Abstract
Lipid droplets (LDs) are ubiquitous organelles that store metabolic energy in the form of neutral lipids (typically triacylglycerols and steryl esters). Beyond being inert energy storage compartments, LDs are dynamic organelles that participate in numerous essential metabolic functions. Cells generate LDs de novo from distinct sub-regions at the endoplasmic reticulum (ER), but what determines sites of LD formation remains a key unanswered question. Here, we review the factors that determine LD formation at the ER, and discuss how they work together to spatially and temporally coordinate LD biogenesis. These factors include lipid synthesis enzymes, assembly proteins, and membrane structural requirements. LDs also make contact with other organelles, and these inter-organelle contacts contribute to defining sites of LD production. Finally, we highlight emerging non-canonical roles for LDs in maintaining cellular homeostasis during stress.
Keywords: lipid droplet, endoplasmic reticulum, fatty acid, lipotoxicity, metabolon
1. Introduction
Eukaryotic cells have achieved a high level of organizational complexity by spatially restricting metabolic reactions within subcellular compartments (Hurtley, 2009). Such spatial compartmentalization is essential for regulation of flux between metabolic pathways and protecting the cell from toxic metabolic intermediates (William, 2010). Lipid droplets (LDs), organelles with a unique architecture and found in most cells, form a nexus of energy metabolism that help maintain cellular homeostasis (Fujimoto and Ohsaki, 2006; Olzmann and Carvalho, 2019; Walther and Farese, 2012). Each LD consists of a neutral lipid core mainly of triacylglycerols (TAG) and steryl esters (SE), encapsulated within an ER-derived phospholipid monolayer surface (Fujimoto and Parton, 2011). LDs are dynamically regulated in response to physiological conditions. Their lipid composition, size, and number vary depending on the cell type, nutrient availability, and metabolic state (Thiam and Beller, 2017). This variability, along with selective binding of different proteins and enzymes to their surface, reflects the functional heterogeneity of LDs (Bersuker et al., 2018; Kory et al., 2016; Prévost et al., 2018).
LD formation is a multistep process (Walther et al., 2017). Newly synthesized neutral lipids produced by mostly ER-resident enzymes are deposited between the phospholipid leaflets, where they coalesce into lenses (Choudhary et al., 2015). Once a critical size is reached, a nascent LD buds towards the cytoplasm. Many factors probably contribute to defining the sites on the ER surface where LD budding occurs, as well as its directionality. First, enzymes of lipid synthesis that promote LD formation can be localized, and these may also partition onto LDs during stages of droplet biogenesis. Structural proteins are also necessary as assembly factors that support the nascent lens as it grows (Cartwright et al., 2015; Choudhary et al., 2015). Additionally, specific phospholipids such as phosphatidic acid (PA) in the membrane can accommodate local perturbations to normal ER shape that promote the remodeling necessary to exude a monolayercoated LD from the ER bilayer (Ben M’barek et al., 2017; Thiam et al., 2013).
Recent work demonstrates that LDs do not work in isolation but are well-connected organelles that make contact with other cellular compartments (Valm et al., 2017). During their growth and maturation, LDs maintain intimate contact with the ER as a source of neutral lipids (Jacquier et al., 2011). Under different metabolic conditions, LDs also establish contacts with many other organelles that can coordinate their breakdown (Barbosa and Siniossoglou, 2017; Gao and Goodman, 2015; Schuldiner and Bohnert, 2017). Remarkably, these inter-organelle contacts also contribute to LD biogenesis, as they can serve as specialized platforms that maintain cellular homeostasis by controlling lipid storage, utilization, and signaling.
Recent studies provide new insights into the structural and mechanistic basis for LD biogenesis at the ER, as well as how LD formation can be spatially positioned in the context of cellular metabolic status (Barbosa and Siniossoglou, 2016; Hariri et al., 2018; Teixeira et al., 2017). These studies reveal previously unappreciated levels of spatial organization to LD production. In this review, we summarize and discuss the current understanding of how different factors work together to contribute to the compartmentalization of LD biogenesis. We also discuss newly characterized tethering proteins that connect LDs to other cellular compartments and spatially regulate LD sub-cellular distributions to promote adaptations to stress. Finally, we highlight key questions remaining in the field of LD biology that future studies need to address.
2. Compartmentalization of LD synthesis enzymes
Although the order of events underlying LD formation are becoming understood, much is still unknown about the precise molecular mechanisms that govern LD biogenesis (Olzmann and Carvalho, 2019). A cascade of enzymatic reactions in the ER synthesizes neutral lipids that are likely deposited among the acyl chains of the bilayer and are laterally mobile in this hydrophobic environment (Coleman and Lee, 2004). While the most prominent neutral lipids that form LD cores are TAGs and SEs, other lipids such as retinyl esters, squalene, ether lipids, and acylceramides accumulate in LDs under different conditions and in different cell types (Bartz et al., 2007; Blaner et al., 2009; Senkal et al., 2017; Spanova et al., 2012; Ta et al., 2012). LDs can remain attached to the ER, forming stable bridges (ER-LD contact sites) or necks (Jacquier et al., 2011; Salo et al., 2016). ER-LD contacts are regularly observed by high-resolution imaging techniques, and it is unclear to what extent droplets separate entirely from the ER (Wilfling et al., 2014).
As major energy-storage compartments for the cell, LDs are highly conserved throughout evolution and have been observed in both prokaryotes and eukaryotes. Bacteria, yeast, plants, and mammals all have the capacity to synthesize TAG and form LDs (Alvarez and Steinbüchel, 2002; Wältermann et al., 2005). Lipid synthesis enzymes and the general architecture of LD metabolic pathways are highly conserved among eukaryotes (Coleman and Lee, 2004; Rajakumari et al., 2008; Santos and Riezman, 2012). In unicellular eukaryotes such as the yeast Saccharomyces cerevisiae, LDs are easily detected which facilitates genetic screens for defects in LD biogenesis and morphology (Fei et al., 2008; Szymanski et al., 2007). In addition to their role in energy homeostasis, LD biogenesis is critical for yeast homeostasis especially during periods of nutritional excess or lipid stress.
How early steps of neutral lipid synthesis are spatially compartmentalized within the vast ER network remains a key unanswered question in LD biology. More than 25 different enzymes and isoenzymes are required to synthesize neutral lipids, and these are localized to various sub-cellular compartments and the cytoplasm (Coleman and Lee, 2004). As a consequence, interorganelle membrane contact sites are emerging as metabolic scaffolds that promote the metabolic cross-talk among these enzymes as enzymatic assemblies (so-called metabolons) that regulate substrate sharing and availability (Srere, 1987). Below we discuss what is known about enzyme segregation in the context of LD biogenesis.
2.1. Enzymes of TAG synthesis
The steps of TAG biosynthesis were identified several decades ago (Kennedy, 1957, 1961). Since then, considerable knowledge has been gained about the enzymes and regulators that are involved in these reactions. De novo TAG synthesis is catalyzed by four consecutive enzymes: glycerol-phosphate acyltransferase (GPAT), acyl-glycerol-phosphate acyltransferase (AGPAT), phosphatidic acid phosphohydrolase (PAP), and acyl-CoA:diacylglycerol acyltransferase (DGAT) (Coleman and Lee, 2004). Multiple isoforms of TAG synthesis enzymes appear to catalyze the same biochemical reaction suggesting that each isoenzyme may play a distinct functional role in regulating TAG production (Coleman and Lee, 2004; Yen et al., 2008). While these enzymes have been identified, relatively little is known about how they are coordinated and physically organized within subcellular locales. TAG synthesis occurs primarily in the ER under ambient conditions; however, an elevated influx of fatty acids can induce ER-localized synthesis enzymes to re-localize to LDs which act as metabolic platforms for the local generation of TAG (Athenstaedt and Daum, 1997; Brasaemle et al., 2004; Goodman, 2009; Jacquier et al., 2011; Kuerschner et al., 2008; Stone et al., 2006). Proteomics analysis of LD fractions isolated from Drosophila S2 cells revealed that at least one isoenzyme catalyzing each step of TAG biosynthesis (e.g. GPAT4, AGPAT3, PAP) can re-localize from the ER to LDs (Wilfling et al., 2013). This study showed that moving TAG synthesis enzymes to the LD surface contributed to the formation of a distinct LD sub-population that can expand in size (Wilfling et al., 2013). This implies that targeting specific enzymes to LDs can promote their growth and maturation.
