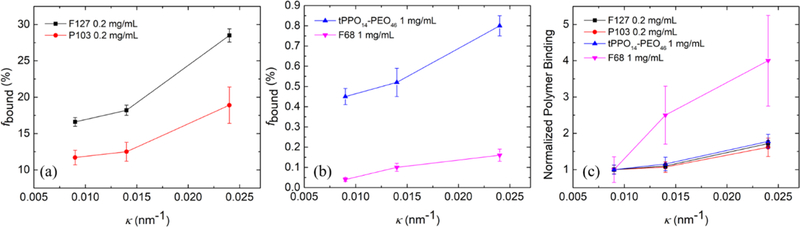
Lipid bilayer curvature can be simply manipulated by altering the liposome size. Our previous work on quantifying polymer binding to lipid bilayers was based on liposomes with an intermediate extrusion pore radius of 50 nm.25 Herein, two additional sizes of extrusion pore radius (25 and 100 nm) were used in order to obtain liposomes with three different curvatures, corresponding to κ = 0.009, 0.014, and 0.024 nm−1, calculated as κ = 1/Rh. F127 and P103 were selected as representative strongly binding polymers, whereas tPPO14–PEO46 and F68 were weakly binding polymers.25 The binding percentages of the strongly and weakly interacting polymers with increasing lipid bilayer curvature are shown in Figure 5a,b, respectively. The bilayer curvature, the diffusion coefficients of free and bound polymers used for fitting, and the resulting polymer binding percentage are summarized in Table S3. The binding of all four polymers increases as the lipid bilayer curvature increases. These results have also been normalized to the binding at the smallest curvature, as shown in Figure 5c. It can be seen that the normalized binding as a function of curvature for all polymer species except F68 collapse onto a consistent increasing trend with κ, i.e., higher curvature results in greater polymer binding. When the bilayer curvature is less than 0.014 nm−1 (corresponding to an extrusion pore radius of 50 nm), polymer binding (except F68) is almost independent of the curvature. This finding validates the rationale for using liposomes with an extrusion pore radius of at least 50 nm as models to mimic the flat configuration of real cell membranes, as assumed in previous studies of polymer−membrane association.14,15,24,69 The inconsistent binding trend of F68 can be attributed to its extremely weak interactions with liposomes, as the binding percentage at highest curvature is only 0.2%; the inherently weak association of F68 with liposomes gives rise to large uncertainty in quantifying the polymer binding. We speculate that the higher polymer binding at higher bilayer curvature may be associated with the existence of greater space in the outer leaflet of the lipid bilayer with higher curvature. Interaction with polymers could mitigate the thermodynamic penalty associated with such inferior packing.
Figure 5.
Polymer binding percentage of (a) two strongly bound polymer species, and (b) two weakly bound polymer species as a function of POPC lipid bilayer curvature. (c) Binding trend of four polymer species as a function of POPC lipid bilayer curvature, normalized to their binding to the liposome with the smallest curvature.
