Abstract
Oral squamous cell carcinoma (OSCC) is capable of constructing a favorable immune escape environment through interactions of cells with cells and of cells with the environment. Programmed death ligand-1 (PD-L1) is a well-recognized inhibitor of anti-tumor immunity that plays an important role in tumor immune escape. However, the molecular mechanisms regulating PD-L1 expression are not yet fully understood. In this study, to investigate the role of protein kinase D3 (PKD3) in the regulation of PD-L1 expression, the expression and correlation of PKD3 and PD-L1 were first analyzed by the immunostaining of human OSCC tissue sections, cell experiments and TCGA gene expression databases. The expression levels of PKD3 and PD-L1 were found to be significantly higher in OSCC cells than in normal tissues or cells. In addition, the expression levels of PKD3 and PD-L1 were found to be significantly positively correlated. Subsequently, it was found that the levsel of PD-L1 expression decreased following the silencing of PKD3 and that the ability of interferon (IFN)-γ to induce PD-L1 expression was also decreased in OSCC. The opposite phenomenon occurred following the overexpression of PKD3. It was also found that the phosphorylation of signal transducer and activator of transcription (STAT)1/STAT3 was reduced by the knockdown of PKD3 in OSCC. Moreover, the expression level of PD-L1 was decreased after the use of siRNA to knockdown STAT1 or STAT3. On the whole, the findings of this study confirm that PKD3 regulates the expression of PD-L1 induced by IFN-γ by regulating the phosphorylation of STAT1/STAT3. These findings broaden the understanding of the biological function of PKD3, suggesting that PKD is a potential therapeutic target for OSCC.
Keywords: oral squamous cell carcinoma, protein kinase D3, programmed death ligand-1, interferon-γ, signal transducer and activator of transcription 1/3
Introduction
Oral squamous cell carcinoma (OSCC) is the most common malignant tumor of the head and neck region. It is associated with rapid growth, strong invasiveness, early cervical lymph node metastasis and a high rate of metastasis. Approximately 90% of oral cancers are squamous cell carcinoma or one of its variants (1,2). It is currently one of the leading causes of cancer-related mortality. Despite recent advances in research and therapies, such as chemotherapy, radiotherapy and immunotherapy in particular, the overall mortality rate of patients with OSCC has remained constant over the past few decades, at approximately 50% (3,4).
Tumor immune escape and chronic inflammation in the tumor microenvironment are two important features necessary for tumorigenesis and cancer progression. Programmed death ligand-1 (PD-L1), a ligand for the programmed cell death protein 1 (PD-1) immunosuppressive checkpoint, can be induced in tumors by their exposure to inflammatory factors in the tumor microenvironment, leading to immune escape. PD-L1 protein expression in tumor cells is upregulated upon their stimulation with interleukin (IL)-1, IL-6, tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ), which are located in the tumor microenvironment (5). Of these effectors, IFN-γ is the most effective inducer of PD-L1 expression (6). Recent studies have suggested that signaling molecules affecting the cell cycle, proliferation, apoptosis and survival [including mitogen-activated protein kinase (MAPK), nuclear factor-κB (NF-κB), phosphatidylinositol 3-kinase (PI3K) and Janus kinase (JAK)/signal transducer and activator of transcription (STAT)] are involved in the regulation of PD-L1 expression (6-9). Notably, OSCC usually exhibits host immunosuppression and cytogenetic alterations in tumor cells. The detailed understanding of the mechanisms through which PD-L1 expression is regulated will facilitate the identification of pathways that inhibit PD-L1 function and modulate cancer cell-responsive immune responses.
The protein kinase D (PKD) family consists of 3 serine/threonine kinases (termed PKCμ/PKD1, PKD2 and PKCν/PKD3). They are extremely important regulators of diverse biological processes involved in cell proliferation, cell migration, differentiation, apoptosis, cardiac contraction, cardiac hypertrophy, angiogenesis, tumorigenesis, epithelial-to-mesenchymal transition and immune regulation (10-16). The PKD subtypes can be localized to the plasma membrane and the Golgi complex, and it has also been reported that they can shuttle to the nucleus, as in the case of PKD3 (17). In recent decades, studies on the functions and mechanisms of PKD have mainly focused on PKD1 and PKD2. However, little is known about the function of PKD3, particularly its mechanisms of action. There is increasing evidence to suggest that PKD3 is connected to multiple pathways involved in oncogenic signaling, such as protein kinase B (AKT), extracellular signal-regulated kinase 1/2 (ERK1/2), NF-κB, STAT1 and STAT3 (16,18,19). These signals can also trigger the expression of PD-L1 in tumor cells. Previously, the authors' research group found that PKD2 exerted a certain regulatory effect on the expression of PD-L1 (11). However, the mechanisms through which PKD3, as an oncogene, regulates PD-L1 expression in OSCC cells remain unknown.
In this study, the role of PKD3 in the tumorigenesis and progression of OSCC was examined. The results suggest that PKD3 expression is elevated in OSCC and that PKD3 regulates PD-L1 expression via STAT1 and STAT3. The findings of this study suggest that PKD may be a promising therapeutic target for OSCC and broaden the current understanding of the molecular mechanisms and function of PKD3 in cancer progression.
Materials and methods
Isolation of PDLCs and cell culture
Human oral normal periodontal ligament cells (PDLCs) were obtained from premolar teeth without inflammation and caries, which were extracted for orthodontic treatment at the West China Hospital of Stomatology of Sichuan University. All donors were healthy and written informed consent was obtained from each donor prior to tooth extraction. The extracted teeth were rinsed and placed in phosphate-buffered saline (PBS) supplemented with 1,000 IU/ml penicillin and 1,000 μg/ml streptomycin (HyClone). The remaining procedures were performed according to a previously described protocol (20). Periodontal tissues were from the middle third of the root, were cut into 1-2 mm2 sections and placed in culture flasks for cell culture in RPMI-1640 medium (Gibco; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA), 100 IU/ml penicillin and 100 μg/ml streptomycin. The present study was approved by the West China Hospital of Stomatology Institutional Review Board.
The dysplastic oral keratinocyte (DOK) cell line and 4 OSCC cell lines (Cal-27, HSC-4, HSC-3 and SCC25) were purchased from the American Type Culture Collection (ATCC). The SCC25 cells were cultured in Dulbecco's modified Eagle's medium/nutrient Mixture F12 (DMEM/F12; Gibco; Thermo Fisher Scientific) with 400 ng/ml hydrocortisone and 10% FBS (Sigma-Aldrich; Merck KGaA). The other cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific) supplemented with 10% FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin. The human normal oral epithelial keratinocytes (HOK) were purchased from ScienCell Research Laboratories, Inc. and were cultured in keratinocyte serum-free medium (KSFM) supplemented with recombinant human epidermal growth factor (5 ng/ml) and bovine pituitary extract (50 μg/ml) (Gibco; Thermo Fisher Scientific). The cells were maintained in a humidified 5% CO2 atmosphere at 37̊C.
