Abstract
Tamoxifen (TAM) resistance is a major challenge in the treatment of estrogen receptor-positive (ER+) breast cancer. To date, to the best of our knowledge, there are only a few studies available examining the response of patients with TAM-resistant breast cancer to chemotherapy, and the guidelines do not specify recommended drugs for these patients. In the present study, TAM-resistant cells were shown to exhibit increased proliferation and invasion compared with the parent cells, and the increased expression of c-MYC was demonstrated to play an important role in TAM resistance. Furthermore, the TAM-resistant cells were significantly more sensitive to cisplatin compared with the parent cells, and the silencing of c-MYC expression desensitized the cells to cisplatin through the inhibition of the cell cycle. An increased c-MYC expression was observed in 28 pairs of primary and metastatic tumors from patients treated with TAM, and the clinical remission rate of cisplatin-based chemotherapy was significantly higher compared with other chemotherapy-based regimens in 122 patients with TAM resistant breast cancer. Taken together, the data of the present study demonstrated that although c-MYC was involved in TAM resistance, it increased the sensitivity of ER+ breast cancer to cisplatin. Thus, cisplatin may be a preferred chemotherapeutic agent for the treatment of patients with TAM-resistant breast cancer, particularly in patients where the rapid control of disease progression is required.
Keywords: c-MYC, tamoxifen resistance, chemotherapy, cisplatin, breast cancer
Introduction
Breast cancer is the most common malignant tumor affecting women (1), and ~70% of breast cancer cases are considered estrogen receptor-positive (ER+) (2-4). In patients with ER+ breast cancer, tumor growth and survival are primarily dependent on the activation of the ER signaling pathway (5-7). Tamoxifen (TAM), which prevents the activation of ER signaling and its downstream pathways by blocking estrogen binding to ER, is the most frequently used endocrine drug in clinical practice (8-10). However, ~30-40% of responsive tumors eventually acquire TAM resistance, which remains a major clinical challenge (11-15).
However, other endocrine therapies with or without targeted therapy may be recommended for patients who develop TAM resistance (16,17). Chemotherapy can control the progression of breast cancer and may prolong the survival times of patients that exhibit rapid disease progression or visceral crisis (18). In a previous study by the authors, and in other studies, it was demonstrated that tumor cells usually exhibit more rapid growth rates and increased invasiveness following the development of TAM resistance (19-21), which may explain why some patients may experience rapid tumor progression and multiple visceral metastases during TAM treatment and, thus, require chemotherapy. However, to date, at least to the best of our knowledge, there are only a few studies available investigating the responses of TAM-resistant patients to chemotherapeutic drugs, and there are no guidelines for the recommended chemotherapeutic regimen.
c-MYC is one of the most upregulated oncogenes in several different types of cancer, and has been reported to play varying roles in different molecular subtypes of breast cancer (22,23). In the present study, TAM resistance and the sensitivities of TAM-resistant cells to various chemotherapeutic agents were examined. The results revealed that although c-MYC plays a pivotal role in TAM resistance, it improves cisplatin sensitivity in ER+ breast cancer. Thus, patients with rapid disease progression during TAM treatment may respond favorably to cisplatin, and a high c-MYC expression may be used as a predictive marker.
Materials and methods
Reagents and antibodies
4-Hydroxytamoxifen (TAM) was purchased from Sigma-Aldrich; Merck KGaA. Doxorubicin, paclitaxel and cisplatin were purchased from Beijing Solarbio Science & Technology Co., Ltd. Antibodies against Erα (cat. no. 8644S), human epidermal growth factor receptor 2 (HER2) (cat. no. 2242S), GAPDH (cat. no. 5174S) and AKT (cat. no. 9272S) were purchased from Cell Signaling Technology, Inc. Antibodies against c-MYC (cat. no. ab32072), cyclin D1 (cat. no. ab226977, phospho (p)-AKT (Ser473) (cat. no. ab81283) and E-cadherin (cat. no. ab133597) were purchased from Abcam. Antibodies against p21 (cat. no. 27296-1-AP) and β-catenin (cat. no. 51067-2-AP) were purchased from ProteinTech Group, Inc., and an antibody against vimentin (cat. no. BS1491) was purchased from Bioworld Technology, Inc.
Cell lines and cell culture
The human breast cancer cell lines, MCF-7 and T47D, were purchased from the American Type Culture Collection. The TAM-resistant cell lines, MCF-7R and T47DR, were generated by exposing parental MCF-7 and T47D cells to progressively increasing concentrations of TAM (up to 5 µM) over a duration of ~8 months as previously described (24,25). All cell lines were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Beyotime Institute of Biotechnology) at 37°C in an humidified incubator with 5% CO2.
Cell viability assay
Cell viability was measured using a Cell Counting kit-8 assay (CCK-8; Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Briefly, 5×103 cells/well were seeded in a 96-well plate in 100 µl medium. After 24 h, the cells were treated with the different drugs (TAM: 5, 10, 11, 12, 13, 14, 15, 16, 18, 20 and 25 µM; doxorubicin: 0.25, 0.5, 1, 2, 4, 8 and 16 µM; paclitaxel: 0.25, 0.5, 1, 2, 4, 8, 16, 32 and 64 nM; cisplatin: 2, 4, 8, 16, 32, 64, 128 and 256 µM) (5 wells for each drug) for 24-72 h. Subsequently, 10 µl CCK-8 solution was added to the medium and the cells were incubated for a further 2 h at 37°C. Optical density (OD) values were measured using a digital spectrophotometer (Bio-Rad Laboratories, Inc.) at a wavelength of 450 nm, and cell viability rates were expressed as a percentage relative to the corresponding control cells. The IC50 value was determined from the concentration-effect curve. After the data were log-transformed, the specific value was obtained through the curve parameter equation, and the viability of the control cells was considered as 100% (26).
Cell invasion assays
The MCF7, T47D, MCF7R or T47DR cells were seeded in the upper chamber of a Transwell insert (3×105 cells/well; EMD Millipore) coated with Matrigel (1:7.5) in 200 µl of serum-free medium. Supplemented RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added to the lower chamber. The cell lines were incubated for 24-48 h at 37°C, and the cells were fixed with 4% paraformaldehyde (Wuhan Boster Biological Technology, Ltd.) for 15 min at room temperature and stained with 0.5% crystal violet (Wuhan Boster Biological Technology, Ltd.) for 5 min at room temperature. the number of migrating cells was determined in 5 random fields at an inverted light microscope (magnification, ×200; TE2000-U; Nikon Corp.).
Cell cycle analysis
The proportion of cells in the S phase of the cell cycle was analyzed by flow cytometry using a standard protocol. Briefly, the cells were seeded in 6-well plates (2×105 cells/well) and cultured for 24 h. The cells were then harvested and washed twice with cold PBS, fixed with 70% ethanol at 4°C for 1 h, incubated with Rnase for 30 min at 37°C and stained with propidium iodide for 30 min at room temperature (cat. no. P4170; Sigma Aldrich; Merck KGaA). Cell cycle distribution was analyzed by flow cytometry using a FACSVantage SE instrument (BD Biosciences).
Reverse transcription-quantitative PCR
Total cellular RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Reverse transcription and quantitative PCR were performed using the PrimeScript RT Master mix kit (Takara Bio, Inc.) and SYBR Pre-mix Ex Taq™ II (Takara Bio, Inc.) according to the manufacturer's protocols. GAPDH was used as the reference gene, and the thermocycling conditions were as follows: 2 min at 95°C, followed by 39 cycles at 95°C for 30 sec, 30 sec at 56.5°C and 20 sec at 72°C. Relative gene expression was normalized to GAPDH and calculated using the 2−∆∆Cq method (27). The sequences of primers used for amplifying GAPDH, c-MYC, multidrug resistance-related protein (MRP), multidrug resistance protein (MDR) and breast cancer resistance protein (BCPR) are presented in Table SI.