The final step of TAG biosynthesis is catalyzed by DGAT enzymes which covalently link fatty acyl-CoAs (FA-CoA) to diacylglycerols (DAG) (Cases et al., 1998; Lehner and Kuksis, 1993). Mammals encode two DGAT enzymes, DGAT1 and DGAT2, which differ significantly in their structure, sub-cellular localization, and substrate preference. It was recently shown that DGAT1 and DGAT2 can largely compensate for each other for TAG storage (Chitraju et al., 2019). DGAT1 is localized exclusively to the ER, whereas DGAT2 is localized to the ER and LDs (Villanueva et al., 2009; Yen et al., 2008). DGAT1 can esterify a variety of substrates; therefore, its localization to the ER has been linked to its role in detoxifying excess lipids thus protecting the ER from the lipotoxic effects of high-fat diets and preventing ER (Chitraju et al., 2019; Nguyen et al., 2017). DGAT2, however, appears to be the major enzyme of TAG synthesis from de novo-synthesized fatty acids (Irshad et al., 2017; Wurie et al., 2012). DGAT2 may re-localize from the ER to the LDs upon lipid loading (Kuerschner et al., 2008; McFie and Stone, 2011). The yeast ortholog of DGAT2, Dga1 migrates from the ER to LDs as cells accumulate TAG in late exponential phase, although it is not clear if this enzyme is more or less active there (Gao et al., 2017; Markgraf et al., 2014). Such re-localization of enzymes to LDs may facilitate efficient substrate channeling within localized multi-enzyme complexes.
DGATs can functionally interact with other lipid synthesis enzymes. C. elegans provides evidence for an interaction between DGAT2 and acyl-CoA synthase FAT1P at the ER-LD contact presumably to coordinate local TAG synthesis and deposition into LDs (Xu et al., 2012). Furthermore, recent work revealed that LD-localized DGAT2 can form a complex with ACSL5, an acyl-CoA synthase, and ceramide synthases on the surface of LDs to catalyze the formation of acyl-ceramides that are sequestered in LDs (Senkal et al., 2017). Consistent with this, Dga1 was also involved in generating acyl-ceramides in yeast (Voynova et al., 2012). This could be a mechanism which allows for ceramide storage and detoxification. This study also showed that stored acyl-ceramides were mobilized when fatty acid biosynthesis is inhibited by the addition of cerulenin (Voynova et al., 2012). As acyl-ceramides are not well-studied, the purpose of their mobilization is not completely clear.
2.2. Enzymes of SE synthesis
In contrast to TAG production, the synthesis of SEs and their role in LD biogenesis and regulation have been less extensively studied. The ratio of TAG to SE in LDs differ depending on the cell types and growth conditions. White adipocytes, for example, contain mostly TAG-rich LDs, whereas macrophages have mainly SE LDs (Wang, 2015). Under normal growth conditions, the budding yeast Saccharomyces cerevisiae accumulates nearly equal amounts of TAGs and SEs that can be typically stored within the same LDs. In contrast, a study conducted in the fission yeast Schizosaccharomyces pombe showed that SE-rich LDs form preferentially at the cell periphery or cortical ER, whereas TAG-rich LDs form on the nuclear ER where DAG pools seem to concentrate (Meyers et al., 2016). What regulates the spatial distribution of the neutral lipids within these LD population remains unclear.
Several enzymes in the SE biosynthesis pathway have been observed to localize to both the ER and LDs, suggesting SE production may be possible at both cellular compartments, but also may serve to regulate SE production (Goodman, 2009). The squalene epoxidase Erg1 is normally observed at the ER by fluorescent microscopy, but is also detected in the LD proteome in both yeast and mammalian cells (Leber et al., 1998; Yamamoto and Bloch, 1970). Surprisingly, this LD localization was found to inhibit Erg1 enzymatic activity, possibly by sequestering it away from functional partners which remain bound to the ER (Ono et al., 1980; Shibata et al., 2001). On the other hand, the lanosterol synthase Erg7, which also localizes to these two compartments, is active mainly on LDs rather than the ER (Milla et al., 2002). In addition to Erg1 and Erg7, several other enzymes in the sterol biosynthesis pathway preferentially localize to the ER or LDs. This dual localization may serve as a mechanism to regulate SE production by restricting their access to binding partners or metabolic intermediates.
Taken together, these and other studies indicate that differential targeting of TAG and SE enzymes to LDs is an evolutionary conserved mechanism which likely plays a fundamental role in regulating lipid storage and diversifying LD functions, supporting the concept that LDs are active metabolic hubs that balance cellular lipid composition.
3. Biophysical mechanisms for ER-mediated LD formation
LDs can form and sprout from the ER bilayer surface, but the precise mechanism by which newly synthesized neutral lipids are channeled into nascent LDs is unknown. Modelling studies reveal that, at low concentrations, TAG molecules are positioned in phospholipid membranes with their glycerol backbones facing the membrane-water interface (Duelund et al., 2013). These studies showed that the solubility of TAG in membranes was lowered in the presence of cholesterol, suggesting that local changes in membrane lipid composition may play an important role in regulating TAG deposition into nascent LDs (Spooner and Small, 1987). At high concentrations of TAG (5 – 10 mol%), molecules coalesce to form oil lenses within the two phospholipid monolayers (Duelund et al., 2013; Khandelia et al., 2010). This phenomenon, demixing, serves to minimize surface tension or the energy cost associated with unfavorable interactions between the two unmixable phases, neutral lipids and phospholipid bilayers (Roux and Loewith, 2017; Thiam et al., 2013).
The phospholipid composition of membranes determines their biophysical properties. Direct measurements of surface tension of synthetic liposomes with different lipid composition showed that membrane regions with higher surface tension favor LD growth and budding (Ben M’barek et al., 2017). The molecular geometry of lipids that enrich locally at the LD/ER interface of growing LDs (bridge, neck, or pore) may determine the stability of this connection, affecting LD size and budding efficiency. Lipids with negative curvature (such as DAG, PE, PA, or cholesterol) would stabilize the connection between the ER and the LD monolayer, whereas the presence of excess positively curved lipids such as PI and lysophospholipids disrupts the stability of this association and would favor pinching-off or detachment of LDs. Notably, the neutral lipid content of LDs may dictate the phospholipid requirements for budding at least in vitro. For example, it was shown that PC alone can promote budding of TAG but not SE LDs (Ben M’barek et al., 2017).
To regulate LD emergence from the ER, cells may regulate membrane surface tension by altering the local composition of ER phospholipids (Chorlay and Thiam, 2018; Choudhary et al., 2018). For example, increasing lyso-PC in the ER through genetic manipulation led to increased LD nucleation and budding in vivo. Conversely, increasing squalene content (which lowers surface tension in vitro) reduced LD emergence (Ben M’barek et al., 2017). Thus, locally modulating ER membrane phospholipid composition may be a mechanism by which cells control LD size, number, and budding efficiency. The local surface tension of ER membranes is also important to determine the directionality of LD budding (Chorlay and Thiam, 2018; Chorlay et al., 2019). In most cases, LD budding occurs towards the cytoplasm and this appears to be regulated in part by the asymmetry in monolayer tension of the ER membrane.
While evidence is mounting that LD formation is influenced by membrane lipid composition, less is known about how the ER architecture may affect this process. The ER network is composed of flat membrane sheets and highly curved tubules that are maintained by different curvature-inducing and -stabilizing proteins such as reticulons and atlastins (Hu et al., 2009; Voeltz et al., 2006). Intriguingly, mutations in ER shaping proteins that alter ER architecture also result in altered LD morphology and size (Klemm et al., 2013). Many of these mutations give rise to hereditary spastic paraplegias, a group of inherited neurologic disorders characterized deformations in the ER structure in neurons (Blackstone, 2012; Chiurchiù et al., 2014). To what extent defects in LDs associated with altered ER structure contribute to disease pathologies is unclear.