Patients and clinical samples
The present study included OSCC tissues specimens from 34 patients with OSCC who underwent partial or total surgical resection at West China Hospital of Stomatology from 2014 to 2016. The 34 patients with OSCC enrolled in this study had not received radiotherapy or chemotherapy prior to surgical resection. The clinical information of the patients is presented in Table I. Clinically normal oral mucosa specimens (>2 cm at a distance from the edge of the tumor mass) and primary cancer tissues were collected by surgical resection. The clinical samples were confirmed by two experienced pathologists. This study was approved by the West China Hospital of Stomatology Institutional Review Board and written informed consent was obtained from each patient.
Table I.
Clinical characteristics of the 34 patients with OSCC.
| Patient characteristics (n=34) | No. of patients | Percentage (%) |
|---|---|---|
| Age (years) | ||
| <57 | 16 | 47.1 |
| >57 | 18 | 52.9 |
| Sex | ||
| Male | 26 | 76.5 |
| Female | 8 | 23.5 |
| N-regional lymph node | ||
| Negative | 20 | 58.8 |
| Positive | 14 | 41.2 |
| Histological grade | ||
| Grade 1 | 12 | 35.3 |
| Grade 2 | 18 | 52.9 |
| Grade 3 | 4 | 11.8 |
| TNM stage | ||
| I-II | 15 | 44.1 |
| III-IV | 19 | 55.9 |
OSCC, oral squamous cell carcinoma; TNM, tumor-node-metastasis.
Plasmids and transfection
The Hu-shRNA PKD3 and control shRNA plasmids were obtained from GeneCopoeia. The Hu-shRNA construct contains the human PKD3 gene-specific sequence (GCT CCT ACT TTC TGT GAT TAC) shRNA expression vector psi-LVRU6GP. The plasmid containing GCT TCG CGC CGT AGT CTT A (scrambled) was used as a control. A PKD3 overexpression plasmid and the corresponding control plasmid were also purchased from GeneCopoeia. All plasmids were transfected into the cells using Lipofectamine 2000 reagent according to the manufacturer's instructions. The DOK, Cal-27 and HSC-4 cells were seeded into 6 well plates, and transfection was carried out at 60-80% confluency. Hu-shRNA PKD3 and control shRNA plasmids were transfected into Cal-27 and HSC-4. DOK stably expressing PKD3 protein was established by transfecting the PKD3 overexpression plasmid. After 24 h, the cells were cultured in 0.2 μg/ml puromycin, and PKD3 expression levels were detected by western blot analysis.
Transient gene knockdown with siRNA
STAT1 siRNA (5′-CAC GAG ACC AAU GGU GUG GdT dT-3′; 5′-CCA CAC CAU UGG UCU CGU GdT dT-3′) was used to knockdown STAT1 expression, as previously described (21). In addition, STAT3 siRNA (5′-AAC AUC UGC CUA GAU CGG CUA dTdT-3′; 3′-dTd TGU AGA CGG AUC UAG CCG AU-5′) was synthesized by Dharmacon Research (22). A scrambled sequence (5′-UUC UCC GAA CGU GUC ACG UTT-3′; 5′-ACG UGA CAC GUU CGG AGA ATT-3′) was used as a negative control. STAT1/3 siRNA was transfected into the Cal-27 and HSC-4 cells using Lipofectamine 2000 reagent according to the manufacturer's instructions. The knockdown efficiency was assessed at 72 h following transfection by western blot analysis.
Cell lysates and western blot analysis
The cells were seeded at a density of 2×105 cells per well in 6-well plates. Following overnight incubation, the medium was replaced with maintenance medium containing 20 ng/ml of cytokines, such as IL-1 and IL-6, TNF-α and recombinant human IFN-γ (R&D Systems). The 'wild' group represented untreated cells. The cells in the MIX group were treated with 20 ng/ml of IL-1β, IL-6, TNF-α and IFN-γ. Subsequently, the cells were washed 3 times with ice-cold PBS and lysed with cell lysis buffer (50 mmol/l Tris-HCl at pH 7.4, 5 mmol/l EDTA, 150 mmol/l NaCl, 0.5% Nonidet P-40, 0.5 mmol/l PMSF, 0.5 mmol/l DTT and protease inhibitor cocktail) for 30 min at 4°C. The supernatant was collected by centrifugation at 13,000 × g for 10 min. Lysates were used for western blot analysis as previously described (11). The protein concentration was measured with BCA protein assay reagent (Beyotime Institute of Biotechnology). Approximately 20 μg of protein was separated by 8% SDS-PAGE and blotted onto PVDF membranes (Bio-Rad Laboratories, Inc.). After blocking in 5% non-fat milk for 1 h at room temperature, the membranes were incubated with primary antibodies on a shaker at 4̊C overnight, followed by incubation with horseradish peroxidase (HRP) conjugated anti-mouse or anti-rabbit immunoglobulin G (IgG; cat. no. 7076 or 7074; 1:2,000; Cell Signaling Technology, Inc.) for 1.5 h at 37°C. The protein bands were visualized using an enhanced chemiluminescence (ECL) substrate kit (Millipore, Inc.) with the ECL western blotting system (Bio-Rad Laboratories, Inc.). The primary antibodies used for western blot were as follows: PD-L1 (cat. no. 13684; 1:1,000) and PKD3 (cat. no. 5655; 1:1,000) from Cell Signaling Technology; and STAT1 (cat. no. ab109320; 1:10,000), STAT3 (cat. no. ab68153; 1:2,000), phospho-STAT1(S727) (cat. no. ab109461; 1:5,000), phospho-STAT1(Y701) (cat. no. ab29045; 1:1,000), phospho-STAT3(S727) (cat. no. ab32143; 1:5,000), phospho-STAT3(Y705) (cat. no. ab76315; 1:10,000) and anti-GAPDH (cat. no. ab128915; 1:20,000) from Abcam. Phostag SDS-PAGE (Wako) was performed according to a previously described protocol (23). Semi-quantitative analysis was performed by densitometry using Gel-Pro 32 software (version 3.1, Media Cybernetics, Bethesda).
Immunofluorescence and flow cytometry
The cells were seeded on coverslips and allowed to attach overnight. They were then washed twice with PBS for 5 min at room temperature, fixed with 4% paraformaldehyde for 15 min at room temperature, and then blocked in blocking buffer (1X PBS, 5% normal goat serum, 0.3% Triton X-100™) for 60 min. Primary antibodies were prepared by their dilution according to the datasheet guidelines in antibody dilution buffer (1X PBS, 1% BSA, 0.3% Triton X-100™). The primary antibodies included a polyclonal anti-PKD3 antibody (cat. no. ab252982; 1:40; Abcam) for indirect immunofluorescence and a PE-conjugated antibody for human PD-L1 (MIH1)(cat. no. 12-5983-42; 1:20; eBioscience) for direct immunofluorescence. Following the removal of the blocking solution, the cells were incubated overnight at 4̊C and then washed 3 times in PBS for 5 min each. The cells were then incubated with fluorochrome-conjugated secondary antibody (FITC-conjugated goat anti-rabbit IgG, cat. no. F-2765; 1:100; Invitrogen; Thermo Fisher Scientific) diluted in antibody dilution buffer for 1 h at room temperature in the dark. The slides were then rinsed in PBS and coverslipped with Prolong® Gold Anti-Fade Reagent with DAPI. Imaging was performed using a fluorescence microscope. Flow cytometry was performed according to a previously described (5) using a PE-conjugated antibody for human PD-L1 (MIH1) (cat. no. 12-5983-42; 1:20; eBioscience).
