Abstract
Acute myelogenous leukaemia (AML) is associated with risk factors that are largely unknown and with a heterogeneous response to treatment. Here, we provide a comprehensive quantitative understanding of AML proteomic heterogeneities and hallmarks by using the AML proteome atlas, a proteomics database that we have newly derived from MetaGalaxy analyses, for the proteomic profiling of 205 AML patients and 111 leukaemia cell lines. The analysis of the dataset revealed 154 functional patterns based on common molecular pathways, 11 constellations of correlated functional patterns, and 13 signatures that stratify the patients’ outcomes. We find limited overlap between proteomics data and both cytogenetics and genetic mutations, and also that leukaemia cell lines show limited proteomic similarities with cells from AML patients, suggesting that a deeper focus on patient-derived samples is needed to gain disease-relevant insights. The AML proteome atlas provides a knowledge base for proteomic patterns in AML, a guide to leukaemia cell-line selection, and a broadly applicable computational approach for quantifying the heterogeneities of protein expression and proteomic hallmarks in AML.
In their seminal manuscripts, Hanahan and Weinberg defined 6 (ref. 1) and later 10 (ref. 2) ‘hallmarks of cancer’ that all malignancies must achieve. Although all malignancies share the same hallmarks, between-patient heterogeneities complicate the response of patients to therapy and subject them to varied outcomes. A comprehensive understanding of the biological heterogeneities and their clinical consequences can therefore greatly facilitate treatment recommendations and the development and personalization of targeted therapies.
This is particularly the case in adult acute myelogeneous leukaemia (AML), a deadly hematological disease known for its biological and clinical heterogeneity. AML patients present with rapidly accumulating cancerous myeloid cells in the bone marrow, leading to hematopoietic insufficiency, infection and anemia, which leads to a 5-year survival rate of only 24%3. In addition to prognostic guidance from clinical features like age and performance status, conventional cytogenetics of recurrent whole or partial chromosome losses and translocations events have so far yielded the most significant prognostic information, which stratifies AML patients into favorable, intermediate or unfavorable risk groups4–6. The sequencing of AML genomes has defined a limited set of mutations that are commonly found7,8, complementing the knowledge gained from conventional cytogenetics to provide a more complete understanding of the genetic changes that underlie AML. Beyond genomic sequences and genetic expressions9, AML blasts are affected by the microenvironment, through direct or indirect contact with mesenchymal stem cells, exposure to cytokines and chemokines, and stimulation by the hypoxic milieu that exists within the AML marrow. The biology of the AML cell is therefore a combination of the existing genetic and epigenetic changes and the environment the cell lives in. The possible combinations of changes at each of these levels produce a seemingly infinite number of states and abnormalities. This extreme degree of heterogeneity creates a paradox: regardless of what events have combined to cause a given case of AML, the events must always satisfy the same “hallmarks” using the circuitry and machinery existing in all leukemic cells. This heterogeneity raises the biological question of how such diverse combinations of events meet the “hallmarks” obligation, and how to therapeutically target individual events that are occurring in various combinations.
Since the unifying consequence of all these genetic, epigenetic and environmental events occurs predominantly at the level of proteins and within the signaling networks that they function in, we hypothesized that despite the genetic diversity, AML can be characterized by a more succinct series of protein expression signatures based on the common circuitry of the cell. We further hypothesized that expression patterns observed at the protein level would provide a roadmap of how the leukemic cell is achieving the “hallmarks,” and that this would enable the rational selection of targeted therapies for use in combination on an individualized basis.
To address these hypotheses, we investigated the proteomic heterogeneities of AML patients and define protein expression signatures using Reverse Phase Protein Array (RPPA). RPPA profiles both protein levels and post-translational modifications including phosphorylation, methylation and activation by cleavage with high sensitivity10,11. The technique is particularly preferable for this study due to the limited amount of material available in clinical samples, which would make mass spectrometry infeasible12. The technique has been successfully applied to study multiple types of tumors in basic, pre-clinical and clinical research11,13–16, and it has been incorporated into cancer clinical trials to monitor signaling molecules and pharmacodynamics17–19.
In contrast to previous proteomic profiling studies20,21 that used individual proteins indiscriminately to identify patient clusters, in this study we developed and applied a computational procedure that integrates biological context into pattern discovery and adopts a hierarchical structure to organize patterns. This procedure, termed “MetaGalaxy analysis”, consists of three steps: (1) combining individual proteins into groups of functionally related proteins (such a group is called a “functional group”); (2) characterizing the expression patterns within each functional group (such a pattern is called a “functional pattern”); (3) determining constellations of correlated functional patterns and identifying patient subpopulations with similar constellation signatures. In this multi-layer structure, the heterogeneity of individual protein expressions gives rise to the heterogeneity of functional patterns, and these functional patterns coalesce and form various signaling and functional constellations. The unique and selected combinations of constellations that characterize specific AML patient subpopulations and determine the patients’ response to therapy are therefore quantitative hallmarks (called “signatures”) in AML. Characterizing and understanding these quantitative AML hallmarks can significantly accelerate the identification and development of new therapeutic targets, aid in the design and testing of new combinatorial therapies, and provide new insight into the wiring of pathways in AML.
