Abstract
Neoadjuvant radiation is standard of care for locally advanced rectal cancer. Response to radiation is highly variable and directly linked with survival. However, there currently are no validated biomarkers or molecular targets to predict or improve radiation response, which would help develop personalized treatment and ideally targeted therapies. Here, we identified a novel biomarker, coenzyme A synthase (COASY), whose mRNA expression was consistently elevated in radioresistant human rectal cancers. This observation was validated in independent patient cohorts and further confirmed in colorectal cancer cell lines. Importantly, genetic overexpression and knockdown yielded radioresistant and sensitive phenotypes, respectively, in vitro and in vivo. COASY-knockdown xenografts were more vulnerable to radiation, showing delayed tumor growth, decreased proliferation, and increased apoptosis. Mechanistically, COASY protein directly interacted with the PI3K regulatory subunit PI3K-P85α, which increased AKT and mTOR phosphorylation, enhancing cell survival. Furthermore, shRNA COASY knockdown disrupted downstream PI3K pathway activation and also hindered DNA double-strand break repair, which both led to improved radiosensitivity. Collectively, this work reveals for the first time, the biological relevance of COASY as a predictive rectal cancer biomarker for radiation response, and offers mechanistic evidence to support COASY as a potential therapeutic target.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer and third most common cause of cancer-related death in the United States, accounting for more than 50,000 deaths each year (1). Recent studies have shown a rising incidence in rectal cancer, particularly in the young (2), with worse survival. Rectal cancer presents a complex clinical challenge requiring multimodality therapy and life-altering surgery to provide the best chance of cure. Neoadjuvant chemoradiation therapy (nCRT) followed by surgery decreases local recurrence (3,4) and is considered standard of care for locally advanced rectal cancer (5,6). The response to nCRT is highly variable, and oncologic outcome is directly associated with histopathologically graded response (7). Approximately 25% of patients will not have any residual cancer cells after neoadjuvant chemoradiation (8,9) and not surprisingly, these patients have the best rates of cure. Unfortunately, there are limited biologic predictors of response to therapy (10,11) that help inform treatments or guide personalized care. In addition, there no identified pathways of specific genes that have been successfully targeted in a clinical setting to enhance radiation sensitivity. There is clearly a need to decipher biological and mechanistic factors that enhance or hinder tumor response as a springboard to increasing treatment efficacy and developing new therapies.
Using our previously established mRNA microarray data (12), we identified differently expressed genes according to response to therapy as defined by the American Joint Commission on Cancer (AJCC) and the American College of Pathologists. Statistical analyses highlighted a potential marker, the COASY (Coenzyme A synthase) gene that strongly predicted rectal cancer radioresistance and correlated with rectal cancer AJCC response scores.
COASY is located on chromosome 17 and encodes the 564-amino acid Coenzyme A synthase (COASY protein), a mitochondrial bi-functional enzyme that has two catalytic domains, phosphopantetheine adenylyltransferase (PPAT) and dephospho-CoA kinase (DPCK); and is strongly activated by phospholipids (13). It mediates the final two stages of de novo Coenzyme A (CoA) synthesis from pantothenic acid (vitamin B5) in mammalian cells (14). CoA and its derivate are involved in multiple cellular metabolic pathways including pyruvate oxidation, fatty acid synthesis, cell cycle progression and cell death (for review (15)). Mutation of the COASY gene has been reported in neurological diseases such as the Neurodegeneration with Brain Iron Accumulation (NBIA) where it defines a key event for the disease progression by altering the mitochondrial function (16). Thus, COASY and its associated protein are necessary for cell survival and tissue homeostasis, but they have not been previously been linked to neoplasia.
In the current study, we define and further validate COASY as a predictive marker for rectal cancer radiation sensitivity and resistance in humans. In addition, we validate our clinical observations in both empiric in vitro and in vivo models. Lastly, we describe and confirm that COASY mechanistically mediates rectal cancer radiation resistance via the PI3K signaling pathway activation and enhanced DNA repair.
MATERIALS AND METHODS
Patients
Fresh frozen biopsies utilized for the transcriptomic analysis were from patients treated between 2006 and 2009 at Cleveland Clinic Main Campus in Cleveland, Ohio. Patients with middle- or lower-third rectal cancers included in this study, who met clinical criteria for nCRT, underwent pretreatment biopsy of the tumor via proctoscopy after investigators obtained informed written consent. Clinical criteria for treatment included patients with stage II or III disease according to National Comprehensive Cancer Network guidelines (http://www.nccn.org/professionals/physician_gls/pdf/rectal.pdf). Patient charts were reviewed for documentation of completion of long-course nCRT, and recording the clinical variables and demographics. The standard nCRT regimen included 50.40 Gy delivered in 25 fractions with 5-fluorouracil delivered as a radiation sensitizer. Patients generally underwent surgery with curative intent approximately 8–12 weeks after completion of nCRT. All surgeries were done by Cleveland Clinic colorectal surgeons adhering to oncologic principles of total mesorectal excision. Resected surgical specimens underwent pathological staging and were scored for treatment response according the College of American Pathologists (CAP) guidelines.
Pathologic Evaluation
Pretreatment rectal cancer biopsies were confirmed to contain adenocarcinoma with at least 60% of the specimen containing tumor before inclusion for microarray experiments. Post-treatment responses were scored according to the American Joint Committee on Cancer (AJCC) criteria (17): AJCC 0, complete response, defined as the lack of viable cancer cells; AJCC 1, moderate response, defined as single cells or small groups of cancer cells; AJCC 2, minimal response, defined as residual cancer outgrown by fibrosis; and AJCC 3 non-response, defined as minimal or no tumor response. Scores were assigned by a board-certified pathologist with specialized expertise in gastrointestinal pathology.
Cell lines
Six CRC cell lines used in this study LS411N (RRID:CVCL 1385), HCT116 (RRID:CVCL_0291), SW620 (RRID:CVCL_0547), SW837 (RRID:CVCL_1729), SW480 (RRID:CVCL_0546), and RKO (RRID:CVCL_0504) were purchased from American Type Culture Collection (ATCC). The HCT116, SW620, SW837, SW480 and RKO cell lines were verified by Short Tandem Repeat (STR) analysis and cultivated in modified Dulbecco’s Eagle’s medium (DMEM)-supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
HRT-18 cell line (RRID:CVCL_2514) was purchased from Sigma Aldrich and cultured in RPMI/1640 medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. The HRT-18 cell lines was also verified by STR analysis. All experiments were done with cell lines cultivated less than 20 passages since their procurement. Both HRT-18 and RKO cell lines were transduced with two lentiviral constructs containing different shRNAs targeting COASY mRNA (shRNA#1 and shRNA#2) as well as a non-targeting control (NT) shRNA (Origene). Cell were selected by adding puromycin in the culture media. To overexpress COASY, cells were transfected with the expression vector (pCMV3-COASY-GFP) or empty vector (pCMV3-GFP) from Sino Biological then cultured under hygromycin selection pressure.
Microarray Analysis, RT-qPCR and Gene Set Enrichment
After mRNA extraction, transcriptome analysis from 33 fresh-frozen rectal adenocarcinoma biopsies was performed as described previously (12) using the Illumina platform Human‐6 v2 (GSE133057). The COASY expression was verified by RT-qPCR using forward primer 5’- AGCCTTGAGGTTTCAGCCTGG- 3’; reverse primer 5’- AAGAACCTCAAACGTGGCCT- 3’; and normalized by two reference genes : β-ACTIN forward primer 5’- AGAAAATCTGGCACCACACC- 3’; reverse primer 5’-AGAGGCGTACAGGGATAGCA- 3’; and PUM1 forward primer 5’-TGTACTTACGAAGAGTTGCGATGTG- 3’; reverse primer 5’- CCAGGCCAGCGGAAGAT- 3’. The thermal profile used was 95°C for 15 seconds followed by 60°C for 1minute, repeated for 40 cycles.
In silico analyses
To establish a heatmap for transcriptomic analysis, we selected and clustered differentially expressed genes by using a cutoff of a 2-fold change (positive or negative) and p<0.0001 between samples with COASYLow compared to COASYHigh. Underexpressed genes are represented in green and overexpressed genes are represented in red. From the 660 differentially expressed genes, an enrichment map was created using Cytoscape software (RRID:SCR_003032) (18). The in silico analysis of differentially expressed genes was completed by a Gene Set Enrichment Assay (GSEA - RRID:SCR_003199) (19,20) performed with Java GSEA desktop software by using 1000 gene set permutations and the remaining default settings. The gene set used was REACTOME. Only geneset with FDR<0.25 and p<0.05 were considered as significant.
To analyze the relative expression of COASY, we examined Oncomine database (RRID:SCR_007834 - www.oncomine.org) in four independent patient cohorts with colorectal cancer (Gaedcke, Ki, Hong and Jorissen) as well as seven independent patients cohort with head and neck, pancreas, bladder, prostate, myeloma and lung cancers (Estilla, Logsdon, Sanchez-Carbayo, Welsh, Zhan, Selemat, Stearman, respectively). In each of the dataset, the COASY expression was dichotomized into lower-than-median and higher-than-median expression groups based on the Log2 median-centered intensity of COASY mRNA.
In vivo studies
To investigate the effect of COASY on tumor radioresistance, a total of 1×106 viable rectal cancer cells stably transfected with shRNA against COASY or shRNA control (RKONT, RKOshRNA#1, RKOshRNA#2) were resuspended in 200 μL of matrigel and injected subcutaneously in the flank of a 6-week old NSG mice (RRID:IMSR_ARC:NSG) obtained from The Jackson Laboratory. Once the tumor volume reached a size of 200 mm3, treatment by radiotherapy was initiated. Mice were anesthetized by isoflurane inhalation then 2 Gy radiotherapy was delivered by a PANTAK XRAD 320 (Precision X-Ray, North Branford, CT) per day during 5 consecutive days at a 1 Gy/min dose rate (10 Gy total). Tumor volume was recorded every two days using calipers. Mice were euthanized at day 21 and xenografts were excised and snap frozen in Optimal Cutting Temperature compound (OCT-Tissue-Tek) and kept at −80°C for future immunostaining.
Immunohistochemistry
All tissues were retrieved under pathologic supervision with IRB approval at Cleveland Clinic. Routine immunohistochemistry for COASY (Abcam, Cat# ab227272, 1:500 dilution) from paraffin embedded sections were completed. Antigen unmasking solution (Vector laboratories, Cat# H300) antigen retrieval method was used.
Clonogenic assay
RKO and HRT-18 rectal cancer cell lines knocked down or overexpressing COASY were used for clonogenic survival assay as previously described (21).