Reverse transcription-quantitative PCR (RT-qPCR)
For RT-qPCR, total RNA was collected from the cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific), and reverse transcription reactions were performed using PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd.). The resulting cDNA was then subjected to qPCR analysis using SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.) and the ABI 7500 real-time PCR machine (Applied Biosystems). The qPCR conditions were as follows: Initial denaturation at 95̊C for 30 sec, followed by 40 cycles of 95̊C for 5 sec and 60̊C for 30 sec. The relative expression values of the targeted genes were calculated using the comparative Cq (2−ΔΔCq) method. The primers (forward and reverse, respectively) used for RT-qPCR included PD-L1 (forward, 5′-CAA TGT GAC CAG CAC ACT GAG AA-3′ and reverse, 5′-GGC ATA ATA AGA TGG CTC CCA GAA-3′) and GAPDH (forward, 5′-ACA ACT TTG GTA TCG TGG AAG G-3′ and reverse, 5′-GCC ATC ACG CCA CAG TT TC-3′). The relative mRNA expression of PD-L1 was normal-ized to that of GAPDH.
Immunohistochemical staining and analysis
The OSCC tissues were fixed with 10% neutral formalin and embedded in paraffin. A series of 5-μm-thick slices was cut, dewaxed with xylene, and rehydrated with a series of graded ethanol solutions. The tissue slices were boiled for 20 min in citrate solution (10 mmol/l, pH 6.0). After cooling, the slices were immersed in 0.3% hydrogen peroxide solution for 15 min to block endogenous peroxidase activity. The slices were rinsed in PBS for 5 min and blocked with 5% BSA solution at room temperature for 20 min. Subsequently, the sections were incubated overnight with rabbit anti-human PD-L1 polyclonal antibody (cat. no. 13684; 1:200; Cell Signaling Technology) or rabbit anti-human PKD3 polyclonal antibody (cat. no. ab252982; 1:40; Abcam) at 4°C. The following day, according to the manufacturer's instructions of ChemMate™ EnVision™ Detection kit (Genetech), the sections were incubated with HRP-labeled goat anti-mouse or -rabbit secondary antibodies. Diaminobenzidine (DAB) was used for color development, and hematoxylin was used to stain the nuclei for 2 min at room temperature. Finally, the slices were dehydrated, cleaned and mounted.
ImageJ 1.48v software was used for analysis. The staining intensity was evaluated according to the following ratings: 0, no staining; 1, weak staining; 2, medium staining; 3, strong staining. The H-score was calculated as follows: H-score = 0 × (% negative tumor cells) + 1 × (% weak staining) + 2 × (% medium staining) + 3 × (% strong staining). The H-scores ranged from 0 (100% negative tumor cells) to 300 (100% strong staining of tumor cells).
The Cancer Genome Atlas (TCGA) data retrieval and analysis
UCSC Xena (http://xena.ucsc.edu/welcome-to-ucsc-xena/) was used to download RNAseq data for queried genes. The gene expression data of 564 head and neck squamous cell carcinoma (HNSCC) samples were obtained. Linear regression curve fitting analysis was performed using GraphPad Prism software. TCGA data were correlated using the Spearman's r test.
Statistical analysis
All data were statistically analyzed using GraphPad Prism software (version 6; GraphPad Software, Inc.) using one-way analysis of variance followed by the post hoc Tukey's multiple comparisons test. The correlation between PKD3 and PD-L1 expression was analyzed using Spearman's correlation analysis. All statistical results with a P-value <0.05 were considered to be significant.
Results
Expression and localization of PD-L1 and PKD3 in non-tumor and OSCC cell lines
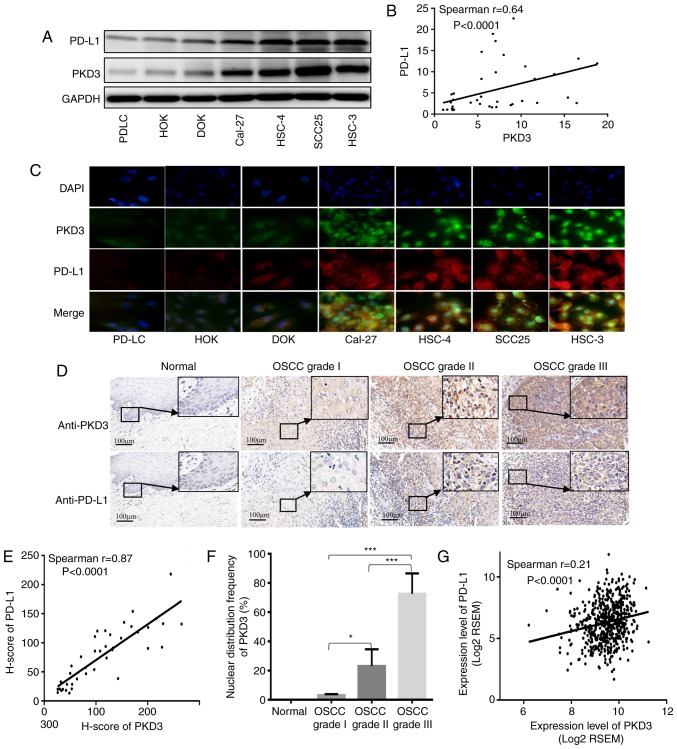
The expression levels of PKD3 and PD-L1 in non-tumor and OSCC cell lines were first measured by western blot analysis and immunofluorescence. As shown in Fig. 1A and C, the expression levels of PKD3 and PD-L1 were higher in the cancer cells than in the non-cancer cells. In addition, the expression pattern of the immunosuppressive protein, PD-L1, correlated well with the expression pattern of PKD3 (Fig. 1B). Subsequently, the localization of PKD3 and PD-L1 expression was analyzed by double fluorescence staining. From non-tumor cells to tumor cells, PKD3 exhibited a progressively increasing nuclear distribution that ranged from only in the cytoplasm, to evenly in the cytoplasm and nucleus, to mainly in the nucleus. However, PD-L1 exhibited an intracellular distribution that contrasted with that of PKD3. PD-L1 exhibited progressively enhanced plasma membrane distribution ranging from evenly in the cytoplasm and nucleus to mainly in the plasma membrane. In addition to the increased accumulation of PKD3 in the nucleus, the staining of PD-L1 at the cell membrane gradually increased. Taken together, the data indicated that PKD3 and PD-L1 are upregulated and highly positively correlated in OSCC, suggesting that PKD3 plays an important role in the regulation of the expression of the immunosuppressive protein, PD-L1.
Figure 1.