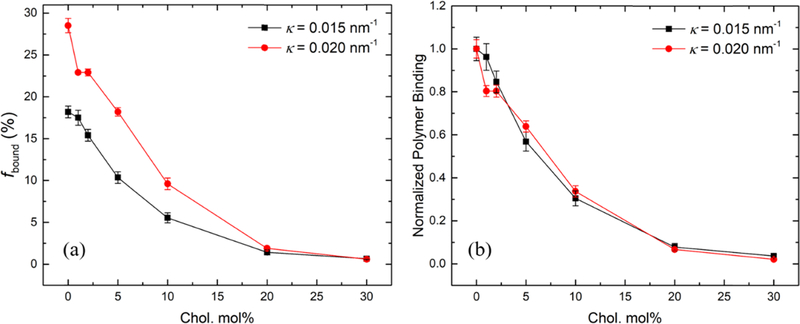
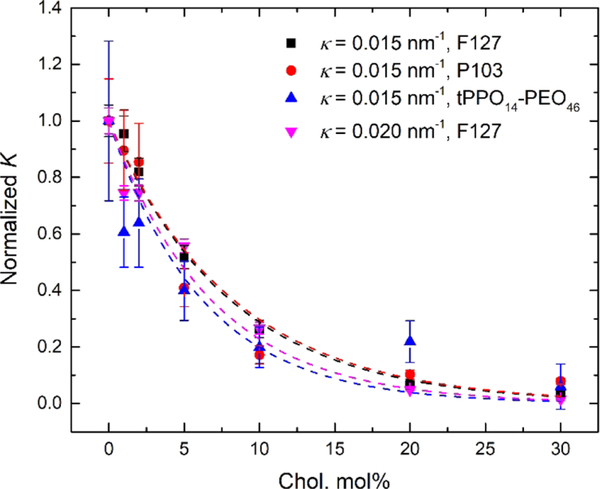
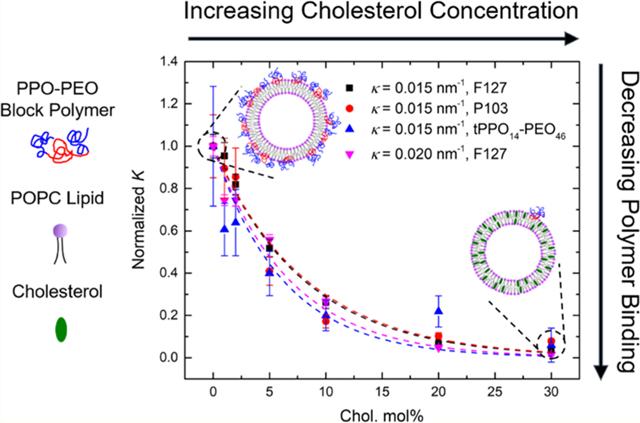
We have demonstrated that higher cholesterol molar percentage in the lipid bilayer results in less polymer binding, whereas higher bilayer curvature results in more polymer binding. To investigate whether the cholesterol weakening effect on polymer binding can be mitigated by higher curvature of the lipid bilayers, we further measured the binding of F127 to lipid bilayers with a higher curvature (i.e., extruded through a pore radius of 25 nm) as a function of cholesterol molar percentage (Figure 6a). Compared with its binding to the bilayer with an intermediate curvature (i.e., extruded through a pore radius of 50 nm), F127 shows significantly more binding at higher curvature until the cholesterol molar percentage reaches 20 mol %. At 20 and 30 mol % cholesterol, the binding percentage at both curvatures drops below 2% and shows no significant difference. Note that the liposomes at these high cholesterol levels exhibit more similar curvatures (κ = 0.018 and 0.014 nm−1) because the curvatures of the liposomes extruded through the 25 nm pores decreased with increased cholesterol (Table 2). For brevity, the average curvature of the liposomes extruded through the same pore radius is shown in Figure 6 (i.e., 0.015 nm−1 for liposomes extruded through the 50 nm pore radius and 0.020 nm−1 for the 25 nm pore radius). Polymer binding at both curvatures as a function of the cholesterol molar percentage was further normalized to their original binding with neat POPC liposomes (i.e., no cholesterol addition). As shown in Figure 6b, the two curves overlap with each other and follow the same decreasing trend as the cholesterol molar percentage increases. This indicates that the cholesterol weakening effect on polymer binding has little dependence on the lipid bilayer curvature. The diffusion coefficients of free and bound polymers used for fitting and the resulting polymer binding percentage at higher curvature (i.e., 0.020 nm−1) are summarized in Table S4.
Figure 6.
(a) Polymer binding percentage and (b) normalized polymer binding of 0.2 mg/mL F127 to POPC/cholesterol liposomes with κ = 0.015 nm−1 (black) and κ = 0.020 nm−1 (red) as a function of the cholesterol molar percentage.
