Abstract
Objectives
This review fills the paucity of information on K. pneumoniae as a nosocomial pathogen by providing pooled data on epidemiological risk factors, resistant trends and profiles and resistant and virulent genes of this organism in Asia.
Methods
Exhaustive search was conducted using PubMed, Web of Science, and Google scholar for most studies addressing the prevalence, risk factors, drug resistant-mediated genes and/or virulent factors of K. pneumoniae in Asia. Data extracted for meta-analysis were analyzed using comprehensive meta-analysis version 3. Trends data for the isolation rate and resistance rates were entered into Excel spread sheet and the results were presented in graphs.
Results
The prevalence rate of drug resistance in K. pneumoniae were; amikacin (40.8%) [95% CI 31.9–50.4], aztreonam (73.3%) [95% CI 59.9–83.4], ceftazidime (75.7%) [95% CI 65.4–83.6], ciprofloxacin (59.8%) [95% CI 48.6–70.1], colistin (2.9%) [95% CI 1.8–4.4], cefotaxime (79.2%) [95% CI 68.0–87.2], cefepime (72.6) [95% CI 57.7–83.8] and imipenem (65.6%) [95% CI 30.8–89.0]. TEM (39.5%) [95% CI 15.4–70.1], SHV-11 (41.8%) [95% CI 16.2–72.6] and KPC-2 (14.6%) [95% CI 6.0–31.4] were some of the resistance mediated genes observed in this study. The most virulent factors utilized by K. pneumoniae are; hypermucoviscous phenotype and mucoviscosity-related genes, genes for biosynthesis of lipopolysaccharide, iron uptake and transport genes and finally, adhesive genes.
Conclusion
It can be concluded that, antimicrobial resistant in K. pneumoniae is a clear and present danger in Asia which needs strong surveillance to curb this menace. It is very important for public healthcare departments to monitor and report changes in antimicrobial-resistant isolates.
Keywords: Klebsiella pneumoniae, Antibiotic resistance profiles, Resistant genes, Virulent genes, Risk factors, Asia
Background
The problem of antibiotic resistance has become an albatross on the neck of clinicians, veterinarians and other infection control agents in their quest to treat and prevent infections caused by microorganisms that were once thought to have been eradicated with antimicrobials. These organisms or superbugs are returning in new forms resistant to almost all clinically important antimicrobials. Unfortunately, the pharmaceutical pipeline merely does not have enough new medicines to maintain pace with drug-resistant bacterial infections [1]. Klebsiella pneumoniae is one of such clinically significant organisms that have acquired much public health concern. Klebsiella pneumoniae is a significant Enterobacteriaceae considered as one of the opportunistic pathogens causing broad spectra of diseases and showing increasingly frequent acquisition of resistance to antibiotics.
According to Shiri et al. [2], this organism accounts for about one-third of all Gram-negative infections such as urinary tract infections, cystitis, pneumonia, surgical wound infections, endocarditis and septicemia. It also causes necrotizing pneumonia, pyogenic liver abscesses and endogenous endophthalmitis [3]. High mortality rates, extended hospitalization, coupled with high cost are often associated with infections caused by this organism [4]. The drastic rise in the incidence of multidrug-resistant (MDR) and extremely drug-resistant (XDR) pathogens belonging to the Enterobacteriaceae group is a major economic problem as these pathogens are prevalent natural residents of human and animal microbiome. Despites its numerous clinical importance, there is still paucity of information on K. pneumoniae.
This review was therefore designed to determine the antibiotic-resistant profiles of Klebsiella pneumonia as a nosocomial pathogen and focuses on some differences between classical and non-classical subtypes, antimicrobial resistance-mediated genes, some virulent factors of this organism, and some epidemiological risk factors through a systematic review and meta-analysis. This review also looked at some trends in the isolation and resistance rates of K. pneumoniae using China as the target country.
Methods
Search strategies
Exhaustive search was conducted using PubMed, Web of Science, and Google scholar for most studies addressing the prevalence and/or the molecular epidemiology of drug resistant strains of K. pneumoniae in some selected countries in Asia. The search filtered articles among the years of 2005 to 2019. The applied keywords included Klebsiella pneumoniae, antibiotic resistance, resistant genes, virulent genes, epidemiological risk factors and Asia.
Inclusion and exclusion criteria
The original published articles on the prevalence of drug resistant strains of K. pneumoniae from hospital-acquired infections in some selected countries in Asia were considered. Before an article will be considered useful to this study, its antibiotic susceptibility testing should use reference standard methods and recommendations by the Clinical and Laboratory Standards Institute (CLSI) for drug susceptibility testing of K. pneumoniae against most commonly used antimicrobial agents. Due to the following reasons, some studies were excluded from this studies; articles not following CLSI recommended drug susceptibility testing methods, case reports, meta analyses or systematic reviews, letters to editor, review articles, non-English, and duplicate publication.
Data extraction
For the meta-analysis, information extracted from each article were authors’ name, the publication time, year of study, number of samples, antimicrobial drug resistant profiles of K. pneumoniae, resistance-mediated genes, and virulent genes.
Statistical analysis
The comprehensive Meta-analysis software version 3 was used to analyze the data. Because of the heterogeneity among studies, random effects models was used and tested with the Cochrane Q test. For trends in antimicrobial resistance and isolation rates, data were entered into excel spread sheet and graphs were plotted based on the available data.
