Abstract
Histone deacetylase inhibitors (HDACI) are promising anticancer agents and their use in combination with conventional anticancer drugs is currently under investigation. We previously reported cell line–specific upregulation of ABCG2, a multidrug resistance transporter shown to control oral bioavailability and CNS penetration, by the HDACI romidepsin, although the precise mechanism in a particular cell line remains to be determined. The aryl hydrocarbon receptor (AhR) is a ligand-dependent transcription factor that can be activated by numerous environmental contaminants and has been shown to be a client protein of heat shock protein 90 (Hsp90). A xenobiotic response element was defined in the ABCG2 promoter and was shown to mediate AhR signaling. Activated AhR was found to be associated with the ABCG2 promoter only in cell line models that respond to romidepsin with ABCG2 upregulation. Our data suggest that romidepsin acetylated Hsp70 and inhibited the chaperone function of Hsp90, thereby allowing the dissociation of AhR from Hsp90. The dissociation of AhR from Hsp90 may be a prerequisite for the differential upregulation of ABCG2 by romidepsin. Increasing our understanding of the mechanism(s) governing differential upregulation of ABCG2 in response to romidepsin could provide an insight into strategies needed to tackle resistance to HDACIs in cancer therapeutics.
Introduction
ABCG2 is a ubiquitous ATP-binding cassette (ABC) multidrug resistance transporter that plays a significant role in normal tissue protection, stem cell maintenance, and clinical pharmacology (1). Its overexpression confers resistance in cancer cell lines to a variety of cancer chemotherapeutic agents such as mitoxantrone, topotecan, and methotrexate (2–7). However, little is known about the mechanisms underlying its regulation.
Histone deacetylase inhibitors (HDACI) such as romidepsin (also known as FK228 or depsipeptide) and vorinostat (suberoylanilide hydroxamic acid or SAHA) were found to be potent anticancer agents, capable of inducing cell cycle arrest and apoptosis in cancer cells (8). A number of HDACIs are currently in clinical trials as both monotherapy and in combination therapy because these compounds have been shown to potentiate the cytotoxic effects of other anticancer drugs in experimental models. We and others have shown that multidrug resistance transporters (including ABCG2 and MDR1) can be upregulated on treatment with various HDACIs (9–13), potentially hindering drug response in combination therapy. Therefore, a better understanding about the mechanism for activation of these transporters on HDACI treatment could help develop strategies for the prevention of drug resistance.
The aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor that belongs to the basic helix-loop-helix/Per-ARNT-Sim (bHLH-PAS) family (14). It regulates the transcription of genes encoding xenobiotic metabolizing enzymes and also mediates the toxic effects caused by environmental carcinogens such as dioxins and polycyclic aromatic hydrocarbons. In the unliganded state, the AhR is associated with 2 molecules of the chaperone heat shock protein 90 (Hsp90), an immunophilin-like protein XAP2, and the Hsp90-interacting acidic protein p23 in the cytoplasm. Ligand binding initiates a cascade of events leading to AhR translocation to the nucleus, release of Hsp90, and dimerization of AhR with nuclear translocator (Arnt). The ligand-bound AhR–Arnt complex subsequently recognizes and binds to its cognate binding site with a DNA sequence of 5′-TA/TGCGTG-3′ (15), termed the AhR response element (or xenobiotic response element; XRE), located in the promoter region of responsive genes, thereby modulating gene transcription (16). Its ligands include many natural and synthetic compounds, some of which, such as polyhalogenated aromatic hydrocarbons and polycyclic aromatic hydrocarbons, are important environmental contaminants.
Hsp90 is an ATP-dependent molecular chaperone that controls the intracellular trafficking and folding of diverse cellular proteins, particularly those involved in signal transduction, cell cycle regulation, and survival (17, 18). It plays a key role in the conformational maturation of oncogenic signaling proteins including HER-2/ErbB2, Akt, and Raf-1 (19–21). AhR is a known Hsp90 client protein. The association with Hsp90 is required for the AhR to assume a conformation that is optimal for ligand binding (22). Reversible acetylation is an important posttranslational modification of Hsp90 that is known to impair the chaperone function of Hsp90 (23–25). This has been shown to cause dissociation of a number of client proteins such as Her-2 & c-Raf (25, 26), causing their polyubiquitination and proteasomal degradation. Importantly, Hsp90 acetylation has been detected in cells treated with various HDACIs including romidepsin and vorinostat (27, 28). However, in the case of AhR, the effect of HDACI treatment on chaperone association of AhR with Hsp90, as well as the levels of AhR, has not been determined. Moreover, within the nucleus, the precise mechanisms mediating the dissociation of AhR from Hsp90 and the formation of a heterodimer with Arnt is still not fully understood.
In the present study, we sought to evaluate the involvement of the AhR pathway in the activation of ABCG2 by romidepsin. Our data show that romidepsin disrupted the chaperone binding of Hsp90 with AhR through Hsp70 acetylation and subsequently promoted the binding of AhR–Arnt with the XRE element on the ABCG2 promoter. In cell lines that did not respond to romidepsin treatment with ABCG2 upregulation, Hsp70 acetylation was not observed and AhR remained bound to Hsp90, thereby blocking the AhR-mediated ABCG2 upregulation.
Materials and Methods
Tissue culture
Human colon cancer cell line S1, a derivative of LS180, has been described previously (5). MCF-7, SF295, and SW620 cells were chosen and obtained from the National Cancer Institute Tumor Drug Screen for this study. The cell lines were maintained in Iscove’s modified Eagle’s medium (S1 and MCF-7) or RPMI medium (H460, SF295, and SW620) supplemented with 10% FBS, 100 units/mL streptomycin sulfate, and 100 units/mL penicillin G sulfate, and incubated at 37°C in 5% CO2.
Drug treatment
To test the effect of HDAC inhibition on ABCG2 expression, cells were treated with 10 ng/mL of romidepsin (also known as depsipeptide, FR901228, or NSC630176; Developmental Therapeutics Program, National Cancer Institute) supplemented with 5 μg/mL of verapamil (Sigma) for 24 hours. Verapamil was added to prevent the efflux of romidepsin mediated by P-glycoprotein upregulation. Stock solutions of romidepsin and verapamil were dissolved in DMSO and water, respectively. For the study of the AhR pathway in the regulation of ABCG2, benzo(a)pyrene [B(a)P; Sigma] was used as the AhR agonist whereas resveratrol (Sigma), kaempferol (Sigma), and salicylamide (Sigma) were used as AhR antagonists.
RNA interference
To construct a small interference hairpin-loop (sh) silencing vector against the human AhR, 2 complementary oligonucleotides were synthesized, annealed, and liganded into the BglII and EcoRI site of the pU6 vector (a kind gift from Dr Chen Yang Chao, School of Biomedical Sciences, Chinese University of Hong Kong, Hong Kong). The sequences were 5′-GGTTTCAGCAGTCTGATGT-CttcaagagaGACATCAGACTGCTGAAACCCTTTTT-3′ (forward) and 5′-AAAAAGGGTTTCAGCAGTCT-GATGTCtctcttgaaGACATCAGACTGCTGAAACC-3′ (reverse), which contained the oligonucleotide encoding a 20-mer hairpin sequence specific to the human AhR mRNA (designed by using the siRNA Selection Program at the Whitehead Institute for Biomedical Research (http://jura.wi.mit.edu/siRNAext/; ref. 29), a ttcaagaga loop sequence separating the 2 complements, and a TTTTT terminator at the 3′-end. The various cell lines were transfected with the backbone vector (pU6) or the shRNA vector specific against AhR (pU6-AhR) by using Lipofectamine 2000 (Invitrogen) before evaluation for ABCG2 regulation by romidepsin. Silencing efficiency was assessed at the protein level by immunoblot analysis.
Reverse transcription and real-time PCR
Total RNA was isolated by using the Trizol regent (Invitrogen). RNA (1 μg) was reverse transcribed by using Tetro reverse transcriptase (Bioline). Quantitative real-time PCR was performed to quantify the change of ABCG2, MDR1, p21, and CYP1A1 transcript expressions by using the KAPA SYBR FAST qPCR Kit (Kapa-Biosystems) in a LightCycler 480 Instrument I (Roche Applied Science). The human glyceraldehyde 3 phosphate dehydrogenase (GAPDH) RNA was amplified in parallel as the internal control. Sequences of the specific primers used are listed in Table 1. PCRs were performed at 95°C for 10 minutes, followed by 50 cycles of 95°C for 10 seconds and 60°C for 10 seconds. Fluorescent signal was acquired at the end of the elongation step of every PCR cycle (72°C for 10 seconds) to monitor the increasing amount of amplified DNA. ΔCt was calculated by subtracting the Ct of GAPDH from the Ct of ABCG2, MDR1, p21, or CYP1A1. ΔΔCt was then calculated by subtracting the ΔCt of the parental cells from the ΔCt of the corresponding resistant cells. Fold change of gene expression was calculated by the equation 2−ΔΔCt.