Expression and correlation of PKD3 and PD-L1 in OSCC. (A) The expression levels of PKD3 and PD-L1 were examined by western blot analysis in various oral epithelium-derived tumor cells and non-tumor cells. (B) Correlation analysis between PKD3 and PD-L1; P<0.0001, Spearman's r=0.64. (C) Expression and localization of PKD3 and PD-L1 by immunofluorescence analysis. (D) The expression levels of PKD3 and PD-L1 were analyzed by immunohistochemistry in normal tissue and tumor specimens. (E) Linear regression analysis of immunostained PKD3 and PD-L1 in human OSCC tissues with H-scores; P<0.0001, Spearman's r=0.87. (F) Nuclear distribution of PKD3 in normal and tumor tissues. (G) Linear regression analysis was performed using the expression data of PKD3 and PD-L1 in HNSCC from TCGA. P<0.0001, Spearman's r=0.21. *P<0.05; ***P<0.001. PKD3, protein kinase D3; OSCC, oral squamous cell carcinoma; PD-L1, programmed death ligand-1.
Expression and localization of PD-L1 and PKD3 in OSCC tissues
The expression and localization of PD-L1 and PKD3 in were then analyzed 26 normal tissue and 34 tumor specimens. More frequent and intense PKD3 and PD-L1 staining was observed in OSCC tissues (Fig. 1D). In addition, it was found that the nuclear localization of PKD3 was significantly associated with tumor grade. PKD3 nuclear staining was not observed in the normal tissues, whereas the grade I tumors exhibited 3.23% PKD3-positive nuclear staining, grade II tumors exhibited 23.14% positive staining, and grade III tumors exhibited 72.65% positive staining (P<0.01; Fig. 1F). The immunohistochemical score (H-score) was then calculated. To determine whether PKD3 affects the level of the immunosuppressive protein, PD-L1, linear trend and Spearman's rank correlation coefficient tests were performed on the H-score. It was found that a significant positive correlation existed between PKD3 and PD-L1 (P<0.0001, r=0.87; Fig. 1E). The expression of PKD3 and PD-L1 was also analyzed in a large number of HNSCC samples using the TCGA gene expression database. It was found that the expression levels of PKD3 and PD-L1 were significantly higher in the OSCC than in the normal tissues. In addition, the expression level of PKD3 gradually increased with an increase in the tumor grade (Fig. S1). Indeed, it was found that PKD3 significantly and positively correlated with PD-L1 (P<0.0001, r=0.21; Fig. 1G). These results indicate that PKD3 may be involved in the regulation of PD-L1 expression and that the nuclear accumulation of PKD3 may play a role in the pathogenesis of OSCC.
PD-L1 expression is induced by inflammatory factors
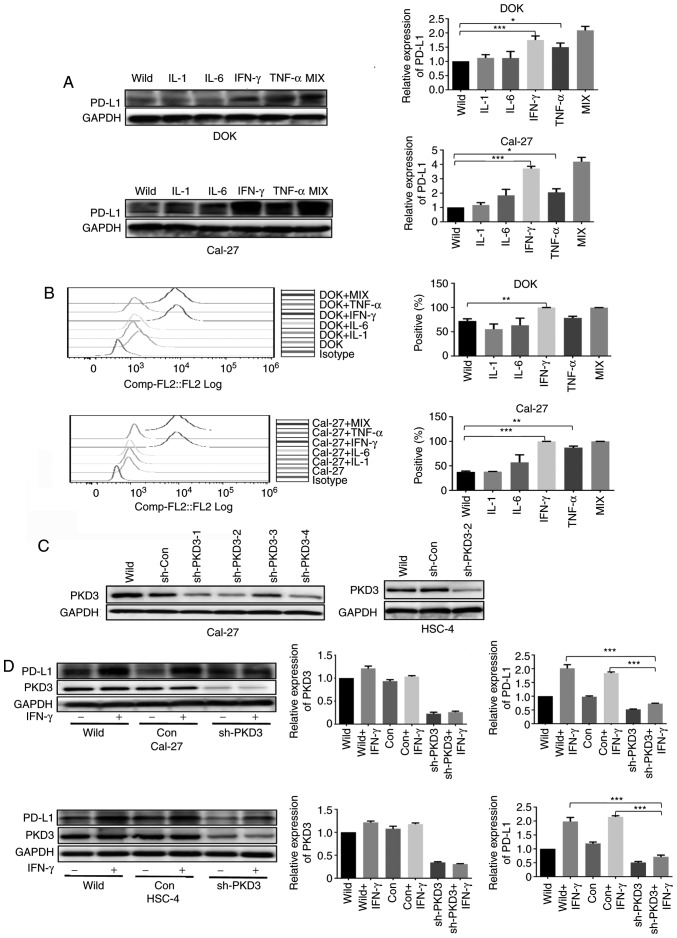
To examine the effects of inflammatory factors on the expression of PD-L1, the DOK and Cal-27 cell lines were treated with 20 ng/ml of IL-1β, IL-6, TNF-α and IFN-γ for 24 h. The expression of PD-L1 was determined by western blot analysis and flow cytometry. It was found that PD-L1 protein expression was upregulated upon the stimulation of the cells with IL-1β, IL-6, TNF-α and IFN-γ, with IFN-γ exhibiting the most potent effect (Fig. 2A and B).
Figure 2.
IFN-γ-induced PD-L1 expression is dependent on PKD3. (A and B) Inflammatory factors increased the expression of PD-L1 in OSCC cells. (C) Cal-27 and HSC-4 cell lines were transfected with a plasmid containing either a PKD3-specific shRNA or a control shRNA. Positive clones were selected with puromycin for 2 weeks. PKD3 expression was examined by western blot analysis. (D) The PD-L1 protein expression levels of Cal-27 and HSC-4 were determined by western blot analysis. Cells transfected with PKD3 shRNA and cells transfected with control shRNA were cultured in the presence or absence of IFN-γ (20 ng/ml) for 24 h. The bar graphs present densitometric analysis of the changes in the levels of PKD3 and PD-L1. Data are presented as the means ± SD (n=3). *P<0.05; **P<0.01; ***P<0.001. IFN-γ, interferon-γ; PD-L1, programmed death ligand-1; PKD3, protein kinase D3; OSCC, oral squamous cell carcinoma.
PKD3 is required for IFN-γ-mediated PD-L1 expression
To determine whether PKD3 is involved in the IFN-γ-induced upregulation of PD-L1 expression in OSCC, a loss-of-function method was used to investigate the biological relevance of PKD3 in the IFN-γ-induced expression of PD-L1. First, shRNA was used to silence the expression of PKD3. The silencing efficiency was confirmed at the protein level by western blot analysis (Fig. 2C). Additional results revealed that PKD3 knockdown decreased the expression of PD-L1. Even with exposure to IFN-γ, PD-L1 expression was only slightly elevated (Fig. 2D). Moreover, the overexpression of PKD3 in the DOK cells significantly increased the expression of PD-L1 induced by IFN-γ (P<0.001, Fig. 3C and D). However, the expression level of PD-L1 was only slightly elevated without IFN-γ treatment (Fig. 3A and B). These data thus suggest that PKD3 is responsible for IFN-γ-mediated PD-L1 expression.