Western blot analysis
Cells were lysed using RIPA lysis buffer with phenyl-methanesulfonyl fluoride (Wuhan Boster Biological Technology, Ltd.), and the protein concentration was determined using a bicinchoninic acid assay (Beyotime Institute of Biotechnology). A total of 50 µg protein was loaded on a 10% SDS gel and resolved using SDS-PAGE. The resolved proteins were transferred to PVDF membranes. Non-specific binding sites were blocked by incubating the membranes with 5% non-fat milk, after which the membranes were incubated overnight at 4°C with the primary antibodies: ERα (1:1,000); HER2 (1:1,000); AKT (1:1,000); p-AKT (1:1,000); c-MYC (1:1,000); cyclin D1 (1:1,000); vimentin (1:1,000); E-cadherin (1:1,000); p21 (1:500); β-catenin (1:500); GAPDH (1:1,000) Membranes were subsequently incubated with a horseradish peroxidase-conjugated anti-rabbit/mouse immunoglobulin G secondary antibody (1:1,000; cat. no. BA1075 and BA1051; Wuhan Boster Biological Technology, Ltd.). Signals were visualized using enhanced chemiluminescence reagent (EMD Millipore). Densitometry analysis was performed using ImageJ and normalized to the respective expression of GAPDH.
Small interfering (si)RNA transfection
siRNAs targeting c-MYC mRNA and a control siRNA were purchased from Shanghai GenePharma Co., Ltd. siRNAs were transfected into the cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The efficiency of RNA-interference was assessed using RT-qPCR 24-36 h following transfection. Cell lysates were collected 48-72 h following transfection and used for western blot analysis. The sequences of the siRNAs targeting c-MYC mRNA are presented in Table SII.
Patient and specimen selection
A total of 178 consecutive patients with recurrent and/or metastatic ER+ breast cancer, treated at the Breast Cancer Center of Chongqing at The First Affiliated Hospital of Chongqing Medical University between July, 2012 and July, 2017, were recruited for the present study. The outcomes of endocrine therapy were investigated for all patients, and 122 patients with TAM resistance were identified (Table I). At the time of diagnosis of recurrent and/or metastatic disease, 125 patients (70.2%) had received various chemotherapeutic regimens, whereas the remaining 53 patients continued to receive endocrine therapy. The Response Evaluation Criteria in Solid Tumors (RECIST) guidelines (version 1.1) (28) were utilized to assess chemotherapy responses as follows: Partial response (PR), a reduction of all target lesion diameters of ≥30%; progressive disease (PD), growth of all target lesion diameters by ≥20%; stable disease (SD), neither a sufficient reduction to be classified as PR, nor sufficient growth to be classified as PD (29). A total of 28 paired, archived paraffin-embedded breast cancer specimens (primary and metastatic breast cancer tissues) from the aforementioned TAM-resistant patients were obtained from the Clinical Diagnostic Pathology Center of Chongqing Medical University (Chongqing, China). Detailed information pertaining to the specimens is presented in Table SIII. The present study was approved by the Ethics Committee of Chongqing Medical University and written informed consent was obtained from all patients.
Table I.
Clinicopathological features of tamoxifen-resistant patients (n=122).
| Parameters | No. (%) |
|---|---|
| Age (years) | |
| <30 | 3 (2.5) |
| 30-40 | 36 (29.5) |
| 40-50 | 69 (56.6) |
| ≥50 | 14 (11.4) |
| Time of metastasis (months) | |
| <24 | 59 (48.4) |
| ≥24 | 63 (51.6) |
| Surgical procedure | |
| Mastectomy | 103 (84.4) |
| Breast-conserving | 19 (15.6) |
| Subtypes of Cancer | |
| Ductal | 115 (94.4) |
| Lobular | 4 (3.2) |
| Others | 3 (2.4) |
| TNM staging | |
| I | 10 (8.2) |
| IIA | 29 (23.8) |
| IIB | 42 (34.4) |
| IIIA | 21 (17.2) |
| IIIB | 10 (8.2) |
| IIIC | 2 (1.6) |
| Unknown | 8 (6.6) |
| Recurrent or/and metastatic site | |
| Breast | 9 (7.4) |
| Chest wall | 27 (22.1) |
| Lymph nodea | 21 (17.2) |
| Boneb | 26 (21.3) |
| Brainc | 3 (2.5) |
| Liverc | 13 (10.7) |
| Lungc | 16 (13.1) |
| Multi-visceral metastasisd | 7 (5.7) |
Irrespective of the local situation;
irrespective of the local situation and lymph node;
irrespective of the local situation, lymph node and bone;
including two parts of lung, liver and brain.
Immunohistochemistry (IHC)
IHC staining was performed as previously described (19). For histochemical analysis, Image-Pro Plus version 6.0 (Media Cybernetics, Inc.) was used to assess the percentage of positive cells. The detailed steps were as follows: Deparaffinized specimens were then sectioned (4-µm-thick slices). Following antigen repair and blocking for non-specific binding, the slices were incubated with specific primary antibodies against c-MYC (1:200; cat. no. ab32072, Abcam) overnight at 4°C. Subsequently, the sections were treated with HRP-conjugated goat anti-rabbit IgG secondary antibody (1:200; cat. no. TA140003; OriGene Technologies, Inc.) for 30 min at room temperature. After staining with diaminobenzidine (OriGene Technologies, Inc.) and hematoxylin for 5 sec at room temperature, images were captured using a Nikon Eclipse 80i microscope (magnification, ×200; Nikon Corp.). The mean OD in 5 randomly selected areas (MOD=IOD/area) was used to evaluate the levels of protein expression. The c-MYC staining intensities in tumor tissues were scored as follows: 0, no staining; 1, weak staining; 2, intermediate staining; 3, strong staining; and 4, very strong staining. Finally, the IHC data were quantified by multiplying the staining intensity by the proportion of positive cells as previously described (30).
Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA)
The microarray analysis data of MCF-7 cells and TAM-resistant cell lines were downloaded from GEO (accession no. GSE26459). c-MYC expression in breast cancer tissues, para-cancerous tissues and in different molecular subtypes of breast cancer were obtained from TCGA, and survival curves based on c-MYC expression in breast cancers were obtained from Kaplan-Meier plotter: c-MYC expression data in tumors and follow-up information for patients with ER-positive breast cancer treated with adjuvant TAM were also downloaded from GEO (accession nos. GSE6532, n=87; and GSE9195, n=77), and the log-rank test was then used to assess the association between c-MYC expression and survival in breast cancer patients.
Statistical analysis
SPSS version 22.0 (SPSS Inc.) was used for the data analysis. Data are presented as the means ± standard deviation of 3 repeats. A one-way ANOVA followed by Tukey's post hoc test was used to compare differences among multiple groups. A χ2 test was used to evaluate clinical response to chemotherapy in patients with recurrent and/or metastatic breast cancer. A value of P<0.05 was considered to indicate a statistically significant difference.