Figures 4c and 6b reveal similar trends for relative polymer binding with an increasing cholesterol molar percentage, independent of polymer species and lipid bilayer curvature. A rather straightforward argument can rationalize both the exponential form of the decay and the fact that the normalized curves for quite different polymers collapse onto a single curve. The polymer binding percentage, fbound, can be converted to an equilibrium binding constant K as follows,
| (5) |
where K is defined as the ratio of the concentration of bound polymers ([bound]) to that of free polymers ([free]), assuming that the concentration of lipid binding sites remains unchanged and thereby is incorporated into K. K is related to the standard free-energy change of the binding process, ΔG°, where R is the gas constant and T is the temperature. For a given polymer, we can define three free energies, corresponding to three possible states: free in solution, bound to 100% lipid, and bound to 100% cholesterol (a hypothetical limit). If the binding to a mixed lipid/cholesterol layer is linear in the cholesterol composition x, analogous to an assumption of “ideal mixing” in the layer, then ΔG° will vary linearly with the cholesterol molar percentage. This assumption is therefore sufficient to explain the single-exponential decay observed (see the derivation in the Supporting Information for more details). Then, if we make a further assumption that the incremental free-energy cost to bind a more hydrophilic polymer compared with a more hydrophobic polymer is the same for pure lipid and for pure cholesterol, then the exponential decay constant A is independent of polymer. K for the various polymer species interacting with liposomes with different curvatures is plotted as a function of x in Figure 7. Each curve is fit to a single-exponential decay model
| (6) |
and the fitted values of parameter A are summarized in Table S5. Within the uncertainty of these fits all data can be accounted for by a single value of A. This confirms the assumption that the difference of the standard free energy of a polymer bound to a liposome composed of 100 mol % POPC compared with a hypothetical liposome composed of 100 mol % cholesterol changes little with variation of the polymer species or bilayer curvature.
Figure 7.
Single-exponential decay model fitted to the normalized K of various polymer species bound to POPC/cholesterol liposomes with different curvatures as a function of the cholesterol molar percentage.
CONCLUSIONS
We have investigated how cholesterol concentration and lipid bilayer curvature affect the interaction of both strongly and weakly binding PPO–PEO block copolymers with POPC and DOPC liposomes. Copolymer binding to liposomes was probed by PFG-NMR, which can resolve the relative amounts of free and liposome-bound polymers. The role of cholesterol was studied by quantifying polymer binding at various cholesterol molar percentages. For POPC, the copolymer binding decays exponentially with the cholesterol composition, by a factor of 10 at 30 mol % cholesterol relative to the pure POPC bilayer. This could be due to an increase in the membrane bending modulus and/or hydrophobicity. Polymer binding also decreases as the bilayer curvature decreases, most notably between liposomes with extrusion radii of 25 and 50 nm, with less variation as the size increases to 100 nm. This may be due to the creation of a greater void space in the outer leaflet of the lipid bilayer with a higher curvature, which is mitigated by the presence of the hydrophobic PPO blocks. All data obtained with the weakly and strongly associating PPO–PEO block copolymers at different bilayer curvatures and with varying cholesterol content can be reduced to a single-exponential curve of the normalized equilibrium binding constant K versus the cholesterol content. This superposition can be rationalized with only two plausible assumptions that the free-energy change of binding is linear in cholesterol content and that the increment in free energy to bind a more hydrophilic polymer than a more hydrophobic one is independent of the membrane composition. In summary, these results demonstrate that cholesterol in the lipid bilayer plays a dominant role in controlling the interactions of liposomes with both weakly and strongly adsorbing PPO–PEO block polymers. This finding establishes a key factor in understanding how Pluronics/poloxamers impact cellular activity in living systems.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Karen J. Haman for synthesizing the diblock polymer and Mihee Kim for useful discussions. We also acknowledge Letitia J. Yao for help with the NMR experiments. NMR instrumentation was supported by the Office of the Vice President of Research, College of Science and Engineering, and the Department of Chemistry at the University of Minnesota. The cryo-TEM images were recorded in the Characterization Facility, University of Minnesota, which receives partial support from NSF through the MRSEC program under Award Number DMR-1420013. This study was funded by the National Institutes of Health (grant R01HL122323).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.langmuir.9b00572.