4. Proteins involved in LD biogenesis
While lipid biosynthesis in the ER is a key factor in controlling LD nucleation and budding, nascent LDs soon become enriched with assembly proteins that facilitate LD biogenesis. Among such proteins are the fat storage-inducing transmembrane (FIT) proteins and seipin.
FIT proteins.
FIT proteins comprise an evolutionary conserved family of proteins that facilitate LD budding in cells (Choudhary et al., 2015; Hayes et al., 2017). Mammals have two proteins, FIT1 and FIT2, while yeast has two FIT2 homologs, Scs3 and Ytf2. Depletion of FIT proteins causes accumulation of neutral lipids and LD lenses that fail to grow in yeast, worms, and mammals, highlighting their role in LD production (Choudhary et al., 2015). However, the molecular mechanism of how FIT proteins promote LD budding is not well understood. Purified FIT2 can bind to TAG and DAG (Gross et al., 2011). Deletion of FIT2 in yeast causes accumulation of DAG in the ER and lowering ER DAG levels is sufficient to rescue LD budding in a FIT2 knock-out. This implicates FIT2 in the metabolism of DAG, a lipid that supports negative membrane curvature (Moir et al., 2012). One possibility is that FIT2 may sense and regulate the local concentration of DAG thereby regulates lens formation. Therefore, FIT2 proteins may promote LD emergence from the ER by reducing DAG levels that typically accumulate at LD biogenesis sites (Choudhary et al., 2018).
Evidence for the involvement of FIT proteins in phospholipid metabolism suggests another possible mechanism by which FITs may promote lens formation. Comprehensive analysis of FIT proteins showed that they contain a sequence corresponding to the active site of lipid phosphatase / phosphotransferase enzymes which remove phosphate or transfer phosphatecontaining head groups between lipids (Hayes et al., 2017). Single amino-acid substitution at the putative active site abolished the ability of FITs to rescue the phenotypes associated with FIT deletion, implying some degree of phosphatase activity that is important for function (Choudhary et al., 2015; Hayes et al., 2017). In agreement with this, loss of FIT2 in yeast also caused defects in targeting of Opi1, a major transcriptional regulator of phospholipid metabolism (Hayes et al., 2017).
Seipin.
Seipin is another important protein for LD assembly. Loss of seipin results in severe lipodystrophy in patients, as originally described by Seip and Berardinelli in the 1950s (Berardinelli, 1954; Seip, 1959). It is conserved in animals, plants, and fungi (Magré et al., 2001; Szymanski et al., 2007b). Seipin is an ER integral membrane protein, the bulk of which resides in the ER lumen with the ends facing the cytoplasm (Lundin et al., 2006). Despite its ER localization, seipin is not uniformly distributed across the ER but forms distinct foci, a sub-population of which concentrate at ER-LD contacts. Seipin is also recruited to nascent droplets as they mature and is among the first proteins observed at LD lenses (Fei et al., 2008; Szymanski et al., 2007; Wang et al., 2016). A role of seipin in LD function was first suggested by genome-wide screens in yeast, where genetic ablation of seipin gene SEI1 (formerly FLD1) induced the formation of morphologically irregular (eg. small or “super-sized”) LDs that could be visualized by fluorescence microscopy (Fei et al., 2008; Szymanski et al., 2007). The ratio of supersized to small clustered droplets was increased if phospholipid synthesis was slow (inositol starvation), suggesting an inability of the ER to regulate flow of phospholipids into nascent droplets or prevent droplet fusion in the absence of seipin (Fei et al., 2011). The LD-ER clusters in the null strain also prevented segregation of LDs to buds (Wolinski et al., 2011).
Seipin plays important roles in both droplet assembly and droplet maintenance through ER contacts. LD assembly can be observed in yeast by inducing an enzyme that generates neutral lipid. Droplets in seipin-null cells are slow to form, with neutral lipids instead accumulating in the ER. While single growing spherical LDs form in the wild type, clusters of tiny isolated lipid drops with accompanying ER fragments are initially observed in the knockout strain (Cartwright et al., 2015). In cultured S2 cells, a short exposure of cells to oleic acid results in small lipid-containing structures in the ER which can be visualized by the fluorescently-labeled LD targeting peptide LiveDrop (Wang et al., 2016). In wild-type cells these soon convert to larger BODIPY-stained puncta. In the seipin-null strain however, the BODIPY puncta are much less efficiently produced from the LiveDrop particles, which continue to multiply (Wang et al., 2016). In both yeast and animal systems, therefore, seipin promotes the development of nascent droplets.
Besides a role in droplet formation, seipin is essential for maintaining ER-LD contacts (Grippa et al., 2015; Salo et al., 2016, 2019). In wild-type cultured cells, the ER-LD contact, characterized by an electron-dense plaque between the two organelles, is fairly uniform in area and morphology. The junctions in seipin-null cells, by contrast, are more heterogeneous in size and shape, often much larger, and some droplets lacked connections with the ER entirely, a situation not usually observed in wild type cells (Salo et al., 2016). The composition of the ER-LD junction is also different in seipin-null cells, with PA, a bilayer-destabilizing lipid, accumulating at this site (Fei et al., 2011; Grippa et al., 2015; Han et al., 2015; Wolinski et al., 2015). In line with this, droplets released from the ER in the absence of seipin are also observed in yeast cells, a cell system in which LDs are thought to be permanently attached to the ER, and some droplets are even observed in the nucleus (Cartwright et al., 2015; Wolinski et al., 2015). Indeed, nuclear droplets are very rare in yeast, although seipin-catalyzed droplet formation into the nucleoplasm, in which droplets remain tethered to the inner nuclear membrane, can occur (Romanauska and Köhler, 2018). Protein trafficking to droplets also is disrupted in the absence of seipin. Some lipases target inefficiently to droplets in seipin-null yeast cells in an older study (Wolinski et al., 2011), and ACSL3, an ER enzyme that concentrates around droplets and on their surface, failed to traffic to droplets in cultured cells, as shown more recently (Salo et al., 2016). Proper lipid trafficking to droplets also depends on seipin (Salo et al., 2019). C12-BODIPY, a fluorescent fatty acid analogue, failed to become esterified and incorporate into established LDs, although it could traffic to newly developing ones (Salo et al., 2016). The facilitation of neutral lipid trafficking from ER to LDs by seipin maintains LD size homogeneity and prevents transfer between droplets resulting in size heterogeneity (Salo et al., 2019). The conclusion is that seipin is not only required for the initial steps of budding of LDs from the ER, but is also essential for maintaining functional junctions which permit trafficking of neutral lipids and at least a subset of proteins to established droplets.
Structural studies on purified seipin are ongoing to determine its function. Seipin (Sei1, formerly Fld1) was first purified from yeast and shown to be a homo-oligomer of approximately nine subunits that formed a toroid (Binns et al., 2010). Similarly, human seipin was purified and estimated to form dodecamers based on atomic force microscopy (Sim et al., 2013). Recently published cryo-EM structures of the luminal domains from human and fly (the transmembrane domains and cytosolic-facing termini could not be resolved) reveal the protein to consist of 11 and 12 seipin subunits, respectively (Sui et al., 2018; Yan et al., 2018); unpublished data on yeast Sei1 shows it to be a 10-mer, suggesting that the absolute number of subunits is not important for function. The seipin oligomer displays radial symmetry with an aperture in the middle. Both transmembrane domains of each subunit, although not visualized, emanate from the luminal domains at their periphery, i.e., distal from the central aperture.