Figure 3.
PKD3 overexpression enhances IFN-γ-induced PD-L1 expression in DOK cells. (A and B) The levels of PKD3 and PD-L1 protein were examined by western blot analysis and immunofluorescence. The PKD3 overexpression plasmid was transfected into DOK cells using Lipofectamine 2000. (C and D) Cells transfected with the PKD3 overexpression plasmid and cells transfected with control shRNA were cultured in the presence or absence of IFN-γ (20 ng/ml) for 24 h. The expression levels of PKD3 and PD-L1 were determined by western blot analysis. In addition, the cell surface expression of PD-L1 was analyzed by flow cytometry. Data are presented as the means ± SD (n=3). ***P<0.001. PKD3, protein kinase D3; PD-L1, programmed death ligand-1; IFN-γ, interferon-γ.
IFN-γ activates PKD3, STAT1 and STAT3 in OSCC cell lines
The findings of this study have thus far demonstrated that PKD3 is involved in the expression of PD-L1 induced by IFN-γ. To investigate the IFN-γ-mediated signal transduction events, the activation status of PKD3 was first examined by using phostag SDS-PAGE immunoblot analysis of cells exposed to IFN-γ (Fig. 4A). IFN-γ induced a marked increase in PKD3 phosphorylation as early as 1 h. The TCGA gene expression database was then used to analyze the major signaling pathway members involved in the regulation of PD-L1 expression in a large number of HNSCC samples, such as MAPK, NF-κB, PI3K and STAT1/3. It was found that only the expression of STAT1/3 was significantly associated with that of PD-L1 and PKD3 (Fig. S2). Moreover, numerous studies have indicated that STAT1 and STAT3 play a key role in regulating the expression of PD-L1 in HNSCC (6,9,24-26). As expected, IFN-γ stimulation activated STAT1 and STAT3, with their phosphorylation apparent after 1 h in all 3 cell lines (Fig. 4A and B). In addition, the PD-L1 mRNA levels were measured by RT-qPCR. As shown in Fig. 4C, the PD-L1 mRNA level peaked at 2 h. Moreover, the level of PD-L1 protein was gradually increased after 6 h in the presence of IFN-γ (Fig. 4A). Thus, these results indicate that IFN-γ induces the activation of PKD3, STAT1 and STAT3 in OSCC, and that the activation of PKD3, STAT1 and STAT3 chronologically follows the increase in the mRNA and protein levels of PD-L1.
Figure 4.
IFN-γ induces the activation of related factors regulating PD-L1 expression in OSCC cells. (A) Cancer cells were stimulated with IFN-γ (20 ng/ml) for the indicated periods of time. The activation of related factors regulating PD-L1 expression was analyzed by western blot analysis. (B) The line graphs show densitometric analysis of changes in the abundance of phosphorylated substrates and total proteins normalized to GAPDH to account for loading variability. (C) PD-L1 mRNA expression was assayed by RT-qPCR. The cells were treated as described in (A). Data are presented as the means ± SD (n=3). IFN-γ, interferon-γ; PKD3, protein kinase D3; PD-L1, programmed death ligand-1.
PKD3 is a key kinase for the STAT1 and STAT3 regulation of PD-L1 expression
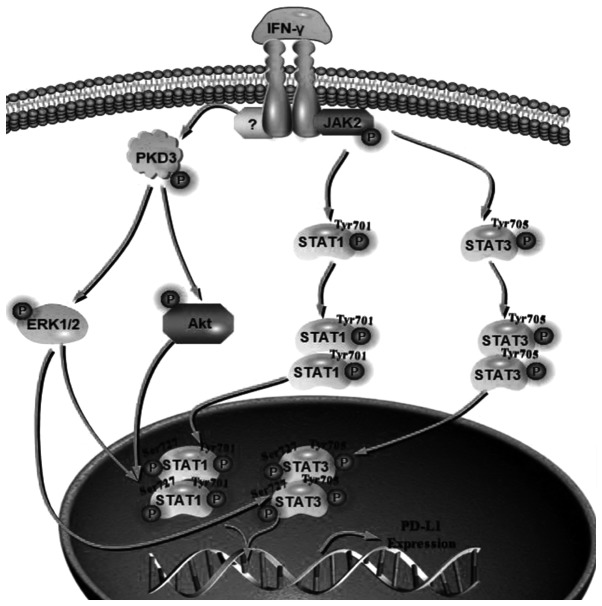
The results presented above provide evidence of an association between the expression of PKD3 and the signaling of IFN-γ-induced PD-L1 expression in OSCC. To further elucidate the role of PKD3 in the IFN-γ-mediated induction of PD-L1 expression, it was then determined whether the IFN-γ-induced activation of STAT1 and STAT3 in OSCC cell lines following exposure to IFN-γ was dependent on PKD3. For this purpose, the phosphorylation levels of STAT1 and STAT3 were examined after 1 h of treatment with or without IFN-γ. It was found that the knockdown of PKD3 significantly decreased the phosphorylation levels of STAT1 and STAT3 at Ser727 in OSCC cell lines (P<0.001, Fig. 5A). However, their phosphorylation levels at tyrosine residues were not markedly altered (Fig. S3). Subsequently, the relative contribution of STAT1 and STAT3 to IFN-γ-mediated PD-L1 expression was examined. The corresponding siRNAs effectively knocked down the expression of STAT1 and STAT3 in OSCC cell lines. The knockdown of STAT1 significantly abrogated the IFN-γ-induced PD-L1 upregulation at the protein level (P<0.001, Fig. 5B). However, IFN-γ was still able to induce an increase in PD-L1 expression after STAT3 silencing, although the elevated level was decreased (Fig. 5C). Taken together, these results indicate that activation of STAT1 and STAT3 is required for IFN-γ-mediated PD-L1 expression and that PKD3 is a key kinase regulating the activation of STAT1 and STAT3, as shown in Fig. 6.
Figure 5.
PKD3 is a key kinase for the regulation of PD-L1 expression by the cytokines, STAT1 and STAT3. (A) The indicated cell lines were transfected with PKD3-shRNA or control shRNA and treated with or without IFN-γ (20 ng/ml). The phosphorylation of STAT1 and STAT3 was assessed by western blot analysis. Data are presented as the means ± SD (n=3). (B and C) The cells were transfected with STAT1 siRNA or control siRNA for 48 h and then stimulated with IFN-γ for 24 h. In addition, the cells were transfected with STAT3-siRNA or control siRNA for 48 h and then stimulated with IFN-γ. Expression of PD-L1 was analyzed by western blot analysis. Data are presented as the means ± SD (n=3). *P<0.05; **P<0.01 and ***P<0.001. IFN-γ, interferon-γ; PKD3, protein kinase D3; PD-L1, programmed death ligand-1.
Figure 6.