Summary of liposome radii and the percentage of liposome surface area within multilamellar liposomes measured from cryo-TEM, summary of the binding results of F127, P103, tPPO14–PEO46, and F68 to POPC/cholesterol liposomes at different cholesterol molar percentages, polymer binding percentage and normalized binding of F127 to DOPC/cholesterol and to POPC/cholesterol liposomes at different cholesterol molar percentages, summary of the binding results of F127, P103, tPPO14–PEO46, and F68 to POPC liposomes at different curvatures, summary of the binding results of F127 to POPC/cholesterol liposomes with a higher curvature of 0.020 nm−1 at different cholesterol molar percentages, fitting results of the decay parameter A, and derivation of the equilibrium binding constant K as a function of cholesterol molar percentage x (PDF)
The authors declare no competing financial interest.
J.M.M. is on the scientific advisory board of and holds zero value equity shares in Phrixus Pharmaceuticals Inc., a company developing novel therapeutics for heart failure and DMD, and this is actively managed by the UMN Office of Institutional Compliance.
REFERENCES
- (1).Pacifico CR; Lundsted LG; Vaughn TH Flake Form Nonionic Detergents. Soap Sanit. Chem 1950, 26, 40–43. [Google Scholar]
- (2).Vaughn TH; Suter HR; Lundsted LG; Kramer MG Properties of Some Newly Developed Nonionic Detergents. J. Am. Oil Chem. Soc 1951, 28, 294–299. [Google Scholar]
- (3).Lundsted LG Polyoxyalkylene Compounds. US2674619A, 1954. [Google Scholar]
- (4).Yasuda S; Townsend D; Michele DE; Favre EG; Day SM; Metzger JM Dystrophic Heart Failure Blocked by Membrane Sealant Poloxamer. Nature 2005, 436, 1025–1029. [DOI] [PubMed] [Google Scholar]
- (5).Houang EM; Haman KJ; Filareto A; Perlingeiro RC; Bates FS; Lowe DA; Metzger JM Membrane-Stabilizing Copolymers Confer Marked Protection to Dystrophic Skeletal Muscle in Vivo. Mol. Ther.-Methods Clin. Dev 2015, 2, No. 15042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bartos JA; Matsuura TR; Tsangaris A; Olson M; McKnite SH; Rees JN; Haman K; Shekar KC; Riess ML; Bates FS; Metzger JM; Yannopoulos D. Intracoronary Poloxamer 188 Prevents Reperfusion Injury in a Porcine Model of ST-Segment Elevation Myocardial Infarction. JACC 2016, 1, 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Venne A; Li S; Mandeville R; Kabanov A; Alakhov V. Hypersensitizing Effect of Pluronic L61 on Cytotoxic Activity, Transport, and Subcellular Distribution of Doxorubicin in Multiple Drug-Resistant Cells. Cancer Res. 1996, 56, 3626–3629. [PubMed] [Google Scholar]
- (8).Valle JW; Lawrance J; Brewer J; Clayton A; Corrie P; Alakhov V; Ranson M. Phase A II, Window Study of SP1049C as First-Line Therapy in Inoperable Metastatic Adenocarcinoma of the Oesophagus. J. Clin. Oncol 2004, 22, 4195. [Google Scholar]
- (9).Batrakova EV; Kabanov AV Pluronic Block Copolymers: Evolution of Drug Delivery Concept from Inert Nanocarriers to Biological Response Modifiers. J. Controlled Release 2008, 130, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kabanov AV; Batrakova EV; Alakhov VY Pluronic Block Copolymers as Novel Polymer Therapeutics for Drug and Gene Delivery. J. Controlled Release 2002, 82, 189–212. [DOI] [PubMed] [Google Scholar]