Each subunit of the luminal domains of human, fly, and yeast seipin consist of one to three alpha-helices close to the central aperture with a sandwich of two beta-sheets comprising the rest of the domain ((Sui et al., 2018; Yan et al., 2018) and unpublished results). The structural features resemble lipid-binding domains of proteins such as NPC2, in which the lipid is enclosed by the beta sandwich. The human protein can bind anionic phospholipids including PA, although the physiological importance of this has not yet been determined. In the fly and human structures, the alpha helices are hydrophobic and are modeled to intimately interact with the ER, perhaps sensing and being stabilized by a neutral lipid lens at the earliest stage of droplet formation and thereafter stabilizing the growing droplet. Unlike the fly and human forms, the seipin alpha-helices in yeast are not hydrophobic; the sensing function could be provided by a hydrophobic domain of a binding partner, the best candidate of which is Ldb16 (Grippa et al., 2015; Wang et al., 2014a).
Seipin shows genetic interactions with several other proteins involved in lipid metabolism, which provides insights into its role in LD biogenesis and crosstalk with other organelles. Yeast seipin was shown to interact with Pex30, a protein involved in peroxisomal assembly. In line with this, LDs and peroxisomes are now known to emanate from the same ER sub-domains, implying roles for seipin in crosstalk with other cellular machinery involved in peroxisome biogenesis (Joshi et al., 2018; Pagac et al., 2016; Wang et al., 2018). Thus, the regulation of lipid droplets (which store esterified fatty acids) and peroxisomal formation (peroxisomes are the only organelle in yeast to oxidize fatty acids) appear to be functionally linked.
Finally, the role of seipin in LD formation must be reconciled with its role in promoting adipogenesis, a transcriptional pathway; PPARγ signaling is key in this process, and it is disrupted in seipin-deficient cells (Chen et al., 2009; Payne et al., 2008). A key may be glycerolipid synthesis. The absence of seipin results in an increase in 3-glycerolphosphate acyltransferase (GPAT) activity, resulting in elevated PA (Pagac et al., 2016). Interestingly, an increase in GPAT enzyme suppressed adipogenesis, suggesting that a lipid, possibly PA, is inhibiting PPARγ signaling, tying a loss of normal lipid droplet assembly with adipogenesis. However, several other models exist to link seipin with adipogenesis, including a role in ER calcium homeostasis (Bi et al., 2014), actin cytoskeleton remodeling (Yang et al., 2014), and prevention of inflammation (Qiu et al., 2013). Dissection of primary from secondary effects, and establishing causation, will be paramount in understanding the role of seipin in this developmental process.
5. Collaboration of LDs with the ER and other organelles at junctions
Numerous studies indicate that LDs are not randomly distributed within the cytoplasm, and their organization reflects the metabolic status of the cell. This intracellular LD positioning is often facilitated by LDs making direct contact with other organelles to promote lipid exchange and metabolic interactions with these compartments. These interactions also provide metabolic adaptations to stress. For example, LD-mitochondria or LD-peroxisomes contacts allow LDs to donate fatty acids to these organelles for energy production through oxidation (Binns et al., 2006; Chang et al., 2019; Joshi et al., 2018; Rambold et al., 2015; Wang et al., 2011). Compartmentalizing neutral lipid metabolism at LDs within the ER network may regulate the efficiency of distinct metabolic pathways (Coleman, 2019). Indeed the organization of neutral lipid synthesis enzymes during ER-localized LD production bears striking resemblance to the formation of sequential enzymatic assemblies, or what Paul Srere defined as metabolons, which can enhance metabolic efficiency by channeling substrates, thereby regulating steady-state flux and the availability of intermediate metabolites that participate in competing reactions (Ovádi and Srere, 1999; Srere, 1987). Although this notion has been demonstrated in several examples in vitro and in artificially engineered synthetic biological systems (Avalos et al., 2013; Chen and Silver, 2012; Dueber et al., 2009; Lee et al., 2012), it remains unclear to what extent this organization exists at a cellular level, let alone its metabolic consequences.
Another physiological outcome for organizing neutral lipid metabolism within the ER network may be promoting cellular adaptations to stress to maintain ER homeostasis and prevent lipotoxicity. In response to an influx of fatty acids, LD biogenesis is normally up-regulated and fatty acids, which can act as detergents, are sequestered into TAG in LDs (Figure 1). In mammalian cells, saturated fatty acids such as palmitate (C16:0), can become incorporated into ER-resident glycerophospholipids (phosphatidylcholine, for example) in the absence of LD function, causing defects in membrane fluidity and compromise ER structure and function if LD biogenesis is disturbed (Piccolis et al., 2019). The addition of exogenous fatty acids also leads to defects in ER structure and function in yeast, and this is worsened if LD biogenesis is perturbed (Hariri et al., 2019).
Figure 1. Metabolic fates of fatty acids.
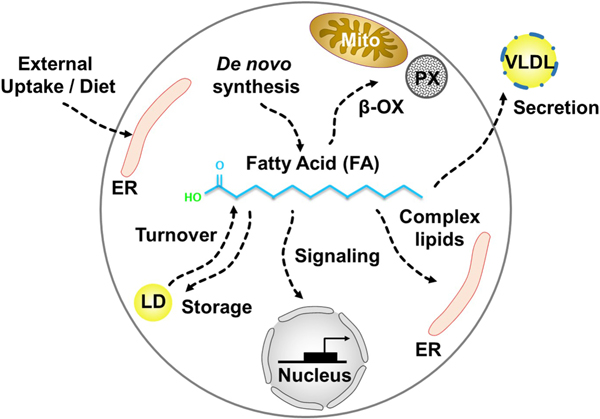
FAs synthesized de novo or taken up from the external cellular environment have multiple metabolic fates. These include incorporation into membrane phospholipids and complex lipids, degradation and energy generation by oxidation, storage in lipid droplets, esterification to proteins, signaling and regulation of transcription factors, or secretion.
6. LDs as stress response factors that promote cellular homeostasis
To combat lipotoxicity, cells express numerous fatty acyl-CoA ligases that can esterify fatty acids and actively promote their incorporation into membranes and neutral lipids. Defects in the incorporation of toxic fatty acids through the ablation or pharmacological inhibition of the DGAT enzymes can cause fatty acid accumulation in the ER bilayer leading to ER dysfunction, protein mis-folding, and the activation of the Unfolded Protein Response (UPR) pathway (Borradaile et al., 2006; Wei et al., 2006). Consistent with this, inducing ER stress stimulates LD formation in yeast, underscoring the functional relationship between ER homeostasis and LD biogenesis (Fei et al., 2009). Remarkably, in yeast fatty acid detoxification itself may be spatially compartmentalized to distinct sub-regions of the ER network. When cultured in oleate-containing medium, the yeast fatty acyl-CoA ligase Faa1 interacts with ER-vacuole tether Mdm1 and promotes LD biogenesis at the nuclear ER-vacuole junction (NVJ) (Hariri et al., 2018). As a consequence, Mdm1 is required to maintain ER homeostasis in the presence of exogenous fatty acids, and loss of Mdm1 leads to defects in ER morphology and sensitivity to lipotoxic fatty acids (Hariri et al., 2019). Similarly, Mdm1-deficient yeast also manifest defects in the incorporation of fatty acids into LDs at the NVJ, suggesting that yeast can spatially localize LD biogenesis at the NVJ to maintain ER lipid homeostasis.
Cells also express an arsenal of proteins that efficiently process or dilute saturated fatty acids. In mammalian cells, recent CRISPR-based genetic screens have revealed factors necessary for protection against palmitate-induced cell death. These include proteins necessary for the production of unsaturated fatty acids (ACSL4, SCAP, AGPAT1, and CEPT1), the synthesis of which can dampen palmitate toxicity (Zhu et al., 2019). Moreover, addition of palmitate to DGAT-deficient cells is lethal, but can be blunted by co-incubation with the unsaturated FA oleate. Interestingly, loss of ACSL3 was hyper-protective against palmitate toxicity, likely because this fatty acyl-CoA ligase primarily activates saturated FAs so that they can be incorporated into downstream lipids (Zhu et al., 2019). This study also identified a poorly characterized protein called CHP1 that regulates glycerol-3-phosphate acyltransferase 4 (GPAT4) that is involved in the initial stages of TAG biosynthesis. A parallel independent study conducting both transcriptomic and lipidomic analysis revealed that elevated palmitate influx causes increased saturated FA incorporation into several glycerophospholipids including PA and lysophospholipids (Piccolis et al., 2019). This work also showed that depletion of the E3 ubiquitin ligase domaincontaining protein RNF213 was protective against palmitate lipotoxicity. Although its function is unknown, RNF213 mutations are linked to Moyamoya vascular disease, and RNF213 may regulate some aspect of fatty acid desaturation (Piccolis et al., 2019). Collectively, these studies indicate that the cell has many ways to suppress saturated fatty acid incorporation into lipids, the failure of which leads to potent cell death.