A predictive model for IFN-γ induction of PD-L1 expression via activation of PKD3-STAT1/STAT3. Due to IFN-γ stimulation, JAK2 is activated and STAT1/STAT3 is phosphorylated at tyrosine, which regulates the dimerization and nuclear translocation of STATs. IFN-γ stimulation can also activate PKD3 and then AKT and ERK. These factors can regulate the phosphorylation of STAT1/STAT3 at Ser727, which is essential for the DNA binding and transcriptional activity of STAT1/3. IFN-γ, interferon-γ; PKD3, protein kinase D3; PD-L1, programmed death ligand-1.
Discussion
The PKD family, as a class of evolutionarily conserved serine/threonine kinases, has been implicated in diverse biological processes, such as cell proliferation, cell migration, differentiation, apoptosis, tumorigenesis, epithelial-to-mesenchymal transition and immune regulation (10-15,17,18). In this study, the role of a lesser-known member of the PKD family, PKD3, in the IFN-γ-induced expression of the immunosuppressive protein, PD-L1, in OSCC was investigated.
Based on the data from western blot analysis and immunofluorescence, PKD3 expression was found to be significantly upregulated in malignant cell lines relative to its expression in non-tumor cells. The increase in the nuclear distribution of PKD3 in malignant tumor cells was evident. However, the specific mechanisms underlying the nuclear accumulation of PKD3 have not been fully elucidated. Moreover, a statistically significant correlation was also found between PKD3 and PD-L1. The immunohistochemical findings were also consistent with the cell experiments. The results of immunohistochemistry and TCGA analysis indicated that the expression level of PKD3 and the frequency and intensity of nuclear staining gradually increased with the increase in the tumor grade. These findings suggest a potential role of PKD3 in the progression of OSCC. These results are consistent with those of previous studies in several types of cancer, which supports the conclusion obtained herein that abnormal PKD3 expression and localization promote cancer progression (18,27,28).
The expression of PD-L1 in tumor cells is mainly regulated by two mechanisms. The external mechanism, such as IFN-γ secreted by natural killer cells and tumor-infiltrating lymphocytes in the tumor microenvironment (7,29-31), can strongly induce the expression of PD-L1 in tumor cells. The intrinsic mechanism may be present in the constitutive carcinogenic signaling pathway of tumor cells. It is now becoming clear that the tumor microenvironment is critical for cancer progression. In addition, the pathogenesis of cancer is largely dependent on its interaction with microenvironmental components. There is increasing evidence to indicate that the induction of PD-L1 expression by inflammatory factors (such as IL-1, IL-6, TNF-α and IFN-γ) in the tumor microenvironment may be one of the most important factors affecting the efficacy of tumor immunotherapy (32). Of these, IFN-γ has the most potent inducing effect on PD-L1 expression. IFN-γ is a multifunctional cytokine produced by natural killer cells, T cells and macrophages. The infiltration of these cells can upregulate the expression of PD-L1 in tumor cells and can protect tumors from immune attack (31,33-35). Previous studies have indicated that IFN-γ can significantly induce tumor progression in some cases (36-38). In some clinical trials, IFN-γ treatment has been found to exert a negative effect on the prognosis of certain patients (39). Taken together, IFN-γ is deemed to have a double-edged sword effect in anti-tumor immunity, upregulating antigen processing machinery (APM) components and human leukocyte antigen (HLA) class I expression in tumor cells. However, it is also one of the most effective inducers of PD-L1. Therefore, it is crucial to elucidate the molecular mechanism underlying the induction of PD-L1 expression by IFN-γ. In HNSCC, the expression of PKD3 is significantly increased and gradually increases with an increase in the tumor grade. PKD3 may be a constitutive oncogenic signal in tumor cells, and internal and external mechanisms may co-regulate the expression of PD-L1.
One of the main findings of this study is the observation that PKD3 is involved in the IFN-γ-mediated upregulation of PD-L1 expression in OSCC. It was found that PKD3 knockdown in tumor cells reduced the level of PD-L1 induced by IFN-γ and that the overexpression of PKD3 in non-tumor cells significantly increased the level of PD-L1 induced by IFN-γ. Previous studies had demonstrated that PD-L1 expression in HNSCC is mainly affected by the activation of the STAT1/STAT3 pathway (9,24,25). To determine whether STAT1/STAT3 is involved in the regulation of PD-L1 expression by PKD3, in this study, the TCGA gene expression database was used to analyze the main signaling pathway members involved in the regulation of PD-L1 expression, such as NF-κB, MAPK, PI3K and STAT1/3 (6-9,24-26,29,40). It was found that only the expression of STAT1/3 was significantly associated with that of PD-L1 and PKD3. This provides direction for future studies. Indeed, it was found that IFN-γ induced a significant increase in the STAT1 and STAT3 phosphorylation levels. However, a significant decrease was only observed in the phosphorylation level of STAT1 and STAT3 at the Ser727 site after PKD3 silencing, and there was no significant change in the phosphorylation level at tyrosine. The activation of STATs involves dimerization, nuclear trans-location, DNA binding and transcriptional activation, which requires the phosphorylation of the Ser727 site and tyrosine. The phosphorylation of tyrosine regulates the dimerization and nuclear translocation of STATs, which is essential for the activation of the JAK-STAT signaling pathway. However, the phosphorylation of STAT1/3 at Ser727 is essential for the optimal DNA binding and transcriptional activity of STAT1/3 (41,42). Previous studies have demonstrated that the inhibition of AKT activation can lead to the downregulation of the IFN-γ-mediated phosphorylation of STAT1 at Ser727, which not only downregulates the expression of the STAT1 target genes, CXCL9 and CXCL10, but also downregulates the expression of PD-L1 (7). In addition, ERK can directly interact with STAT1/STAT3 to phosphorylate Ser727, increasing the viability and growth rate of cells (41-50). AKT and ERK, as downstream targets of PKD3, regulate the growth and survival of cancer cells (18). Taken together, these findings suggest that the activation of STAT1/STAT3 at the Ser727 site contributes to tumor development. These results indicate that PKD3 regulates IFN-γ-induced PD-L1 expression by regulating the phosphorylation of STAT1/STAT3 (Fig. 6).
In conclusion, PKD3, as a serine/threonine kinase, is involved in the regulation of STAT1/STAT3 activation. The findings of this study shed light on the function of PKD3 in the development of OSCC. It was demonstrated that in human OSCC, PKD3 participates in the regulation of PD-L1 expression by modulating the activation of STAT1/STAT3, thereby providing a theoretical basis for the combination of PKD inhibition and immunotherapy of human OSCC.
Supplementary Data
Acknowledgments
The authors would like to thank the patients, researchers, and staff involved in this study.
Abbreviations
- PKD3
protein kinase D3
- OSCC
oral squamous cell carcinoma
- HNSCC
head and neck squamous cell carcinoma
- PD-L1
programmed death ligand-1
- PD-1
programmed cell death protein 1
- IFN-γ
interferon-γ
- STAT
signal transducer and activator of transcription
- TCGA
The Cancer Genome Atlas
- IL
interleukin
- TNF
tumor necrosis factor
- NF-κB
nuclear factor-κB
- MAPK
mitogen-activated protein kinase
- PI3K
phosphatidylinositol 3-kinase
- AKT
protein kinase B
- ERK
extracellular signal-regulated kinase
- APM
antigen processing machinery
- HLA
human leukocyte antigen
Funding
This study was supported by National Natural Science Foundations of China (grant nos. 81802717 and 81372892).