- (11).Alakhov V; Klinski E; Li S; Pietrzynski G; Venne A; Batrakova E; Bronitch T; Kabanov A. Block Copolymer-Based Formulation of Doxorubicin. From Cell Screen to Clinical Trials. Colloids Surf., B 1999, 16, 113–134. [Google Scholar]
- (12).Kim M; Haman KJ; Houang EM; Zhang W; Yannopoulos D; Metzger JM; Bates FS; Hackel BJ PEO–PPO Diblock Copolymers Protect Myoblasts from Hypo-Osmotic Stress In Vitro Dependent on Copolymer Size, Composition, and Architecture. Biomacromolecules 2017, 18, 2090–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Houang EM; Haman KJ; Kim M; Zhang W; Lowe DA; Sham YY; Lodge TP; Hackel BJ; Bates FS; Metzger JM Chemical End Group Modified Diblock Copolymers Elucidate Anchor and Chain Mechanism of Membrane Stabilization. Mol. Pharmaceutics 2017, 14, 2333–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Cheng C-Y; Wang J-Y; Kausik R; Lee KYC; Han S. Nature of Interactions between PEO-PPO-PEO Triblock Copolymers and Lipid Membranes: (II) Role of Hydration Dynamics Revealed by Dynamic Nuclear Polarization. Biomacromolecules 2012, 13, 2624–2633. [DOI] [PubMed] [Google Scholar]
- (15).Wang J-Y; Marks J; Lee KYC Nature of Interactions between PEO-PPO-PEO Triblock Copolymers and Lipid Membranes: (I) Effect of Polymer Hydrophobicity on Its Ability to Protect Liposomes from Peroxidation. Biomacromolecules 2012, 13, 2616–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang J-Y; Chin J; Marks JD; Lee KYC Effects of PEO–PPO–PEO Triblock Copolymers on Phospholipid Membrane Integrity under Osmotic Stress. Langmuir 2010, 26, 12953–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wang J; Segatori L; Biswal SL Probing the Association of Triblock Copolymers with Supported Lipid Membranes Using Microcantilevers. Soft Matter 2014, 10, 6417. [DOI] [PubMed] [Google Scholar]
- (18).Firestone MA; Wolf AC; Seifert S. Small-Angle X-ray Scattering Study of the Interaction of Poly(ethylene Oxide)-bPoly(propylene Oxide)-b-Poly(ethylene Oxide) Triblock Copolymers with Lipid Bilayers. Biomacromolecules 2003, 4, 1539–1549. [DOI] [PubMed] [Google Scholar]
- (19).Firestone MA; Seifert S. Interaction of Nonionic PEO–PPO Diblock Copolymers with Lipid Bilayers. Biomacromolecules 2005, 6, 2678–2687. [DOI] [PubMed] [Google Scholar]
- (20).Lee B; Firestone MA Electron Density Mapping of Triblock Copolymers Associated with Model Biomembranes: Insights into Conformational States and Effect on Bilayer Structure. Biomacromolecules 2008, 9, 1541–1550. [DOI] [PubMed] [Google Scholar]
- (21).Hezaveh S; Samanta S; De Nicola A; Milano G; Roccatano D. Understanding the Interaction of Block Copolymers with DMPC Lipid Bilayer Using Coarse-Grained Molecular Dynamics Simulations. J. Phys. Chem. B 2012, 116, 14333–14345. [DOI] [PubMed] [Google Scholar]
- (22).Rabbel H; Werner M; Sommer J-U Interactions of Amphiphilic Triblock Copolymers with Lipid Membranes: Modes of Interaction and Effect on Permeability Examined by Generic Monte Carlo Simulations. Macromolecules 2015, 48, 4724–4732. [Google Scholar]