Results
Proliferation and invasion are increased in TAM-resistant cells
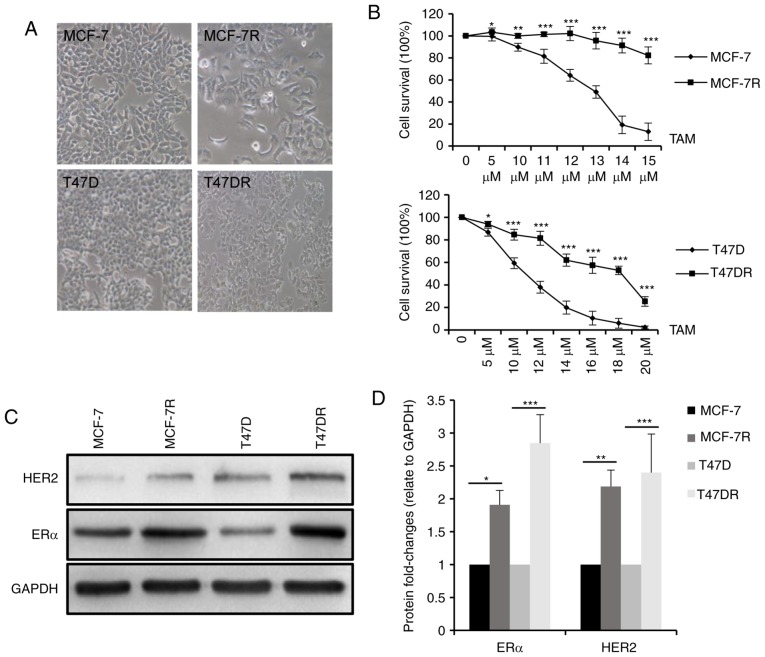
In agreement with a previous study (19) by the authors, the morphology of the MCF-7R and T47DR cells exhibited a flatter and more polygonal shape compared with that of the parental control cells, and the cells were more mesenchymal phenotypically (Fig. 1A). To validate TAM resistance in these established cells, MCF-7R and T47DR cells, and their parental cells were treated with various concentrations of TAM and the cell survival rates were measured compared with a negative control. As shown in Fig. 1B, survival was higher in the MCF-7R and T47DR cells compared with the respective parental cells treated with the same concentration of TAM. The IC50 values of the 4 cell lines to TAM were 12.53±0.95 µM for the MCF-7, 26.93±1.76 µM for the MCF-7R, 9.29±1.23 µM for the T47D and 17.27±1.16 µM for the T47D-7R cells (data not shown). In addition, the protein expression levels of ERα and HER2 were significantly higher in the TAM-resistant cells (Fig. 1C and D), consistent with previous preclinical models and clinical observations (24,31-33).
Figure 1.
Establishment of TAM-resistant breast cancer cells. (A) TAM-resistant cells exhibited a more elongated, spindle-shaped morphology compared with the respective parental cells. Magnification, ×200. (B) Survival of TAM-resistant cells and the respective parental cells was evaluated using a Cell Counting kit-8 assay following treatment with various concentrations of TAM for 24 h. (C) Western blot analysis (D) and densitometry analysis of ERα and HER2 expression in MCF-7, MCF-7R, T47D and T47DR cells. Data are presented as the means ± standard deviation of the mean of 3 repeats. *P<0.05, **P<0.01, ***P<0.001 vs. respective parental cells. TAM, tamoxifen.
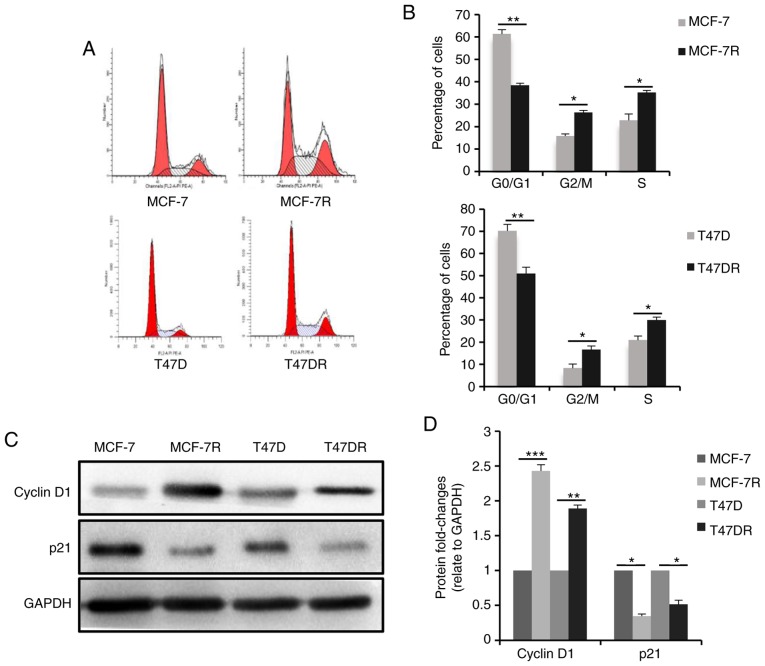
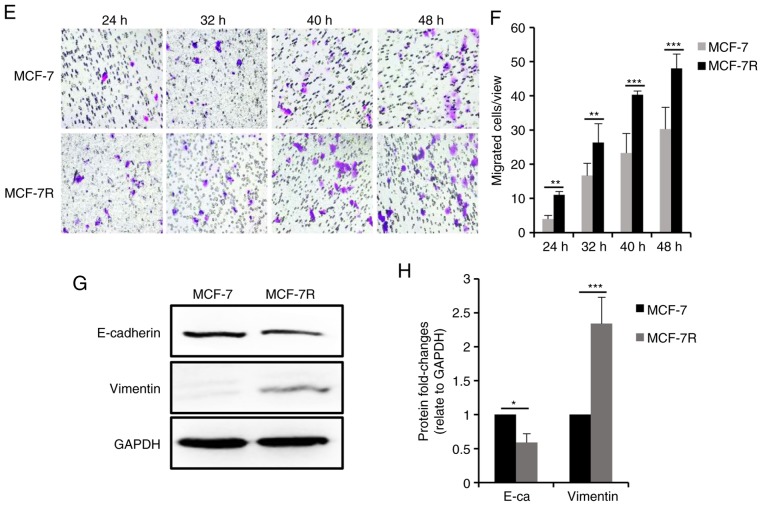
To evaluate the proliferation of the TAM-resistant cells, cell cycle progression was analyzed by flow cytometry, and the results revealed that the percentage of cells in the G0/G1 phase was decreased and the percentage of cells in the S phase was increased significantly in the TAM-resistant cells (Fig. 2A and B), suggesting that proliferation of the TAM-resistant cells was increased. Consistently, the expression of the cell cycle checkpoint-associated protein, p21waf1, decreased significantly in the TAM-resistant cells, whereas the expression of cyclin D1 was increased (Fig. 2C and D). Furthermore, the TAM-resistant cells were significantly more invasive compared with the parental cells (Figs. 2E and F, and S1A and B). E-cadherin protein expression was decreased and vimentin expression was increased in the TAM-resistant cells compared with the respective parental cells (Figs. 2G and H, and S1C and D), suggesting the cells has undergone epithelial-mesenchymal transition, consistent with the morphology of resistant cells.
Figure 2.
Characteristics of TAM-resistant cells. (A) Distribution of cells in different phases of the cell cycle in TAM-sensitive and TAM-resistant cells detected by flow cytometry. (B) Bar charts representing the percentage of cells in the G0/G1, G2/M or S phase. (C) Western blot analysis and (D) densitometric analysis of cyclin D1 and p21 expression in TAM-sensitive and TAM-resistant cells. (E) Representative images of successfully invaded MCF-7 and MCF-7R cells and (F) quantitative analysis of invasion measured using a Transwell invasion assay at different times. Magnification, ×200. (G) Western blot analysis and (H) densitometric analysis of E-cadherin and vimentin expression in TAM-sensitive and TAM-resistant cells. Data are presented as the means ± standard deviation of mean of three repeats. *P<0.05, **P<0.01, ***P<0.001 vs. respective parental cells. TAM, tamoxifen.