Availability of data and materials
All data are included in this article and its supplementary information files.
Authors' contributions
PZ, YF and XZ designed the outline of the study. BC and JC designed the study, conducted experiments and wrote the manuscript. JC and ML supervised the study, contributed to data acquisition and revised the manuscript. LW, HC, YK and JW collected and analyzed the data. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
This study was approved by the West China Hospital of Stomatology Institutional Review Board (approval no. WCHSIRB-D-2013-039). Written informed consent was obtained from each donor and patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest.
References
- 1.Sathiyasekar AC, Chandrasekar P, Pakash A, Kumar KUG, Jaishlal MS. Overview of immunology of oral squamous cell carcinoma. J Pharm Bioallied Sci. 2016;8(Suppl 1):S8–S12. doi: 10.4103/0975-7406.191974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Lin YM, Sung WW, Hsieh MJ, Tsai SC, Lai HW, Yang SM, Shen KH, Chen MK, Lee H, Yeh KT, Chen CJ. High PD-L1 expression correlates with metastasis and poor prognosis in oral squamous cell carcinoma. PLoS One. 2015;10:e0142656. doi: 10.1371/journal.pone.0142656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferris RL. Immunology and immunotherapy of head and neck cancer. J Clin Oncol. 2015;33:3293–3304. doi: 10.1200/JCO.2015.61.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu W, Lu L, Feng Y, Chen J, Li Y, Kong X, Chen S, Li X, Chen Q, Zhang P. Inflammation promotes oral squamous carcinoma immune evasion via induced programmed death ligand-1 surface expression. Oncol Lett. 2013;5:1519–1526. doi: 10.3892/ol.2013.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51:221–228. doi: 10.1016/j.oraloncology.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Gao Y, Yang J, Cai Y, Fu S, Zhang N, Fu X, Li L. IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer. 2018;143:931–943. doi: 10.1002/ijc.31357. [DOI] [PubMed] [Google Scholar]
- 8.Moon JW, Kong SK, Kim BS, Kim HJ, Lim H, Noh K, Kim Y, Choi JW, Lee JH, Kim YS. IFNγ induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci Rep. 2017;7:17810. doi: 10.1038/s41598-017-18132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bu L, Yu G, Wu L, Mao L, Deng WW, Liu JF, Kulkarni AB, Zhang WF, Zhang L, Sun ZJ. STAT3 induces immunosuppression by upregulating PD-1/PD-L1 in HNSCC. J Dent Res. 2017;96:1027–1034. doi: 10.1177/0022034517712435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy A, Ye J, Deng F, Wang QJ. Protein kinase D signaling in cancer: A friend or foe? Biochim Biophys Acta Rev Cancer. 2017;1868:283–294. doi: 10.1016/j.bbcan.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Feng Y, Lu L, Wang H, Dai L, Li Y, Zhang P. Interferon-γ-induced PD-L1 surface expression on human oral squamous carcinoma via PKD2 signal pathway. Immunobiology. 2012;217:385–393. doi: 10.1016/j.imbio.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 12.LaValle CR, George KM, Sharlow ER, Lazo JS, Wipf P, Wang QJ. Protein kinase D as a potential new target for cancer therapy. Biochim Biophys Acta. 2010;1806:183–192. doi: 10.1016/j.bbcan.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans IM, Zachary IC. Protein kinase D in vascular biology and angiogenesis. IUBMB Life. 2011;63:258–263. doi: 10.1002/iub.456. [DOI] [PubMed] [Google Scholar]
- 14.Zhang T, Braun U, Leitges M. PKD3 deficiency causes alterations in microtubule dynamics during the cell cycle. Cell Cycle. 2016;15:1844–1854. doi: 10.1080/15384101.2016.1188237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durand N, Borges S, Storz P. Protein kinase D enzymes as regulators of EMT and cancer cell invasion. J Clin Med. 2016;5:E20. doi: 10.3390/jcm5020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker J, Falconer AM, Wilkinson DJ, Europe-Finner GN, Litherland GJ, Rowan AD. Protein kinase D3 modulates MMP1 and MMP13 expression in human chondrocytes. PLoS One. 2018;13:e0195864. doi: 10.1371/journal.pone.0195864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozengurt E, Rey O, Waldron RT. Protein kinase D signaling. J Biol Chem. 2005;280:13205–13208. doi: 10.1074/jbc.R500002200. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Deng F, Singh SV, Wang QJ. Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCε/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 2008;68:3844–3853. doi: 10.1158/0008-5472.CAN-07-5156. [DOI] [PubMed] [Google Scholar]
- 19.Zou Z, Zeng F, Xu W, et al. PKD2 and PKD3 Promote Prostate Cancer Cell Invasion by Modulating NF-κB- And HDAC1-mediated Expression and Activation of uPA. J Cell Sci. 2012;125:4800–4811. doi: 10.1242/jcs.106542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Wang CM, Zhang P, Wang X, Chen J, Yang J, Lu W, Zhou W, Yuan W, Feng Y. Expression of programmed death 1 ligand 1 on periodontal tissue cells as a possible protective feedback mechanism against periodontal tissue destruction. Mol Med Rep. 2016;13:2423–2430. doi: 10.3892/mmr.2016.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin W, Choe WH, Hiasa Y, Kamegaya Y, Blackard JT, Schmidt EV, Chung RT. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology. 2005;128:1034–1041. doi: 10.1053/j.gastro.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu W, Steinberg SF. Phos-tag SDS-PAGE resolves agonist-and isoform-specific activation patterns for PKD2 and PKD3 in cardiomyocytes and cardiac fibroblasts. J Mol Cell Cardiol. 2016;99:14–22. doi: 10.1016/j.yjmcc.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C, Cao W, Ren Z, Tang Y, Zhang C, Yang R, Chen Y, Liu Z, Peng C, Wang L, et al. GDNF secreted by nerves enhances PD-L1 expression via JAK2-STAT1 signaling activation in HNSCC. Oncoimmunology. 2017;6:e1353860. doi: 10.1080/2162402X.2017.1353860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, Seethala RR, Freeman GJ, Ferris RL. Identification of the cell-intrinsic and-extrinsic pathways downstream of EGFR and IFNγ that induce PD-L1 expression in head and neck cancer. Cancer Res. 2016;76:1031–1043. doi: 10.1158/0008-5472.CAN-15-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann J, Hoesli R, Michmerhuizen N, Devenport SN, Ludwig ML, Vandenberg TR, Matovina C, Jawad N, Mierzwa M, Shuman AG, et al. Surveilling the potential for precision medicine-driven PD-1/PD-L1-targeted therapy in HNSCC. J Cancer. 