In yeast, failure to store fatty acids in LDs alters the functions of numerous cellular pathways. LD-deficient yeast (in which the enzymes necessary for TAG and SE synthesis are genetically ablated) are alive, but exhibit defects in macro-autophagy and the formation of the autophagophore (Velázquez et al., 2016). This could be caused by general defects in ER homeostasis, the source organelle for autophagophore biogenesis. Additionally, LDs may be required to supply fatty acids necessary for building the autophagophore membrane. Consistent with this, LD-deficient yeast also exhibit defects in sporulation because LDs donate fatty acids necessary to build the prospore membrane (Hsu et al., 2017). These studies suggests that fatty acids serve as general building blocks for membrane synthesis, and the failure of LDs to provide these fatty acids may perturb the cell’s ability to mount stress response during times of nutrient shortage.
7. Organelle tethers in spatially-organized LD biogenesis
How do cells organize their LDs to ensure their maintenance and efficient mobilization when the need arises? In yeast facing carbon starvation, LDs are produced on the surface of the nuclear envelope and accumulate next to the vacuole (eg. yeast lysosome), clustering them at an inter-organelle contact site known as the nucleus-vacuole junction (NVJ). Indeed, the NVJ becomes a hotspot for LD biogenesis itself, and accumulates numerous lipid synthesis enzymes including the PA phosphatase Pah1 that promotes the local synthesis of DAG, the precursor to TAG (Adeyo et al., 2011; Barbosa et al., 2015). Recent studies also identified specific proteins that decorate NVJ-associated LDs. These include Lipid Droplet Organizing (LDO) proteins Ldo16 and Ldo45, which decorate the LD monolayer surface and are required for protein Pdr16 to be recruited to LDs (Eisenberg-Bord et al., 2017; Teixeira et al., 2017). Remarkably, Ldo16 and Ldo45 are isoforms and products of the same genetic ORF in yeast. Both Ldo45 and Ldo16 are expressed in exponentially growing yeast, whereas expression levels of Ldo45 are diminished in stationary phase, indicating the expression level of these proteins is metabolically regulated. In humans, a protein termed promethin that has strong structural homology to LDOs was also found to associate with LDs and co-purify with seipin (Castro et al., 2019).
Another protein that associates with LDs at the NVJ is Mdm1 (Henne et al., 2015). Mdm1 binds LDs via its Phox-Associated (PXA) domain, allowing this protein to simultaneously establish tri-organelle contacts between ER, LD, and vacuole and thus promote LD clustering at the ER-vacuole interface (Hariri et al., 2018, 2019). Beyond its role as a molecular tether, Mdm1 also interacts with the fatty acyl-CoA ligase Faa1, and that is sufficient to recruit Faa1 to sites of LD biogenesis in yeast to promote the formation of a local fatty acid pool that facilitates LD growth. Consistent with this, loss of either MDM1 or FAA1 causes similar defects in fatty acid activation and sensitizes yeast to lipotoxicity, suggesting that Mdm1 coordinates local fatty acid activation at the NVJ to facilitate fatty acid incorporation into TAG (Hariri et al., 2019).
8. Yeast lipophagy and ER homeostasis
Why do yeast LDs accumulate at the NVJ during nutritional stress? One model posits that these LDs will eventually be delivered into the vacuole lumen for breakdown via a micro-autophagy process known as micro-lipophagy (Seo et al., 2017; Toulmay and Prinz, 2013; Wang et al., 2014b). Yeast grown into late stationary phase eventually deliver their LDs into the vacuole lumen, but this process is only observed after multiple days of low-nutrient subsistence (Barbosa and Siniossoglou, 2016). LD clustering at the NVJ actually occurs early in the transition from exponential growth into stationary phase, a time-frame known as the diauxic shift when cell growth slows and yeast transition from glycolysis into aerobic respiration (Barbosa et al., 2015; Hariri et al., 2018). This metabolic transition requires drastic changes in metabolic signaling at the vacuole/lysosome, the organelle from which the TORC1 complex regulates cell growth and division (Murley et al., 2017). Yeast undergoing diauxic shift also exhibit slowed membrane synthesis and increased mitochondrial biogenesis, which may lead to a buildup and/or re-allocation of cellular fatty acids as phospholipid production is altered. Thus, an intriguing hypothesis is that LD biogenesis at the NVJ provides a metabolic platform to efficiently process fatty acids which can accumulate in this metabolic transition phase. These NVJ-associated LDs may also execute specialized signaling functions with the vacuole/lysosome that allow it to metabolically remodel itself in preparation for stationary phase subsistence. Collectively, NVJ-associated LDs may play signaling as well as lipid storage roles that promote the metabolic transition into low-nutrient subsistence. Their biogenesis may also protect the cell from FA-induced lipotoxicity during the onset of stationary phase, as these LDs will sequester away otherwise toxic FAs, which can ultimately be deposited into the vacuole via micro-lipophagy during long-term starvation, or mobilized by cytoplasmic lipases if growth is resumed.
9. Current questions and future perspectives
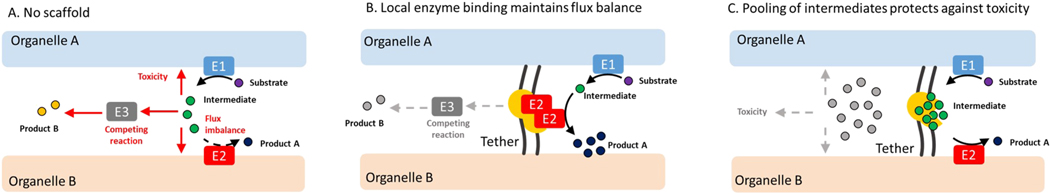
Despite recent advancements, several important question remain concerning the initial stages of LD formation. It is unclear how the many neutral lipid synthesis enzymes organize and work together in response to different metabolic cues to promote droplet formation, and what triggers their dispersion during times of low LD growth. Specific roles for various scaffolding proteins or ER-LD tethers in coordinating LD formation at inter-organelle junctions also remain largely unclear. We propose that inter-organelle tethers and scaffolds constitute an important and still unexplored mechanism that cells use to regulate metabolic efficiency, through binding enzymes and pooling lipid intermediates at sites of organelle biogenesis (Figure 2). These organizing factors become critical for shunting common lipid intermediates such as DAG, which is shared between neutral lipid and phospholipid synthesis pathways, into specific destinations such as LD lens formation. How LD-associated proteins like seipin, FIT proteins, Ldo45/16/promethin, and Mdm1/Snx14 contribute to lipid processing and LD production to maintain cellular lipid balance are unclear. To answer these questions new tools for tracking lipid movements and methods to detect transient protein assemblies will be needed. A molecular understanding of how LD metabolism is spatially organized on a subcellular level will be an exciting challenge for future investigations. New functions for LDs in signaling and switching metabolic pathways on and off will also emerge. High-resolution structural information from intact LDs inside cells will likely yield many insights into these processes.
Figure 2. Potential mechanisms for organelle tethers / scaffolds in regulating metabolic organization.
In the absence of compartmentalization cells face many challenges. Enzyme availability affects flux through metabolic pathways. Toxic intermediates accumulate in membranes and cause toxicity. Intermediates enter into competing reactions diluting the final product yield (A). Scaffolds or tethers recruit metabolic enzyme complexes which enhances pathway efficiency (B). Scaffolds can also bind and sequester intermediates which enhances their stability and prevents their entry into competing reactions. Increasing local concentration of intermediates also enhances metabolic efficiency (C).
Highlights.