Annexin V/PI assay
RKO and HRT-18 cells were cultured in 6-well culture plates at 8–10×105/well in 3 mL of appropriate medium for 24 hours before irradiation. Cells were harvested 72 hours after irradiation and stained with Annexin V-FITC/PI before being analyzed by flow cytometry. Values represent the mean (± SE) of the sums of Annexin V+/PI-, Annexin V-/PI+ and Annexin V+/PI+.
Co-Immunoprecipitation and LC-MS/MS analysis
Whole-cell protein was extracted from RKO cell lines using Pierce IP Lysis Buffer (Thermo Scientific) supplemented with cocktail of anti-protease and anti-phosphatase. For each conditions, 3 mg of protein were immunoprecipitated with COASY (Santa cruz, Cat# sc-393812, 1:1000 dilution), P70S6K1 (Cell Signaling, Cat# 9202S, 1:1000 dilution, RRID:AB_331676) or PI3K-P85α (Cell signaling, Cat# 4292S, 1:1000 dilution, RRID:AB_329869) antibodies using protein G magnetic beads. The elution products were separated by SDS-PAGE 4–20% (BioRad). For the protein digestion, the bands were cut from the gel as closely as possible from the gel, washed/destained in 50% ethanol, 5% acetic acid and then dehydrated in acetonitrile. The bands were then reduced with Dithiothreitol and alkylated with iodoacetamide prior to the in-gel digestion. All bands were digested in-gel using trypsin, by adding 5 μL 10 ng/μL chymotrypsin in 50 mM ammonium bicarbonate and incubating overnight at room temperature to achieve complete digestion. The peptides that were formed were extracted from the polyacrylamide in two aliquots of 30 μL of 50% acetonitrile with 5% formic acid. These extracts were combined and evaporated to <10 μL in a Speedvac and then resuspended in 1% acetic acid to make up a final volume of ~30 μL for LC-MS analysis.
The LC-MS system used a Thermo Scientific Orbitrap Elite mass spectrometer system. The HPLC column was a Dionex 15 cm x 75 μm id Acclaim Pepmap C18, 2 μm, 100 Å reversed-phase capillary chromatography column. Five microliters volumes of the extract were injected and the peptides were eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.3 μL/min were introduced into the source of the mass spectrometer on-line. The microelectrospray ion source is operated at 2.5 kV. The digest was analyzed using the data dependent multitask capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans. The data were analyzed by using all CID spectra collected in the experiment to search the human UniProtKB sequence database (RRID:SCR_004426) with the search program Mascot (Mascot, RRID:SCR_014322) and more specifically against the sequence of Myd88 with the program Sequest.
Immunoblot
Protein concentrations were measured using the Bio-Rad Protein Assay Reagent. 20 μg of protein was loaded and separated on a 4–20% gradient Tris-HCl gel. Membranes were then incubated in primary antibody overnight at 4°C: COASY (Abcam, Cat# ab129012, 1:1000 dilution, RRID:AB_329869); γ-H2AX (Cell Signaling, Cat# 2577S, 1:500 dilution, RRID:AB_2118010); DNA-PKcs (Santa Cruz, Cat# sc-390698, 1:1000 dilution); MRE11 (Cell Signaling, Cat# 4895S, 1:1000 dilution, RRID:AB_2145100); Ku70 (Santa Cruz, Cat# sc-17789, 1:1000 dilution, RRID:AB_628454); RAD51 (Cell Signaling, Cat# 8875S 1:1000 dilution, RRID:AB_2721109); β-ACTIN (Santa Cruz, Cat# sc-47778, 1:2000 dilution, RRID:AB_2714189). The membranes were then washed and blotted with a horseradish peroxidase-conjugated secondary antibody (Invitrogen, Cat# A16072, 1:2000 dilution, RRID:AB_2534745; Thermo Fisher, Cat# 65–6120, 1:5000 dilution, RRID:AB_2533967) for 1 hour.
Phospho-AKT (ser473) and mTOR (ser2448) probing were done using the PI3K protein array (Cell Signaling, Cat# 7323) according the manufacturer instructions. The array were developed and imaged on a ChemiDoc™ Touch Imaging System (Biorad, RRID:SCR_014210). Images were then analyzed with Image Lab 3.0.1 (BioRad).
Statistics
All data are represented as mean ± SE. Statistical significance was determined by unpaired, 2-tailed non-parametric t-test; ANOVA test; Wilcoxon test; Receiver-Operatoring Characteristics analyses or Pearson correlation according the experiments. The p-values and the type of statistical analysis performed are described in the figure legends. In the figures, a p-values < 0.05 were considered statistically significant. These statistical analyses were performed using GraphPad Prism, version 7 (GraphPad Software, RRID:SCR_002798). A Kolmogorov–Smirnov-like statistic was used for enrichment score determination during GSEA analysis. For GSEA analysis p-values < 0.05 and a false discovery rate (FDR) < 0.25 was considered to be statistically significant.
Study approval
De-identified patient samples and clinical information for this study were retrieved from a prospectively maintained, single institution, Institutional Review Board-approved CRC biobank (IRB#4134). The experiments using mice was approved by the Cleveland Clinic Institutional Animal Care and Use Committee (IACUC Ref #2015–1516) and conducted in accordance with the National Institutes of Health guide for the care and use of laboratory animals.
RESULTS
COASY is overexpressed in human rectal cancers and increased levels are associated with worse response to nCRT
Thirty-three pretreatment biopsies were prospectively collected from patients with clinical stage II or III rectal adenocarcinoma who underwent nCRT. After completion of nCRT, patients underwent proctectomy as treatment of their disease and the resected specimens were evaluated by a gastrointestinal pathologist. A response to treatment score was assigned according to the American Joint Committee on Cancer (AJCC) criteria ranging from AJCC 0–3 (see Materials and Methods). To identify potential predictive biomarkers of nCRT response, an mRNA microarray analysis was done on pretreatment specimens and compared to post-treatment AJCC scores (GSE133057) (12). All 4 groups were compared for differentially expressed genes. Furthermore, the best responders (AJCC 0) were compared to all other groups (AJCC 1-2-3); the worst responders (AJCC 3) were compared to all other groups (AJCC 0-1-2). Differentially expressed genes were identified for each comparison. An ANOVA analysis identified 145 genes differentially expressed between the four AJCC scores (Figure 1A). Using a Wilcoxon rank sum test, 170 differentially expressed genes were found between AJCC 0 (complete responders) and AJCC 1-2-3; and 218 genes were differentially expressed between the worst responders (AJCC 3) and all patients with any other responses (AJCC 0-1-2) (Figure 1A). After cross-referencing the three resultant lists of differentially expressed genes for overlap, only 3 genes were identified on all lists, being consistently differentially expressed between the clinically defined groups: COASY, SPATA20, and PUSL1. Based on a literature review, we decided to focus on the biology of COASY, as it has been implicated in the generation of CoA, an essential co-enzyme required for over 4% of all cellular enzyme reactions (22).
Figure 1. COASY expression is a predictive biomarker for rectal cancer patient tumor response to neoadjuvant chemoradiation.
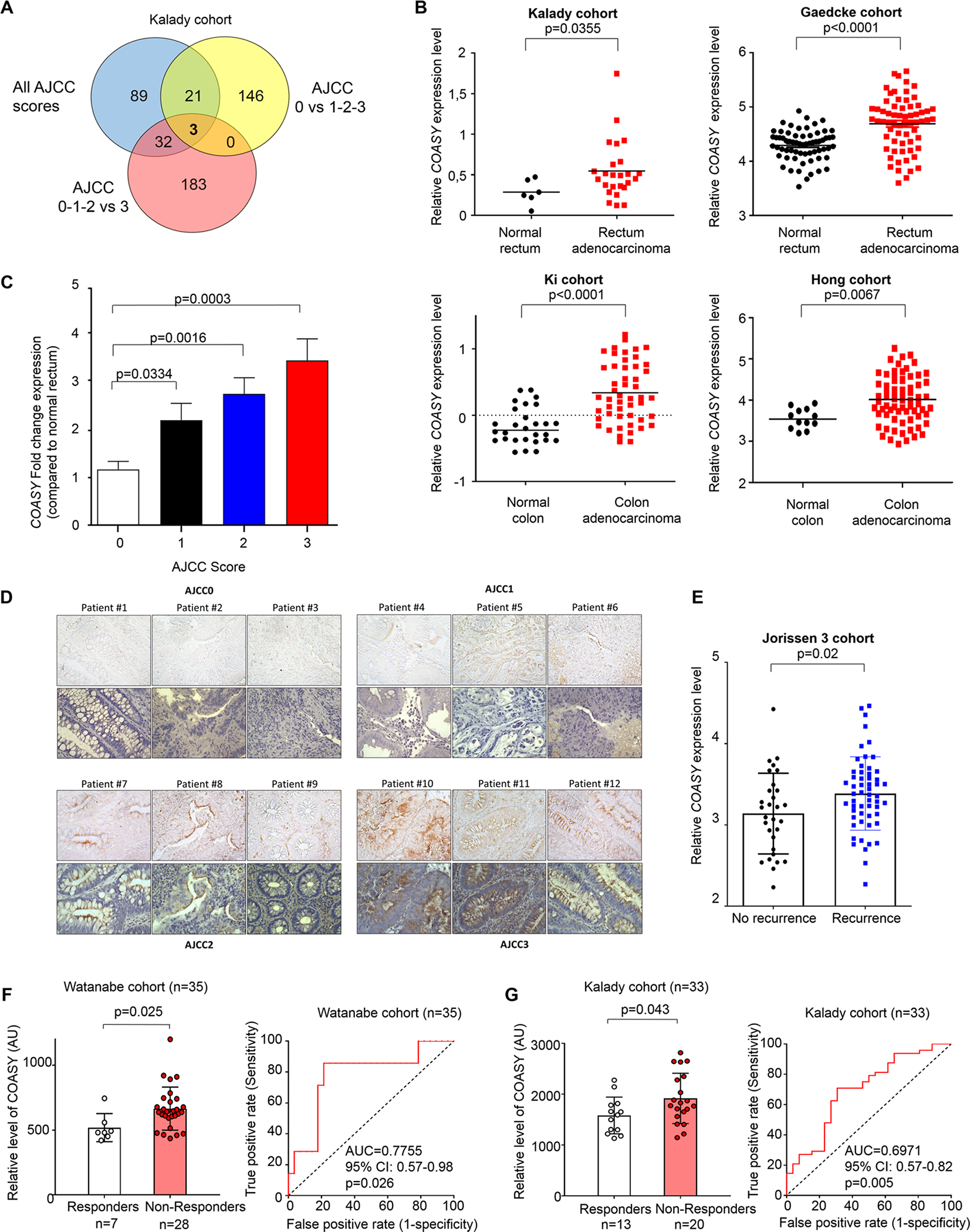
A) The Venn diagram displays the number of genes differentially expressed between all scores (ANOVA test), between complete responders compared to all others (AJCC 0 vs 1-2-3), and non-responders compared to all others (AJCC 0-1-2 vs 3) (Wilcoxon test); (n=33, Kalady cohort). COASY is one of 3 genes differentially expressed in all comparisons. B) Comparative analysis of relative COASY expression level between normal tissue and colorectal cancer in Kalady cohort (upper left) and three independent cohorts (Hong, Gaedcke and Ki cohorts) available on Oncomine Database. Bold bar represents the mean. Statistics were performed using a Mann-Whitney test. C) RT-qPCR of COASY gene expression from 33 patients according to their respective AJCC tumor regression scores. Results were normalized by two references genes (β-ACTIN and PUM1). Data are represented as mean ± SE. Statistics were performed using a Mann-Whitney test. D) Immunohistochemistry analysis of COASY on paraffin-embedded pre-treatment rectal adenocarcinoma patient samples. Three patients were studied per each AJCC response score. For each group, top and bottom panel show the staining without and with hematoxylin counter staining, respectively. E) Comparative analysis of relative COASY expression level using the Jorissen 3 database comparing CRC patients with or without recurrence within 5 years. Statistics were performed using a Mann-Whitney test. F) COASY expression level measured by microarray in rectal cancer pretreatment biopsies and analyzed according to response to nCRT as a retrospective validation cohort (Watanabe et al., 2006. GSE3493) (left panel). A Receiver-Operating Characteristic (ROC) analysis showing the sensitivity and specificity of COASY expression to discriminate responders from non-responders in the Watanabe cohort G) By means of comparison, the same analysis is shown for the Kalady cohort. COASY expression level measured by microarray in rectal cancer pretreatment biopsies and analyzed according to response to nCRT. Data are represented as mean ± SE. Statistics were performed using a Mann-Whitney test (left panel). A ROC analysis showing the sensitivity and specificity of COASY expression to discriminate responders from non-responders right panel).