2017;8:332–344. doi: 10.7150/jca.17547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H, Xu M, Chi X, Yan Q, Wang Y, Xu W, Zhuang K, Li A, Liu S. Higher PKD3 expression in hepatocellular carcinoma (HCC) tissues predicts poorer prognosis for HCC patients. Clin Res Hepatol Gastroenterol. 2017;41:554–563. doi: 10.1016/j.clinre.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Huck B, Duss S, Hausser A, Olayioye MA. Elevated protein kinase D3 (PKD3) expression supports proliferation of triple-negative breast cancer cells and contributes to mTORC1-S6K1 pathway activation. J Biol Chem. 2014;289:3138–3147. doi: 10.1074/jbc.M113.502633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mimura K, The JL, Okayama H, Shiraishi K, Kua LF, Koh V, Smoot DT, Ashktorab H, Oike T, Suzuki Y, et al. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 2018;109:43–53. doi: 10.1111/cas.13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassilakopoulou M, Avgeris M, Velcheti V, Kotoula V, Rampias T, Chatzopoulos K, Perisanidis C, Kontos CK, Giotakis AI, Scorilas A, et al. Evaluation of PD-L1 expression and associated tumor-infiltrating lymphocytes in laryngeal squamous cell carcinoma. Clin Cancer Res. 2016;22:704–713. doi: 10.1158/1078-0432.CCR-15-1543. [DOI] [PubMed] [Google Scholar]
- 31.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, Chen L. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127ra137. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, Wu X, Ma J, Zhou M, Li X, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:10. doi: 10.1186/s12943-018-0928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaCasse CJ, Janikashvili N, Larmonier CB, Alizadeh D, Hanke N, Kartchner J, Situ E, Centuori S, Har-Noy M, Bonnotte B, et al. Th-1 lymphocytes induce dendritic cell tumor killing activity by an IFN-γ-dependent mechanism. J Immunol. 2011;187:6310–6317. doi: 10.4049/jimmunol.1101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maraskovsky E, Chen W, Shortman K. IL-2 and IFN-gamma are two necessary lymphokines in the development of cytolytic T cells. J Immunol. 1989;143:1210–1214. [PubMed] [Google Scholar]
- 35.Chen L, Tourvieille B, Burns GF, Bach FH, Mathieu-Mahul D, Sasportes M, Bensussan A. Interferon: A cytotoxic T lymphocyte differentiation signal. Eur J Immunol. 1986;16:767–770. doi: 10.1002/eji.1830160709. [DOI] [PubMed] [Google Scholar]
- 36.Xiao M, Wang C, Zhang J, Li Z, Zhao X, Qin Z. IFNgamma promotes papilloma development by up-regulating Th17-associated inflammation. Cancer Res. 2009;69:2010–2017. doi: 10.1158/0008-5472.CAN-08-3479. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda M, Nakamoto Y, Suzuki S, Kurata T, Kaneko SJ. Interferon-gamma-mediated hepatocarcinogenesis in mice treated with diethylnitrosamine. Lab Invest. 2005;85:655–663. doi: 10.1038/labinvest.3700257. [DOI] [PubMed] [Google Scholar]
- 38.Hanada T, Kobayashi T, Chinen T, Saeki K, Takaki H, Koga K, Minoda Y, Sanada T, Yoshioka T, Mimata H, et al. IFNgamma-dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice. J Exp Med. 2006;203:1391–1397. doi: 10.1084/jem.20060436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alberts DS, Marth C, Alvarez RD, Johnson G, Bidzinski M, Kardatzke DR, Bradford WZ, Loutit J, Kirn DH, Clouser MC, et al. Randomized phase 3 trial of interferon gamma-1b plus standard carboplatin/paclitaxel versus carboplatin/paclitaxel alone for first-line treatment of advanced ovarian and primary peritoneal carcinomas: Results from a prospectively designed analysis of progression-free survival. Gynecol Oncol. 2008;109:174–181. doi: 10.1016/j.ygyno.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Zhang J, Zhang Z, Guo Y, Wu Y, Wang R, Wang L, Mao S, Yao X. Overexpression of indoleamine 2,3-dioxygenase 1 promotes epithelial-mesenchymal transition by activation of the IL-6/STAT3/PD-L1 pathway in bladder cancer. Transl Oncol. 2019;12:485–492. doi: 10.1016/j.tranon.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 42.Aziz MH, Manoharan HT, Church DR, Dreckschmidt NE, Zhong W, Oberley TD, Wilding G, Verma AK. Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Cancer Res. 2007;67:8828–8838. doi: 10.1158/0008-5472.CAN-07-1604. [DOI] [PubMed] [Google Scholar]
- 43.Sakaguchi M, Oka M, Iwasaki T, Fukami Y, Nishigori C. Role and regulation of STAT3 phosphorylation at Ser727 in melanocytes and melanoma cells. J Invest Dermatol. 2012;132:1877–1885. doi: 10.1038/jid.2012.45. [DOI] [PubMed] [Google Scholar]
- 44.Gough DJ, Koetz L, Levy DE. The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and Ras-mediated transformation. PLoS One. 2013;8:e83395. doi: 10.1371/journal.pone.0083395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Decker T, Kovarik P. Transcription factor activity of STAT proteins: Structural requirements and regulation by phosphorylation and interacting proteins. Cell Mol Life Sci. 1999;55:1535–1546. doi: 10.1007/s000180050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang Y, Zhong L, Lin M, Zhou X, Jing H, Ying M, Luo P, Yang B, He Q. MEK/ERK dependent activation of STAT1 mediates dasatinib-induced differentiation of acute myeloid leukemia. PLoS One. 2013;8:e66915. doi: 10.1371/journal.pone.0066915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nitulescu II, Meyer SC, Wen QJ, Crispino JD, Lemieux ME, Levine RL, Pelish HE, Shair MD. Mediator kinase phosphorylation of STAT1 S727 promotes growth of neoplasms with JAK-STAT activation. EBioMedicine. 2017;26:112–125. doi: 10.1016/j.ebiom.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goren I, Pfeilschifter J, Frank S. Determination of leptin signaling pathways in human and murine keratinocytes. Biochem Bioph Res Commun. 2003;303:1080–1085. doi: 10.1016/S0006-291X(03)00480-7. [DOI] [PubMed] [Google Scholar]
- 49.Kondo K, Shaim H, Thompson PA, Burger JA, Keating M, Estrov Z, Harris D, Kim E, Ferrajoli A, Daher M, et al. Ibrutinib modulates the immunosuppressive CLL microenvironment through STAT3-mediated suppression of regulatory B-cell function and inhibition of the PD-1/PD-L1 pathway. Leukemia. 2018;32:960–970. doi: 10.1038/leu.2017.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou L, Schandené L, Mordvinov VA, Chatelain P, Pradier O, Goldman M, Stordeur P. Trapidil inhibits monocyte CD40 expression by preventing IFN-γ-induced STAT1 S727 phosphorylation. Int Immunopharmacol. 2004;4:863–871. doi: 10.1016/j.intimp.2004.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in this article and its supplementary information files.