Lipid droplets biogenesis is spatially compartmentalized within specialized domains of the ER network
Several factors drive the formation of lipid droplets including enzymes, lipids, and structural proteins
Lipid droplets collaborate with subcellular organelles via membrane contact sites
Lipid droplets are active metabolic hubs that balance cellular lipid composition
Acknowledgements:
Support is gratefully acknowledged from the NIH (R01GM084210 to JMG and R35GM119768 to WMH), American Diabetes Association (7–13-BS-055 to JMG), American Heart Association (16GRNT27540010 to JMG), The Welch Foundation (I-1936 to MLR and I1873 to WMH), and The Searle Foundation (SSP-2016–1482 to WMH).
Abbreviations:
- LD
lipid droplet
- ER
endoplasmic reticulum
- TAG
triacylglycerol
- DAG
diacylglycerol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, and Goodman JM (2011). The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez HM, and Steinbüchel A (2002). Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 60, 367–376. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, and Daum G (1997). Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J. Bacteriol. 179, 7611–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos JL, Fink GR, and Stephanopoulos G (2013). Compartmentalization of metabolic pathways in yeast mitochondria improves production of branched chain alcohols. Nat. Biotechnol. 31, 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, and Siniossoglou S (2016). Spatial distribution of lipid droplets during starvation: Implications for lipophagy. Commun. Integr. Biol. 9:4, DOI: 10.1080/19420889.2016.1183854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, and Siniossoglou S (2017). Function of lipid droplet-organelle interactions in lipid homeostasis. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1459–1468. [DOI] [PubMed] [Google Scholar]
- Barbosa AD, Sembongi H, Su W-M, Abreu S, Reggiori F, Carman GM, and Siniossoglou S (2015). Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell 26, 3641–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R, Li W-H, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RGW, Liu P, and Chapman KD (2007). Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 48, 837–847. [DOI] [PubMed] [Google Scholar]
- Ben M’barek K, Ajjaji D, Chorlay A, Vanni S, Forêt L, and Thiam AR (2017). ER Membrane Phospholipids and Surface Tension Control Cellular Lipid Droplet Formation. Dev. Cell 41, 591–604.e7. [DOI] [PubMed] [Google Scholar]
- Berardinelli W (1954). An undiagnosed endocrinometabolic syndrome: report of 2 cases. J. Clin. Endocrinol. Metab. 14, 193–204. [DOI] [PubMed] [Google Scholar]
- Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA, Nomura DK, and Olzmann JA (2018). A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Dev. Cell 44, 97–112.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Wang W, Liu Z, Huang X, Jiang Q, Liu G, Wang Y, and Huang X (2014). Seipin promotes adipose tissue fat storage through the ER Ca2+-ATPase SERCA. Cell Metab. 19, 861–871. [DOI] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RGW, and Goodman JM (2006). An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D, Lee S, Hilton CL, Jiang Q-X, and Goodman JM (2010). Seipin is a discrete homooligomer. Biochemistry 49, 10747–10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C (2012). Cellular pathways of hereditary spastic paraplegia. Annu. Rev. Neurosci. 35, 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaner WS, O’Byrne SM, Wongsiriroj N, Kluwe J, D’Ambrosio DM, Jiang H, Schwabe RF, Hillman EMC, Piantedosi R, and Libien J (2009). Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim. Biophys. Acta 1791, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradaile NM, Han X, Harp JD, Gale SE, Ory DS, and Schaffer JE (2006). Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J. Lipid Res. 47, 2726–2737. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Dolios G, Shapiro L, and Wang R (2004). Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279, 46835–46842. [DOI] [PubMed] [Google Scholar]
- Cartwright BR, Binns DD, Hilton CL, Han S, Gao Q, and Goodman JM (2015). Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell 26, 726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Smith SJ, Zheng Y-W, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, et al. (1998). Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. 95, 13018–13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro IG, Eisenberg-Bord M, Persiani E, Rochford JJ, Schuldiner M, and Bohnert M (2019). Promethin Is a Conserved Seipin Partner Protein. Cells 2019, 8(3), 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-L, Weigel AV, Ioannou MS, Pasolli HA, Xu CS, Peale DR, Shtengel G, Freeman M, Hess HF, Blackstone C, et al. (2019). Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. J. Cell Biol. jcb.201902061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AH, and Silver PA (2012). Designing biological compartmentalization. Trends Cell Biol. 22, 662–670. [DOI] [PubMed] [Google Scholar]
- Chen W, Yechoor VK, Chang BH-J, Li MV, March KL, and Chan L (2009). The human lipodystrophy gene product Berardinelli-Seip congenital lipodystrophy 2/seipin plays a key role in adipocyte differentiation. Endocrinology 150, 4552–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitraju C, Walther TC, and Farese RV (2019). The triglyceride synthesis enzymes DGAT1 and DGAT2 have distinct and overlapping functions in adipocytes. J. Lipid Res. 60, 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurchiù V, Maccarrone M, and Orlacchio A (2014). The Role of Reticulons in Neurodegenerative Diseases. Neuromolecular Med. 16, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlay A, and Thiam AR (2018). An Asymmetry in Monolayer Tension Regulates Lipid Droplet Budding Direction. Biophys. J. 114, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlay A, Monticelli L, Ferreira JV, M’barek KB, Ajjaji D, Wang S, Johnson E, Beck R, Omrane M, Beller M, et al. (2019). Membrane Asymmetry Imposes Directionality on Lipid Droplet Emergence from the ER. Dev Cell. 50(1):25–42.e7. [DOI] [PubMed] [Google Scholar]
- Choudhary V, Ojha N, Golden A, and Prinz WA (2015). A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J Cell Biol 211, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V, Golani G, Joshi AS, Cottier S, Schneiter R, Prinz WA, and Kozlov MM (2018). Architecture of Lipid Droplets in Endoplasmic Reticulum Is Determined by Phospholipid Intrinsic Curvature. Curr. Biol. CB 28, 915–926.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA (2019). It takes a village: channeling fatty acid metabolism and triacylglycerol formation via protein interactomes. J. Lipid Res. 60(3):490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, and Lee DP (2004). Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43, 134–176. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KLJ, and Keasling JD (2009). Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759. [DOI] [PubMed] [Google Scholar]
- Duelund L, Jensen GV, Hannibal-Bach HK, Ejsing CS, Pedersen JS, Pakkanen KI, and Ipsen JH (2013). Composition, structure and properties of POPC-triolein mixtures. Evidence of triglyceride domains in phospholipid bilayers. Biochim. Biophys. Acta 1828, 1909–1917. [DOI] [PubMed] [Google Scholar]
- Eisenberg-Bord M, Mari M, Weill U, Rosenfeld-Gur E, Moldavski O, Castro IG, Soni KG, Harpaz N, Levine TP, Futerman AH, et al. (2017). Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J Cell Biol. 217(1):269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, and Yang H (2008). Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Wang H, Fu X, Bielby C, and Yang H (2009). Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem. J. 424, 61–67. [DOI] [PubMed] [Google Scholar]
- Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, et al. (2011). A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7, e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, and Ohsaki Y (2006). Cytoplasmic lipid droplets: rediscovery of an old structure as a unique platform. Ann. N. Y. Acad. Sci. 1086, 104–115. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, and Parton RG (2011). Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, and Goodman JM (2015). The lipid droplet—a well-connected organelle. Front. Cell Dev. Biol. 3(49). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Binns DD, Kinch LN, Grishin NV, Ortiz N, Chen X, and Goodman JM (2017). Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J Cell Biol 216, 3199–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JM (2009). Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50, 2148–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa A, Buxó L, Mora G, Funaya C, Idrissi F-Z, Mancuso F, Gomez R, Muntanyà J, Sabidó E, and Carvalho P (2015). The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DA, Zhan C, and Silver DL (2011). Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. 108, 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Binns DD, Chang Y-F, and Goodman JM (2015). Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(ΔNterm) only in combination with Ldb16p. BMC Cell Biol. 16, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri H, Rogers S, Ugrankar R, Liu YL, Feathers JR, and Henne WM (2018). Lipid droplet biogenesis is spatially coordinated at ER–vacuole contacts under nutritional stress. EMBO Rep. 19, 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri H, Speer N, Bowerman J, Rogers S, Fu G, Reetz E, Datta S, Feathers JR, Ugrankar R, Nicastro D, et al. (2019). Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J Cell Biol. 218(4):1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M, Choudhary V, Ojha N, Shin JJ, Han G-S, Carman GM, Loewen CJ, Prinz WA, and Levine T (2017). Fat storage-inducing transmembrane (FIT or FITM) proteins are related to lipid phosphatase/phosphotransferase enzymes. Microb. Cell Graz Austria 5, 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Zhu L, Balogi Z, Stefan C, Pleiss JA, and Emr SD (2015). Mdm1/Snx13 is a novel ER–endolysosomal interorganelle tethering protein. J. Cell Biol. 210, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T-H, Chen R-H, Cheng Y-H, and Wang C-W (2017). Lipid droplets are central organelles for meiosis II progression during yeast sporulation. Mol. Biol. Cell 28, 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu P-P, Voss C, Rismanchi N, Prinz WA, Rapoport TA, and Blackstone C (2009). A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtley S (2009). Spatial cell biology. Location, location, location. Introduction. Science 326, 1205. [DOI] [PubMed] [Google Scholar]
- Irshad Z, Dimitri F, Christian M, and Zammit VA (2017). Diacylglycerol acyltransferase 2 links glucose utilization to fatty acid oxidation in the brown adipocytes. J. Lipid Res. 58, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, and Schneiter R (2011). Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci 124, 2424–2437. [DOI] [PubMed] [Google Scholar]
- Joshi AS, Nebenfuehr B, Choudhary V, Satpute-Krishnan P, Levine TP, Golden A, and Prinz WA (2018). Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 9(1):2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EP (1957). Metabolism of Lipides. Annu. Rev. Biochem. 26, 119–148. [DOI] [PubMed] [Google Scholar]
- Kennedy EP (1961). Biosynthesis of complex lipids. Fed. Proc. 20, 934–940. [PubMed] [Google Scholar]
- Khandelia H, Duelund L, Pakkanen KI, and Ipsen JH (2010). Triglyceride Blisters in Lipid Bilayers: Implications for Lipid Droplet Biogenesis and the Mobile Lipid Signal in Cancer Cell Membranes. PLOS ONE 5(9), e12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm RW, Norton JP, Cole RA, Li CS, Park SH, Crane MM, Li L, Jin D, Boye-Doe A, Liu TY, et al. (2013). A Conserved Role for Atlastin GTPases in Regulating Lipid Droplet Size. Cell Rep. 3, 1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, Farese RV, and Walther TC (2016). Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol. 26, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerschner L, Moessinger C, and Thiele C (2008). Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic Cph. Den. 9, 338–352. [DOI] [PubMed] [Google Scholar]
- Leber R, Landl K, Zinser E, Ahorn H, Spök A, Kohlwein SD, Turnowsky F, and Daum G (1998). Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol. Biol. Cell 9, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, DeLoache WC, and Dueber JE (2012). Spatial organization of enzymes for metabolic engineering. Metab. Eng. 14, 242–251. [DOI] [PubMed] [Google Scholar]
- Lehner R, and Kuksis A (1993). Triacylglycerol synthesis by an sn-1,2(2,3)-diacylglycerol transacylase from rat intestinal microsomes. J. Biol. Chem. 268, 8781–8786. [PubMed] [Google Scholar]
- Lundin C, Nordström R, Wagner K, Windpassinger C, Andersson H, von Heijne G, and Nilsson I (2006). Membrane topology of the human seipin protein. FEBS Lett. 580, 2281–2284. [DOI] [PubMed] [Google Scholar]
- Magré J, Delépine M, Khallouf E, Gedde-Dahl T, Van Maldergem L, Sobel E, Papp J, Meier M, Mégarbané A, Bachy A, et al. (2001). Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28, 365–370. [DOI] [PubMed] [Google Scholar]
- Markgraf DF, Klemm RW, Junker M, Hannibal-Bach HK, Ejsing CS, and Rapoport TA (2014). An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Rep. 6, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFie PJ, and Stone SJ (2011). A fluorescent assay to quantitatively measure in vitro acyl CoA:diacylglycerol acyltransferase activity. J. Lipid Res. 52, 1760–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers A, del Rio ZP, Beaver RA, Morris RM, Weiskittel TM, Alshibli AK, Mannik J, Morrell-Falvey J, and Dalhaimer P (2016). Lipid Droplets Form from Distinct Regions of the Cell in the Fission Yeast Schizosaccharomyces pombe. Traffic 17, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla P, Athenstaedt K, Viola F, Oliaro-Bosso S, Kohlwein SD, Daum G, and Balliano G (2002). Yeast Oxidosqualene Cyclase (Erg7p) Is a Major Component of Lipid Particles. J. Biol. Chem. 277, 2406–2412. [DOI] [PubMed] [Google Scholar]
- Moir RD, Gross DA, Silver DL, and Willis IM (2012). SCS3 and YFT2 link transcription of phospholipid biosynthetic genes to ER stress and the UPR. PLoS Genet. 8, e1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A, Yamada J, Niles BJ, Toulmay A, Prinz WA, Powers T, and Nunnari J (2017). Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling. J. Cell Biol. 216, 2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, Nomura DK, and Olzmann JA (2017). DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev. Cell 42, 9–21.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, and Carvalho P (2019). Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Takahashi K, Odani S, Konno H, and Imai Y (1980). Purification of squalene epoxidase from rat liver microsomes. Biochem. Biophys. Res. Commun. 96, 522–528. [DOI] [PubMed] [Google Scholar]
- Ovádi J, and Srere PA (1999). Macromolecular Compartmentation and Channeling In International Review of Cytology, Walter H, Brooks DE, and Srere PA, eds. (Academic Press; ), pp. 255–280. [DOI] [PubMed] [Google Scholar]
- Pagac M, Cooper DE, Qi Y, Lukmantara IE, Mak HY, Wu Z, Tian Y, Liu Z, Lei M, Du X, et al. (2016). SEIPIN Regulates Lipid Droplet Expansion and Adipocyte Development by Modulating the Activity of Glycerol-3-phosphate Acyltransferase. Cell Rep. 17, 1546–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne VA, Grimsey N, Tuthill A, Virtue S, Gray SL, Dalla Nora E, Semple RK, O’Rahilly S, and Rochford JJ (2008). The human lipodystrophy gene BSCL2/seipin may be essential for normal adipocyte differentiation. Diabetes 57, 2055–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolis M, Bond LM, Kampmann M, Pulimeno P, Chitraju C, Jayson CBK, Vaites LP, Boland S, Lai ZW, Gabriel KR, et al. (2019). Probing the Global Cellular Responses to Lipotoxicity Caused by Saturated Fatty Acids. Mol. Cell. 74(1):32–44.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost C, Sharp ME, Kory N, Lin Q, Voth GA, Farese RV, and Walther TC (2018). Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev. Cell 44, 7386.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Wee K, Takeda K, Lim X, Sugii S, Radda GK, and Han W (2013). Suppression of adipogenesis by pathogenic seipin mutant is associated with inflammatory response. PloS One 8, e57874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Grillitsch K, and Daum G (2008). Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid Res. 47, 157–171. [DOI] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, and Lippincott-Schwartz J (2015). Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanauska A, and Köhler A (2018). The Inner Nuclear Membrane Is a Metabolically Active Territory that Generates Nuclear Lipid Droplets. Cell 174, 700–715.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, and Loewith R (2017). Tensing Up for Lipid Droplet Formation. Dev. Cell 41, 571–572. [DOI] [PubMed] [Google Scholar]
- Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magré J, Thiele C, Hölttä-Vuori M, Jokitalo E, et al. (2016). Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35, 2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Li S, Vihinen H, Hölttä-Vuori M, Szkalisity A, Horvath P, Belevich I, Peränen J, Thiele C, Somerharju P, et al. (2019). Seipin Facilitates Triglyceride Flow to Lipid Droplet and Counteracts Droplet Ripening via Endoplasmic Reticulum Contact. Dev Cell. pii: S1534–5807(19)30388–0. [DOI] [PubMed] [Google Scholar]