- (23).Zhirnov AE; Pavlov DN; Demina TV; Badun GA; Grozdova ID; Melik-Nubarov NS Effect of the Structure of Ethylene Oxide-Propylene Oxide Block Copolymers on Their Interaction with Biological Membranes. Polym. Sci., Ser. A 2006, 48, 1202–1210. [Google Scholar]
- (24).Johnsson M; Silvander M; Karlsson G; Edwards K. Effect of PEO–PPO–PEO Triblock Copolymers on Structure and Stability of Phosphatidylcholine Liposomes. Langmuir 1999, 15, 6314–6325. [Google Scholar]
- (25).Zhang W; Haman KJ; Metzger JM; Hackel BJ; Bates FS; Lodge TP Quantifying Binding of Ethylene Oxide−Propylene Oxide Block Copolymers with Lipid Bilayers. Langmuir 2017, 33, 12624–12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhirnov AE; Demina TV; Krylova OO; Grozdova ID; Melik-Nubarov NS Lipid Composition Determines Interaction of Liposome Membranes with Pluronic L61. Biochim. Biophys. Acta, Bioenerg 2005, 1720, 73–83. [DOI] [PubMed] [Google Scholar]
- (27).van Meer G. Lipid Traffic in Animal Cells. Annu. Rev. Cell Biol 1989, 5, 247–275. [DOI] [PubMed] [Google Scholar]
- (28).Shrestha R; Anderson CM; Cardenas AE; Elber R; Webb LJ Direct Measurement of the Effect of Cholesterol and 6Ketocholestanol on the Membrane Dipole Electric Field Using Vibrational Stark Effect Spectroscopy Coupled with Molecular Dynamics Simulations. J. Phys. Chem. B 2017, 121, 3424–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chiu SW; Jakobsson E; Scott HL Combined Monte Carlo and Molecular Dynamics Simulation of Hydrated Lipid-Cholesterol Lipid Bilayers at Low Cholesterol Concentration. Biophys. J 2001, 80, 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Huang C-H Roles of Carbonyl Oxygens at the Bilayer Interface in Phospholipid−sterol Interaction. Nature 1976, 259, 242–244. [DOI] [PubMed] [Google Scholar]
- (31).Yeagl PL. Cholesterol and Related Sterols The Membranes of Cells; Elsevie; r, 2016; pp 189–218. [Google Scholar]
- (32).Presti FT The Role of Cholesterol in Regulating Membrane Fluidity In Membrane Fluidity in Biology; Aloia RC, Boggs JM, Eds.; Academic Press: Orlando, 1985; pp 97–146. [Google Scholar]
- (33).Finegold L, Ed. Cholesterol in Membrane Models; CRC Press: Boca Raton, 1993. [Google Scholar]
- (34).Mouritsen OG; Jørgensen K. Dynamical Order and Disorder in Lipid Bilayers. Chem. Phys. Lipids 1994, 73, 3–25. [DOI] [PubMed] [Google Scholar]
- (35).Khelashvili G; Pabst G; Harries D. Cholesterol Orientation and Tilt Modulus in DMPC Bilayers. J. Phys. Chem. B 2010, 114, 7524–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kucerka N; Nieh M-P; Katsaras J. Fluid Phase Lipid Areaš and Bilayer Thicknesses of Commonly Used Phosphatidylcholines as a Function of Temperature. Biochim. Biophys. Acta, Bioenerg 2011, 1808, 2761–2771. [DOI] [PubMed] [Google Scholar]