Results
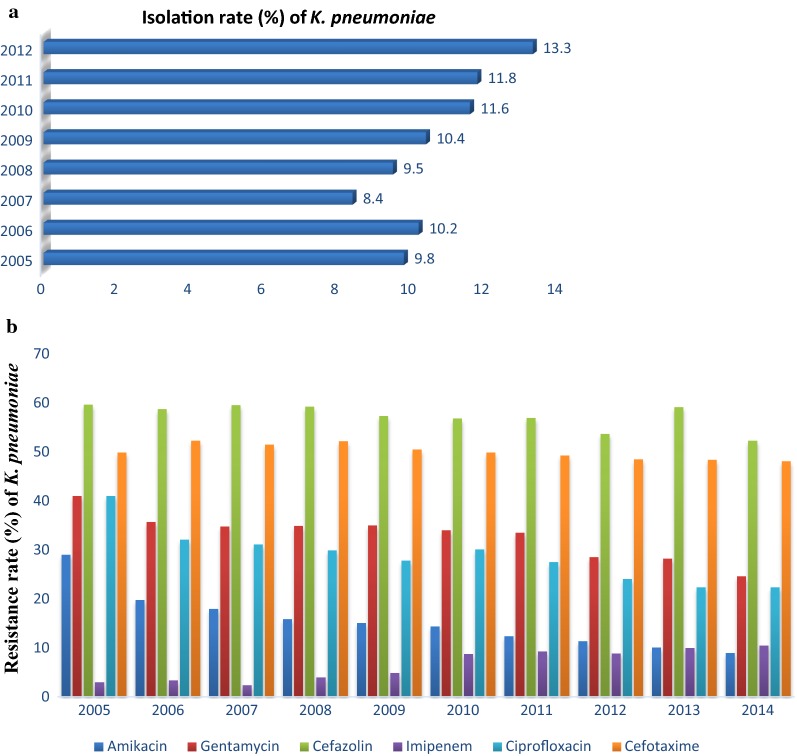
From the literature search, a total of 143 studies or articles were screened from PubMed, Web of Science, and Google scholar. After the removal of duplicates and evaluation of titles and abstract, 64 full text scripts were evaluated. After secondary full text evaluation, 20 studies or articles (Table 1) addressing the prevalence, drug resistant-mediated genes and/or virulent factors in K. pneumoniae in Asia were selected for the final meta-analysis. From Fig. 1, it can be seen that there is an increasing trend in the isolation rate of K. pneumoniae (from 9.8% in 2005 to 13.3% in 2012) in China. Interestingly, the isolation rate decreased in 2007 but increased in the preceding years. In contrast, the resistant trends of K. pneumoniae in China was not congruent to the isolation rate as there were decreasing resistance trends from 2005 to 2014. Imipenem recorded the lowest resistance rate but its resistance trends tend to increase steadily from 2005 to 2014. From Table 2, it can be seen that K. pneumoniae has a great resistance rate to most of the commonly used antimicrobials. Cefotaxime recorded the highest prevalence (79.2%) followed by aztreonam (73.3%) and cefepime (72.6%). Colistin recorded the lowest resistance rate of 2.9%. The individual studies as seen in Table 3, details the number of isolates which were resistant to these antimicrobials. These numbers in terms of percentage resistance to the various antimicrobials ranges from ‘’no isolate’’ (0%) to ‘’all isolates’’ (100%). From Table 4, it can be seen that K. pneumoniae harbor some genes that confers most of its resistance properties. In this review, the selected resistant-mediated genes were in the decreasing order of CTX-M-1 (41.9%), SHV-11 (41.8%), TEM (39.5%), CTX-M-15 (35.3%), KPC-2 (14.6%) and NDM-1 (6.7%). The genes, Mag, Armp, ArmpA2, allS (hypermucoviscous phenotype and mucoviscosity-related genes); wabG, uge, wcaG (biosynthesis of lipopolysaccharide genes); iutA, icuA, iroN, iroB, ybtA, irp2, kfu, entB (iron uptake and transport genes) and Cf29a, fimH, mrkD (Adhesion genes) are all some virulent factors that are used by K. pneumoniae to cause various harm or infections (Table 5).
Table 1.
Details of articles included in the meta-analysis
| Ref nos. | Refs | Publication year | Enrolment time | Province/country | No. of K.P isolates (N) | Type of K.P strain used (n) |
|---|---|---|---|---|---|---|
| [30] | Liu et al. | 2019 | 2013–2017 | Anhui/China | 106 | CRKP (106) |
| [39] | Tian et al. | 2018 | 2016–2017 | Shanghai/China | 170 | CRKP (170) |
| [40] | Zhao et al. | 2019 | 2015–2016 | Anhui/China | 63 | CRKP (63) |
| [41] | Meng et al. | 2019 | 2014–2015 | Central China | 142 | CRKP (142) |
| [42] | Kim et al. | 2019 | 2016–2017 | South Korea | 579 | KP (579) |
| [43] | Xu et al. | 2019 | 2013–2015 | Dalian/China | 30 | ESBL-P KP (30) |
| [44] | Guo et al. | 2017 | 2009–2014 | Henan/China | 8203 | KP (8203) |
| [29] | Dong et al. | 2018 | 2011–2014 | Beijing/China | 146 | CRKP (52) |
| [45] | Cha et al. | 2018 | 2010–2014 | Seoul/Korea | 260 | ESBL:AmpC-KP (54) |
| [46] | Alizade et al. | 2018 | 2014–2015 | Kerman/Iran | 103 | K.P (103) |
| [47] | Lu et al. | 2018 | 2015–2016 | Sichuan/China | 112 | HvCoR-KP (5) |
| [48] | van Dorp et al. | 2019 | 2016–2017 | Beijing/China | 100 | CRKp (100) |
| [49] | Shanker et al. | 2018 | 2015 | India | HvKP | |
| [50] | Huang et al. | 2018 | 2012–2014 | Taipei/Taiwan China | 63 | HvCR-KP (63) |
| [51] | Abrar et al. | 2019 | 2014–2017 | Lahore/Pakistan | 124 | KP (124) |
| [52] | Mitra et al. | 2019 | 2012–2014 | Kolkata/India | 55 | KP (55) |
| [53] | Gautam et al. | 2019 | 2014–2016 | New Delhi, Chandigarh, Vellore, Puducherry/India | 304 | ESBL-KP and Non-ESBL-KP (304) |
| [54] | Mansury et al. | 2016 | 2012–2013 | Shiraz/Iran | 38 | ESBL KP (38) |
| [55] | Heidary et al. | 2017 | 2013–2014 | Tehran/Iran | 117 | K.P (117) |
| [56] | Ma et al. | 2015 | 2012–2014 | Taiwan China | 760 | CnSKP (760) |
HvCoR-KP, hypervirulent colistin-resistant Klebsiella pneumonia; CRKP, carbapenem-resistant Klebsiella pneumoniae; hvKP, hypervirulent Klebsiella pneumoniae; HvCR-KP, hypervirulent carbapenem-resistant Klebsiella pneumoniae; CnSKP, carbapenem non-susceptible Klebsiella pneumoniae; K.P, Klebsiella pneumoniae; ESBL-P KP, extended spectrum β-lactamase producing Klebsiella pneumoniae
Fig. 1.