Table 1.
Sequence of the primers used for real-time PCR analysis
| Gene | Primer sequence |
|---|---|
| ABCG2 | Forward: 5′-CAATGGGATCATGAAACCTG-3′ Reverse: 5′-GAGGCTGATGAATGGAGAA-3′ |
| MDR1 | Forward: 5′-CTGTCAGTGTATTTTCAATGTTTCG-3′ Reverse: 5′-AATTTTGTCACCAATTCCTTCATTA-3′ |
| p21 | Forward: 5′-GAGCGATGGAACTTCGACTTT-3′ Reverse: 5′-GGGCTTCCTCTTGGAGAAGAT-3′ |
| CYP1A1 | Forward: 5′-CCATGACCAGAAGCTATGGGT-3′ Reverse: 5′-GCTCTCAAGCACCTAAGAGCG-3′ |
| GAPDH | Forward: 5′-ACCACAGTCCATGCCATCAC-3′ Reverse: 5′-TCCACCACCCTGTTGCTGTA-3′ |
Luciferase reporter assays
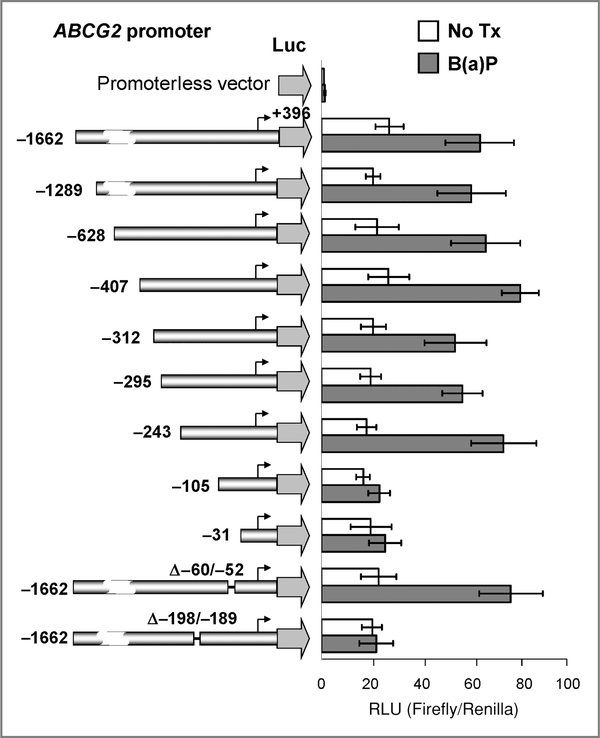
A series of human ABCG2 promoter constructs with progressive deletions at the 5′-end has been previously described (30). Two new reporter constructs were prepared with the putative XRE sequences deleted from the full-length ABCG2 promoter (Fig. 2; Δ−60/−52 and Δ−198/−189). The ABCG2 promoter/firefly luciferase fusion genes (200 ng DNA) were transfected in S1 cells by using Fugene 6 (Roche). The pGL3-Basic (promoterless) plasmid, encoding firefly luciferase (Promega), was used to determine the basal levels. In each experiment, the phRG-Basic plasmid (50 ng), encoding Renilla luciferase (Promega), was cotransfected for normalization purposes. Luminescence was measured 48 hours after transfection by the Dual-Luciferase Reporter Assay System (Promega). In experiments involving drug treatment, the drugs were added 24 hours after transfection. Reporter activity was normalized by calculating the ratio of firefly/Renilla values. Results were expressed as mean ± SD of duplicate measurements from 3 independent transfections.
Figure 2.
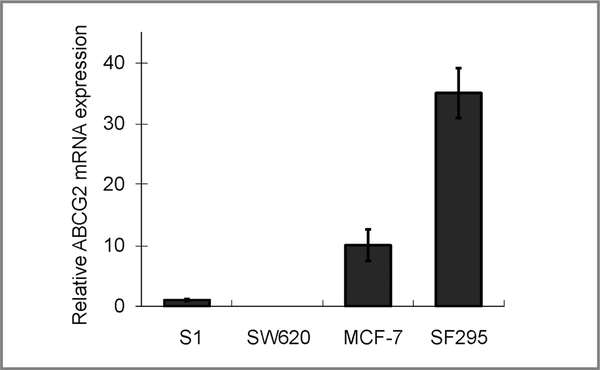
Real-time PCR analysis comparing the basal ABCG2 mRNA expression in the various cell lines used in this study (S1, SW620, MCF-7, and SF295). Relative ABCG2 transcript expression was shown after normalization with GAPDH (the level in S1cells was set as 1 for comparison). The level in SW620 was found to be nearly undetectable.
Western blot analysis
AhR repressor (AhRR) protein expression was examined in S1, MCF-7, HepG2, SW620, A549, SF295, and HeLa cells treated with or without romidepsin or B(a)P. Cells were lysed in buffer containing 0.2 mol/L Tris (pH 7.4), 1% Triton X-100, 0.02% β-mercaptoethanol, and complete mini EDTA-free protease inhibitor cocktail tablets (Roche), followed by sonication for homogenization. Whole cell lysates prepared were separated by SDS-PAGE and subjected to immunoblot analysis with the respective antibodies for AhRR (ab85666; Abcam) and GAPDH (American Research Products). The antigen–antibody complex was analyzed with an Odyssey (Li-Cor) infrared imaging system after incubation with a 1:10,000 dilution of goat anti-mouse (IRDye800CW) or anti-rabbit (IRDye680) secondary antibody (Li-Cor).
Coimmunoprecipitation
For coimmunoprecipitation experiments, cells were lysed in IP buffer (50 mmol/L Tris-HCl, pH 8.0; 150 mmol/L NaCl; 1% Triton X-100; and protease inhibitor cocktail tablets), homogenized by sonication, and centrifuged at 13,000 rpm for 2 minutes at 4°C. Lysates (500 μg) were precleared for 1 hour at 4°C then incubated with rabbit polyclonal anti-Hsp90 (SPA-846; Stressgen), rabbit polyclonal anti-acetyl lysine antibody (ab21623; Abcam) or mouse monoclonal anti-AhR (RPT9; Abcam) antibody overnight at 4°C. Protein and antibody were then incubated with ImmunoPure Plus Immunobilized Protein A (Pierce) for 2 hours at 4°C. Samples were washed 5 times with 500 μL of IP buffer. The Protein A pellet was resuspended in 35 μL of 1 × sample loading buffer and heated at 100°C for 5 minutes before being resolved by 12% SDS–polyacrylamide gels and immunoblotted with mouse monoclonal antibodies for Hsp90 (AC88; Abcam), Hsp70 (5A5; Abcam), AhR (RPT1; Abcam), or Bcl-2 (100/D5; Abcam). Results were determined by chemiluminescence detection with an Odyssey (Li-Cor) infrared imaging system as described earlier.
ChIP assays
Chromatin-immunoprecipitation (ChIP) was performed by a ChIP assay kit (Upstate Biotechnology) according to the manufacturer’s instructions, with some modifications. Briefly, proteins were cross-linked with DNA in the cultured cells by using 1% formaldehyde for 10 minutes at 37°C and quenched with 0.125 mol/L glycine for 5 minutes at room temperature. The cells were then rinsed in ice-cold PBS containing 5 mmol/L sodium butyrate (Sigma), scraped, and resuspended in a lysis buffer (Active Motif) with the addition of complete protease inhibitor cocktail (Roche). The DNA–protein complexes were sheared by using an enzymatic shearing kit (Active Motif) under conditions that gave a range of DNA fragments from 200 to 600 bp, as determined by agarose gel electrophoresis. ChIP was then carried out overnight at 4°C with either mouse monoclonal anti-AhR (RPT9; Abcam) or anti-AhRR (Abcam) antibody. The amount of immunoprecipitated DNA was assessed by semiquantitative PCR, using primers spanning the distal region (distal, nt −1,527 to 1,268) or proximal regions (P3, nt −293 to −139) of the ABCG2 promoter (Fig. 3; ref. 12), and compared with the amount of input DNA before immunoprecipitation.