Using the median level of COASY expression from the microarray data, the patient population was then classified as having low or high COASY levels (Table 1). Using a univariate analysis, the low and high COASY groups were statistically similar in terms of demographics and clinicopathologic characteristics. However, the AJCC tumor regression scores were significantly different between the two groups, demonstrating that patients with higher COASY levels were more likely to have a worse response to radiation, independent of other variables (Table 1).
Table 1:
Patient and tumor characteristics of the entire study cohort, and analyzed according to COASY level.
| Characteristics | All patients (n=33) | Low COASY (n=16) | High COASY (n=17) | p-value* |
|---|---|---|---|---|
| Age | ||||
| <50 | 10 (30.3%) | 5 (15.2%) | 5 (15.2%) | p=0.908 |
| ≥50 | 23 (69.7%) | 11 (33.3%) | 12 (36.3%) | |
| Gender | ||||
| Male | 23 (69.7%) | 10 (30.3%) | 13 (39.4%) | p=0.302 |
| Female | 10 (30.3%) | 6 (18.2%) | 4 (12.1%) | |
| TNM stage | ||||
| 1 | 4 (12.1%) | 2 (6%) | 2 (6%) | p=0.372 |
| 2 | 11 (33.3%) | 6 (18.2%) | 5 (15.2%) | |
| 3 | 15 (45.5%) | 8 (24.2%) | 7 (21.2%) | |
| 4 | 3 (9.1%) | 0 (0%) | 3 (9.1%) | |
| Lymphovascular invasion | ||||
| yes | 10 (30.3%) | 5 (15.2%) | 6 (18.2%) | p=0.805 |
| no | 23 (69.7%) | 11 (33.3%) | 11 (33.3%) | |
| Survival | ||||
| Dead | 11 (33.3%) | 6 (18.2%) | 5 (15.2%) | p=0.622 |
| Alive | 22 (66.7%) | 10 (30.3%) | 12 (36.3%) | |
| Time between nCRT and surgery | ||||
| < 8weeks | 17 (51.5%) | 9 (27.3%) | 8 (24.2%) | p=0.597 |
| ≥ 8weeks | 16 (48.5%) | 7 (21.2%) | 9 (27.3%) | |
| AJCC score | ||||
| 0 | 6 (18.2%) | 6 (18.2%) | 0 (0%) | p=0.020 |
| 1 | 7 (21.2%) | 3 (9.1%) | 4 (12.1%) | |
| 2 | 13 (39.4%) | 6 (18.2%) | 7 (21.2%) | |
| 3 | 7 (21.2%) | 1 (3%) | 6 (18.2%) | |
The p-value is obtained by χ2 test and is considered signifcant when p value <0.05
To further validate the microarray results, RT-qPCR for COASY was performed using all 33 specimens included in the microarray, as well as normal rectal mucosa. COASY was significantly overexpressed in rectal cancer tissue compared to normal tissue from patients in the study population (Kalady cohort) (Figure 1B). This finding was supported by similar results found in three different independent cohorts from the Oncomine database (Hong cohort, Gaedcke cohort, and Ki cohort) (Figure 1B). In addition, COASY was overexpressed in multiple cancer types compared to corresponding normal tissue as shown for cancer of the head and neck, pancreas, bladder, prostate, bone marrow myeloma, and lung using the Oncomine database (Supplementary Figure 1A–G). In our cohort, we also observed a stepwise increase in COASY expression with increasing AJCC score from 0 to 3 (Figure 1C). Comparing COASY levels from complete responders (AJCC 0) to all other patients (AJCC 1-2-3), those with a complete response had significantly lower COASY expression (Supplementary Figure 2A). Similarly, patients with no response to nCRT (AJCC 3) had significantly higher COASY expression compared to all other patients (AJCC 0-1-2) (Supplementary Figure 2B). Receiver operating characteristic (ROC) analysis revealed that COASY expression level had an area under the curve (AUC) of 0.827, indicating a strong predictive power for identifying complete responders (Supplementary Figure 2C). Using COASY expression to identify the worst responders, ROC analysis estimated the AUC at 0.730 indicating good predictive power (Supplementary Figure 2D). To determine if our transcriptomic data translated to protein expression, COASY protein level was measured by immunohistochemistry on paraffin-embedded pretreated human rectal cancer biopsies comparing 3 independent patients per AJCC score. Consistent with COASY gene expression, COASY protein level increased from AJCC 0 to AJCC 3 (Figure 1D). This observation was also confirmed using immunofluoresence of freshly frozen human rectal cancer samples (Supplement Figure 2E).
Furthermore, while the Oncomine database does not specifically contain response to radiotherapy data, interrogation of the Oncomine Jorissen 3 cohort (154 patients) revealed that overexpression of COASY is associated with CRC recurrence, suggesting worse response to treatment (Figure 1E).
To further assess if our findings were reproducible in other cohorts, we tested the ability of COASY expression to predict therapeutic response of rectal cancer by using a microarray gene expression dataset published in 2009 in Cancer Research from Watanabe and collaborators (23) deposited on NCBI (GSE3493). Of note, Watanabe’s data did not provide granularity between AJCC scores, only broadly between responder or non-responder categories. Therefore, as a comparison we analyzed our data the same way: responders (AJCC 0– 1) and non-responders (AJCC 2–3). In the Watanabe cohort of 35 samples, we analyzed the COASY mRNA expression and determined that cancers from non-responder patients harbored a significantly higher levels of COASY expression compared to cancers from responders (Figure 1F left panel). The same difference was observed in our cohort (Figure 1G left panel, p=0.043). These results support independent patient cohort validation for our observations. The ROC analysis on this validation cohort revealed an AUC of 0.775 (p=0.026) demonstrating the capacity of COASY expression level to distinguish responder patients from non-responder patients (Figure 1F right panel). For comparison, the AUC was 0.697 (p=0.005) in our discovery cohort (Figure 1G right panel).
Taken together, these results support that COASY expression level correlates with rectal cancer response to nCRT in a clinical setting and thus could be potentially used as a predictive biomarker.
COASY influences radiosensitivity of rectal cancer cell lines
Considering the correlation of COASY expression and radiation response in the clinical setting, we investigated COASY expression in multiple CRC cell lines to expand the study using in vitro models. COASY mRNA levels were measured by RT-qPCR on 7 different CRC cell lines. Heterogeneity of COASY expression was observed among the cell lines (up to 12.5 fold difference) (Figure 2A). To determine the correlation between COASY expression and response to radiation, we measured the radiation dose required for each cell line to achieve 90% cell death (or 10% cell survival, D10) by clonogenic assays for each cell line. There was a distinct statistically significant positive linear correlation (R2=0.77; p=0.009) between COASY expression and the radiosensitivity index, D10 (Figure 2B).
Figure 2. Elevated COASY levels correlate with increased CRC cell line radioresistance.