Trends of resistance and isolation rate of K. pneumoniae in China. a The isolation rate of K. pneumoniae in China (Data extracted from CHINET surveillance system 2015 which collected data from outpatients and inpatients in 19 big hospitals from 14 provinces. b The resistance rate of K. pneumoniae commonly used antimicrobials in China (Data extracted from CHINET surveillance system 2015 which collected data from outpatients and inpatients in 19 big hospitals from 14 provinces for a period of 10-year [57]
Table 2.
Overall resistance rate of Klebsiella pneumoniae to various antimicrobials
| Subgroups | No. of events/studies | Prevalence of drug resistance, % (CI) | Heterogeneity test | |
|---|---|---|---|---|
| I2 (%) | p value | |||
| Overall resistant to amikacin | 11 | 40.8 (31.9–50.4) | 93.7 | < 0.001 |
| Overall resistant to aztreonam | 9 | 73.3 (59.9–83.4) | 97.4 | < 0.001 |
| Overall resistant to ceftazidime | 10 | 75.7 (65.4–83.6) | 90.5 | < 0.001 |
| Overall resistant to ciprpfloxacin | 11 | 59.8 (48.6–70.1) | 96.4 | < 0.001 |
| Overall resistant to colistin | 5 | 2.9 (1.8–4.4) | 0.0 | 0.5250 |
| Overall resistant to cefotaxime | 8 | 79.2 (68.0–87.2) | 93.8 | < 0.001 |
| Overall resistant to cefepime | 8 | 72.6 (57.7–83.8) | 96.9 | < 0.001 |
| Overall resistant to gentamicin | 9 | 58 (49.2–66.3) | 89.7 | < 0.001 |
| Overall resistant to imipenem | 10 | 65.6 (30.8–89.0) | 99.6 | < 0.001 |
| Overall resistant to levofloxacin | 6 | 54.1 (36.0–71.2) | 92.7 | < 0.001 |
| Overall resistant to meropenem | 11 | 62.7 (31.1–86.2) | 99.5 | < 0.001 |
| Overall resistant to trimethoprim_sulfamethoxazole | 7 | 58.2 (35.5–77.9) | 98.8 | < 0.001 |
CI, confidence interval; n, number of events (drug resistance); N, total number of Klebsiella pneumoniae from the included studies
Table 3.
The prevalence of antimicrobial drug resistance among Klebsiella pneumoniae isolates according to individual studies
| References | N | AMK | ATM | CAZ | CIP | COL | CTX | FEP | GEN | IPM | LVX | MEM | SXT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liu et al. [30] | 106 | 33 | 49 | 106 | 49 | 0 | – | 92 | 33 | 106 | 49 | 106 | 16 |
| Tian et al. [39] | 170 | 99 | 161 | 170 | 107 | 5 | 170 | – | 105 | 163 | 105 | 162 | 139 |
| Zhao et al. [40] | 63 | 48 | 61 | 62 | 59 | – | – | 58 | 50 | 62 | 56 | 63 | – |
| Meng et al. [41] | 142 | 45 | 89 | 78 | 67 | – | – | 81 | 69 | – | 60 | – | 42 |
| Kim et al. [42] | 579 | – | 139 | – | 141 | – | – | – | – | – | – | – | 131 |
| Xu et al. [43] | 30 | 16 | – | 29 | 26 | – | 28 | 24 | – | – | 23 | – | – |
| Guo et al. [44] | 8203 | 2568 | 4323 | 4077 | 4184 | – | 6005 | 2887 | 4651 | 468 | – | 476 | 5357 |
| Dong et al. [29] | 52 | 3 | 47 | 52 | 8 | 0 | – | 51 | 37 | 46 | 4 | 49 | 47 |
| Cha et al. [45] | 54 | – | – | 53 | 51 | – | 52 | 34 | – | 13 | – | 7 | – |
| Mitra et al. [52] | 55 | 45 | 50 | – | 52 | – | 53 | – | 52 | – | – | 29 | 49 |
| Gautam et al. [53] | 304 | 109 | – | 159 | 115 | 8 | 157 | 155 | – | 103 | – | 120 | – |
| Mansury et al. [54] | 38 | 6 | – | – | – | – | 19 | – | 16 | 6 | – | 4 | – |
| Heidary et al. [55] | 117 | 40 | 75 | 73 | – | 5 | 77 | – | 51 | 28 | – | 28 | – |
| Ma et al. [56] | 760 | – | – | – | – | – | – | – | – | 568 | – | 519 | – |
| Sum | 3012 | 4994 | 4859 | 4859 | 18 | 6561 | 3382 | 5064 | 1563 | 297 | 1563 | 5781 | |
| Rate (%) | 40.8 | 73.3 | 75.7 | 59.8 | 2.9 | 79.2 | 72.6 | 58 | 65.6 | 54.1 | 62.7 | 58.2 |
AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; COL, colistin; CTX, cefotaxime; FEP, cefepime; GEN, gentamicin; IPM, imipenem; LVX, levofloxacin; MEM, meropenem; SXT, trimethoprim-sulfamethoxazole
Table 4.