Figure 3.
ABCG2 promoter luciferase reporter assay. Left, schematic representations of the 5′-deletion ABCG2-promoter constructs. The 5′-end of each of the constructs relative to the transcription start site (arrows) is indicated. The pGL3-basic (promoterless) vector, encoding firefly luciferase, was used to determine the basal levels. The last 2 constructs (Δ−60/−52 and Δ−198/−189) were prepared with the putative XRE elements deleted from the full-length ABCG2 promoter. Right, ABCG2 transcriptional activity, with or without B(a)P treatment in S1 cells transiently transfected with the various ABCG2 promoter constructs. The mean reporter activity ± SD [firefly/Renilla luciferase units (RLU)] from 3 independent experiments is presented. The reporter assay was also repeated in S1 cells treated with romidepsin (data not shown). As we have previously noted, romidepsin aberrantly increases the luciferase activity of the Renilla vector (12). Therefore, the specific effect of romidepsin on the XRE could not be confirmed by this assay.
Taq DNA polymerase (Bioline) was used to amplify 1 μL of either the immunoprecipitated DNA by using the specific antibody, the immunoprecipitate by using a normal immunoglobulin G (IgG) control antibody, or a 1:10 dilution of input chromatin. Preliminary experiments were carried out to determine optimal PCR conditions so that the yield of PCR products was dependent on the amount of input DNA (data not shown). The conditions for the PCR reactions were as follows: 94°C for 3 minutes, 30 cycles at 94°C for 1 minute, 55°C for 1 minute, 72°C for 1 minute, and a final extension at 72°C for 7 minutes. PCR products were electrophoresed on 2% agarose gel and stained with ethidium bromide. Band intensity was quantified as mentioned earlier in the text. Fold enrichment in each immunoprecipitation was determined by quantifying the intensities of the PCR product in immunoprecipitated DNA versus input DNA (total chromatin). Only 10% of the total input was used in the PCR reactions and they were run as the “input” lanes in gel electrophoresis. Percentage enrichment was calculated accordingly. ChIP assays were repeated thrice by different chromatin preparations.
Flow cytometry
ABCG2-mediated efflux activity in romidepsin-pretreated cells was determined by flow cytometric assays as described previously (31). Cells after a 24-hour treatment of romidepsin (10 ng/mL) or vorinostat (5 μmol/L) were incubated in 1 μmol/L pheophorbide A (PhA) with or without 10 μmol/L ABCG2-specific inhibitor fumitremorgin C (FTC) in complete medium (phenol red–free RPMI 1640 with 10% FBS) at 37°C in 5% CO2 for 30 minutes. Subsequently, the cells were washed with cold complete medium and then incubated for 1 hour at 37°C in PhA-free medium continuing with FTC to generate the FTC/efflux histogram, or without FTC to generate the efflux histogram. The FTC-inhibitable PhA efflux was determined as the difference in mean fluorescence intensity (ΔMFI) between the FTC/efflux and efflux histograms, which indicates the ABCG2-mediated transport activity. Cells were finally washed with cold Dulbecco’s PBS and placed on ice in the dark until analysis by flow cytometry.
Samples were analyzed on an LSRFortessa Cell Analyzer (BD Biosciences). PhA fluorescence was detected with a 488-nm argon laser and a 670-nm bandpass filter. At least 10,000 events were collected for all flow cytometry studies.
Results
Induction of ABCG2 by romidepsin or B(a)P in selected cancer cell lines
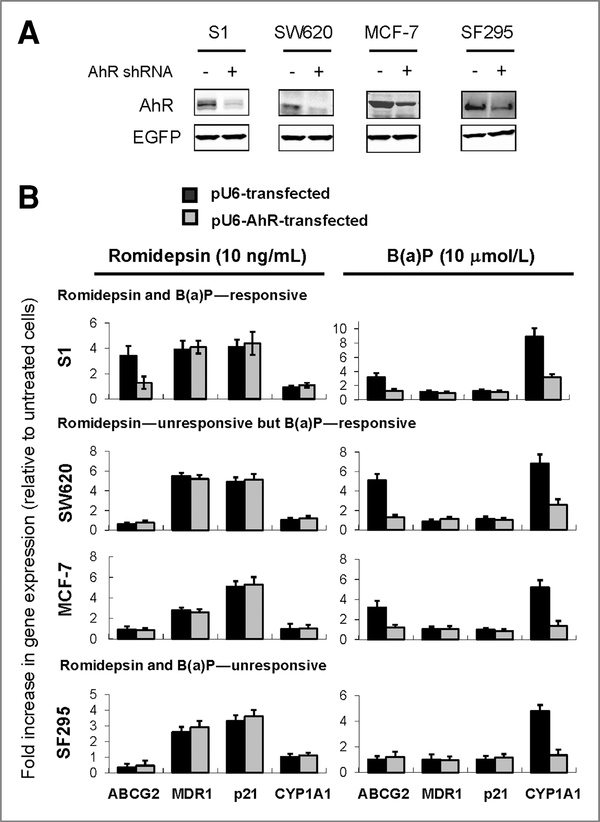
Previously, we showed that romidepsin upregulates ABCG2 in selected cancer cell lines and this coincides with the induction of permissive histone modification marks at the ABCG2 promoter in these cells (12). In our continued effort aiming at understanding the regulation of ABCG2, we pursued an earlier report suggesting that ABCG2 could be regulated by AhR agonists (32). Our result showed that ABCG2 can be upregulated by B(a)P (an AhR agonist; 24 hours) in several cell lines including S1, SW620, and MCF-7 cells (Fig. 1). This upregulation can be reversed in AhR-silenced cells (Fig. 1) or by concomitant treatment with known AhR antagonists [Supplementary Fig. 1; resveratrol (33), kaempferol (34), or salicylamide (35)]. Interestingly, knockdown of AhR (Fig. 1) or AhR antagonists (Supplementary Fig. 1) could also inhibit the upregulation of ABCG2 in S1 by romidepsin. Consistent with our previous findings, SW620, MCF-7, and SF-295 did not respond to romidepsin treatment with ABCG2 upregulation (Fig. 1; ref. 12). Similar results regarding ABCG2 regulation were also obtained for other HDACIs including vorinostat and panobinostat (LBH-589; Table 2). Of note, the cell lines were chosen to cover a range of basal ABCG2 expression levels (Fig. 2). Although ABCG2 levels in S1 and SW620 were marginally detectable, MCF-7 and SF295 express much higher basal levels of ABCG2. As controls for comparison, the upregulation of MDR1 and p21, 2 genes commonly found to be elevated by HDACIs, was not affected by AhR knockdown (Fig. 1) or AhR antagonists (Supplementary Fig. 1) in all cell lines tested. Moreover, both MDR1 and p21 levels were not changed by B(a)P, suggesting that the AhR-related ABCG2 upregulation observed is specific. It is also interesting to note that a typical AhR-responsive gene, CYP1A1, was not affected by romidepsin treatment in all cell lines tested (Fig. 1).
Figure 1.
Real-time PCR analysis showing the absolute requirement for AhR in the upregulation of ABCG2 by romidepsin or B(a)P. A, effective knockdown of AhR by pU6-AhR was confirmed by immunoblot analysis. Enhanced green fluorescent protein (EGFP) represents the loading control for the cotransfections. B, the cells were treated with either romidepsin or B(a)P for 24 hours, with or without the prior knockdown of AhR (48 hours after transfection). The cell lines tested can be divided into 3 categories according to the upregulation of ABCG2 on the drug treatment: romidepsin and B(a)P–-responsive (S1); romidepsin–-nonresponsive but B (a)P–-responsive (SW620 & MCF-7); and romidepsin and B(a)P—nonresponsive (SF295). MDR1 and p21 were also measured because they were found to be universally upregulated by HDACIs. Expression level of a typical AhR-responsive gene CYP1A1 was also evaluated. EGFP (from cotransfection) was used as the internal control for normalization. The data shown represent the fold increase in the levels of the various mRNAs relative to the no-treatment control after normalization with EGFP. Mean ± SD from 3 independent experiments is shown.
Table 2.