c-MYC expression is increased in TAM-resistant cells and c-MYC knockdown enhances sensitivity to TAM
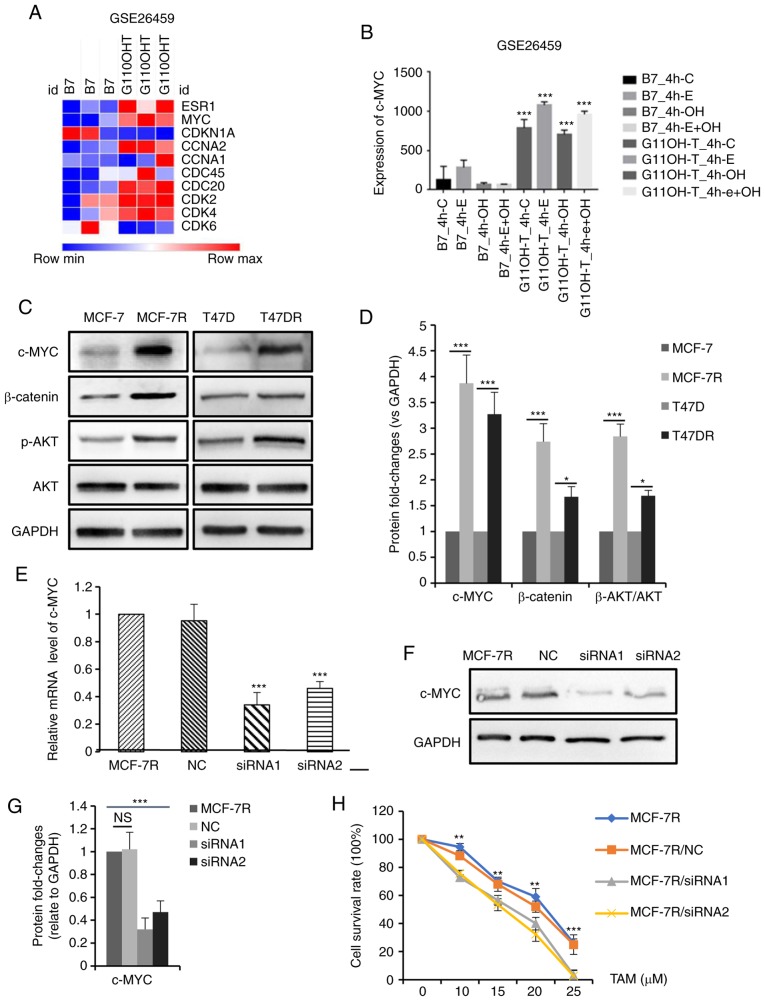
To explore the potential mechanisms through which TAM resistance is acquired, microarray data from TAM-resistant cells were downloaded from GEO (accession no. GSE26459) and used to identify TAM resistance-associated genes. A heatmap depicting the expression of proliferation-related genes in the TAM-resistant cells relative to the parental controls is presented in Fig. 3A. c-MYC expression was found to be abnormally high in the TAM-resistant cells (P<0.001, Fig. 3B). c-MYC is one of the primary target genes of the ER signaling pathway and possesses similar functions to ER (34-36). The expression of c-MYC was observed in the TAM-resistant cells used in the present study (Fig. 3C and D). Consistently, the activation of PI3K/AKT and β-catenin signaling in TAM-resistant cells was also confirmed in the cells used (Fig. 3C and D). To further examine the role of c-MYC in TAM resistance, the siRNA-mediated knockdown of c-MYC in the MCF-7R cells (Fig. 3E-G) was shown to enhance the sensitivity of MCF-7R cells to TAM (Fig. 3H). The TAM IC50 values of the MCF-7R cells prior to and following the knockdown of c-MYC were 20.26±0.76 µM for the MCF-7R, 18.88±0.69 µM for the MCF-7R/NC,13.95±1.82 µM for the MCF-7R/siRNA1 and 13.71±1.63 µM for the MCF-7R/siRNA2 cells (data not shown). Taken together, these data demonstrate that c-MYC plays a critical role in TAM resistance in vitro.
Figure 3.
c-MYC expression is increased in TAM-resistant breast cancer cells, and the knockdown of c-MYC increases TAM sensitivity. (A) Heatmap depicting the fold-changes of proliferation-related genes in TAM-sensitive and TAM-resistant cells. (B) Bar graph representing the expression levels of c-MYC in TAM-sensitive and TAM-resistant cells. (C) Western blot analysis and (D) densitometric analysis of c-MYC, β-catenin, p-AKT and AKT expression in TAM-sensitive and TAM-resistant cells. Efficiency of siRNA in knocking down c-MYC expression in MCF-7R cells was evaluated by (E) reverse transcription-quantitative PCR and (F and G) western blot analysis. (H) Cellular viability was evaluated in MCF-7R cells transfected with siRNA and treated with TAM for 24 h (**P<0.01, ***P<0.001, compared to siRNA). Data are presented as the means ± standard deviation of mean of 3 repeats. *P<0.05, **P<0.01, ***P<0.001. TAM, tamoxifen; siRNA, small interfering RNA; NC, negative control; p-, phospho-.
c-MYC expression is associated with TAM resistance in patients with breast cancer
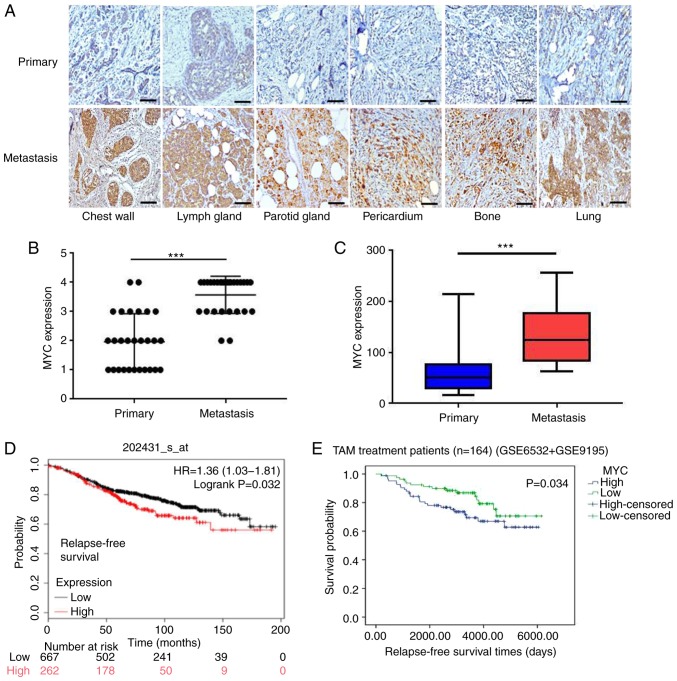
To further confirm the association between c-MYC and clinical TAM-resistance in patients with breast cancer, 28 pairs of primary and recurrent/metastatic tumor specimens from the same patients who underwent adjuvant TAM treatment were collected. As shown in Fig. 4A, c-MYC expression in the primary tumors was weaker, but was increased significantly in the metastatic lesions following the development of TAM in the same patients (all P<0.001; Fig. 4B and C), consistent with the results observed in TAM-resistant cell lines.
Figure 4.
Higher c-MYC expression levels are closely associated with TAM resistance in breast cancer patients. (A-C) Representative images of intensity and quantification of immunohistochemistry staining of c-MYC expression in paired primary and metastatic breast cancer tissues following treatment with TAM. Magnification, ×200. (D) Higher levels of c-MYC expression were associated with a less favorable recurrence-free survival in patients with breast cancer who received endocrine therapy based on publicly available data. (E) Higher expression levels of c-MYC predicted long-term disease outcomes following adjuvant TAM therapy. ***P<0.001. TAM, tamoxifen. Scale bar, 100 µm.
To assess c-MYC expression in breast cancer, TCGA was used for bioinformatics analysis. As shown in Fig. S2A and B, c-MYC expression was relatively low in breast cancer tissues compared with normal tissues (P<0.05); however, c-MYC was highly expressed in basal subtypes (endocrine therapy-resistant), which indicated an association with the therapeutic efficacy of TAM. Furthermore, Kaplan-Meier survival analysis revealed that a higher c-MYC expression was associated with a shorter distant metastasis-free survival in patients with breast cancer (P=0.011; Fig. S2C), and its expression was also associated with a poor relapse-free survival in patients with breast cancer that received endocrine therapy (P=0.032; Fig. 4D). In addition, a high c-MYC expression predicted an improved post-progression survival of patients with ER+ breast cancer (P=0.023), but not of the patients with ER-negative breast cancer (P=0.27; Fig. S2D and E).