The prevalence of some resistant-mediated genes among Klebsiella pneumoniae isolates
| Subgroups | No. of events/studies | Prevalence of resistant gene % (CI) |
n/N | Heterogeneity Test | |
|---|---|---|---|---|---|
| I2 (%) | P-value | ||||
| Overall prevalence of TEM genes | 5 | 39.5 (15.4–70.1) | 241/484 | 96.6 | < 0.001 |
| Overall prevalence of SHV-11 genes | 7 | 41.8 (16.2–72.6) | 350/1117 | 98.1 | < 0.001 |
|
Overall prevalence of CTX-M-1 Genes |
5 | 41.9 (21.6–65.4) | 229/826 | 95.7 | < 0.001 |
| Overall prevalence of CTX-M-15 genes | 5 | 35.3 (17.1–58.9) | 153/548 | 95.0 | < 0.001 |
| Overall prevalence of KPC-2 genes | 6 | 14.6 (6.0–31.4) | 301/2031 | 97.1 | < 0.001 |
| Overall prevalence of NDM-1 genes | 5 | 6.7 (1.7–23.4) | 108/1452 | 95.6 | < 0.001 |
CI, confidence interval; n, number of events (resistance-mediated genes); N, total number of Klebsiella pneumoniae from the included studies
Table 5.
Some selected virulent factors encoded by different strains of Klebsiella pneumoniae
| Virulence factors | References |
|---|---|
|
Hypermucoviscous phenotype and mucoviscosity-related genes magA rmpA rmpA2 allS |
[30, 42, 47–50] |
|
Biosynthesis of lipopolysaccharide wabG uge wcaG |
[42] |
|
Iron uptake and transport iutA icuA iroN iroB ybtA irp2 kfu entB |
[30, 42, 47–50] |
|
Adhesion Cf29a fimH mrkD |
[42, 47, 49, 50] |
Klebsiella pneumoniae: the classical and other subtypes
Klebsiella pneumoniae can be broadly classified into two subtypes; classical Klebsiella pneumoniae (cKp) and non-classical Klebsiella pneumoniae (ncKp). The antimicrobial resistance profiles and the virulence profiles of these strains vary with the former tagged as notorious [3, 5]. Notwithstanding, several clones of these ncKp have also been implicated in causing severe and difficult to threat infections due to their continuous mutation and the acquisition of plasmids and transposons which carries resistant and virulent genes. This has led to the emergences of strains such as hypervirulent Klebsiella pneumoniae (hvKp) or hypermucoviscous Klebsiella pneumoniae (HMKP). This strain was first identified in Eastern part of Asia and has since spread worldwide [6]. This subtype is non-resistant to most of the commonly used antimicrobials such as colistin and carbapenems. But the recent reports of carbapenem-resistant hvKp strains which belong to the sequence types 11 (ST11) [7], ST25 and ST65 [8] poses a major clinical concern.
HvKp strains can cause serious infections in both immunocompetent, diseased and healthy young individuals [9]. This hvKp is known to habour (i) sidephore; predominant of which is aerobactin which is concomitant with hypermucoviscosity, (ii) virulent factors such as; K1, K2, K20 capsular types, rmpA and rmpA2 mucoid-regulator genes [10]. The horizontal transfer of these plasmids and transposons has led to the multidrug resistance (MDR) and the extremely drug resistance (XDR) nature of most of these subtypes. The high prevalence rate of MDR and XDR K. pneumoniae subtypes reflects a multifactorial dissemination processes that include but not limited to: the spread of high risk global multi-resistant genetic lineage [11]; acquisition of successful multi-resistant plasmids; and acquisition of resistant genes located on successful transposons. Klebsiella is a major source of carbapenem resistance worldwide by the dissemination of its plasmids which is facilitated by high genetic transfer (HGT) to other species. Spread of these extended-spectrum β-lactamase (ESBL) and Carbapenemase-encoding plasmids poses a major threat, as acquisition of these plasmids turn bacteria into MDR and XDR. In China, the most dominant MDR KPC-producing clone is the ST11. Once the blaKPC-2 gene is introduces into a certain location, especially in a hospital setup, under antibiotic selection pressure, further dissemination of this gene may occur which may lead to MDR and XDR strains. Hypermucoviscous K. pneumoniae (HMKP) strain types which are sporadically distributed in Asia and the Middle East are the NDM-producing isolates [12, 13]. Liu et al. [14], had reported the first outbreak of CR-HMKP strains which harbored blaNDM-1 gene, shared the same pulsotypes (PT) and belong to the same Sequence types (ST).
Epidemiological risk factors associated with K. pneumoniae colonization and infections
It is believed that several factors can cause the colonization of K. pneumoniae in a community as well as in a hospital setting. The cases of K. pneumoniae infections vary from country to country. In a study by Ling et al. [15], it was reported that, Chinese people had a colonization rate of 66.0% compared to Malay (14.3%), Indian (7.9%) and others nationals (11.8%). This is an indication that infections caused by K. pneumoniae can be locality specific although there can be some form of intercontinental similarities.