Summary of real-time PCR analysis in cells treated with romidepsin (10 ng/mL), vorinostat (5 μmol/L), or panobinostat (100 nmol/L)
| Cells | ABCG2 | MDR1 |
|---|---|---|
| S1 | ||
| No treatment | 1 | 1 |
| Romidepsin-treated | 8.2 | 1.9 |
| Vorinostat-treated | 6.5 | 2.2 |
| Panobinostat-treated | 7.6 | 2.3 |
| SW620 | ||
| No treatment | ND | 1 |
| Romidepsin-treated | ND | 4.8 |
| Vorinostat-treated | ND | 5.4 |
| Panobinostat-treated | ND | 4.9 |
| SF295 | ||
| No treatment | 1 | 1 |
| Romidepsin-treated | 0.23 | 1.8 |
| Vorinostat-treated | 0.2 | 1.9 |
| Panobinostat-treated | 0.23 | 1.9 |
NOTE: Real-time PCR analysis was performed by the Universal Probe Library (UPL) assays (Roche). The specific probes used for ABCG2 and MDR1 are #56 and #49, respectively. Relative ABCG2 or MDR1 expression was shown after normalization with GAPDH.
Abbreviation: ND, not detectable.
An XRE cis-element is required for the activation of ABCG2 promoter activity by B(a)P
Because AhR was potentially involved in romidepsin-mediated upregulation of ABCG2 in S1 cells (Fig. 1), the role of AhR in ABCG2 regulation was further examined. A series of 5′-deletion reporter gene constructs were generated from the ABCG2 promoter region with the 3′-end terminating at +396 bp (ref. 30; Fig. 3). Using a transient transfection assay system in S1 cells, all of the ABCG2 promoter-luciferase constructs showed good activities above the pGL3-Basic background (Fig. 3). Reporter activity of all constructs was found to be activated on B(a)P treatment (24 hours), except the 2 shortest 5′-deleted promoter constructs (−105/+396 and −31/+ 396). Using the PROMO virtual library for the identification of putative transcription factor binding sites, 2 putative XRE elements were predicted on the ABCG2 proximal promoter (“−60 to −52” and “−198 to −189”) based on the TRANSFAC database (36). B(a)P-mediated upregulation of ABCG2 promoter activity could be abolished in the −198/−189-deleted reporter construct (Fig. 3), suggesting that this XRE region is important for ABCG2 regulation. Moreover, activation of the ABCG2 promoter activity was also abolished in AhR-silenced cells (Supplementary Fig. 2). The reporter assay was also repeated in S1 cells treated with romidepsin (data not shown). As we have previously noted, romidepsin aberrantly increases the luciferase activity of the Renilla vector (12). Therefore, the specific effect of romidepsin on the XRE could not be confirmed by this assay.
Romidepsin increases the association of AhR with XRE at the ABCG2 promoter in the responsive cell line
An important step in the AhR pathway is the formation of the AhR–Arnt heterodimer and the subsequent binding of this dimer to the XRE of its target gene promoter to regulate transcriptional activity. To find out whether this process can differentiate the regulation of ABCG2 by romidepsin, we examined the ability of romidepsin to affect the association of the AhR to the ABCG2 proximal promoter by ChIP analysis. ChIP assays measure the interaction of protein with DNA in a given chromatin region in vivo (37). Differential histone H3 hyperacetylation in the proximal ABCG2 promoter has been shown in romidepsin-treated cells with ABCG2 upregulation, indicating the activation of transcription (12).
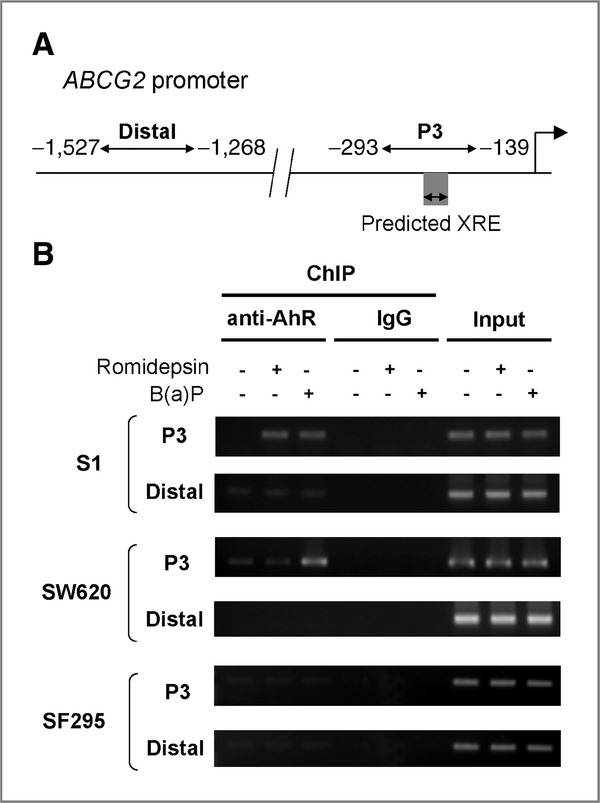
DNA purified after immunoprecipitation with anti-AhR antibody was evaluated by PCR by using primers targeting the proximal (nt −293 to −139) or distal regions (nt −1,527 to −1,268) of the ABCG2 promoter (Fig. 4A). The relative amount of promoter-associated AhR was quantified by measuring the PCR band intensity in a molecular imager followed by normalization with the input. Serial dilutions of the input DNA were amplified by PCR to determine the linearity of the PCR reactions. The levels of PCR amplifications for all ChIP samples were within this linear range. No signal was obtained from immunoprecipitated samples if normal IgG was used (Fig. 4B). The putative XRE element (nt −198 to −189) of the ABCG2 promoter was located within the proximal promoter fragment evaluated by ChIP (Fig. 4A). Romidepsin treatment (4 hours) increased the association of AhR with the proximal ABCG2 promoter in the romidepsin-responsive S1 cells that upregulate ABCG2 (Fig. 4B), but not in SW620 and SF295 cells. On the contrary, an increase in AhR binding to the proximal ABCG2 promoter following B(a)P was observed in both S1 and SW620 cells but not in SF295 cells (4 hours), consistent with the upregulation of ABCG2 in these cell lines. In contrast, there was negligible association of AhR with the distal ABCG2 promoter, confirming the specific binding of AhR to the XRE after romidepsin or B(a)P treatment in the corresponding respective cell lines (Fig. 4B).
Figure 4.
ChIP assay showing that the ABCG2 promoter was enriched with AhR in romidepsin- or B(a)P-treated responsive cell lines. A, a schematic representation of the ABCG2 promoter studied by ChIP analysis. Two sets of primers were used for amplification of the proximal (P3) and distal ABCG2 promoter. Nucleotide positions are numbered relative to the major transcription start site (+1) defined in the published DNA sequence (Genbank accession no. AF151530). Big arrowhead, transcription start site. B, ChIP analyses targeting the proximal and distal region of the ABCG2 promoter in romidepsin or B(a)P-treated cells (4 hours), using antibody against AhR. Input, DNA isolated from the lysate before immunoprecipitation (only 10% of the total chromatin was used for the PCR reactions and it was run as the “input” in the gel electrophoresis); IgG, ChIP using normal IgG for immunoprecipitation. A representative set of result from 3 independent immunoprecipitations is shown.
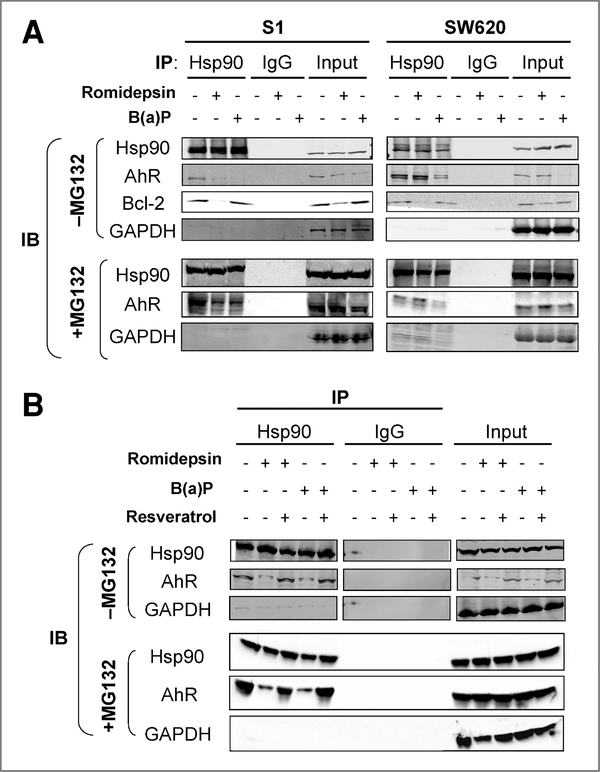
Romidepsin decreased the binding of AhR to Hsp90 in the responsive S1 cells, probably by inducing Hsp70 acetylation
Hsp90 is a chaperone protein that is important in maintaining its client protein in a functional conformation for its biological function. Although somewhat controversial, previous reports have shown that the class I HDAC inhibitor romidepsin induced acetylation of Hsp90, which inhibits the ATP binding and chaperone association of Hsp90 with its client proteins, including Her-2, Akt, and c-Raf, leading to their polyubiquitylation and degradation by the 20S proteasome (23, 25, 38). AhR is a known Hsp90 client protein and it is normally stabilized by the chaperone activity of Hsp90. Because AhR was found to be involved in the upregulation of ABCG2 by romidepsin, additional experiments were performed to find whether romidepsin alters the ability of Hsp90 to interact with AhR, thereby regulating AhR stability. S1 and SW620 cells were treated with 10 ng/mL romidepsin (+5 μg/mL verapamil) or 10 μmol/L B(a)P for 16 hours. Hsp90 was immunoprecipitated with Hsp90 antibody and its associated proteins were determined by immunoblot analysis.