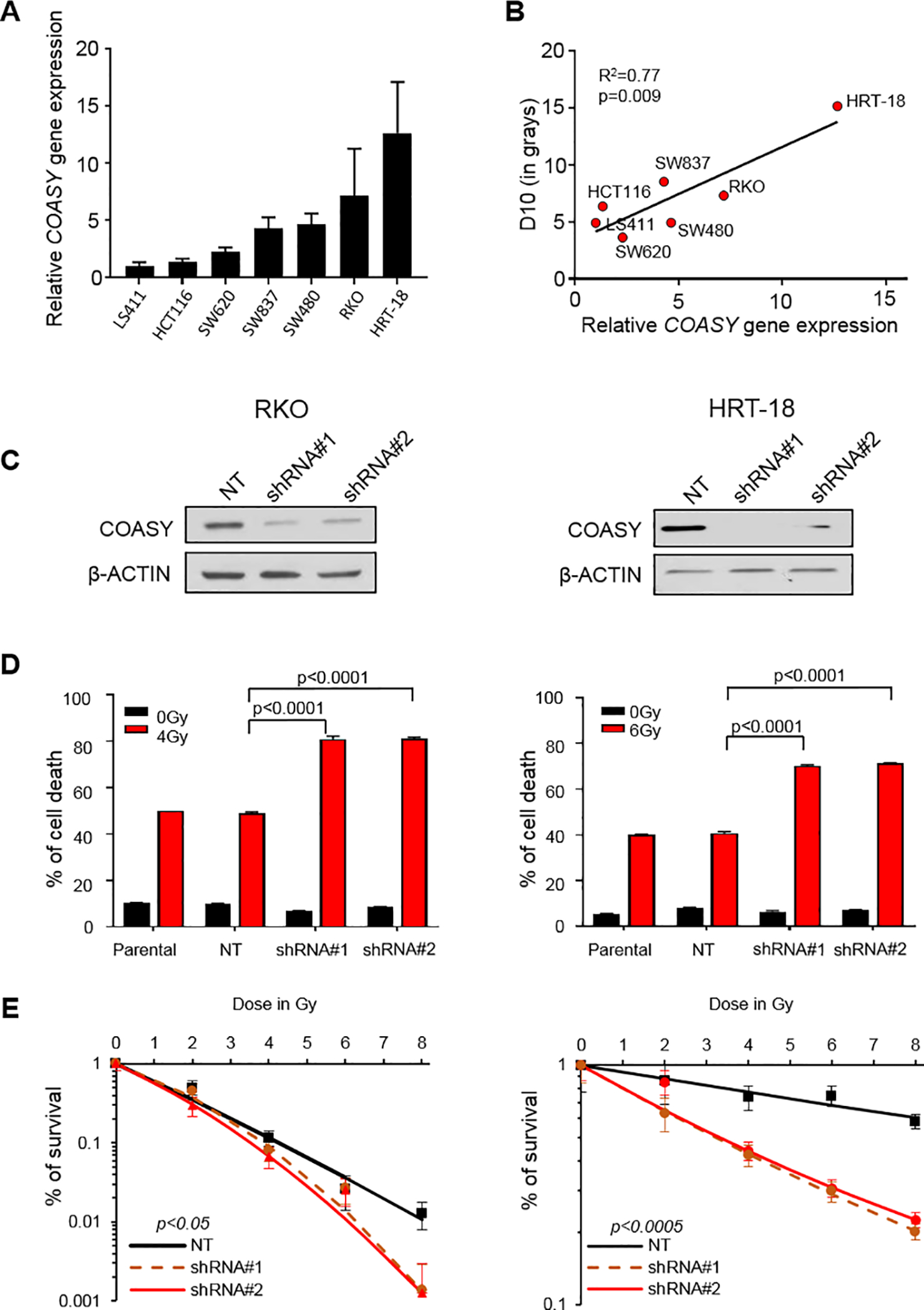
A) The expression of COASY was measured by RT-qPCR and normalized by β-ACTIN in 7 CRC cell lines. B) Correlation of COASY expression to radiation doses required to obtain 10% survival (D10), determined by clonogenic assays in the previously mentioned 7 CRC cell lines. C) Two different shRNA constructs (shRNA#1 and shRNA#2) targeted against COASY mRNA were stably transduced in two CRC cell lines (RKO and HRT-18) via lentiviral infection and compared to a control non-targeting shRNA (NT). COASY knockdown was observed by immunoblot. D) Flow cytometry was performed to determine the levels of apoptosis by AnnexinV/PI in parental and shRNA cell lines 72 hours after 4 Gy or 6 Gy irradiation in RKO and HRT-18, respectively. The percentage of cell death included the cells that were AnnexinV+/PI-; AnnexinV-/PI+ and AnnexinV+/PI+. Data are represented as mean ± SE. Statistical analysis was done using Mann-Whitney test. E) Clonogenic survival assays for various doses of irradiation. The percentage of cell survival is shown as a function of the irradiation dose in black (NT), dashed orange (shRNA#1), and red lines (shRNA#2) for RKO and HRT-18. ANOVA was performed to measure the interaction between the irradiation dose, the cell lines, and the survival percentage; p values are shown
Next, we tested if COASY gene knockdown could alter the radiation response. For that, we used two different lentivirus-mediated short hairpin RNAs (shRNAs) to stably knockdown COASY gene expression in the two CRC cell lines having the strongest COASY expression in our panel: RKO and HRT-18 (Figure 2C). We did not observe a significant effect of COASY knock down on cellular growth in vitro at basal level in RKO and HRT-18 cell lines (Supplementary Figure 3A–B). Briefly, both cell lines were exposed to their respective IC50 irradiation dose (4 Gy for RKO; 6 Gy for HRT-18) and cell death was quantified by Annexin V/PI. For both cell lines, 50% cell death was observed in the parental cell lines and in the non-targeting shRNA control cell lines (NT). However, the same irradiation dose delivered to the knockdown lines achieved 80% cell death (Figure 2D). To analyze the effect of COASY knockdown in a different way, cell lines were subjected to clonogenic survival assay using various doses of irradiation as reported by Emons et al (24). For both lines, the shRNA knockdowns experienced a decrease in survival compared to the parental cell lines and non-targeting shRNA (Figure 2E). The opposite phenomenon was observed in COASY overexpressing lines using a stable expression vector (Supplementary Figure 3C–D). Taken together, these data demonstrated that COASY influences radiosensitivity in vitro, and the effect exists across multiple cell lines.
Gene expression analysis and gene set enrichment identify distinct signatures and signaling pathways associated with COASY.
To identify genes or pathways associated with COASY-mediated resistance, patient samples were dichotomized into COASYLow (bottom 25% of the cohort for COASY expression n=8) and COASYHigh (top 25% of the cohort for COASY expression n=8). On gene expression analysis, using a cut off of at least two-fold difference and a p<0.0001, 660 genes were differentially expressed between the low and high groups (Supplementary Table 1). As shown with the heat map, the majority of the differentially expressed genes associated with high COASY levels are downregulated compared to their expression in the low COASY group (Figure 3A). To understand which classes of genes are associated with COASY expression, the list of 660 genes was processed through the Gene Ontology (GO) database. In the COASYHigh group, 67 GO were downregulated including cell death, regulation of cell differentiation, regulation of gene expression, and immune response. Only 4 GO were upregulated in the COASYHigh group: response to oxidative stress, positive regulation of phosphorus metabolism, ion homeostasis, and monovalent inorganic anion homeostasis (Figure 3B). This analysis was completed by Gene Set Enrichment Analysis (GSEA) comparing expression of all genes in COASYLow cancers compared to COASYHigh cancers (Figure 3C). Among the REACTOME gene sets significantly enriched in COASYHigh phenotype, gene sets associated with PI3K/AKT activation, EGFR signaling, and PDGF signaling in cancer were present (p=0.03 FDR=0.21; p=0.003 FDR=0.13, p=0.001 FDR=0.13, respectively).
Figure 3. Gene expression analysis and gene set enrichment identify distinct signatures and signaling pathways associated with COASY.
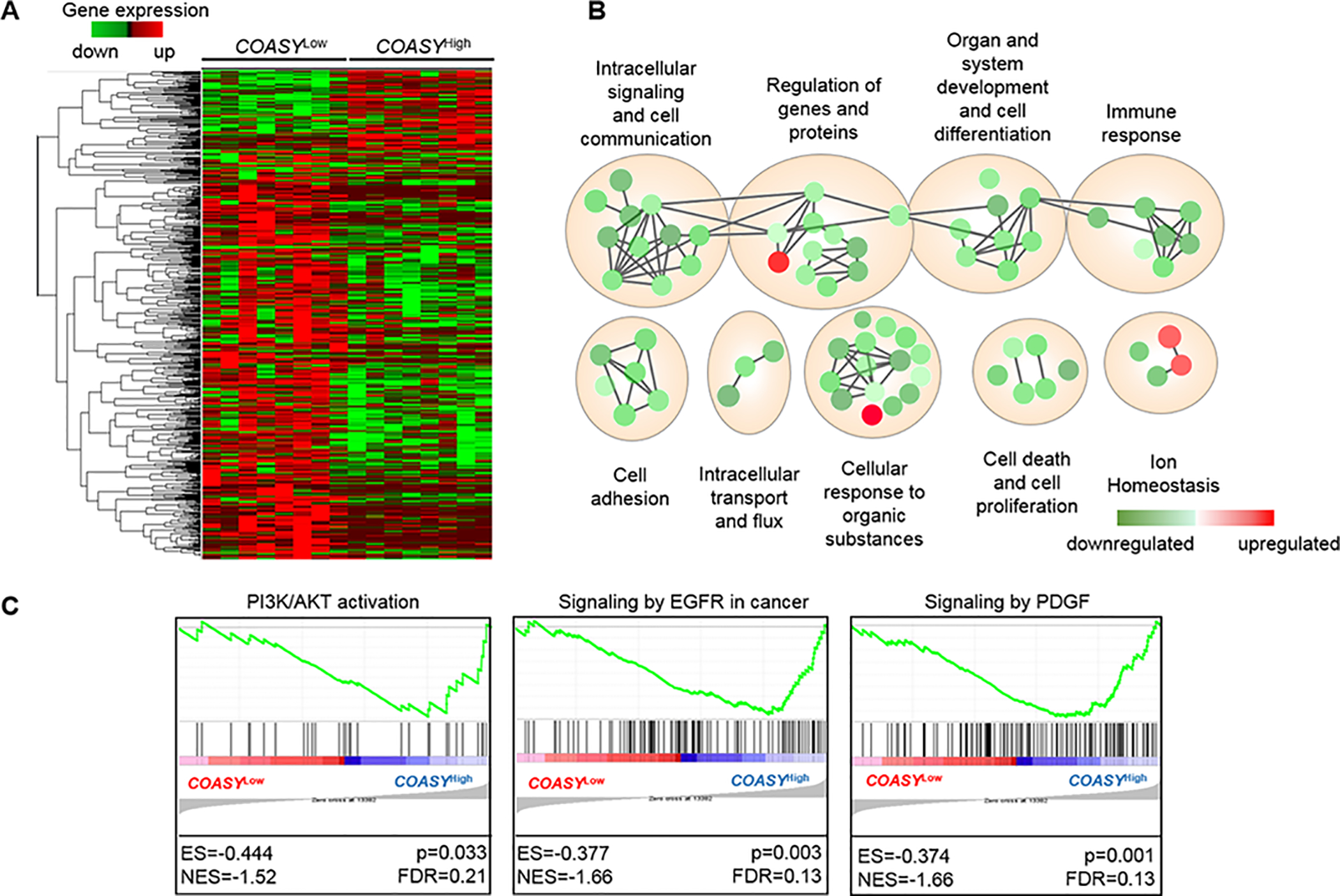
A) Heatmap from hierarchical clustering of 660 differentially expressed genes comparing two groups: COASYLow (bottom 25% of the cohort for COASY expression; n=8) to COASYHigh (Top 25% of the cohort for COASY expression; n=8. B) Gene clusters from COASYHigh hierarchical classification were subjected to Gene Ontology Analysis, and the most enriched term for each cluster was determined using the q-value from the FDR test. Green nodes represent downregulated gene families; red nodes represent upregulated gene families. Size of nodes represent the number of genes present in the cluster and color intensity represents the p-value. C) Gene Set Enrichment Assay (GSEA) made from microarray data of the Kalady cohort comparing two phenotypes, COASYLow and COASYHigh, with the REACTOME dataset. ES, enrichment score; NES, normalized enrichment score, FDR false discovery rate.