Some epidemiological risk factors associated with K. pneumoniae infections may include socio-demographic factors such as gender, age, hospitalization status, source of domestic water (river, rain, well, bottled, piped, boiled), companion animals (cats, dogs, birds), livestock (chicken, ducks, pigs, cow or water buffalo), malnutrition, co-morbidity and the use and misuse of detergents and antiseptics. In Asia, it has been reported that exposure to health care facility and history of previous overseas hospitalization (OR: 33.667; 95% CI 4.539–259.700) is one of the notifiable risk factors associated with K. pneumoniae colonization but this had been disagreed by Ling et al. [15], who iterated that persons with no history of overseas travel and overseas hospitalization are also at risk of K. pneumoniae colonization and infections, hence, suggesting that K. pneumoniae is a persistent organism in our community setting.
Also, admission to an ICU facility (OR: 11.899; 95% CI 4.986–28.399), antimicrobial exposure; particularly carbapenems and fluoroquinolones [14, 16], hematology patients and patients with immunodeficiency are all high risk factors for K. pneumoniae colonization and infections. Carrier levels in hospitalized patients are significantly higher, with reported rates of 77% in the stool, 19% in the pharynx, and 42% on the hands [3]. The higher rates of colonization are primarily related to the increasing use of antibiotics [3, 17, 18]. The increase in colonization rate of K. pneumoniae as observed in these clinical samples is of epidemiological important because, Klebsiella nosocomial infection was four times higher in stool carriers compared with non-carriers [19]. In a study in Taiwan, antibiotic use (e.g. ampicillin or amoxicillin) within the last 30 days was associated with an increased risk of liver abscess [20], suggesting that an increasing exposure to antimicrobials and the last period of antimicrobial administration is a major risk factor for K. pneumoniae colonization and infection.
In a study by Saleem et al. [21], it was reported that some risk factors associated with K. pneumoniae sepsis and mortality in a neonate intensive care units in Pakistan were; extremely low birth weight (p = 0.01, OR 6.1, 95% CI 0.8–44.4), being a male (p = 0.06, OR 9.2, 95% CI 1.3–66.9), severe thrombocytopenia (p = 0.07, OR 3.9, 95% CI 1.2–13.0), and failure to achieve microbiological clearance (p < 0.001, OR 19.6, 95% CI 4.0–98.0). The above listed factors can in combination or singly predispose individuals to K. pneumoniae colonization and infections.
Discussions
Klebsiella pneumoniae is rapidly becoming known for its resistance properties to most of the last-line antibiotics that are usually used. It is especially problematic in hospitals, where it causes a range of acute infections. The increasing trends in the isolation rate of K. pneumoniae is of much concern. Economically developed areas such as China have a more advanced medical system which may increase the chance of exposure to antibiotics and this will increase the possibility of bacterial resistance. In China, the higher population density may also have increased the isolation rate among the population. In this review, although there is an increasing trend in the isolation rate, their resistance rates were not in tandem as this was evident in the decreasing trends over the years. Although, imipenem and meropenem have shown good activity against Enterobacteriaceae [22], the situation observed in this review reiterates the public health implications of K. pneumoniae. In Fig. 1, there is a steady increase in resistance of imipenem over the years and this can be as a result of their increasing use among the populace. Generally, the decreasing resistant rate of K. pneumoniae to most of the antimicrobials in China can be attributed to; (i) the enforcement of taking various actions for prevention of bacterial infection such as separating the pathogen carriers and enforcement of hand sanitization of medical professionals by the government through the Nosocomial Infection Control Committee, (ii) the restriction and control of the use of antibiotics by the Chinese Ministry of Hygiene, which has implemented guidelines for the rational use of antibiotics since 2006.
The global emergence and spread of genes of antimicrobial resistance such as ESBL and carbapenemase genes in K. pneumoniae isolates present a significant danger to public health. This is because carbapenems have long been deemed as the last therapeutic resort or option of antibiotics used to treat diseases and infections caused by multidrug-resistant gram-negative bacteria. The rapid global emergence of K. pneumoniae strains, resistant to almost all β-lactams, including carbapenems as seen in this study shows the organism’s ability to react quickly to selective environmental pressure modifications. The extensive use and misuse of carbapenems is one of the attributable reasons that has led to the evolution of plasmid-mediated carbapenemases, i.e. enzymes that hydrolyze all β-lactams including the last-line carbapenems [23]. Different resistance-mediated genes mediate antimicrobial drug resistance in K. pneumoniae. The high rate of resistance to carbapenems (imipenem and meropenem) observed in this study can be partly be attributed to the presence of some carbapenemase resistant-mediated genes such as blaOXA. bla NDM and blaKPC realized in this study. In K. pneumoniae, the blaKPC genes which confers reduced susceptibility or resistance to nearly all β-lactam antibiotics by various enterobacteria are mostly carried on plasmids. The detection of carbapenemases is important from an epidemiological perspective as they are plasmid-mediated and may be transferred horizontally between different bacterial species [24]. Dissemination of resistant determinants have been recognized as a major challenge in the treatment of bacterial infections worldwide [25]. Also, resistance of K. pneumoniae to cephalosporin (ceftazidime, cefepime and cefotaxime) as seen in this study can also be partly be attributed to the KPC gene because the KPC enzyme hydrolyzes extended-spectrum cephalosporins. This can therefore be used to identify KPC-mediated genes that are resistant to these cephalosporins (e.g. ceftazidime, ceftriaxone, and cefotaxime).
Klebsiella pneumoniae resistance to Aminoglycosides (amikacin and gentamicin) as seen in this review may be as a result of modifications in cell permeability due to alterations in AcrAB-TolC and KpnEF efflux pump systems and due to loss of putative porin, KpnO. Also the disruptions in AcrAB-TolC may increase the susceptibility of K. pneumoniae to gentamicin [26]. The 16S rRNA methylases which are encoded on the plasmids [27] confers resistance to all aminoglycosides. Mutations which confer resistance via target modification can also be a possible attributable reason for the increasing resistance of K. pneumoniae to most aminoglycosides.