In S1 cells, romidepsin or B(a)P treatment decreased the association of AhR with Hsp90, which is believed to have led to the lower protein stability of AhR (Fig. 5A, −MG132). However, in SW620 cells, the association of AhR with Hsp90 could only be decreased by B(a)P but not by romidepsin. In romidepsin-treated SW620 cells, AhR protein stability remained unchanged (Fig. 5A, −MG132, Input). This result suggested that the dissociation of AhR from Hsp90 may be a prerequisite for the upregulation of ABCG2 by romidepsin. Moreover, consistent with the finding that resveratrol (an AhR antagonist) could inhibit ABCG2 upregulation by romidepsin (Supplementary Fig. 1), the association between AhR and Hsp90 was restored in S1 cells treated with romidepsin (16 hours) in the presence of resveratrol (Fig. 5B). A similar dissociation of AhR from Hsp90 was also observed in S1 or MCF-7 cells on B(a)P treatment (Fig. 5A; Supplementary Fig. 3, respectively), but not in the nonresponsive SF295 cells (Supplementary Fig. 3).
Figure 5.
Immunoprecipitation assay showing that romidepsin led to a dissociation of AhR from Hsp90 in the responsive cell lines. A, total cell lysates were prepared from S1 or SW620 cells as described in the Materials and Methods. For each immunoprecipitation, a total of 500 μg of protein was incubated with rabbit anti-Hsp90 or control IgG antibody prebound to Protein A for 2 hours at 4°C. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membrane. Hsp90, AhR, or Bcl-2 in the immunoprecipitate was visualized by the respective antibodies. To ascertain that the observed AhR–Hsp90 dissociation was not because of artifacts attributed by the decreased AhR stability, the immunoprecipitation experiments were carried out side-by-side in cells treated concurrently with or without MG-132. It was noted that similar results were obtained in both cases, except that AhR protein stability was not affected in cells treated with MG-132. B, dissociation of AhR from Hsp90 in the presence of romidepsin or B(a)P was inhibited by resveratrol (an AhR antagonist) in S1 cells. The immunoprecipitation experiment was performed as described in (A). Hsp90 was immunoprecipitated from cell lysates after treatment of romidepsin or B(a)P, or a combination with resveratrol (in the presence or absence of MG132). AhR in the immunoprecipitate was detected by immunoblot analysis as in (A). A representative blot from 3 independent experiments is shown. GAPDH represents the loading control for the immunoprecipitation experiments.
Inhibition of the Hsp90 chaperone function causes degradation of its client proteins via the ubiquitin-proteasome pathway. The protein expression of such a client protein, Bcl-2, and its binding to Hsp90 in the presence of romidepsin was therefore also evaluated as a control. Romidepsin remarkably reduced the binding of Bcl-2 to Hsp90 and decreased the Bcl-2 protein expression in S1 cells (Fig. 5A, −MG132). To ascertain that the observed AhR–Hsp90 dissociation was not because of artifacts attributed by the decreased AhR stability, the immunoprecipitation experiments following romidepsin or B(a)P were repeated in cells treated concurrently with the proteasome inhibitor MG-132 (10 μmol/L; Calbiochem). Although AhR was not degraded, AhR–Hsp90 dissociation was still observed in romidepsin/B(a)P-treated S1 cells, B(a)P-treated SW620 cells, or B(a)P-treated MCF-7 cells (Fig. 5A and B, +MG132 versus −MG132; Supplementary Fig. 3).
It is known that Hsp90 chaperone activity is regulated by reversible acetylation and is controlled by the deacetylase HDAC6 (39). As a class I HDACI, romidepsin has been reported to have only a weak effect on HDAC 6 (27, 28) but it has been shown to disrupt the chaperone function of Hsp90 and induce apoptosis in human non–small cell lung cancer cells (23). Hsp70 is an important cochaperone protein with Hsp90. It was reported that one of the class I HDACs is the deacetylase of Hsp70 and that romidepsin treatment can increase acetylation of Hsp70 (40). Of note, Hsp70 is required for the assembly of Hsp90–client protein complexes (41). More recently, it has been reported that romidepsin may disrupt the chaperone function of Hsp90 indirectly through acetylation of Hsp70 (42). Therefore, immunoprecipitation experiments were also performed to assess the effect of romidepsin and vorinostat on acetylation of Hsp90/Hsp70 and the association between Hsp90 and AhR.
As expected for the pan-HDACI, vorinostat induced hyperacetylation of both Hsp90 and Hsp70 in S1 cells (Fig. 6A). On the contrary, romidepsin cannot inhibit HDAC6 and therefore it did not cause Hsp90 hyperacetylation but instead only Hsp70 hyperacetylation in S1 cells (Fig. 6A). Consistent with the effect of Hsp90/Hsp70 hyperacetylation on Hsp90 chaperone function and agreeing with the result in Figure 5A, romidepsin or vorinostat treatment (4 hours) was found to deplete the binding of AhR to Hsp90 in S1 cells (Fig. 6B). Interestingly, no Hsp70 and Hsp90 acetylation was observed in the nonresponsive SW620 cells after exposing to romidepsin or vorinostat (Fig. 6A), which is believed to account for the unchanged association between AhR and Hsp90 (Fig. 6B). As a control, B(a)P treatment was able to dissociate AhR from Hsp90 in both S1 and SW620 cells (Fig. 6B), consistent with the well-established biological effect of AhR ligand binding (16).
Figure 6.
Immunoprecipitation assay showing that romidepsin decreased the binding of AhR to Hsp90 in S1 cells by inducing Hsp70 acetylation. A, comparison of the effects of romidepsin or vorinostat on Hsp90/Hsp70 acetylation in S1 and SW620 cells. In S1 cells, vorinostat increased both Hsp90 and Hsp70 acetylation but romidepsin only induced Hsp70 acetylation. On the contrary, no Hsp90/Hsp70 acetylation was detected in SW620 cells treated with either vorinostat or romidepsin. Cell lysates immunoprecipitated with anti-acetyl Lysine antibody were subjected to immunoblot analysis by anti-Hsp90 or anti-Hsp70 antibody. The levels of Hsp90 and Hsp70 in the whole cell lysate not subjected to immunoprecipitation were used as the loading controls. B, cell lysates from S1 and SW620 cells were prepared as in Figure 5A and immunoprecipitated with anti-AhR antibody. Hsp90 or Hsp70 in the immunoprecipitate was visualized by the respective antibodies. A representative blot from 3 independent immunoprecipitations is shown. GAPDH represents the loading control for the immunoprecipitation experiments.
Romidepsin or B(a)P did not increase AhRR expression or its binding to the ABCG2 promoter in the nonresponsive cell lines
The AhR repressor (AhRR) has been shown to inhibit AhR signaling through a proposed negative feedback mechanism involving induction of its constitution expression and competition with AhR for dimerization to Arnt and subsequently binding to AhR regulatory elements (XRE; ref. 43). To determine whether ABCG2 upregulation was constrained by AhRR in the nonresponsive cell lines, AhRR protein expression and their association with the ABCG2 promoter were evaluated by immunoblot and ChIP analysis, respectively. Romidepsin or B(a)P (16 hours) decreased AhRR expression in HepG2 cells but had no detectable effect in other cell lines tested (Fig. 7A). In SW620 (the romidepsin nonresponsive and B(a)P responsive cell line), no association between AhRR and the ABCG2 promoter can be detected under either treatment with romidepsin or B(a)P (Fig. 7B). These results rule out the involvement of AhRR in the lack of ABCG2 upregulation in the nonresponsive cell lines.