COASY-induced radioresistance is mediated by activation of the PI3K/AKT/mTOR pathway
Similar to ligand stimulation, it is well known that exposure to ionizing radiation induces activation of EGFR, secretion of PDGF, and triggers activation of the downstream PI3K/AKT pathway in different types of cancers (25). The PI3K/AKT pathway is a major contributor to radioresistance by promoting cell survival after irradiation (26). Previous studies have shown physical interactions between the COASY, PI3K-P85α, and P70S6K1 proteins (27–29) leading to modulation of PI3K/AKT pathway activation in normal, non-cancerous cells. Thus, we hypothesized that COASY modulates the PI3K/AKT pathway activation after irradiation in cancer cells. Using Liquid Chromatography followed by Mass Spec (LC-MS/MS), the presence the COASY peptide (504)DGLSEAAAQSR(514) was detected in PI3K-P85α and P70S6K1 immunoprecipitated samples, thus proving a physical interaction between COASY and PI3K pathway members (Figure 4A). To provide additional evidence, we co-stained COASY and PI3K-P85α protein by immunofluorescence and observed an overlap indicating co-localization (Figure 4B).
Figure 4. COASY-induced radioresistance is mediated by modulation of the PI3K/AKT/mTOR pathway.
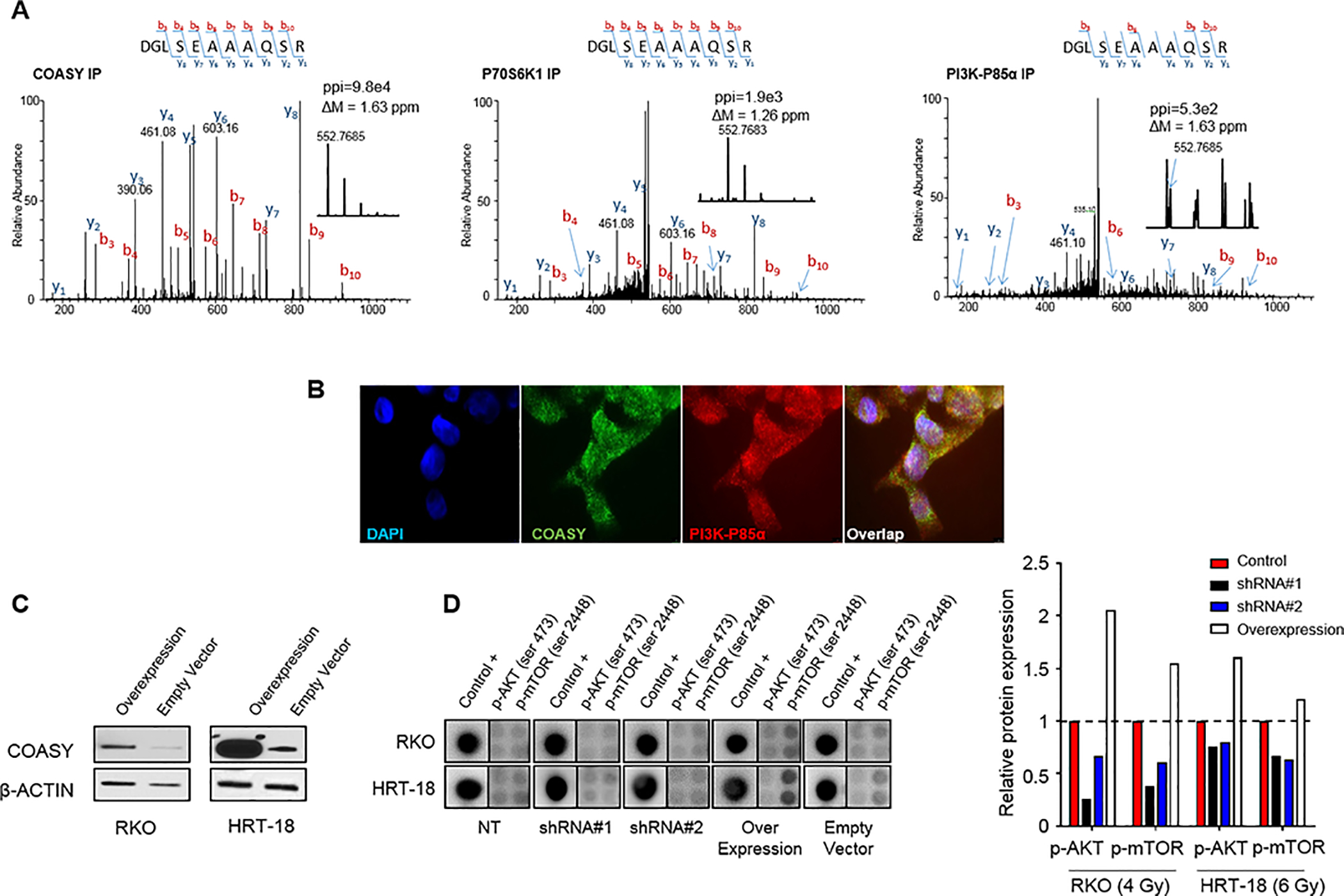
A) Using RKO, endogenous PI3K-P85α and P70S6K1 were co-immunoprecipitated in a complex with COASY using a specific antibody from exponentially growing cells. Protein G magnetic beads alone were incubated with the same cell extract and used as a control for nonspecific binding. The immunoprecipitated proteins were fractionated on an SDS-Page gel, and LC-MS/MS. The MS/MS spectra for the COASY tryptic peptide, (504)DGLSEAAAQSR(514), is shown from the COASY IP, P70S6K1 IP, and PI3K-P85α IP. ppi stands for the peptide peak intensities. B) Co-staining analysis of COASY and PI3K-P85α protein in RKO cell line by immunofluorescence (100x). C) Immunoblotting was used to detect COASY expression in RKO and HRT-18 cell lines transfected with either a control vector (empty vector) or a COASY overexpression vector (overexpression). D) PI3K protein arrays were done 24h after irradiation (4 Gy for RKO and 6 Gy for HRT-18) comparing the knockdown cell line for COASY (shRNA#1 and shRNA#2) and overexpressing cell lines to their respective control transfected with non-targeting shRNA (NT) or an empty vector. Histograms correspond to the signal intensity quantification with background subtracted and normalized by the 3 internal positive controls.
To determine whether COASY modulates the radiation-induced activation of the PI3K pathway, p-AKT and p-mTOR levels were measured in stable COASY knockdown (shRNA#1 and shRNA#2) and NT shRNA cell lines 24 hours after irradiation. Additionally, stable COASY overexpressing cell lines established by transduction of a COASY expression vector, and controls (empty vector) were also included in the assay (Figure 4C). Twenty-four hours after irradiation (4 Gy and 6 Gy for RKO and HRT-18, respectively), p-AKT (ser473) and p-mTOR (ser2448) levels were determined. We found that p-AKT and p-mTOR were lower in the two shRNA conditions compared to their non-targeting control conditions in both cancer cell lines. Conversely, when COASY was overexpressed, p-AKT and p-mTOR levels were strongly increased compared to the control condition (Figure 4D). In conclusion, COASY directly affected the level of radiation-induced activation of PI3K pathway in CRC cell lines suggesting a potential mechanism for radioresistance.
COASY enhances DNA repair efficiency
Since the PI3K/AKT/mTOR pathway is known to directly regulate DNA damage repair in cancer cells, alterations of this pathway can be contextually connected to DNA repair defects and radioresistance (30). As ionizing radiation confers lethality through the induction of DNA double-strand breaks (DSB), the role of COASY in DNA damage repair was assessed. The efficiency of DNA damage repair was measured using a γ-H2AX foci formation assay 24 hours after irradiation (4 Gy and 6 Gy for RKO and HRT-18 cell lines, respectively). In both cell lines, there was a significantly higher content of residual DSB in the shRNA COASY cell populations compared to their NT shRNA control (Figure 5A). These results demonstrate that COASY downregulation yields a decrease in DNA repair efficiency.
Figure 5. COASY enhances DNA repair ability.
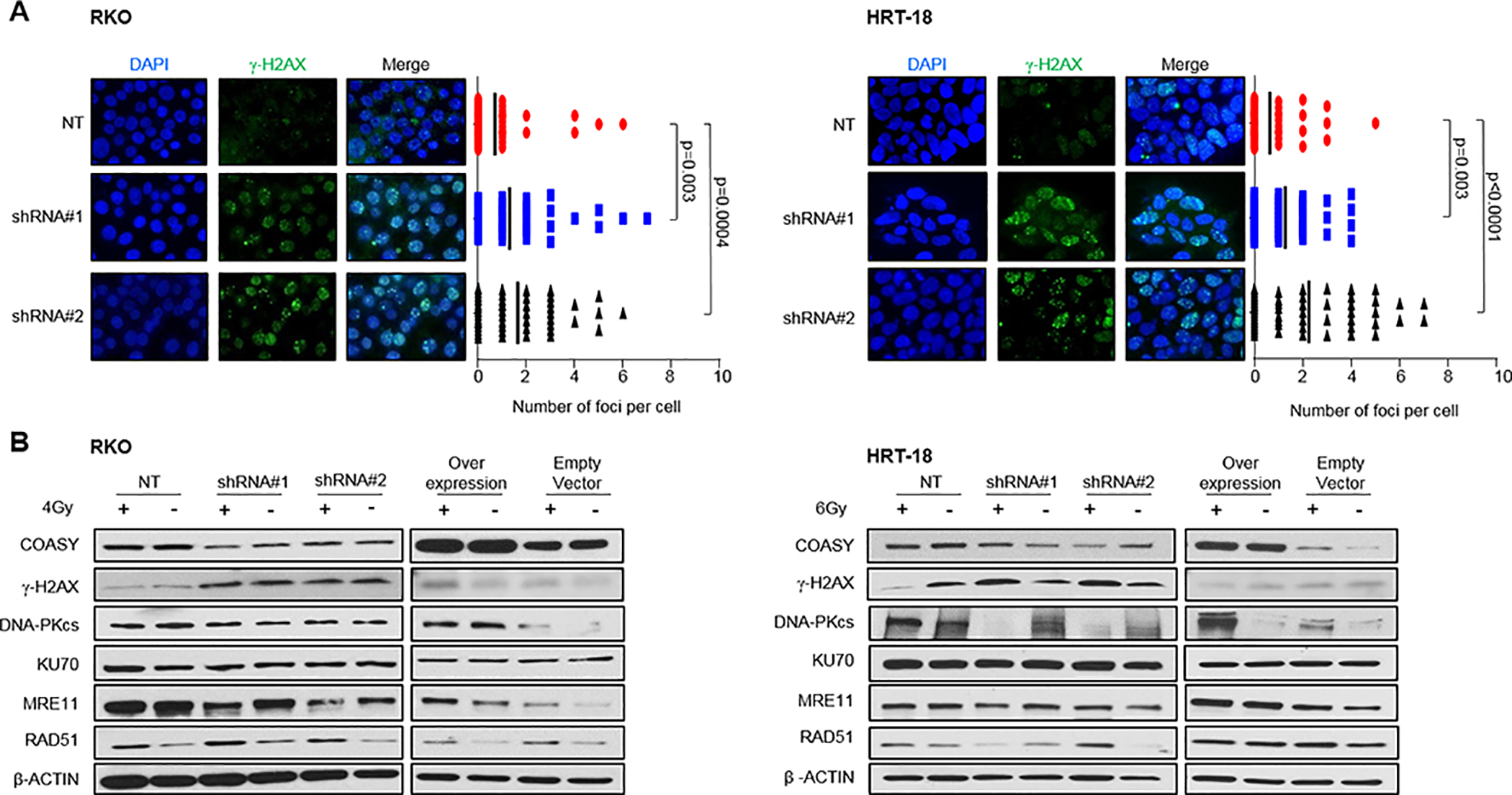
A) COASY knockdown and non-targeting irradiated cells were evaluated for DSB number by measuring γ-H2AX 24 hours after irradiation with 4 Gy and 6 Gy for RKO and HRT-18, respectively. For each cell line, the left panel shows representative pictures of γ-H2AX staining by immunofluorescence. The right panel box‐plot shows the distribution of cells counted in each cell population (n=50). Bold lines represent the mean number of foci per cell. Statistical analyses were done using Mann-Whitney test. B) COASY shRNAs and NT shRNA from RKO and HRT-18 cell lines were treated with 4 Gy and 6 Gy of irradiation, respectively. After 24 hours, lysates were prepared and analyzed for COASY, γ-H2AX, DNA-PKcs, KU70, MRE11, RAD51 and normalized by β-ACTIN via immunoblot.