To determine whether c-MYC expression may serve as a direct predictor of favorable outcomes following adjuvant TAM therapy, tumor gene expression levels and corresponding follow-up data from patients with ER+ breast cancer treated with TAM as an adjuvant were downloaded from GEO (accession nos. GSE6532 and GSE9195). The results revealed that an increased c-MYC expression was associated with a reduced relapse-free survival (P=0.034; Fig. 4E), suggesting that c-MYC expression was closely associated with clinical TAM resistance.
c-MYC affects the sensitivity of cisplatin by regulating cell cycle progression
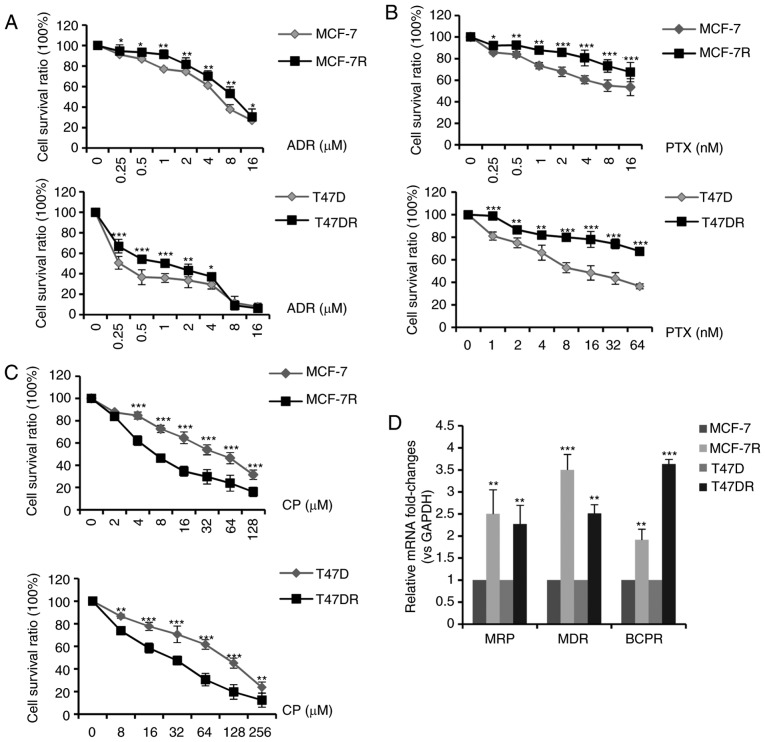
According to standard clinical chemotherapy, 3 major chemotherapeutic drugs (doxorubicin, paclitaxel and cisplatin) were evaluated to determine the alterations in chemosensitivity following the development of TAM resistance. As shown in Fig. 5A-C, the MCF-7R and T47DR cells exhibited an increased resistance to doxorubicin and paclitaxel; however, they exhibited a greater sensitivity to cisplatin, in comparison with the parental cells. The IC50 values of ADR, PTX and CP to the MCF-7, MCF-7R, T47D and T47DR cells were 5.30±0.85, 8.37±1.05, 1.36±0.36, 1.94±0.26 µM for ADR, 14.10±1.63, 57.78±3.85, 16.20±2.37, 102.89±5.37 nM for PTX, and 42.37±3.35, 20.76±2.86, 84.32±4.35, 25.76±3.24 µM for CP, respectively (data not shown). RT-qPCR assays revealed that the mRNA expression levels of genes associated with multidrug resistance (MRP, MDR and BCPR) were significantly higher in the TAM-resistant cells compared with the parental cells (Fig. 5D), which may partially explain the increase in resistance to doxorubicin and paclitaxel.
Figure 5.
Sensitivity of TAM-resistant cells to different chemotherapeutic drugs. TAM-sensitive and TAM-resistant cells were treated with increasing concentrations of (A) ADR for 24 h, (B) PTX for 72 h or (C) CP for 24 h. (D) mRNA expression levels of multidrug-resistance genes in TAM-sensitive and TAM-resistant cells. Data are presented as the means ± standard deviation of the mean of 3 repeats. *P<0.05, *P<0.01, ***P<0.001 vs. respective parental cells. TAM, tamoxifen; ADR, doxorubicin; PTX, paclitaxel; CP, cisplatin.
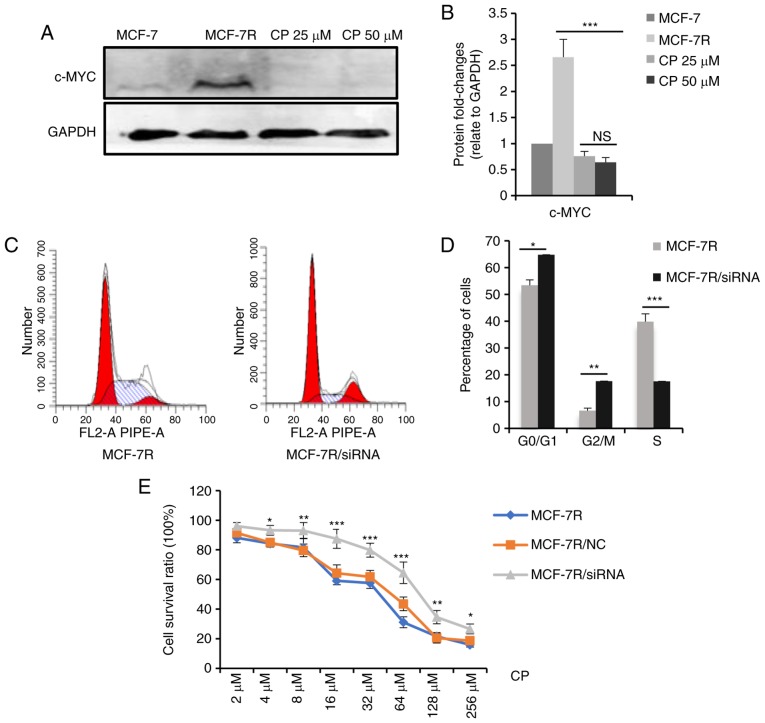
Thus, the mechanisms underlying increased cisplatin sensitivity were examined. It is hypothesized that cisplatin-induced DNA lesions are most cytotoxic in the S phase of the cell cycle (37-39) and that c-MYC accelerates the G1/S phase transition (40,41). In the present study, it was hypothesized that a high c-MYC expression increased cisplatin sensitivity by increasing the proportion of cells in the S phase. To test this hypothesis, the MCF-7R cells were treated with a medium concentration of cisplatin for 24 h, and the dead cells were washed away. Western blot analysis was performed to evaluate c-MYC protein expression levels in the residual adherent cells. As shown in Fig. 6A and B, c-MYC protein expression significantly decreased in the surviving cells following cisplatin treatment, indicating that the cells with a high c-MYC expression were sensitized to cisplatin-induced cell death. Furthermore, the percentage of MCF-7R cells in the S phase decreased significantly following the knockdown of c-MYC expression (Fig. 6C and D), and c-MYC knockdown desensitized the MCF-7R cells to cisplatin (Fig. 6E). The IC50 values of the MCF-7R cells to cisplatin prior to and following knockdown of c-MYC were 24.07±2.25 µM for the MCF-7R, 28.89±3.02 µM for the MCF-7R/NC and 54.23±3.62 µM for the MCF-7R/siRNA cells (data not shown). Collectively, these results suggest that a high c-MYC expression in TAM-resistant cells increased cisplatin sensitivity by regulating the cell cycle.
Figure 6.
c-MYC affects the sensitivity of cisplatin through regulating the cell cycle. (A) MCF-7R cells were treated with 0, 25 or 50 µM cisplatin for 24 h. After removing the dead cells, cells were lysed. c-MYC expression levels in the indicated cells were determined by western blot analysis. (B) Densitometric analysis of c-MYC in the western blots shown in (A). (C) c-MYC expression was knocked down in MCF-7R cells and flow cytometric analysis was performed to evaluate proportion of cells in the S-phase. (D) Bar chart representing the percentage of MCF-7R cells in the G0/G1, G2/M or S phase, prior to and following c-MYC/siRNA transfection. (E) Cell-viability of MCF-7R with c-MYC knocked down compared with MCF-7R and MCF-7R/NC cells, after treatment of cells with different doses of cisplatin for 24 h (*P<0.05, **P<0.01, ***P<0.001, compared to NC only). Data are presented as the means ± standard deviation of the mean of 3 repeats. *P<0.05, **P<0.01, ***P<0.001. TAM, tamoxifen; siRNA, small interfering; NC, negative control.