Figure 7.
Immunoblot and ChIP analyses showing that AhRR was not involved in the lack of ABCG2 upregulation in SW620 cells treated with romidepsin or B(a)P. A, immunoblot analysis showing the cellular level of AhRR in various cell lines indicated after treatment of romidepsin or B(a)P. HepG2 and HeLa cells were known to express detectable protein level of AhRR, therefore they were included for comparison. GAPDH was used as a loading control. No activation of AhRR was detected in all the cell lines tested. B, ChIP analyses targeting the proximal and distal region of the ABCG2 promoter in romidepsin- or B(a)P-treated SW620 cells, using anti-AhRR antibody. Input, DNA isolated from the lysate before immunoprecipitation (only 10% of the total chromatin was used for the PCR reactions and it was run as the “input” in the gel electrophoresis); IgG, ChIP using normal IgG for immunoprecipitation. A representative gel image from 2 independent immunoprecipitations is shown.
Romidepsin increased ABCG2 efflux activity only in the responsive S1 cells
Consistent with the PCR analysis (Fig. 1; Table 2), increased PhA efflux (as measured by ΔMFI) was also noted in romidepsin or vorinostat-treated S1 cells (Fig. 8). On the contrary, PhA efflux remained unchanged or was slightly decreased in SW620 and SF295 cells, respectively, on romidepsin or vorinostat treatment (Fig. 8).
Figure 8.
Drug efflux assay showing the increase in ABCG2-mediated efflux by romidepsin or vorinostat treatment in the S1 cells. Cells were incubated with the fluorescent substrate PhA with or without 10 μmol/L FTC (ABCG2-specific inhibitor) for 30 minutes at 37°C, washed, and then allowed to efflux for 1 hour at 37°C in substrate-free medium continuing with (dashed lines, FTC/Efflux histogram) or without (solid lines, Efflux histogram) FTC. Representative histograms from 3 independent experiments are shown. ΔMFI was determined as the difference in mean fluorescence intensity between the FTC/efflux and efflux histograms, which indicates the ABCG2-mediated transport activity.
Discussion
This study extends our earlier report about the modulation of the ABCG2 gene by HDACIs. We found that romidepsin increased ABCG2 expression in selected cancer cell lines and this modulation was accompanied by histone modifications such as an increase in histone acetylation at the ABCG2 promoter (12). Unlike MDR1, the induction of ABCG2 following romidepsin exposure was found to be cell line specific (12). Although differential display (44) or microarray analyses (45) identified only 2% to 10% of expressed genes as sensitive to HDACIs, we have found a gene responding to HDAC inhibition in some cell lines but not in others. The upregulation of ABCG2 by HDACIs may lead to drug resistance when used in combination with substrate drugs. Therefore, understanding the differential effects by romidepsin could not only provide the guidance for the avoidance of HDACI-induced drug resistance, but also potentially provide insight into the induction of therapeutically important genes by HDAC inhibition.
The involvement of the AhR pathway in the upregulation of ABCG2 has been suggested in studies showing that known AhR agonists, including 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and B(a)P, elevated the levels of ABCG2 mRNA and protein expression, resulting in increased transport of ABCG2 substrates (33, 46). In fact, during the preparation of this manuscript, it was reported that AhR is a transcriptional activator of ABCG2 (47). In our continued investigation into the molecular mechanisms that regulate ABCG2, we confirmed and extended these findings, observing that ABCG2 was differentially upregulated in cancer cell lines after treatment with B(a)P, which could be inhibited by concomitant treatment with AhR antagonists (resveratrol, kaempferol, and salicylamide) or by AhR knock-down. Upregulation of ABCG2 following romidepsin could also be abolished by chemically antagonizing AhR or genetic silencing (Fig. 1; Supplementary Fig. 1). To this end, romidepsin is unlikely to be a direct AhR ligand because the typical AhR-responsive gene CYP1A1 was not affected in all cell lines tested (Fig. 1). As additional controls, the universal upregulation of MDR1 and p21 by romidepsin in our study was not affected by chemical antagonists or AhR knockdown (Fig. 1; Supplementary Fig. 1), suggesting that the observed AhR-mediated ABCG2 upregulation should be specific. An XRE consensus sequence was identified and subsequently confirmed at the ABCG2 promoter close to the transcription start site by a reporter gene assay (Fig. 3; Supplementary Fig. 2), which is consistent with that reported recently by Tan and colleagues (47). Thus, we sought to elucidate the mechanisms for this differential effect by romidepsin on ABCG2 regulation.
The formation of the AhR–Arnt heterodimer and its subsequent binding to the XRE sequence at its target genes is critical for the activation of the AhR pathway. To study the cell line–specific upregulation of ABCG2 by romidepsin, we examined the association of AhR with the XRE on the ABCG2 promoter. Results from ChIP analyses showed that treatment with romidepsin or B(a)P induced the association of the AhR to XRE harbored in the ABCG2 promoter only in the responsive cells (Fig. 4), which is consistent with the cell line–dependent activation of the AhR pathway by drug treatment.
Hsp90 is an important molecular chaperone playing a critical role in maintaining the stability and function of its client proteins (17, 18), and Hsp90 acetylation has been shown to impair the chaperone function of Hsp90 (23) and target the client proteins for degradation (48, 49). Hsp70 is an important cochaperone protein of Hsp90 and is required for the assembly of Hsp90–client protein complexes (41). It has been reported that romidepsin may disrupt the chaperone function of Hsp90 indirectly through acetylation of Hsp70 (42). Because the chaperone function of Hsp90 also determines AhR protein stability, AhR protein expression was also assessed. Concurrent with release of AhR and translocation to the nucleus, AhR degradation also occurs following B(a)P exposure (50). We observed this phenomenon following B(a)P exposure in both S1 and SW620 cells. Similarly, our data showed that AhR degrades substantially after 16-hour exposure to romidepsin in S1 cells (Fig. 5A, −MG132, Input). In contrast, the Hsp90 chaperone was found to protect AhR from degradation in the romidepsin nonresponsive SW620 cells (Fig. 5A, −MG132, Input). Importantly, the AhR–Hsp90 dissociation was still observed in romidepsin/B(a)P-responsive cell lines when the proteasome inhibitor MG-132 was used to stabilize the AhR protein (Fig. 5A, +MG132 versus −MG132). These results suggest that the observed lower AhR/Arnt/XRE binding in the responsive cells is not due to a decrease in the cellular level of the AhR protein. Agonist-induced degradation of the AhR protein is known to occur and it does not compromise the induction of AhR-regulated genes; rather, it is probably a regulated proteolytic event to keep the gene induction under control (51).
Romidepsin and vorinostat were found to cause Hsp90/Hsp70 acetylation only in the responsive cell line S1 but not in the nonresponsive SW620 cells (Fig. 6A). It has been recently shown that Hsp90 dissociation from the AhR is essential for the formation of the AhR–Arnt heterodimer (52), which binds to the XRE of target genes and initiates transcription. In S1 cells, in which the dissociation of Hsp90 and AhR was observed on romidepsin or vorinostat treatment, there was a remarkable increase in the association between AhR and Hsp70 (Fig. 6B). Because Hsp70 may also promote disaggregation and protein degradation through dissociation of Hsp90 and its client proteins (53), this could serve to prevent the excessive upregulation of ABCG2. In our model system (Fig. 5A, −MG132), romidepsin disrupts the association of AhR from Hsp90 and leads to degradation of AhR. However, induction of Hsp70 was not observed after romidepsin treatment in either S1 (responsive) or SW620 (unresponsive) cells (Fig. 6A, lysate). Further, the IC50s of romidepsin in S1 and SW620 were found to be similar (approximately 0.5 ng/mL in both cell lines; ref. 54), suggesting that the differential Hsp90/Hsp70 acetylation on romidepsin treatment in S1 and SW620 cells apparently did not impact HDACI sensitivity. Although Hsp90/Hsp70 acetylation may be the critical determinant for the differential upregulation of ABCG2 by romidepsin, the role of other cochaperone proteins such as p23 and XIAP should not be excluded and deserves further investigation. A better understanding of the markers or cell phenotypes that make some cells responsive to HDACI with ABCG2 upregulation is needed to design strategies to modulate ABCG2 by the AhR pathway.