The low prevalence of Polymyxin (Colistin) resistance in this review makes a lot of sense because of their restricted use in human medicine dating back between the 1980s and 2000s, due to their recognized toxicity. According to Falagas and Kasiakou [28], Polymyxin works by disrupting the membrane integrity through displacement of cations (Ca2+/Mg2+) in the outer membrane, by binding to the negatively charged lipopolysaccharides (LPS) which leads to cell lysis. Undoubtedly, the presence of resistance determinants will allow K. pneumoniae strains to survive the barrage of antibiotics used in treatment of hospital infections.
According to a report by Dong et al. [29], K. pneumoniae utilize an array of virulence factors to colonize and propagate in a host cell. These include at least (a) surface antigen, particularly capsular polysaccharide (CPS, K antigen); (b) siderophores responsible for binding ferric iron secreted by the host’s iron-binding proteins; and (c) adherence variables responsible for binding to host cell surfaces, such as fimbriae type 1 and type 3, and non-fimbrial adherence proteins.
Hypermucoviscous phenotype and mucoviscosity-related genes, genes for biosynthesis of lipopolysaccharide, iron uptake and transport genes and Adhesive genes are all virulent factors that are employed by K. pneumoniae strains in pathogenesis. Iron is a key component for K. pneumoniae’s survival. As free iron is scarce in host plasma, K. Pneumoniae acquires iron predominantly through the secretion of siderophores; molecules with a greater iron affinity than the host transport proteins [30]. Among the siderophores secreted by K. pneumoniae, aerobactin is considers the most important virulent factor [31, 32], as it can cause severe infection by assisting in the transport of the organism from the intestinal tract to various tissues and also the multiplication of these organisms in the tissues.
The rmpA and rmpA2 are Plasmid-carried genes, which contribute to the enhancement of capsular production. Also, the MagA gene is an important gene used by K. pneumoniae strains to demonstrate an extraordinarily high resistance to human serum and phagocytosis. This gene can be used as a molecular marker for quick diagnosis and can also be useful in tracing the roots of emerging infectious diseases caused by K. pneumoniae. According to Fang et al. [33], the MagA protein could be a good candidate for new drug targets.
It has been shown that fimbriae contribute to diseases of the urinary tract [34], mediates the development of biofilms and also it is involve in the adherence of the organism to medical devices. Therefore, expression of these genes could improve K. pneumoniae’s adhesive ability to respiratory epithelial cells and also to surfaces of other medical devices such as ventilators, thereby, increasing their ability to cause ventilator-associated diseases. Fimbria, according to Huang et al. [35] and Stahlhut et al. [36], may be a major factor for biofilm-associated diseases and host entry abilities of K. pneumoniae.
Limitations
Although, very positive results were obtained from this meta-analysis, it is worth noting that there were a number of limitations with this meta-analysis. This study did not utilized data from abstracts, posters and conference proceedings but only full paper publications. Because of the limited number of studies included in this meta-analysis, this study did not assess for publication bias because according to Terrin and Schmid [37] and Thornton and Lee [38], funnel plots and statistical tests for detection of publication bias are unreliable for limited number of studies.
Conclusions
It can be concluded that antimicrobial resistance in K. pneumoniae is a clear and present danger in Asia which needs strong surveillance to curb this menace. Although low resistance rate to colistin and imipenem were recorded in this review, more cautious efforts should be made to develop new line of antimicrobials as resistance to these drugs are surging. It is very important for public healthcare departments to monitor and report the changes in antimicrobial-resistant isolates. This will guide doctors in prescribing the proper antimicrobials in case of resistance gene evolution. It is essential to design a preparedness plan in order to tackle this public health danger. On the other side, control measures must include a multi-faceted strategy coordinated by the domestic health officials in fields.
Acknowledgements
None.
Authors’ contributions
CYE, TS, SL and YW contributed equally in the preparation and reviewing of the manuscript. All authors read and approved the final manuscript.
Funding
None
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Infectious Diseases Society of America. Bad bugs, no drugs. Infectious Diseases Society of America, Alexandria, VA 2004. http://www.idsociety.org/pa/IDSA_Paper4_final_web.pdf.