Patients with TAM-resistant breast cancer exhibit higher remission rates with cisplatin-based chemotherapy
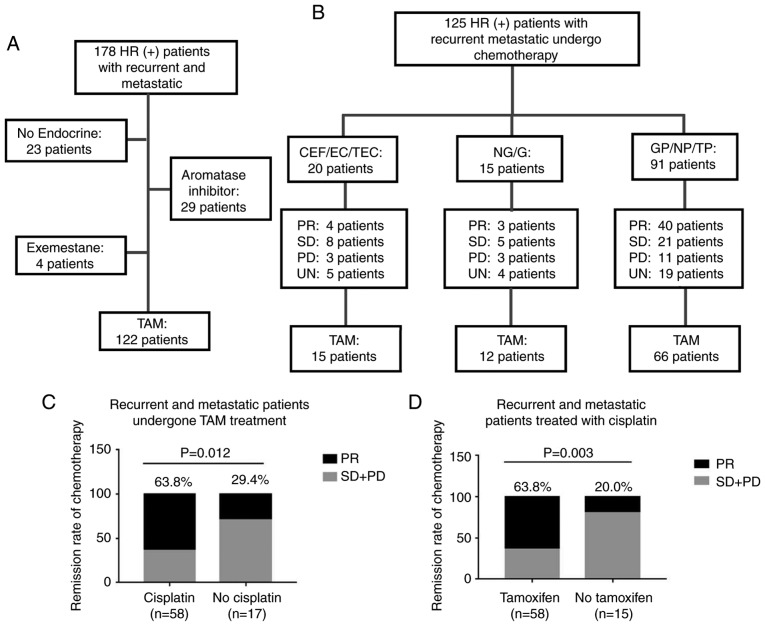
A total of 122 patients were shown to possess TAM-resistant breast cancer among the 178 patients with recurrent and/or metastatic ER+ breast cancer based on their endocrine therapy regimen (Fig. 7A). The median age of the patients at the time of recurrence/metastatic disease was 42.2 years (range, 25-65 years), and the median time from TAM therapy to recurrence and metastasis was 36.0 months (range, 5-132 months). In addition, 23 patients (18.9%) received treatment with TAM for >5 years. The baseline characteristics of the TAM-resistant patients are presented in Table I.
Figure 7.
Patients with TAM-resistance have favorable remission rates in response to cisplatin-based chemotherapy. (A) Summary of endocrine therapies administered to 178 patients with ER+ recurrent and metastatic breast cancer. A total of 122 patients were determined to exhibit TAM resistance. (B) Chemotherapy regimens and responses in 125 patients with ER+ recurrent and metastatic breast cancer. Clinical responses were evaluated in comparison with alteration of target lesions during chemotherapy. (C) Clinical remission rates of TAM-resistant patients administered different chemotherapy regimens. (D) Clinical remission rates of patients administered cisplatin-based chemotherapy following treatment with different endocrine-therapy regimens. C, cyclophosphamide; E, epirubicin; F, fluorouracil; T, paclitaxel; N, vinorelbine; G, gemcitabine; P, cisplatin; PR, partial response; SD, stable disease; PD, progressive disease; UN, unknown; ER+, estrogen receptor-positive; TAM, tamoxifen.
Following diagnosis with tumor recurrence and metastasis, 125 patients (93 with TAM resistance) received chemotherapy. The chemotherapeutic regimens and therapeutic responses are detailed in Fig. 7B. Following the analysis of the clinical remission efficiencies of patients that had undergone different chemotherapy regimens, it was determined that the TAM-resistant patients who received cisplatin-based chemotherapy exhibited improved clinical responses compared with those who received other regimens (P=0.012; Fig. 7C). Furthermore, the patients treated with TAM exhibited higher clinical remission rates when treated with cisplatin-based chemotherapy, compared with patients that had undergone other endocrine treatments (P=0.003; Fig. 7D).
Discussion
Despite the fact that aromatase inhibitors and other endocrine therapy drugs have been successfully used in the treatment of breast cancer (42,43), TAM remains the most widely used therapeutic in clinical practice (9,10,44). A major obstacle of using TAM is that a substantial proportion of patients develop drug resistance, leading to relapse and progression (12,15,45). For these patients, chemotherapy is recommended to control the disease when visceral crisis and rapid tumor progression occurs (18,46). However, in contrast to the large number of studies on the mechanisms underlying the development of TAM resistance (13-15,47,48), to the best of our knowledge, few studies to date have examined the effect of endocrine resistance on sensitivity to chemotherapeutics.
As an oncogene, c-MYC has been shown to play an important role in multiple cellular functions, including proliferation, migration, angiogenesis and differentiation through binding with nuclear DNA (41,49). It has been reported that c-MYC may partially replace the function of ER in estrogen-deprived breast cancer (34,37,50) and that c-MYC signaling may predict responses to endocrine therapy (51,52). Thus, c-MYC is hypothesized to play an important role in TAM-resistant breast cancer. In agreement with the findings of previous studies, the present study also confirmed that c-MYC expression was significantly increased in TAM-resistant cells and clinical tumor samples, and the inhibition of c-MYC expression in MCF-7R cells partly restored TAM sensitivity.
Subsequently, alterations in the sensitivities of cells to various chemotherapeutic agents following the occurrence of TAM resistance was determined, and the results revealed that TAM-resistant cells were significantly more sensitive to cisplatin-induced cell death compared with the parental cell line, consistent with previous literature (53-55). Moreover, previous clinical findings have demonstrated that a high c-MYC expression predicts more favorable clinical responses in patients with cervical cancer treated with cisplatin-based chemotherapy (56), and TAM-resistant triple-negative breast cancer exhibits an increased sensitivity to cisplatin; in addition, c-MYC amplification is the most frequently reported aberration in these tumors (55,57). Therefore, it was hypothesized that c-MYC may enhance sensitivity to cisplatin by regulating the proportion of cells in the S phase in TAM-resistant cells, and the results of the present study confirmed the above hypothesis. The clinical data generated in the present study also demonstrated that TAM-resistant breast cancer patients treated with cisplatin-based chemotherapy had higher clinical remission rates compared with those treated with other chemotherapeutic regimens, further confirming the findings of the in vitro experiments. To the best of our knowledge, the present study is the first to demonstrate that c-MYC plays different roles in treatment with endocrine therapy and chemotherapy in ER+ breast cancer. Furthermore, it is also the first time that the changes of chemosensitivity have been investigated in clinical TAM-resistant patients. In addition, it is noteworthy that the c-MYC downstream genes, CCNA2 and CDK4, also exhibit a high expression in TAM resistance, and it has been reported that both CCNA2 (58) and CDK4 (59) play an important role in TAM resistance. Therefore, the high expression of CCNA2 and CDK4 may also be involved in the sensitivity of TAM-resistant cells cisplatin; further research is warranted to confirm the above mechanisms.
In the present study, TAM-resistant cells also exhibited resistance to doxorubicin and paclitaxel; however, they exhibited an increased sensitivity to cisplatin, in agreement with the findings of a previous study (60). A possible reason underlying this phenomenon may be that upregulated expression levels of multidrug-resistance-associated genes in TAM-resistant cells are involved in the doxorubicin and paclitaxel efflux; thus, the cycle change caused by c-MYC may not increase the sensitivity of these two drugs to TAM-resistant cells (58). In addition, these proteins exhibited less of an effect on cisplatin efflux, as the limited intracellular accumulation of cisplatin is usually the result of reduced uptake (61-63). Furthermore, p21 has been identified as an important effector of c-MYC activity in the cell cycle, and its expression was significantly reduced in the TAM-resistant cells in the present study. Previous studies have revealed that the inhibition of p21 expression sensitized tumor cells to cisplatin (64,65). Thus, it was hypothesized that c-MYC may affect the cell cycle and increase cisplatin sensitivity by regulating p21 expression in TAM-resistant cells, and further experiments are required to examine this hypothesis. In addition, a recent study demonstrated that TAM-resistant breast cancer cells are resistant to DNA-damaging chemotherapy (66), which was inconsistent with the findings of the present and previous studies (53-55,60). The different results observed in the various studies may reflect the diversity in the cell models used, and further research aimed at determining the underlying mechanisms is required due to the complexity of drug resistance.