To understand the dynamics of DNA repair, we investigated the expression of DNA repair proteins responsible for homologous recombination (HR), MRE11 and RAD51, as well as proteins implicated for Non-Homologous End Joining (NHEJ), DNA-PKcs and KU70, 24 hours after irradiation. The level of COASY was not modified after irradiation in either cell line (Figure 5B). Analyzing COASY expression dynamically over time from 1 hour to 24 hours after irradiation confirmed an absence of COASY protein level modification after irradiation (Supplementary Figure 4). The downregulation of COASY via shRNA resulted in an increase in γ-H2AX, signifying increased DSB in both RKO and HRT-18 cell lines, confirming the results seen with fluorescent microscopy. In RKO cell lines, which are HR proficient, the level of DNA-PKcs and KU70 were not affected by irradiation, but MRE11 protein expression decreased with COASY downregulation. In contrast, HRT-18 cells which are HR-deficient and relies on NHEJ as the main DNA repair mechanism, displayed a strong alteration of DNA-PKcs expression that directly correlated with COASY protein level after irradiation (Figure 5B). These findings suggest that the COASY protein affects DNA repair efficiency in both HR and NHEJ proficient cells.
Inhibition of COASY decreases cancer growth in vivo
To further study the relationship between COASY and CRC radiation resistance, we established CRC mouse xenograft models. Using two different shRNAs as well as a non-targeting shRNA in RKO cell line (RKOshRNA#1, RKOshRNA#2 and RKONT), 1×106 cells were injected subcutaneously into the flanks of NSG mice. The tumors were allowed to grow to a size of approximately 200 mm3, and then randomized into two groups for each of the cell lines (control and irradiated). The irradiated group received 2 Gy per day during 5 consecutive days (10 Gy total) as shown in Figure 6A. Tumor size was measured daily for the control mice or every two days for irradiated mice. In the control condition, we did not observed a significant difference in tumor growth comparing the shRNAs to the non-targeting shRNA (NT). However, in the irradiated conditions the two COASY knockdown lines exhibited significantly slower growth kinetics compared to the non-targeted shRNA tumors (Figure 6B) suggesting that COASY knockdown enhances the effect of radiation and impedes tumor growth, but does not inherently affect non-treated tumor growth. Harvested xenografts were analyzed for proliferation and apoptosis via immunostaining for Ki67 and cleaved caspase-3. RKONT tumors demonstrated extensive proliferation and minimal cell death compared to both RKOshRNA#1 and RKOshRNA#2 tumors which displayed decreased proliferation and increased apoptosis (Figure 6C–D). These data demonstrate that COASY expression modulates cancer radiation response in vivo.
Figure 6. Knockdown of COASY induces radiosensitization of CRC in vivo.
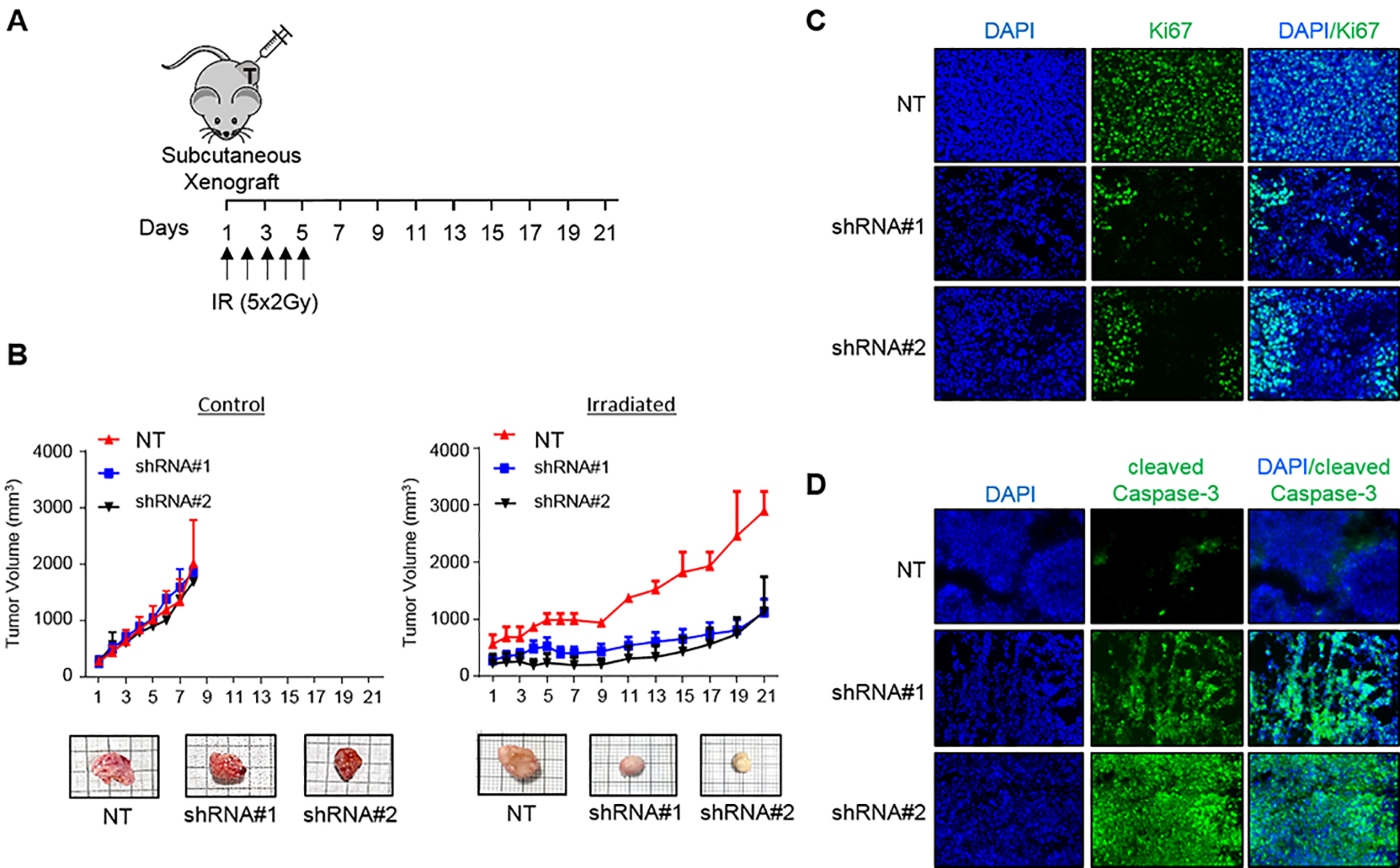
A) Experimental design: NSG mice underwent subcutaneous flank injection with RKONT, RKOshRNA#1, or RKOshRNA#2. When tumors reached approximatively 200 mm3, mice received radiation with 2 Gy using X-beam at 1 Gy/min per day during 5 consecutive days (arrows). B) Tumor growth was monitored in control condition (left panel) or treated with irradiation (right panel) every two days using calipers. Data are represented as mean ± SE (n=3 for control and n=4 for both shRNA groups). Representative images of harvested tumors on day 8 for control and day 21 for tumor treated with irradiation are shown. C) Ki67 immunofluorescence staining on tumors harvested on day 21. Representative pictures of each xenograft cell line are shown. D) Cleaved caspase-3 immunofluorescence staining on tumors harvested on day 21. Representative pictures of each xenograft cell line are shown.
DISCUSSION
In this study, we have demonstrated for the first time a direct link between COASY expression and radiation response in rectal cancer. Mechanistically, COASY expression augments radiation-induced PI3K/AKT pathway activation via physical interaction with PI3K pathway members, subsequently increasing p-AKT and p-mTOR, thus promoting cell survival. Furthermore, COASY facilitates DNA damage repair efficiency and thus cell survival via repair of DNA double strand breaks. Using human cancer samples linked to clinical outcome, we identified COASY as a candidate biomarker for rectal cancer radiation resistance and demonstrated reproducibility in other independent patient validation cohorts as well as in cellular and mouse models. Taken together, this study not only characterizes COASY as a potential biomarker for radiation treatment response in rectal cancer, but also mechanistically provides a possible novel therapeutic strategy.
COASY was originally identified as the bi-functional enzyme that mediates the final two steps of de novo CoA from pantothenic acid in mammalian cells (14). However, the role of COASY remains incompletely defined. Sparse studies have shown non-canonical roles of COASY in embryonic development, lipid metabolism, mitochondrial function and iron metabolism in several models (zebrafish, yeast, and human cells) (31–33). One recent report describes COASY as an important regulator of productive mitosis in several cancer cell lines, supporting its importance in cellular proliferation (34). COASY has not previously been studied in normal colon or rectum, nor colorectal cancer. Importantly, these differential COASY expression patterns were observed in multiple other cancer types with COASY elevated compared to normal corresponding tissue types, suggesting potential broader applications to our findings in rectal cancer.