The romidepsin-induced dissociation of AhR from Hsp90 was inhibited by cotreatment with resveratrol (Fig. 5B). In fact, resveratrol has been proposed to antagonize the induction of AhR genes either by competing for AhR binding with the AhR ligands (55) or by inhibiting the recruitment of the AhR–Arnt heterodimer to the XRE (33). Our data suggest that resveratrol may prevent romidespin-mediated upregulation of ABCG2 by maintaining the association between Hsp90 and AhR.
The negative feedback mechanism involving the AhRR was also examined to determine its role in the lack of ABCG2 upregulation in the nonresponsive cell lines. To this end, the constitutive expression of AhRR was not altered by romidepsin treatment (Fig. 7A). In the romidepsin nonresponsive SW620 cell line, no AhRR binding to the ABCG2 promoter can be detected (Fig. 7B). Therefore, AhRR is not likely to participate in the cell line–specific regulation of ABCG2 by romidepsin treatment.
The biological/functional consequence of the cell line–specific upregulation of ABCG2 was also investigated. Consistent with the ABCG2 mRNA analysis, an increase in ABCG2-mediated efflux was observed in romidepsin- or vorinostat-treated S1 cells (Fig. 8). It is noteworthy that ABCG2 protein expression or FTC-inhibitable efflux was not observed in S1 cells under basal condition. Given that ABCG2 upregulation may lead to treatment failure when HDACI (romidepsin) is used in combination with other chemotherapeutic agents, strategies could be designed to modulate the AhR pathway therapeutically for the circumvention of drug resistance. Moreover, because constitutive overexpression of AhR is commonly observed in several cancer types including lung and esophageal cancers (56, 57), the involvement of this AhR-mediated upregulation of ABCG2 in the acquired multidrug resistance and poor prognosis of these cancers may deserve investigation.
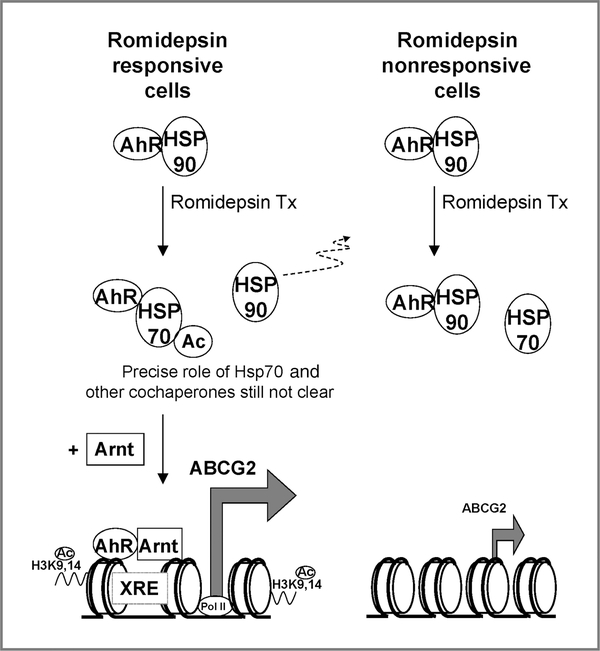
Taken together, the AhR signaling pathway was shown to be involved in the cell line–specific upregulation of ABCG2 by romidepsin treatment. A working model is illustrated in Figure 9. The AhR–Hsp90 interaction seems to represent a novel pathway for regulating ABCG2 expression in cancer cells. Whether other therapeutic genes will be similarly regulated is a subject for further study.
Figure 9.
A working model for the cell line–specific ABCG2 upregulation via the AhR pathway after romidepsin treatment. In romidepsin responsive cell lines, romidepsin treatment caused Hsp70 acetylation and impaired Hsp90 chaperone activity. Although AhR protein expression was decreased because of the inhibition of Hsp90 chaperone activity, AhR–Hsp90 dissociation is a prerequisite for AhR–XRE binding and the subsequent activation of ABCG2. ABCG2 upregulation in these cell lines is also characterized by the enhanced enrichment of acetylated histone H3 (H3 K9,14 Ac) and RNA polymerase II (Pol II) at the ABCG2 promoter (12). Regulation by other cochaperones may also play a role in this cell line–dependent ABCG2 regulation. In contrast, in romidepsin nonresponsive cell lines, although AhR was stabilized by Hsp90 because of the lack of Hsp90/Hsp70 acetylation, the AhR activation was hindered by the constant association between Hsp90 and AhR.
Supplementary Material
Acknowledgments
Grant Support
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and the Seed Research Funding provided by the School of Pharmacy, Chinese University of Hong Kong (K.K.W. To).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interests
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Robey RW, To KK, Polgar O, Dohse M, Fetsch P, Dean M, et al. ABCG2: a perspective. Adv Drug Deliv Rev 2009;61:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allikmets R, Schriml LM, Hutchinson A, Romano-Spica V, Dean M. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res 1998;58:5337–9. [PubMed] [Google Scholar]
- 3.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA 1998;95:15665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maliepaard M, van Gastelen MA, de Jong LA, Pluim D, van Waardenburg RC, Ruevekamp-Helmers MC, et al. Overexpression of the BCRP/MXR/ABCP gene in a topotecan-selected ovarian tumor cell line. Cancer Res 1999;59:4559–63. [PubMed] [Google Scholar]
- 5.Miyake K, Mickley L, Litman T, Zhan Z, Robey R, Cristensen B, et al. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: demonstration of homology to ABC transport genes. Cancer Res 1999;59:8–13. [PubMed] [Google Scholar]
- 6.Robey RW, Medina-Perez WY, Nishiyama K, Lahusen T, Miyake K, Litman T, et al. Overexpression of the ATP-binding cassette half-transporter, ABCG2 (Mxr/BCrp/ABCP1), in flavopiridol-resistant human breast cancer cells. Clin Cancer Res 2001;7:145–52. [PubMed] [Google Scholar]
- 7.Volk EL, Farley KM, Wu Y, Li F, Robey RW, Schneider E. Overexpression of wild-type breast cancer resistance protein mediates methotrexate resistance. Cancer Res 2002;62:5035–40. [PubMed] [Google Scholar]
- 8.Peart MJ, Tainton KM, Ruefli AA, Dear AE, Sedelies KA, O’Reilly LA, et al. Novel mechanisms of apoptosis induced by histone deacetylase inhibitors. Cancer Res 2003;63:4460–71. [PubMed] [Google Scholar]
- 9.Morrow CS, Nakagawa M, Goldsmith ME, Madden MJ, Cowan KH. Reversible transcriptional activation of mdr1 by sodium butyrate treatment of human colon cancer cells. J Biol Chem 1994;269: 10739–46. [PubMed] [Google Scholar]
- 10.Robey RW, Zhan Z, Piekarz RL, Kayastha GL, Fojo T, Bates SE. Increased MDR1 expression in normal and malignant peripheral blood mononuclear cells obtained from patients receiving depsipeptide (FR901228, FK228, NSC630176). Clin Cancer Res 2006;12:1547–55. [DOI] [PubMed] [Google Scholar]
- 11.Tabe Y, Konopleva M, Contractor R, Munsell M, Schober WD, Jin L, et al. Up-regulation of MDR1 and induction of doxorubicin resistance by histone deacetylase inhibitor depsipeptide (FK228) and ATRA in acute promyelocytic leukemia cells. Blood 2006;107:1546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.To KK, Polgar O, Huff LM, Morisaki K, Bates SE. Histone modifications at the ABCG2 promoter following treatment with histone deacetylase inhibitor mirror those in multidrug-resistant cells. Mol Cancer Res 2008;6:151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauswald S, Duque-Afonso J, Wagner MM, Schertl FM, Lübbert M, Peschel C, et al. Histone deacetylase inhibitors induce a very broad, pleiotropic anticancer drug resistance phenotype in acute myeloid leukemia cells by modulation of multiple ABC transporter genes. Clin Cancer Res 2009;15:3705–15. [DOI] [PubMed] [Google Scholar]
- 14.Denison MS, Pandini A, Nagy SR, Baldwin EP, Bonati L. Ligand binding and activation of the Ah receptor. Chem Biol Interact 2002;141:3–24. [DOI] [PubMed] [Google Scholar]
- 15.Denison MS, Fisher JM, Whitlock JP Jr. The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. J Biol Chem 1988;263:17221–4. [PubMed] [Google Scholar]
- 16.Swanson HI. DNA binding and protein interactions of the AHR/ARNT heterodimer that facilitate gene activation. Chem Biol Interact 2002;141:63–76. [DOI] [PubMed] [Google Scholar]
- 17.Neckers L Hsp90 inhibitors as novel cancer chemotherapeutic agents. Trends Mol Med 2002;8:S55–61. [DOI] [PubMed] [Google Scholar]
- 18.Whitesell L, Lindquist SL. Hsp90 and the chaperoning of cancer. Nat Rev Cancer 2005;5:761–72. [DOI] [PubMed] [Google Scholar]
- 19.Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci USA 2000;97:10832–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem 2002;277:39858–66. [DOI] [PubMed] [Google Scholar]