- 2.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 3.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giske CG, Monnet DL, Cars O, Carmeli Y. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob Agents Chemother. 2008;52(3):813–821. doi: 10.1128/AAC.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta A. Hospital-acquired infections in the neonatal intensive care unit-Klebsiella pneumoniae. Semin Perinatol. 2002;26(5):340–345. doi: 10.1053/sper.2002.36267. [DOI] [PubMed] [Google Scholar]
- 6.Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, Jeong BC, Lee SH. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;21(7):483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu DX, Huang YL, Ma JH, Zhou HW, Fang Y, Cai JC, Hu YY, Zhang R. Detection of colistin resistance gene mcr-1 in hypervirulent Klebsiella pneumoniae and Escherichia coli isolates from an infant with diarrhea in China. Antimicrob Agents Chemother. 2016;60(8):5099–5100. doi: 10.1128/AAC.00476-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;1(37):107–112. doi: 10.1016/j.ijid.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 9.Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62(1):1–6. doi: 10.1016/j.diagmicrobio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci. 2015;112(27):E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 12.Shankar C, Nabarro LE, Ragupathi NK, Sethuvel DP, Daniel JL, Veeraraghavan B. Draft genome sequences of three hypervirulent carbapenem-resistant Klebsiella pneumoniae isolates from bacteremia. Genome Announc. 2016;4(6):e01081–e01116. doi: 10.1128/genomeA.01081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mei YF, Liu PP, Wan LG, Liu Y, Wang LH, Wei DD, Deng Q, Cao XW. Virulence and genomic feature of a virulent klebsiella pneumoniae sequence type 14 strain of serotype K2 harboring blaNDM–5 in China. Front Microbiol. 2017;23(8):335. doi: 10.3389/fmicb.2017.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu SW, Chang HJ, Chia JH, Kuo AJ, Wu TL, Lee MH. Outcomes and characteristics of ertapenem-nonsusceptible Klebsiella pneumoniae bacteremia at a university hospital in Northern Taiwan: a matched case-control study. J Microbiol Immunol Infect. 2012;45(2):113–119. doi: 10.1016/j.jmii.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 15.Ling ML, Tee YM, Tan SG, Amin IM, How KB, Tan KY, Lee LC. Risk factors for acquisition of carbapenem resistant Enterobacteriaceae in an acute tertiary care hospital in Singapore. Antimicrobial Resist Infect Control. 2015;4(1):26. doi: 10.1186/s13756-015-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kritsotakis EI, Tsioutis C, Roumbelaki M, Christidou A, Gikas A. Antibiotic use and the risk of carbapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae infection in hospitalized patients: results of a double case–control study. J Antimicrob Chemother. 2011;66(6):1383–1391. doi: 10.1093/jac/dkr116. [DOI] [PubMed] [Google Scholar]
- 17.Pollack M, Nieman R, Reinhardt J, Charache P, Jett M, Hardy JRP. Factors influencing colonisation and antibiotic-resistance patterns of gram-negative bacteria in hospital patients. Lancet. 1972;300(7779):668–671. doi: 10.1016/s0140-6736(72)92084-3. [DOI] [PubMed] [Google Scholar]
- 18.Asensio A, Oliver A, González-Diego P, Baquero F, Perez-Diaz JC, Ros P, Cobo J, Palacios M, Lasheras D, Cantón R. Outbreak of a multiresistant Klebsiella pneumoniae strain in an intensive care unit: antibiotic use as risk factor for colonization and infection. Clin Infect Dis. 2000;30(1):55–60. doi: 10.1086/313590. [DOI] [PubMed] [Google Scholar]
- 19.Selden R, Lee S, Wang WL, Bennett JV, Eickhoff TC. Nosocomial Klebsiella infections: intestinal colonization as a reservoir. Ann Intern Med. 1971;74(5):657–664. doi: 10.7326/0003-4819-74-5-657. [DOI] [PubMed] [Google Scholar]
- 20.Lin YT, Liu CJ, Yeh YC, Chen TJ, Fung CP. Ampicillin and amoxicillin use and the risk of Klebsiella pneumoniae liver abscess in Taiwan. J Infect Dis. 2013;208(2):211–217. doi: 10.1093/infdis/jit157. [DOI] [PubMed] [Google Scholar]
- 21.Saleem AF, Qamar FN, Shahzad H, Qadir M, Zaidi AK. Trends in antibiotic susceptibility and incidence of late-onset Klebsiella pneumoniae neonatal sepsis over a 6-year period in a neonatal intensive care unit in Karachi, Pakistan. Int J Infect Dis. 2013;17(11):e961–e965. doi: 10.1016/j.ijid.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Hoban DJ, Bouchillon SK, Hawser SP, Badal RE. Trends in the frequency of multiple drug-resistant Enterobacteriaceae and their susceptibility to ertapenem, imipenem, and other antimicrobial agents: data from the Study for Monitoring Antimicrobial Resistance Trends 2002 to 2007. Diagn Microbiol Infect Dis. 2010;66(1):78–86. doi: 10.1016/j.diagmicrobio.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 23.MerieQueenan A, Bush K. Carbapenemases: the versatile B-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2010;54(1):177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chew KL, Lin RT, Teo JW. Klebsiella pneumoniae in Singapore: hypervirulent infections and the carbapenemase threat. Front Cell Infect Microbiol. 2017;12(7):515. doi: 10.3389/fcimb.2017.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrade LN, Vitali L, Gaspar GG, Bellissimo-Rodrigues F, Martinez R, Darini AL. Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J Clin Microbiol. 2014;52(7):2530–2535. doi: 10.1128/JCM.00088-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galimand M, Courvalin P, Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003;47(8):2565–2571. doi: 10.1128/AAC.47.8.2565-2571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falagas ME, Kasiakou SK, Saravolatz LD. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis. 2005;40(9):1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 29.Dong N, Zhang R, Liu L, Li R, Lin D, Chan EW, Chen S. Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China. Microbial Genom. 2018;4:2. doi: 10.1099/mgen.0.000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Gu Y, Li X, Liu Y, Ye Y, Guan S, Li J. Identification and Characterization 7 of NDM-1-producing hypervirulent (hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med. 2019;39(2):167–175. doi: 10.3343/alm.2019.39.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. 