In conclusion, the results of the present study suggest that although c-MYC participates in TAM resistance, it improves cisplatin sensitivity in ER+ breast cancer. TAM-resistant patients may respond favorably to cisplatin, and the upregulated expression of c-MYC expression may be used as a predictive marker. However, further research and prospective clinical trials are required to confirm the observations of the present study.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was funded by National Natural Science Foundation of China (grant nos. 81472658, NSFC 81772979, NSFC 81472476 and NSFC 31671481).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
RC designed the study, performed the in vitro experiments and drafted the manuscript. SG and CY participated in the development of methodology of the study and performed the bioinformatics analysis. LS, BZ, KL and LL obtained the clinical data and performed the analysis. GT performed the analysis and interpretation of data. ML and SL participated in design of the study and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics committee of Chongqing Medical University (Chongqing, China). Written informed consent was obtained from all patients.
Patient consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Anderson WF, Katki HA, Rosenberg PS. Incidence of breast cancer in the United States: Current and future trends. J Natl Cancer Inst. 2011;103:1397–1402. doi: 10.1093/jnci/djr257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall E. Breast cancer. Dare to do less Science. 2014;343:1454–1456. doi: 10.1126/science.343.6178.1454. [DOI] [PubMed] [Google Scholar]
- 4.Vargo-Gogola T, Rosen JM. Modelling breast cancer: One size does not fit all. Nat Rev Cancer. 2007;7:659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- 5.Green KA, Carroll JS. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer. 2007;7:713–722. doi: 10.1038/nrc2211. [DOI] [PubMed] [Google Scholar]
- 6.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 7.Patani N, Martin LA. Understanding response and resistance to oestrogen deprivation in ER-positive breast cancer. Mol Cell Endocrinol. 2014;382:683–694. doi: 10.1016/j.mce.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 8.Cuzick J, Sestak I, Pinder SE, Ellis IO, Forsyth S, Bundred NJ, Forbes JF, Bishop H, Fentiman IS, George WD. Effect of tamoxifen and radiotherapy in women with locally excised ductal carcinoma in situ: Long-term results from the UK/ANZ DCIS trial. Lancet Oncol. 2011;12:21–29. doi: 10.1016/S1470-2045(10)70266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan VC. Tamoxifen as the first targeted long-term adjuvant therapy for breast cancer. Endocr Relat Cancer. 2014;21:R235–R246. doi: 10.1530/ERC-14-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briest S, Stearns V. Tamoxifen metabolism and its effect on endocrine treatment of breast cancer. Clin Adv Hematol Oncol. 2009;7:185–192. [PubMed] [Google Scholar]
- 11.Lønning PE. Adjuvant endocrine treatment of early breast cancer. Hematol Oncol Clin North Am. 2007;21:223–238. doi: 10.1016/j.hoc.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Arimidex, Tamoxifen, Alone or in Combination (ATAC) Trialists' Group. Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 13.Raha P, Thomas S, Munster PN. Epigenetic modulation: A novel therapeutic target for overcoming hormonal therapy resistance. Epigenomics. 2011;3:451–470. doi: 10.2217/epi.11.72. [DOI] [PubMed] [Google Scholar]
- 14.Thewes V, Simon R, Schroeter P, Schlotter M, Anzeneder T, Büttner R, Benes V, Sauter G, Burwinkel B, Nicholson RI, et al. Reprogramming of the ERRalpha and ERalpha target gene landscape triggers tamoxifen resistance in breast cancer. Cancer Res. 2015;75:720–731. doi: 10.1158/0008-5472.CAN-14-0652. [DOI] [PubMed] [Google Scholar]
- 15.Baneshi MR, Warner P, Anderson N, Edwards J, Cooke TG, Bartlett JM. Tamoxifen resistance in early breast cancer: Statistical modelling of tissue markers to improve risk prediction. Br J Cancer. 2010;102:1503–1510. doi: 10.1038/sj.bjc.6605627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baselga J, Campone M, Piccart M, Burris HA, III, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner NC, Ro J, André F, Loi S, Verma S, Iwata H, Harbeck N, Loibl S, Huang Bartlett C, Zhang K, et al. Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med. 2015;373:209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 18.Zagouri F, Liakou P, Bartsch R, Peccatori FA, Tsigginou A, Dimitrakakis C, Zografos GC, Dimopoulos MA, Azim HA., Jr Discrepancies between ESMO and NCCN breast cancer guidelines: An appraisal. Breast. 2015;24:513–523. doi: 10.1016/j.breast.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 19.Yuan J, Liu M, Yang L, Tu G, Zhu Q, Chen M, Cheng H, Luo H, Fu W, Li Z, Yang G. Acquisition of epithelial-mesenchymal transition phenotype in the tamoxifen-resistant breast cancer cell: A new role for G protein-coupled estrogen receptor in mediating tamoxifen resistance through cancer-associated fibroblast-derived fibronectin and β1-integrin signaling pathway in tumor cells. Breast Cancer Res. 2015;17:69. doi: 10.1186/s13058-015-0579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bui QT, Im JH, Jeong SB, Kim YM, Lim SC, Kim B, Kang KW. Essential role of Notch4/STAT3 signaling in epithelial-mesenchymal transition of tamoxifen-resistant human breast cancer. Cancer Lett. 2017;390:115–125. doi: 10.1016/j.canlet.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Ward A, Balwierz A, Zhang JD, Küblbeck M, Pawitan Y, Hielscher T, Wiemann S, Sahin Ö. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32:1173–1182. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 22.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fallah Y, Brundage J, Allegakoen P, Shajahan-Haq AN. MYC-driven pathways in breast cancer subtypes. Biomolecules. 2017;7:E53. doi: 10.3390/biom7030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raha P, Thomas S, Thurn KT, Park J, Munster PN. Combined histone deacetylase inhibition and tamoxifen induces apoptosis in tamoxifen-resistant breast cancer models, by reversing Bcl-2 overexpression. Breast Cancer Res. 2015;17:26. doi: 10.1186/s13058-015-0533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi H, Jiang Z, Wang C, Yang Y, Li L, He H, Yu Z. Sensitization of tamoxifen-resistant breast cancer cells by Z-ligustilide through inhibiting autophagy and accumulating DNA damages. Oncotarget. 2017;8:29300–29317. doi: 10.18632/oncotarget.16832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sebaugh JL. Guidelines for accurate EC50/IC50 estimation. Pharm Stat. 2011;10:128–134. doi: 10.1002/pst.426. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 29.Yuan JQ, Wang SM, Tang LL, Mao J, Wu YH, Hai J, Luo SY, Ou HY, Guo L, Liao LQ, et al. Relative dose intensity and therapy efficacy in different breast cancer molecular subtypes: A retrospective study of early stage breast cancer patients treated with neoadjuvant chemotherapy. Breast Cancer Res Treat. 2015;151:405–413. doi: 10.1007/s10549-015-3418-z. [DOI] [PubMed] [Google Scholar]
- 30.Han C, Yang L, Choi HH, Baddour J, Achreja A, Liu Y, Li Y, Li J, Wan G, Huang C, et al. Amplification of USP13 drives ovarian cancer metabolism. Nat Commun. 2016;7:13525. doi: 10.1038/ncomms13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindström LS, Karlsson E, Wilking UM, Johansson U, Hartman J, Lidbrink EK, Hatschek T, Skoog L, Bergh J. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol. 2012;30:2601–2608. doi: 10.1200/JCO.2011.37.2482. [DOI] [PubMed] [Google Scholar]