- Santos AXS, and Riezman H (2012). Yeast as a model system for studying lipid homeostasis and function. FEBS Lett. 586, 2858–2867. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, and Bohnert M (2017). A different kind of love – lipid droplet contact sites. Biochim. Biophys. Acta BBA - Mol. Cell Biol. Lipids 1862, 1188–1196. [DOI] [PubMed] [Google Scholar]
- Seip M (1959). Lipodystrophy and gigantism with associated endocrine manifestations. A new diencephalic syndrome? Acta Paediatr. 48, 555–574. [PubMed] [Google Scholar]
- Senkal CE, Salama MF, Snider AJ, Allopenna JJ, Rana NA, Koller A, Hannun YA, and Obeid LM (2017). Ceramide Is Metabolized to Acylceramide and Stored in Lipid Droplets. Cell Metab. 25, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo AY, Lau P-W, Feliciano D, Sengupta P, Gros MAL, Cinquin B, Larabell CA, and Lippincott-Schwartz J (2017). AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation. ELife 6, e21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Arita M, Misaki Y, Dohmae N, Takio K, Ono T, Inoue K, and Arai H (2001). Supernatant protein factor, which stimulates the conversion of squalene to lanosterol, is a cytosolic squalene transfer protein and enhances cholesterol biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 98, 2244–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim MFM, Talukder MMU, Dennis RJ, O’Rahilly S, Edwardson JM, and Rochford JJ (2013). Analysis of naturally occurring mutations in the human lipodystrophy protein seipin reveals multiple potential pathogenic mechanisms. Diabetologia 56, 2498–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanova M, Zweytick D, Lohner K, Klug L, Leitner E, Hermetter A, and Daum G (2012). Influence of squalene on lipid particle/droplet and membrane organization in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1821, 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner PJR, and Small DM (1987). Effect of free cholesterol on incorporation of triolein in phospholipid bilayers. Biochemistry 26, 5820–5825. [DOI] [PubMed] [Google Scholar]
- Srere PA (1987). Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89–124. [DOI] [PubMed] [Google Scholar]
- Stone SJ, Levin MC, and Farese RV (2006). Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2. J. Biol. Chem. 281, 40273–40282. [DOI] [PubMed] [Google Scholar]
- Sui X, Arlt H, Brock KP, Lai ZW, DiMaio F, Marks DS, Liao M, Farese RV, and Walther TC (2018). Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. Cell Biol. 217, 4080–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li W-P, Agarwal AK, Garg A, Anderson RGW, and Goodman JM (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U. S. A. 104, 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta MT, Kapterian TS, Fei W, Du X, Brown AJ, Dawes IW, and Yang H (2012). Accumulation of squalene is associated with the clustering of lipid droplets. FEBS J. 279, 4231–4244. [DOI] [PubMed] [Google Scholar]
- Teixeira V, Johnsen L, Martínez-Montañés F, Grippa A, Buxó L, Idrissi F-Z, Ejsing CS, and Carvalho P (2017). Regulation of lipid droplets by metabolically controlled Ldo isoforms. J Cell Biol. 217(1):127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, and Beller M (2017). The why, when and how of lipid droplet diversity. J. Cell Sci. 130, 315–324. [DOI] [PubMed] [Google Scholar]
- Thiam AR, Farese RV, and Walther TC (2013). The Biophysics and Cell Biology of Lipid Droplets. Nat. Rev. Mol. Cell Biol. 14, 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, and Prinz WA (2013). Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J Cell Biol 202, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, and Lippincott-Schwartz J (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez AP, Tatsuta T, Ghillebert R, Drescher I, and Graef M (2016). Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 212, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva CJ, Monetti M, Shih M, Zhou P, Watkins SM, Bhanot S, and Farese RV (2009). Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatol. Baltim. Md 50, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, and Rapoport TA (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586. [DOI] [PubMed] [Google Scholar]
- Voynova NS, Vionnet C, Ejsing CS, and Conzelmann A (2012). A novel pathway of ceramide metabolism in Saccharomyces cerevisiae. Biochem. J. 447, 103–114. [DOI] [PubMed] [Google Scholar]
- Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla H-J, Kalscheuer R, Stöveken T, Landenberg PV, et al. (2005). Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol. Microbiol. 55, 750–763. [DOI] [PubMed] [Google Scholar]
- Walther TC, and Farese RV (2012). Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Chung J, and Farese RV (2017). Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 33, 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-W (2015). Lipid droplet dynamics in budding yeast. Cell. Mol. Life Sci. CMLS 72, 2677–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-W, Miao Y-H, and Chang Y-S (2014a). Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127, 1214–1228. [DOI] [PubMed] [Google Scholar]
- Wang C-W, Miao Y-H, and Chang Y-S (2014b). A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol 206, 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sreenivasan U, Hu H, Saladino A, Polster BM, Lund LM, Gong D, Stanley WC, and Sztalryd C (2011). Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52, 2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, et al. (2016). Seipin is required for converting nascent to mature lipid droplets. ELife 5, e16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Idrissi F-Z, Hermansson M, Grippa A, Ejsing CS, and Carvalho P (2018). Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat. Commun. 9, 2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Wang D, Topczewski F, and Pagliassotti MJ (2006). Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am. J. Physiol. Endocrinol. Metab. 291, E275–281. [DOI] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng J-X, Graham M, Christiano R, Fröhlich F, et al. (2013). Triacylglycerol Synthesis Enzymes Mediate Lipid Droplet Growth by Relocalizing from the ER to Lipid Droplets. Dev. Cell 24, 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Thiam AR, Olarte M-J, Wang J, Beck R, Gould TJ, Allgeyer ES, Pincet F, Bewersdorf J, Farese RV, et al. (2014). Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. ELife 3, e01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William M (2010). Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos. Trans. R. Soc. B Biol. Sci. 365, 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H, Kolb D, Hermann S, Koning RI, and Kohlwein SD (2011). A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci 124, 3894–3904. [DOI] [PubMed] [Google Scholar]
- Wolinski H, Hofbauer HF, Hellauer K, Cristobal-Sarramian A, Kolb D, Radulovic M, Knittelfelder OL, Rechberger GN, and Kohlwein SD (2015). Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta 1851, 1450–1464. [DOI] [PubMed] [Google Scholar]
- Wurie HR, Buckett L, and Zammit VA (2012). Diacylglycerol acyltransferase 2 acts upstream of diacylglycerol acyltransferase 1 and utilizes nascent diglycerides and de novo synthesized fatty acids in HepG2 cells. FEBS J. 279, 3033–3047. [DOI] [PubMed] [Google Scholar]
- Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV, and Mak HY (2012). The FATP1–DGAT2 complex facilitates lipid droplet expansion at the ER–lipid droplet interface. J Cell Biol 198, 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, and Bloch K (1970). Studies on Squalene Epoxidase of Rat Liver. J. Biol. Chem. 245, 1670–1674. [PubMed] [Google Scholar]
- Yan R, Qian H, Lukmantara I, Gao M, Du X, Yan N, and Yang H (2018). Human SEIPIN Binds Anionic Phospholipids. Dev. Cell. 47(2):248–256.e4 [DOI] [PubMed] [Google Scholar]
- Yang W, Thein S, Wang X, Bi X, Ericksen RE, Xu F, and Han W (2014). BSCL2/seipin regulates adipogenesis through actin cytoskeleton remodelling. Hum. Mol. Genet. 23, 502–513. [DOI] [PubMed] [Google Scholar]
- Yen C-LE, Stone SJ, Koliwad S, Harris C, and Farese RV (2008). DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 49, 2283–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Puthenveedu SN, Shen Y, La K, Ozlu C, Wang T, Klompstra D, Gultekin Y, Chi J, Fidelin J, et al. (2019). CHP1 Regulates Compartmentalized Glycerolipid Synthesis by Activating GPAT4. Mol. Cell 74(1):45–58.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]