In addition, this is the first description of COASY in a biologic role related to radiation resistance. Radiation therapy induces DNA double strand breaks, which if not repaired, triggers cell death. There has been extensive research on the molecular pathways responsible for rectal cancer response to radiotherapy including apoptosis (35), cell proliferation (36,37), DNA repair (38), autophagy (39), and cell metabolism (40). The PI3K pathway is activated after irradiation and plays a central role in these cellular events (41,42). In CRC, several mutations play a role in tumor initiation, progression, metastasis and response to some therapeutic agents. The most common mutations include APC (80% of CRC), TP53 (40–50%), BRAF/KRAS (40%), PI3KCA (25%) and PTEN (5–14%). Multiple studies have described an overactivation of PI3K pathway in tumors harboring these mutations (43–47). Furthermore, activation of the PI3K/AKT pathway correlates with poor patient outcomes in patients with colorectal cancer (25,48).
We propose that COASY-induced radioresistance results from a direct interaction of COASY with PI3K-P85α, leading to over-activation of the PI3K pathway after irradiation. As a result, the increase of AKT and mTOR phosphorylation promotes the recruitment of DNA-PKcs and MRE11 to the DSB site to facilitate the DNA repair promoting cellular survival (48,49). One of the well-characterized kinases, S6K1 (a key regulatory protein of the PI3K pathway involved in cellular proliferation, apoptosis, transcription and protein synthesis) is also associated with COASY protein (27,29). Interestingly, the S6K1 gene product, p70S6K1, is overexpressed in many cancers, including CRC (50,51). Multiple studies have shown different factors may predict rectal cancer response to radiotherapy such as Rho family members (RAC2), antioxidant family members (GPX2), cell cycle members (TP53), and cell adhesion members (β-catenin) (23,24,52). We quantified the activation of APC/β-catenin pathway, comparing COASY shRNAs to the control cell lines after irradiation. Results revealed COASY-induced radioresistance is not dependent on APC/β-Catenin (Supplementary Figure 5), highlighting that multiple independent mechanisms of radiation resistance likely exist in rectal cancer.
We have no direct experimental evidence yet distinguish the specific role of COASY in association with PI3K-P85α and P70S6K1. However the direct binding the two proteins and the direct correlation of PI3K pathway activation to COASY levels triggers speculation that this interaction is related to allosteric regulation of PI3K enzyme. Several proteins have already been shown to interact with different domains of PI3K-P85α, such as CDC42 (to the Rho-GAP domain), FYN, LYN or PAK4 (SH3 domain) and modulate the PI3K complex enzyme activity (53–55). Although not previously shown in cancer cells, Breus, et al. reported interaction between COASY and the SH2 and SH3 domains of PI3K-P85α in embryonic cells. This interaction was only observed when COASY is phosphorylated on tyrosine residues, suggesting that the interaction depends more on a post-translation modification of COASY rather than a transcription upregulation (Figure 5B, Supplementary Figure 4) (28).
Rectal cancer is treated with multimodality therapy including neoadjuvant radiation, surgery, and chemotherapy. Patients with better response to upfront radiation have significantly improved outcomes including decreased local recurrence and improved overall survival (7,56). Furthermore, there is a changing paradigm in the treatment of rectal cancer that utilizes the clinical response after radiation as a deciding factor regarding surgery. In the subset of patients that do not show any residual tumor on imaging, endoscopy, or clinical examination, surgery may be avoided (57) and thus avoiding procedural morbidity and the need for a colostomy. This so called “watch and wait” or organ preservation approach is gaining momentum as more studies have demonstrated its success (58,59). Unfortunately, only approximately 25% of patients develop a clinical complete response to radiation, limiting applicability of watch and wait. Even for patients without a complete response, improved response scores equate with better outcomes. Thus, the need for personalized care to predict responders, and to improve the percentage of patients with good response, the search for both biomarkers and therapeutic targets for radiation sensitivity have become the Holy Grail for rectal cancer treatment. Although other biomarkers for rectal cancer response have been reported (35–37), few have been validated and none are utilized for clinical decisions.
The fact that this study demonstrates a functional mechanistic role for COASY in radiation resistance provides credence to its role as a predictive biomarker, and provides promise as a therapeutic target. There are currently more than 20 clinical trials evaluating PI3K pathway inhibitors alone or in combination with neoadjuvant chemotherapy for cancer treatment (www.clinicaltrials.gov). Although this approach is promising, the lack of specificity and strong toxicities of these novel agents limit their use. Perhaps manipulation of a more directed target such as COASY via small molecule inhibitors targeting the interaction domain between COASY and PI3K members could overcome the treatment resistance observed in cancer with multiple genetic mutations as well as decreasing clinical side effects.
In summary, we classified COASY as a novel gene with key implications in rectal cancer cell survival after radiation treatment. Since radiation resistance is a major hurdle to cure, these findings have significant clinical implications. As a biomarker, COASY levels directly correlated with the clinical response to radiation in humans, cells, and in vivo models. By delineating the underlying mechanisms and pathways of action, we have identified a new therapeutic target for future study as a potential radiation sensitizing agent.
Supplementary Material
SIGNIFICANCE.
COASY is a novel radiotherapy response modulator in rectal cancer that regulates PI3K activation and DNA repair. Furthermore, COASY levels directly correlate with radiation response and serve as a predictive biomarker.
ACKNOWLEDGMENTS
The authors would like to thanks Dr Samaneh Kamali-Sarvestani for her scientific advices; Dr Patrick Leahy for his computational analysis assistance; Dr Belinda Willard of the proteomic core from Lerner Research Institute and Dr Plesec from anatomic pathology department from Cleveland Clinic. This work was supported by a Cleveland Clinic Velosano grant (MFK); and a grant from the American Society of Colon and Rectal Surgeons (MFK); MFK is supported by NIH R01 CA193359-01, and holds the James Church, MD and Edward DeBartolo Jr. Family Chair in Colorectal Surgery.
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Howlader N, Noone A, Krapcho M, Miller D, Bishop K, Kosary C, et al. SEER Cancer Statistics Review, 1975–2014, National Cancer Institute [Internet] Bethesda, MD; Report No.: based on November 2016 SEER data submission, posted to the SEER web site, April 2017. [Google Scholar]
- 2.Murphy CC, Singal AG, Baron JA, Sandler RS. Decrease in Incidence of Young-Onset Colorectal Cancer Before Recent Increase. Gastroenterology. 2018. December 1;155(6):1716–1719.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauer R, Becker H, Hohenberger W, Rödel C, Wittekind C, Fietkau R, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004. October 21;351(17):1731–40. [DOI] [PubMed] [Google Scholar]
- 4.Sauer R, Liersch T, Merkel S, Fietkau R, Hohenberger W, Hess C, et al. Preoperative Versus Postoperative Chemoradiotherapy for Locally Advanced Rectal Cancer: Results of the German CAO/ARO/AIO-94 Randomized Phase III Trial After a Median Follow-Up of 11 Years. JCO. 2012. June 1;30(16):1926–33. [DOI] [PubMed] [Google Scholar]
- 5.Monson JRT, Weiser MR, Buie WD, Chang GJ, Rafferty JF, Buie WD, et al. Practice parameters for the management of rectal cancer (revised). Dis Colon Rectum. 2013. May;56(5):535–50. [DOI] [PubMed] [Google Scholar]
- 6.Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen Y- J, Ciombor KK, et al. Rectal Cancer, Version 2.2018, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2018. July 1;16(7):874–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mace AG, Pai RK, Stocchi L, Kalady MF. American Joint Committee on Cancer and College of American Pathologists regression grade: a new prognostic factor in rectal cancer. Dis Colon Rectum. 2015. January;58(1):32–44. [DOI] [PubMed] [Google Scholar]
- 8.Janjan NA, Khoo VS, Abbruzzese J, Pazdur R, Dubrow R, Cleary KR, et al. Tumor downstaging and sphincter preservation with preoperative chemoradiation in locally advanced rectal cancer: the M. D. Anderson Cancer Center experience. International Journal of Radiation Oncology*Biology*Physics. 1999. July 15;44(5):1027–38. [DOI] [PubMed] [Google Scholar]
- 9.Pucciarelli S, Toppan P, Friso ML, Russo V, Pasetto L, Urso E, et al. Complete Pathologic Response Following Preoperative Chemoradiation Therapy for Middle to Lower Rectal Cancer Is Not a Prognostic Factor for a Better Outcome. Dis Colon Rectum. 2004. November 1;47(11):1798–807. [DOI] [PubMed] [Google Scholar]
- 10.Karagkounis G, Kalady MF. Molecular Biology: Are We Getting Any Closer to Providing Clinically Useful Information? Clin Colon Rectal Surg. 2017. November;30(5):415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karagkounis G, Thai L, Mace AG, Wiland H, Pai RK, Steele SR, et al. Prognostic Implications of Pathological Response to Neoadjuvant Chemoradiation in Pathologic Stage III Rectal Cancer. Ann Surg. 2018. February 20; [DOI] [PubMed] [Google Scholar]
- 12.Gantt GA, Chen Y, DeJulius K, Mace AG, Barnholtz-Sloan J, Kalady MF. Gene expression profile is associated with chemoradiation resistance in rectal cancer. Colorectal Dis. 2014. January 1;16(1):57–66. [DOI] [PubMed] [Google Scholar]
- 13.Zhyvoloup A, Nemazanyy I, Panasyuk G, Valovka T, Fenton T, Rebholz H, et al. Subcellular localization and regulation of coenzyme A synthase. J Biol Chem. 2003. December 12;278(50):50316–21. [DOI] [PubMed] [Google Scholar]
- 14.Zhyvoloup A, Nemazanyy I, Babich A, Panasyuk G, Pobigailo N, Vudmaska M, et al. Molecular Cloning of CoA Synthase THE MISSING LINK IN CoA BIOSYNTHESIS. J Biol Chem. 2002. June 21;277(25):22107–10. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan B, Sibon OCM. Coenzyme A, more than ‘just’ a metabolic cofactor. Biochemical Society Transactions. 2014. August 1;42(4):1075–9. [DOI] [PubMed] [Google Scholar]