- 21.Schulte TW, Blagosklonny MV, Ingui C, Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem 1995;270:24585–8. [DOI] [PubMed] [Google Scholar]
- 22.Pongratz I, Mason GG, Poellinger L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J Biol Chem 1992;267:13728–34. [PubMed] [Google Scholar]
- 23.Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, et al. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst 2002;94:504–13. [DOI] [PubMed] [Google Scholar]
- 24.Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J, et al. Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or-refractory chronic myelogenous leukemiablast crisis cells. Cancer Res 2003;63:5126–35. [PubMed] [Google Scholar]
- 25.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antiluekemia activity of histone deacetylase inhibitors. J Biol Chem 2005;280:26729–34. [DOI] [PubMed] [Google Scholar]
- 26.Bali P, Pranpat M, Swaby R, Fiskus W, Yamaguchi H, Balasis M, et al. Activity of suberoylanilide hydroxamic acid against human breast cancer cells with amplification of Her-2. Clin Cancer Res 2005;11:6382–9. [DOI] [PubMed] [Google Scholar]
- 27.Blagosklonny MV, Robey R, Sackett DL, Du L, Traganos F, Darzynkiewicz Z, et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol Cancer Ther 2002;1:937–41. [PubMed] [Google Scholar]
- 28.Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res 2002;62:4916–21. [PubMed] [Google Scholar]
- 29.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. siRNA selection server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res 2004;32:W130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.To KK, Zhan Z, Bates SE. Aberrant promoter methylation of the ABCG2 gene in renal carcinoma. Mol Cell Biol 2006;26:8572–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robey RW, Steadman K, Polgar O, Morisaki K, Blayney M, Mistry P, Bates SE. Pheophorbide A is a specific probe for ABCG2 function and inhibition. Cancer Res 2004;64:1242–6. [DOI] [PubMed] [Google Scholar]
- 32.Ebert B, Seidel A, Lampen A. Identification of BCRP as transporter of benzo[a]pyrene conjugates metabolically formed in Caco-2 cells and its induction by Ah-receptor agonists. Carcinogenesis 2005;26:1754–63. [DOI] [PubMed] [Google Scholar]
- 33.Beedanagari SR, Bebenek I, Bui P, Hankinson O. Resveratrol inhibits dioxin-induced expression of human CYP1A1 and CYP1B1 by inhibiting recruitment of the aryl hydrocarbon receptor complex and RNA polymerase II to the regulatory regions of the corresponding genes. Toxicol Sci 2009;110:61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puppala D, Gairola CG, Swanson HI. Identification of kaempferol as an inhibitor of cigarette smoke-induced activation of the aryl hydrocarbon receptor and cell transformation. Carcinogenesis 2007;28:639–47. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald CJ, Ciolino HP, Yeh GC. The drug salicylamide is an antagonist of the aryl hydrocarbon receptor that inhibits signal transduction induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Cancer Res 2004;64:429–34. [DOI] [PubMed] [Google Scholar]
- 36.Farre D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 2003;31:3651–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods 1999;19:425–33. [DOI] [PubMed] [Google Scholar]
- 38.Fuino L, Bali P, Wittmann S, Donapaty S, Guo F, Yamaguchi H, et al. Histone deacetylase inhibitor LAQ824 down regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine and epothilone B. Mol Cancer Ther 2003;10:971–84. [PubMed] [Google Scholar]
- 39.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 2005;18: 601–7. [DOI] [PubMed] [Google Scholar]
- 40.Johnson CA, White DA, Lavender JS, O’Neill LP, Turner BM. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J Biol Chem 2002; 277:9590–7. [DOI] [PubMed] [Google Scholar]
- 41.Pratt WB, Toft DO. Regulation of signaling protein function, trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228:111–33. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Wang SY, Zhang XH, Zhao M, Hou CM, Xu YJ, et al. FK228 inhibits Hsp90 chaperone function in K562 cells via hyperacetylation of Hsp70. Biochem Biophys Res Commun 2007;356: 998–1003. [DOI] [PubMed] [Google Scholar]
- 43.Hahn ME, Allan LL, Sherr DH. Regulation of constitutive and inducible AhR signaling: complex interactions involving the AhR repressor. Biochem Pharmacol 2009;77:485–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr 1996;5:245–53. [PMC free article] [PubMed] [Google Scholar]
- 45.Johnstone R Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 2002;1:287–99. [DOI] [PubMed] [Google Scholar]
- 46.Ebert B, Seidel A, Lampen A. Phytochemicals induce breast cancer resistance protein in Caco-2 cells and enhance the transport of benzo (a)pyrene-3-sulfate. Toxicol Sci 2007;96:227–36. [DOI] [PubMed] [Google Scholar]
- 47.Tan KP, Wang B, Yang M, Boutros PC, Macaulay J, Xu H, et al. Aryl hydrocarbon receptor (AHR) is a transcriptional activator of human breast cancer resistance protein (BCRP/ABCG2). Mol Pharmacol 2010;78:175–85. [DOI] [PubMed] [Google Scholar]
- 48.Isaacs HS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell 2003;3:213–7. [DOI] [PubMed] [Google Scholar]
- 49.Fiskus W, Ren Y, Mohapatra A, Bali P, Mandawat A, Rao R, et al. Hydroxamic acid analogue histone deacetylase inhibitors attenuate estrogen receptor-α levels and transcriptional activity: a result of hyperacetylation and inhibition of chaperone function of heat shock protein 90. Clin Cancer Res 2007;13:4882–90. [DOI] [PubMed] [Google Scholar]
- 50.Lin P, Chang JT, Ko JL, Laio SH, Lo WS. Reduction of androgen receptor expression by benzo(a)pyrene and 7,8-dihydro-9,10-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene in human lung cells. Biochem Pharmacol 2004;67:1523–30. [DOI] [PubMed] [Google Scholar]
- 51.Giannone J, Li W, Probst M, Okey A. Prolonged depletion of AH receptor without alteration of receptor mRNA levels after treatment of cells in culture with 2,3,7,8-tetrachloro-dibenzo-p-dioxin. Biochem Pharmacol 1998;55:489–97. [DOI] [PubMed] [Google Scholar]
- 52.Heid SE, Pollenz RS, Swanson HI. Role of heat shock protein 90 dissociation in mediating agonist-induced activation of the aryl hydrocarbon receptor. Mol Pharmacol 2000;57:82–92. [PubMed] [Google Scholar]
- 53.Wegele H, Muller L, Buchner J. Hsp70 and Hsp90 – a relay team for protein folding. Rev Phyisol Biochem Pharmacol 2004;151:1–44. [DOI] [PubMed] [Google Scholar]
- 54.Robey RW, Zhan Z, Piekarz R, Kayastha GL, Fojo T, Bates SE. Increased MDR1 expression in normal and malignant peripheral blood mononuclear cells obtained from patients receiving depsipeptide (FR901228, FK228, NSC630176). Clin Cancer Res 2006;12:1547–55. [DOI] [PubMed] [Google Scholar]
- 55.Ciolino HP, Daschner PJ, Yeh GC. Resveratrol inhibits transcription of CYP1A1 in vitro by preventing activation of the aryl hydrocarbon receptor. Cancer Res 1998;58:5707–12. [PubMed] [Google Scholar]
- 56.Wang CK, Chang H, Chen PH, Chang JT, Kuo YC, Ko JL, Lin P. Aryl hydrocarbon receptor activation and overexpression upregulated fibroblast growth factor-9 in human lung adenocarcinomas. Int J Cancer 2009;125:807–15. [DOI] [PubMed] [Google Scholar]
- 57.Roth MJ, Wei WQ, Baer J, Abnet CC, Wang GQ, Sternberg LR, et al. Aryl hydrocarbon receptor expression is associated with a family history of upper gastrointestinal tract cancer in a high-risk population exposed to aromatic hydrocarbons. Cancer Epidemiol Biomarkers Prev 2009;18:2391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.