- (37).Henriksen J; Rowat AC; Brief E; Hsueh YW; Thewalt JL; Zuckermann MJ; Ipsen JH Universal Behavior of Membranes with Sterols. Biophys. J 2006, 90, 1639–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Smaby JM; Momsen MM; Brockman HL; Brown RE Phosphatidylcholine Acyl Unsaturation Modulates the Decrease in Interfacial Elasticity Induced by Cholesterol. Biophys. J 1997, 73, 1492–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Rog T; Pasenkiewicz-Gierula M. Cholesterol Effects on the Phosphatidylcholine Bilayer Nonpolar Region: A Molecular Simulation Study. Biophys. J 2001, 81, 2190–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Jedlovszky P; Mezei M. Effect of Cholesterol on the Properties of Phospholipid Membranes. 1. Structural Features. J. Phys. Chem. B 2003, 107, 5311–5321. [DOI] [PubMed] [Google Scholar]
- (41).Marsh D; Smith ICP Interacting Spin Labels as Probes of Molecular Separation within Phospholipid Bilayers. Biochem. Biophys. Res. Commun 1972, 49, 916–922. [DOI] [PubMed] [Google Scholar]
- (42).Kusumi A; Tsuda M; Akino T; Ohnishi S; Terayama Y. Protein-Phospholipid-Cholesterol Interaction in the Photolysis of Invertebrate Rhodopsin. Biochemistry 1983, 22, 1165–1170. [DOI] [PubMed] [Google Scholar]
- (43).Lingwood D; Simons K. Lipid Rafts as a Membrane-Organizing Principle. Science 2010, 327, 46–50. [DOI] [PubMed] [Google Scholar]
- (44).Simons K; Ikonen E. Functional Rafts in Cell Membranes. Nature 1997, 387, 569–572. [DOI] [PubMed] [Google Scholar]
- (45).Veatch SL; Keller SL Seeing Spots: Complex Phase Behavior in Simple Membranes. Biochim. Biophys. Acta, Mol. Cell Res. 2005, 1746, 172–185. [DOI] [PubMed] [Google Scholar]
- (46).Ahmed S; Nikolov Z; Wunder SL Effect of Curvature on Nanoparticle Supported Lipid Bilayers Investigated by Raman Spectroscopy. J. Phys. Chem. B 2011, 115, 13181–13190. [DOI] [PubMed] [Google Scholar]
- (47).Holz M; Heil SR; Sacco A. Temperature-Dependent SelfDiffusion Coefficients of Water and Six Selected Molecular Liquids for Calibration in Accurate 1H NMR PFG Measurements. Phys. Chem. Chem. Phys 2000, 2, 4740–4742. [Google Scholar]
- (48).Li Z; Johnson LM; Ricarte RG; Yao LJ; Hillmyer MA; Bates FS; Lodge TP Enhanced Performance of Blended Polymer Excipients in Delivering a Hydrophobic Drug through the Synergistic Action of Micelles and HPMCAS. Langmuir 2017, 33, 2837–2848. [DOI] [PubMed] [Google Scholar]
- (49).Li Z; Lenk TI; Yao LJ; Bates FS; Lodge TP Maintaining Hydrophobic Drug Supersaturation in a Micelle Corona Reservoir. Macromolecules 2018, 51, 540–551. [Google Scholar]
- (50).So S; Yao LJ; Lodge TP Permeability of Rubbery and Glassy Membranes of Ionic Liquid Filled Polymersome Nanoreactors in Water. J. Phys. Chem. B 2015, 119, 15054–15062. [DOI] [PubMed] [Google Scholar]
- (51).So S; Lodge TP Rate of Molecular Exchange through the Membranes of Ionic Liquid Filled Polymersomes Dispersed in Water. J. Phys. Chem. C 2014, 118, 21140–21147. [Google Scholar]
- (52).Leson A; Hauschild S; Rank A; Neub A; Schubert R; Förster, S.; Mayer, C. Molecular Exchange through Membranes of Poly(2-Vinylpyridine-Block-Ethylene Oxide) Vesicles. Small 2007, 3, 1074–1083. [DOI] [PubMed] [Google Scholar]
- (53).Ding J; Price C; Booth C. Use of Crown Ether in the Anionic Polymerization of Propylene oxide—1. Rate of Polymerization. Eur. Polym. J 1991, 27, 891–894. [Google Scholar]
- (54).Ndoni S; Papadakis CM; Bates FS; Almdal K. Laboratory-scale Setup for Anionic Polymerization under Inert Atmosphere. Rev. Sci. Instrum 1995, 66, 1090–1095. [Google Scholar]