- 32.Cui J, Germer K, Wu T, Wang J, Luo J, Wang SC, Wang Q, Zhang X. Cross-talk between HER2 and MED1 regulates tamoxifen resistance of human breast cancer cells. Cancer Res. 2012;72:5625–5634. doi: 10.1158/0008-5472.CAN-12-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Malerva L, Park J, Zou L, Hu Y, Moradpour Z, Pearlberg J, Sawyer J, Stevens H, Harlow E, LaBaer J. High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc Natl Acad Sci USA. 2011;108:2058–2063. doi: 10.1073/pnas.1018157108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng R, Liu YJ, Cui JW, Yang M, Liu XL, Li P, Wang Z, Zhu LZ, Lu SY, Zou L, et al. Aspirin regulation of c-myc and cyclinD1 proteins to overcome tamoxifen resistance in estrogen receptor-positive breast cancer cells. Oncotarget. 2017;8:30252–30264. doi: 10.18632/oncotarget.16325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 36.Musgrove EA, Sergio CM, Loi S, Inman CK, Anderson LR, Alles MC, Pinese M, Caldon CE, Schütte J, Gardiner-Garden M, et al. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS One. 2008;3:e2987. doi: 10.1371/journal.pone.0002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cepeda V, Fuertes MA, Castilla J, Alonso C, Quevedo C, Pérez JM. Biochemical mechanisms of cisplatin cytotoxicity. Anticancer Agents Med Chem. 2007;7:3–18. doi: 10.2174/187152007779314044. [DOI] [PubMed] [Google Scholar]
- 38.Dobbelstein M, Sørensen CS. Exploiting replicative stress to treat cancer. Nat Rev Drug Discov. 2015;14:405–423. doi: 10.1038/nrd4553. [DOI] [PubMed] [Google Scholar]
- 39.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, Jia L, Li S, Cancer Genome Atlas Research Network. Xie W, Yang D. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–720.e9. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 42.Graham J, Pitz M, Gordon V, Grenier D, Amir E, Niraula S. Clinical predictors of benefit from fulvestrant in advanced breast cancer: A Meta-analysis of randomized controlled trials. Cancer Treat Rev. 2016;45:1–6. doi: 10.1016/j.ctrv.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Ma CX, Reinert T, Chmielewska I, Ellis MJ. Mechanisms of aromatase inhibitor resistance. Nat Rev Cancer. 2015;15:261–275. doi: 10.1038/nrc3920. [DOI] [PubMed] [Google Scholar]
- 44.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Thürlimann B, Senn HJ. Tailoring therapies-improving the management of early breast cancer: St Gallen International Expert Consensus on the primary therapy of early breast cancer 2015. Ann Oncol. 2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, Look MP, Smid M, Veldscholte J, Sleijfer S, Foekens JA, Dorssers LC. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol. 2009;27:542–549. doi: 10.1200/JCO.2008.17.1462. [DOI] [PubMed] [Google Scholar]
- 46.Cardoso F, Costa A, Senkus E, Aapro M, André F, Barrios CH, Bergh J, Bhattacharyya G, Biganzoli L, Cardoso MJ, et al. 3rd ESO-ESMO International Consensus Guidelines for advanced breast cancer (ABC 3) Ann Oncol. 2017;28:3111. doi: 10.1093/annonc/mdx036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin K, Park S, Teo WW, Korangath P, Cho SS, Yoshida T, Győrffy B, Goswami CP, Nakshatri H, Cruz LA, et al. HOXB7 is an ERα cofactor in the activation of HER2 and multiple ER target genes leading to endocrine resistance. Cancer Discov. 2015;5:944–959. doi: 10.1158/2159-8290.CD-15-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manso L, Mourón S, Tress M, Gómez-López G, Morente M, Ciruelos E, Rubio-Camarillo M, Rodriguez-Peralto JL, Pujana MA, Pisano DG, Quintela-Fandino M. Analysis of paired primary-metastatic hormone-receptor positive breast tumors (HRPBC) uncovers potential novel drivers of hormonal resistance. PLoS One. 2016;11:e0155840. doi: 10.1371/journal.pone.0155840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller TW, Balko JM, Ghazoui Z, Dunbier A, Anderson H, Dowsett M, González-Angulo AM, Mills GB, Miller WR, Wu H, et al. A gene expression signature from human breast cancer cells with acquired hormone independence identifies MYC as a mediator of antiestrogen resistance. Clin Cancer Res. 2011;17:2024–2034. doi: 10.1158/1078-0432.CCR-10-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin S, Rishi AK, Reddy KB. Anti-estrogenresistant breast cancer cells are sensitive to cisplatin plus TRAIL treatment. Oncol Rep. 2015;33:1475–1480. doi: 10.3892/or.2015.3721. [DOI] [PubMed] [Google Scholar]
- 54.Yde CW, Gyrd-Hansen M, Lykkesfeldt AE, Issinger OG, Stenvang J. Breast cancer cells with acquired antiestrogen resistance are sensitized to cisplatin-induced cell death. Mol Cancer Ther. 2007;6:1869–1876. doi: 10.1158/1535-7163.MCT-07-0072. [DOI] [PubMed] [Google Scholar]
- 55.Leung EY, Kim JE, Askarian-Amiri M, Joseph WR, McKeage MJ, Baguley BC. Hormone resistance in Two MCF-7 breast cancer cell lines is associated with reduced mTOR signaling, decreased glycolysis, and increased sensitivity to cytotoxic drugs. Front Oncol. 2014;4:221. doi: 10.3389/fonc.2014.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu H, Wu J, Zhang W, Luo H, Shen Z, Cheng H, Zhu X. PKM2 enhances chemosensitivity to cisplatin through interaction with the mTOR pathway in cervical cancer. Sci Rep. 2016;6:30788. doi: 10.1038/srep30788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao T, Han Y, Yu L, Ao S, Li Z, Ji J. CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PLoS One. 2014;9:e91771. doi: 10.1371/journal.pone.0091771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao A, Sun T, Ma G, Cao J, Hu Q, Chen L, Wang Y, Wang Q, Sun J, Wu R, et al. LEM4 confers tamoxifen resistance to breast cancer cells by activating cyclin D-CDK4/6-Rb and ERα pathway. Nat Commun. 2018;9:4180. doi: 10.1038/s41467-018-06309-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kangaspeska S, Hultsch S, Jaiswal A, Edgren H, Mpindi JP, Eldfors S, Brück O, Aittokallio T, Kallioniemi O. Systematic drug screening reveals specific vulnerabilities and co-resistance patterns in endocrine-resistant breast cancer. BMC Cancer. 2016;16:378. doi: 10.1186/s12885-016-2452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang D, Sui M, Zhong W, Huang Y, Fan W. Different administration strategies with paclitaxel induce distinct phenotypes of multidrug resistance in breast cancer cells. Cancer Lett. 2013;335:404–411. doi: 10.1016/j.canlet.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baekelandt MM, Holm R, Nesland JM, Tropé CG, Kristensen GB. P-glycoprotein expression is a marker for chemotherapy resistance and prognosis in advanced ovarian cancer. Anticancer Res. 2000;20:1061–1067. [PubMed] [Google Scholar]
- 63.Wada H, Saikawa Y, Niida Y, Nishimura R, Noguchi T, Matsukawa H, Ichihara T, Koizumi S. Selectively induced high MRP gene expression in multidrug-resistant human HL60 leukemia cells. Exp Hematol. 1999;27:99–109. doi: 10.1016/S0301-472X(98)00027-7. [DOI] [PubMed] [Google Scholar]
- 64.Jacobsen C, Honecker F. Cisplatin resistance in germ cell tumours: Models and mechanisms. Andrology. 2015;3:111–121. doi: 10.1111/andr.299. [DOI] [PubMed] [Google Scholar]
- 65.Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O'Reilly T, Natt F, Hall J, Lane HA, Thomas G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 66.Zhu Y, Liu Y, Zhang C, Chu J, Wu Y, Li Y, Liu J, Li Q, Li S, Shi Q, et al. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat Commun. 2018;9:1595. doi: 10.1038/s41467-018-03951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.