2015;83(8):3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo TA, Olson R, MacDonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun. 2014;82(6):2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang R, Li M, Gregory RL. Bacterial interactions in dental biofilm. Virulence. 2011;2:435–444. doi: 10.4161/viru.2.5.16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahlhut SG, Struve C, Krogfelt KA, Reisner A. Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol Med Microbiol. 2012;65(2):350–359. doi: 10.1111/j.1574-695X.2012.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terrin N, Schmid CH, Lau J. In an empirical evaluation of the funnel plot, researchers could not visually identify publication bias. J Clin Epidemiol. 2005;58(9):894–901. doi: 10.1016/j.jclinepi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Thornton A, Lee P. Publication bias in meta-analysis: its causes and consequences. J Clin Epidemiol. 2000;53(2):207–216. doi: 10.1016/s0895-4356(99)00161-4. [DOI] [PubMed] [Google Scholar]
- 39.Tian D, Pan F, Wang C, Sun Y, Zhang H. Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai. Infection and drug resistance. 2018;11:1935. doi: 10.2147/IDR.S175584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao D, Zuo Y, Wang Z, Li J. Characterize carbapenem-resistant Klebsiella pneumoniae isolates for nosocomial pneumonia and their Gram-negative bacteria neighbors in the respiratory tract. Mol Biol Rep. 2019;46(1):609–616. doi: 10.1007/s11033-018-4515-y. [DOI] [PubMed] [Google Scholar]
- 41.Meng X, Yang J, Duan J, Liu S, Huang X, Wen X, Huang X, Fu C, Li J, Dou Q, Liu Y. Assessing molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae (CR-KP) with MLST and MALDI-TOF in Central China. Sci Rep. 2019;9(1):2271. doi: 10.1038/s41598-018-38295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, Park BY, Choi MH, Yoon EJ, Lee H, Lee KJ, Park YS, Shin JH, Uh Y, Shin KS, Shin JH. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrob Chemother. 2018;74(1):190–199. doi: 10.1093/jac/dky397. [DOI] [PubMed] [Google Scholar]
- 43.Xu H, Huo C, Sun Y, Zhou Y, Xiong Y, Zhao Z, Zhou Q, Sha L, Zhang B, Chen Y. Emergence and molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates harboring blaCTX-M-15 extended-spectrum β-lactamases causing ventilator-associated pneumonia in China. Infect Drug Resist. 2019;12:33. doi: 10.2147/IDR.S189494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo X, Cao Z, Dai Z, Li Y, He X, Hu X, Tian F, Ren Y. Antimicrobial susceptibility and molecular epidemiology of multidrug-resistant Klebsiella pneumoniae in Central China. Jpn J Infect Dis. 2017;70(3):229–234. doi: 10.7883/yoken.JJID.2016.049. [DOI] [PubMed] [Google Scholar]
- 45.Cha MK, Kang CI, Kim SH, Chung DR, Peck KR, Lee NY, Song JH. High prevalence of CTX-M-15-type extended-spectrum β-lactamase among AmpC β-lactamase-producing Klebsiella pneumoniae isolates causing bacteremia in Korea. Microbial Drug Resist. 2018;24(7):1002–1005. doi: 10.1089/mdr.2017.0362. [DOI] [PubMed] [Google Scholar]
- 46.Alizade H, Jajarmi M, Aflatoonian MR, Kalantar-Neyestanaki D, Shoja S, Ghanbarpour R. Comparative prevalence of blaCTX-m-15 gene with virulence genes and serotypes in Klebsiella pneumoniae. Jundishapur J Microbiol. 2018;11:4. [Google Scholar]
- 47.Lu Y, Feng Y, McNally A, Zong Z. The emergence of colistin-resistant hypervirulent Klebsiella pneumoniae in China. Front Microbiol. 2018;9:2568. doi: 10.3389/fmicb.2018.02568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Dorp L, Wang Q, Shaw LP, Acman M, Brynildsrud OB, Eldholm V, Wang R, Gao H, Yin Y, Chen H, Ding C. Rapid phenotypic evolution in multidrug-resistant Klebsiella pneumoniae hospital outbreak strains. Microbial Genom. 2019;5:4. doi: 10.1099/mgen.0.000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shankar C, Veeraraghavan B, Nabarro LE, Ravi R, Ragupathi NK, Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 2018;18(1):6. doi: 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang YH, Chou SH, Liang SW, Ni CE, Lin YT, Huang YW, Yang TC. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73(8):2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 51.Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S. Distribution of bla CTX-M, bla TEM, bla SHV and bla OXA genes in extended-spectrum-β-lactamase-producing clinical isolates: a three-year multi-center study from Lahore, Pakistan. Antimicrobial Resist Infect Control. 2019;8(1):80. doi: 10.1186/s13756-019-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitra S, Mukherjee S, Naha S, Chattopadhyay P, Dutta S, Basu S. Evaluation of co-transfer of plasmid-mediated fluoroquinolone resistance genes and bla NDM gene in Enterobacteriaceae causing neonatal septicaemia. Antimicrobial Resist Infect Control. 2019;8(1):46. doi: 10.1186/s13756-019-0477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vikas Gautam, Thaku Anjana, Sharm Megha, Singh Avinash, Bansal Shruti, Sharma Aditi. Molecular characterization of extended-spectrum β-lactamases among clinical isolates of Escherichia coli & Klebsiella pneumoniae: a multi-centric study from tertiary care hospitals in India. Indian J Med Res. 2019;146:208–215. doi: 10.4103/ijmr.IJMR_172_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mansury D, Motamedifar M, Sarvari J, Shirazi B, Khaledi A. Antibiotic susceptibility pattern and identification of extended spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae from Shiraz, Iran. Iran J Microbiol. 2016;8(1):55. [PMC free article] [PubMed] [Google Scholar]
- 55.Heidary M, Goudarzi H, Hashemi A, Eslami G, Goudarzi M, Chirani AS, Amraei S. Prevalence of quinolone resistance genes in Klebsiella pneumoniae strains isolated from hospitalized patients during 2013–2014. Archiv Pediatr Infect Dis. 2017;5:4. [Google Scholar]
- 56.Ma L, Wang JT, Wu TL, Siu LK, Chuang YC, Lin JC, Lu MC, Lu PL. Emergence of OXA-48-producing Klebsiella pneumoniae in Taiwan. PLoS ONE. 2015;10(9):e0139152. doi: 10.1371/journal.pone.0139152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fupin HU, Zhu D, Wang F, Jiang X, Sun Z, Chen Z, Zhidong HU, Jin LI, Xie Y, Kang M, Yingchun XU. CHINET 2013 surveillance of bacterial resistance in China. Chin J Infect Chemother. 2014;1(5):365–374. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.