- (55).Hillmyer MA; Bates FS Synthesis and Characterization of Model Polyalkane−Poly(ethylene Oxide) Block Copolymers. Macromolecules 1996, 29, 6994–7002. [Google Scholar]
- (56).Van Krevelen DW; Te Nijenhuis K. Properties of Polymers: Their Correlation with Chemical Structure; Their Numerical Estimation and Prediction from Additive Group Contributions, 4th ed; Elsevier, 2009. [Google Scholar]
- (57).Brandrup J; Immergut EH; Grulke EA Polymer Handbook, 4th ed; Wiley, 1999. [Google Scholar]
- (58).Olson F; Hunt CAA; Szoka FCC; Vail WJJ; Papahadjopoulos D. Preparation of Liposomes of Defined Size Distribution by Extrusion through Polycarbonate Membranes. Biochim. Biophys. Acta, Bioenerg 1979, 557, 9–23. [DOI] [PubMed] [Google Scholar]
- (59).MacDonald RC; MacDonald RI; Menco BPM; Takeshita K; Subbarao NK; Hu LR Small-Volume Extrusion Apparatus for Preparation of Large, Unilamellar Vesicles. Biochim. Biophys. Acta 1991, 1061, 297–303. [DOI] [PubMed] [Google Scholar]
- (60).Hope MJ; Bally MB; Webb G; Cullis PR Production of Large Unilamellar Vesicles by a Rapid Extrusion Procedure. Characterization of Size Distribution, Trapped Volume and Ability to Maintain a Membrane Potential. Biochim. Biophys. Acta, Bioenerg 1985, 812, 55–65. [DOI] [PubMed] [Google Scholar]
- (61).Wu DH; Che AD; Johnso CS An Improved DiffusionOrdered Spectroscopy Experiment Incorporating Bipolar-Gradient Pulses. J. Magn. Reson., Ser. A 1995, 115, 260–264. [Google Scholar]
- (62).Batrakova E; Lee S; Li S; Venne A; Alakhov V; Kabanov A. Fundamental Relationships between the Composition of Pluronic Block Copolymers and Their Hypersensitization Effect in MDR Cancer Cells. Pharm. Res 1999, 16, 1373–1379. [DOI] [PubMed] [Google Scholar]
- (63).Alexandridis P; Holzwarth JF; Hatton TA Micellization of Poly(ethylene Oxide)-Poly(propylene Oxide)-Poly(ethylene Oxide) Triblock Copolymers in Aqueous Solutions: Thermodynamics of Copolymer Association. Macromolecules 1994, 27, 2414–2425. [Google Scholar]
- (64).Arriaga LR; Lopez-Montero I; Monroy F; Orts-Gil G; Farago B; Hellweg T Stiffening Effect of Cholesterol on Disordered Lipid Phases: A Combined Neutron Spin Echo + Dynamic Light Scattering Analysis of the Bending Elasticity of Large Unilamellar Vesicles. Biophys. J 2009, 96, 3629–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Coldren B; Van Zanten R; Mackel MJ; Zasadzinski JA; Jung HT From Vesicle Size Distributions to Bilayer Elasticity via Cryo-Transmission and Freeze-Fracture Electron Microscopy. Langmuir 2003, 19, 5632–5639. [Google Scholar]
- (66).Pan J; Mills TT; Tristram-Nagle S; Nagle JF Cholesterol Perturbs Lipid Bilayers Nonuniversally. Phys. Rev. Lett 2008, 100, No. 198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Saad HY; Higuchi WI Water Solubility of Cholesterol. J. Pharm. Sci 1965, 54, 1205–1206. [DOI] [PubMed] [Google Scholar]
- (68).Ralston AW; Hoerr CW The Solubilities of the Normal Saturated Fatty Acids. J. Org. Chem 1942, 07, 546–555. [DOI] [PubMed] [Google Scholar]
- (69).Krylova OO; Pohl P. Ionophoric Activity of Pluronic Block Copolymers. Biochemistry 2004, 43, 3696–3703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.