- 16.Aoun M, Tiranti V. Mitochondria: A crossroads for lipid metabolism defect in neurodegeneration with brain iron accumulation diseases. Int J Biochem Cell Biol. 2015. June;63:25–31. [DOI] [PubMed] [Google Scholar]
- 17.Trakarnsanga A, Gönen M, Shia J, Nash GM, Temple LK, Guillem JG, et al. Comparison of tumor regression grade systems for locally advanced rectal cancer after multimodality treatment. J Natl Cancer Inst. 2014. October;106(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003. November 1;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005. October 25;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mootha VK, Lindgren CM, Eriksson K- F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003. July;34(3):267–73. [DOI] [PubMed] [Google Scholar]
- 21.Ferrandon S, Saultier P, Carras J, Battiston-Montagne P, Alphonse G, Beuve M, et al. Telomere Profiling: Toward Glioblastoma Personalized Medicine. Mol Neurobiol. 2013. February 1;47(1):64–76. [DOI] [PubMed] [Google Scholar]
- 22.Begley TP, Kinsland C, Strauss E. The biosynthesis of coenzyme A in bacteria. Vitam Horm. 2001;61:157–71. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe T, Komuro Y, Kiyomatsu T, Kanazawa T, Kazama Y, Tanaka J, et al. Prediction of Sensitivity of Rectal Cancer Cells in Response to Preoperative Radiotherapy by DNA Microarray Analysis of Gene Expression Profiles. Cancer Res. 2006. April 1;66(7):3370–4. [DOI] [PubMed] [Google Scholar]
- 24.Emons G, Spitzner M, Reineke S, Möller J, Auslander N, Kramer F, et al. Chemoradiotherapy Resistance in Colorectal Cancer Cells is Mediated by Wnt/β-catenin Signaling. Mol Cancer Res. 2017;15(11):1481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toulany M, Rodemann HP. Phosphatidylinositol 3-kinase/Akt signaling as a key mediator of tumor cell responsiveness to radiation. Seminars in Cancer Biology. 2015. December;35:180–90. [DOI] [PubMed] [Google Scholar]
- 26.HEIN AL, OUELLETTE MM, YAN Y. Radiation-induced signaling pathways that promote cancer cell survival (Review). Int J Oncol. 2014. August 20;45(5):1813–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemazanyy I, Panasyuk G, Zhyvoloup A, Panayotou G, Gout IT, Filonenko V. Specific interaction between S6K1 and CoA synthase: a potential link between the mTOR/S6K pathway, CoA biosynthesis and energy metabolism. FEBS Letters. 2004. December 17;578(3):357–62. [DOI] [PubMed] [Google Scholar]
- 28.Breus O, Panasyuk G, Gout IT, Filonenko V, Nemazanyy I. CoA Synthase is in complex with p85αPI3K and affects PI3K signaling pathway. Biochemical and Biophysical Research Communications. 2009. August 7;385(4):581–5. [DOI] [PubMed] [Google Scholar]
- 29.Arif A, Jia J, Willard B, Li X, Fox PL. Multisite Phosphorylation of S6K1 Directs a Kinase Phospho-code that Determines Substrate Selection. Mol Cell. 2018. December 27; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De P, Carlson JH, Leyland-Jones B, Dey N. PI3K-AKT-mTOR Pathway Cooperates with the DNA Damage Repair Pathway: Carcinogenesis in Triple-Negative Breast Cancers and Beyond In: PI3K-mTOR in Cancer and Cancer Therapy [Internet]. Humana Press, Cham; 2016. [cited 2017 Nov 8]. p. 65–108. (Cancer Drug Discovery and Development). [Google Scholar]
- 31.Berti CC, Dallabona C, Lazzaretti M, Dusi S, Tosi E, Tiranti V, et al. Modeling human Coenzyme A synthase mutation in yeast reveals altered mitochondrial function, lipid content and iron metabolism. Microb Cell. 2015. April 6;2(4):126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khatri D, Zizioli D, Tiso N, Facchinello N, Vezzoli S, Gianoncelli A, et al. Down-regulation of coasy, the gene associated with NBIA-VI, reduces Bmp signaling, perturbs dorso-ventral patterning and alters neuronal development in zebrafish. Sci Rep. 2016. 28;6:37660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinghorn KJ, Castillo-Quan JI. Mitochondrial dysfunction and defects in lipid homeostasis as therapeutic targets in neurodegeneration with brain iron accumulation. Rare Dis. 2016;4(1):e1128616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin C- C, Kitagawa M, Tang X, Hou M- H, Wu J, Qu DC, et al. CoA synthase regulates mitotic fidelity via CBP-mediated acetylation. Nature Communications. 2018. March 12;9(1):1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng H, You K, Zhang R, Xi S, Zhang T, Dong J, et al. Predictive value of APAF-1 and COX-2 expression in pathologic complete response to neoadjuvant chemoradiotherapy for patients with locally advanced rectal adenocarcinoma. Oncotarget. 2016. May 2;7(23):35233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voboril R, Weberova-Voborilova J. Constitutive NF-kappaB activity in colorectal cancer cells: impact on radiation-induced NF-kappaB activity, radiosensitivity, and apoptosis. Neoplasma. 2006;53(6):518–23. [PubMed] [Google Scholar]
- 37.Ahmed MA, Selzer E, Dörr W, Jomrich G, Harpain F, Silberhumer GR, et al. Fibroblast growth factor receptor 4 induced resistance to radiation therapy in colorectal cancer. Oncotarget. 2016. September 17;7(43):69976–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho V, Chung L, Revoltar M, Lim SH, Tut T- G, Abubakar A, et al. MRE11 and ATM Expression Levels Predict Rectal Cancer Survival and Their Association with Radiotherapy Response. PLoS One [Internet]. 2016. December 8 [cited 2017 May 12];11(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaanan A, Park JM, Tougeron D, Huang S, Wu T- T, Foster NR, et al. Association of Beclin 1 expression with response to neoadjuvant chemoradiation therapy in patients with locally advanced rectal carcinoma. Int J Cancer. 2015. September 15;137(6):1498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y- E, He H- L, Shiue Y- L, Lee S- W, Lin L- C, Wu T- F, et al. The prognostic impact of lipid biosynthesis-associated markers, HSD17B2 and HMGCS2, in rectal cancer treated with neoadjuvant concurrent chemoradiotherapy. Tumour Biol. 2015. September;36(10):7675–83. [DOI] [PubMed] [Google Scholar]
- 41.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014. April;15(4):243–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016. January;16(1):20–33. [DOI] [PubMed] [Google Scholar]
- 43.Tan BS, Tiong KH, Choo HL, Fei-Lei Chung F, Hii L- W, Tan SH, et al. Mutant p53-R273H mediates cancer cell survival and anoikis resistance through AKT-dependent suppression of BCL2-modifying factor (BMF). Cell Death & Disease. 2015. July;6(7):e1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deming DA, Leystra AA, Nettekoven L, Sievers C, Miller D, Middlebrooks M, et al. PIK3CA and APC mutations are synergistic in the development of intestinal cancers. Oncogene. 2014. April 24;33(17):2245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foley TM, Payne SN, Pasch CA, Yueh AE, Van De Hey DR, Korkos DP, et al. Dual PI3K/mTOR Inhibition in Colorectal Cancers with APC and PIK3CA Mutations. Mol Cancer Res. 2017. February 9;15(3):317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao M, Feng T, Mariadason JM, Tsao CC, Lemos R, Dayyani F, et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin Cancer Res. 2013. February 1;19(3):657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong S, Kim S, Kim HY, Kang M, Jang HH, Lee W. Targeting the PI3K signaling pathway in KRAS mutant colon cancer. Cancer Med. 2015. December 29;5(2):248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toulany M, Lee K- J, Fattah KR, Lin Y- F, Fehrenbacher B, Schaller M, et al. Akt Promotes Post-Irradiation Survival of Human Tumor Cells through Initiation, Progression, and Termination of DNA-PKcs–Dependent DNA Double-Strand Break Repair. Mol Cancer Res. 2012. July 1;10(7):945–57. [DOI] [PubMed] [Google Scholar]
- 49.Deng R, Tang J, Ma J- G, Chen S- P, Xia L- P, Zhou W- J, et al. PKB/Akt promotes DSB repair in cancer cells through upregulating Mre11 expression following ionizing radiation. Oncogene. 2011. February 24;30(8):944–55. [DOI] [PubMed] [Google Scholar]
- 50.Gao N, Flynn DC, Zhang Z, Zhong X- S, Walker V, Liu KJ, et al. G1 cell cycle progression and the expression of G1 cyclins are regulated by PI3K/AKT/mTOR/p70S6K1 signaling in human ovarian cancer cells. American Journal of Physiology - Cell Physiology. 2004. August 1;287(2):C281–91. [DOI] [PubMed] [Google Scholar]
- 51.Lu Q, Wang J, Yu G, Guo T, Hu C, Ren P. Expression and clinical significance of mammalian target of rapamycin/P70 ribosomal protein S6 kinase signaling pathway in human colorectal carcinoma tissue. Oncol Lett. 2015. Jul;10(1):277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kandioler D, Zwrtek R, Ludwig C, Janschek E, Ploner M, Hofbauer F, et al. TP53 Genotype but Not p53 Immunohistochemical Result Predicts Response to Preoperative Short-Term Radiotherapy in Rectal Cancer. Ann Surg. 2002. April;235(4):493–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng Y, Bagrodia S, Cerione RA. Activation of phosphoinositide 3-kinase activity by Cdc42Hs binding to p85. J Biol Chem. 1994. July 22;269(29):18727–30. [PubMed] [Google Scholar]
- 54.Pleiman CM, Hertz WM, Cambier JC. Activation of phosphatidylinositol-3’ kinase by Src-family kinase SH3 binding to the p85 subunit. Science. 1994. March 18;263(5153):1609–12. [DOI] [PubMed] [Google Scholar]
- 55.King H, Thillai K, Whale A, Arumugam P, Eldaly H, Kocher HM, et al. PAK4 interacts with p85 alpha: implications for pancreatic cancer cell migration. Scientific Reports. 2017. February 16;7:42575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huh JW, Kim HC, Kim SH, Park YA, Cho YB, Yun SH, et al. Tumor regression grade as a clinically useful outcome predictor in patients with rectal cancer after preoperative chemoradiotherapy. Surgery. 2018. October 9; [DOI] [PubMed] [Google Scholar]
- 57.Dossa F, Chesney TR, Acuna SA, Baxter NN. A watch-and-wait approach for locally advanced rectal cancer after a clinical complete response following neoadjuvant chemoradiation: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2017;2(7):501–13. [DOI] [PubMed] [Google Scholar]
- 58.Renehan AG, Malcomson L, Emsley R, Gollins S, Maw A, Myint AS, et al. Watch-and-wait approach versus surgical resection after chemoradiotherapy for patients with rectal cancer (the OnCoRe project): a propensity-score matched cohort analysis. The Lancet Oncology. 2016. February 1;17(2):174–83. [DOI] [PubMed] [Google Scholar]
- 59.van der Valk MJM, Hilling DE, Bastiaannet E, Meershoek-Klein Kranenbarg E, Beets GL, Figueiredo NL, et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): an international multicentre registry study. Lancet. 2018. 23;391(10139):2537–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