Results
Proteomic profiling of AML patient samples
Peripheral blood and bone marrow specimens, as well as clinical data, from 511 patients with newly diagnosed AML were collected at the University of Texas M.D. Anderson Cancer Center (MDACC) between September 1999 and March 2007. A proteomic profile was generated for each sample using RPPA11 with 228 strictly validated antibodies (Table S1). For each protein, the expression levels in AML samples were normalized relative to the mean expression of ten cryopreserved bone marrow CD34+ samples from healthy subjects (control samples) printed on the same array: positive values indicate higher expression levels than that in control samples, and vice versa.
The patient samples in this study were derived from two sources, either from peripheral blood (n = 282) or from bone marrow (n = 387); and they were prepared using two approaches, either from fresh materials on the day of collection (n = 292) or from cryopreserved cells at a later time (n = 377). In general, the proteomics of blood-derived samples are similar to that of marrow-derived samples. However, differences were observed between fresh and cryopreserved samples. At the protein level, 71% (162/228) of the proteins show statistically significant differences in expression between fresh and cryopreserved samples (two-sided t-test of p < 0.05, corrected for multiple testing to control the false discovery rate (FDR) at 5% using the Benjamini-Hochberg Procedure (BH corrected)22, Table S1; an example of adhesion-related proteins is shown in Figure S1B). To investigate the expression differences at the pathway level, we first divided all 228 proteins into 31 functional groups based on their functional similarities (see more details in Experimental Procedures), and then clustered all samples based on the expression levels of the proteins in each functional group. We found that 24 of 31 functional groups contain expression patterns that were only seen in cryopreserved samples but not in fresh samples (Figure S1A), whereas 9 of 31 functional groups contain patterns exclusive to fresh samples. Due to this proteomic difference between fresh and cryopreserved samples, we restricted the analysis in this study to the 232 patients whose samples were prepared fresh, of which 205 patients were treated at MDACC and were thus evaluated for outcome. Since fresh samples were uninfluenced by the potential effects of cryopreservation or thawing, they may represent a more accurate picture of the AML biology compared to cryopreserved samples. In addition to sample sources and processing conditions, we checked other factors such as collection time (i.e., duration of cryopreservation) and slide batch but did not detect any noticeable batch effects.
The proteomic profile includes expression measurements of 61 post-translational modifications (including 52 phosphorylation sites, 6 cleaved proteins and 3 histone methylation sites), 47 of which have matching measurements of total proteins. We found that the levels of post-translational modifications in general do not correlate with the levels of total protein expression levels, with Pearson Correlation Coefficient (PCC) < 0.5 in 40 phosphorylation sites and PCC > 0.75 in only 4 sites (Table S2). The lack of correlation between post-translational modifications and total protein levels indicates that we will likely observe signaling activities from RPPA that were previously undiscoverable in mRNA-based gene expression profiling (GEP) studies.
Functional patterning: characterizing heterogeneities in pathways
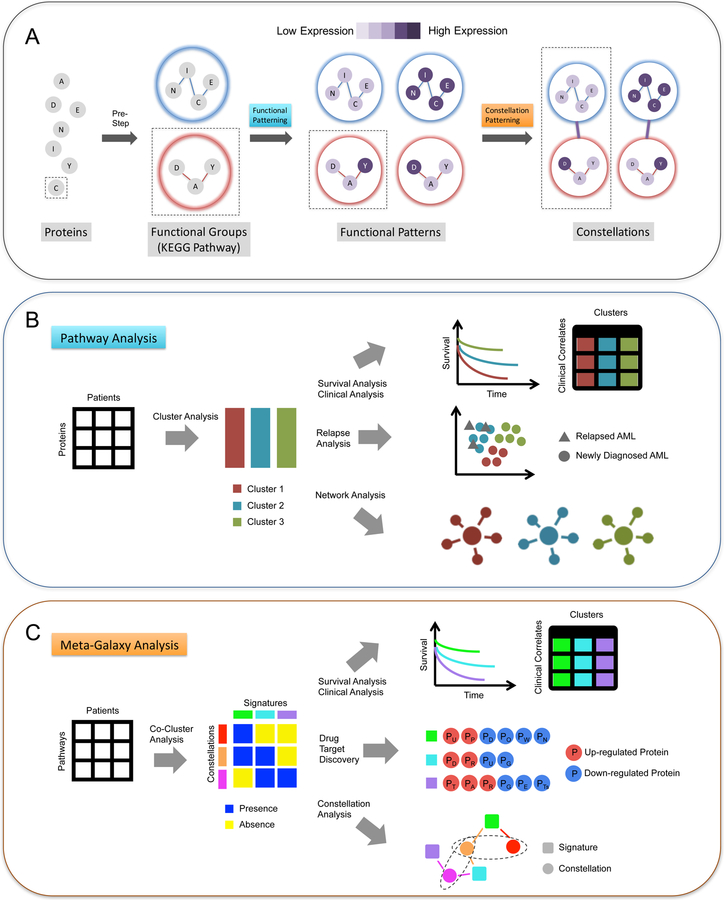
We designed MetaGalaxy analysis (Figure 1) to identify proteomic patterns within the context of functionally related protein networks. As a pre-step to the analysis, we first divided all proteins into 31 functional groups based on their known cellular functionalities and pathway memberships (Table S3). MetaGalaxy analysis proceeds in two steps: (1) identify expression patterns in each functional group and (2) connect functional groups to discovery patterns globally. As the first step of MetaGalaxy analysis, we clustered the AML samples for each functional group based on the expression levels of proteins involved in that functional group. Each resulting cluster is defined as a Functional Pattern. In total, 154 distinct functional patterns were observed in AML patients.
Figure 1. MetaGalaxy analysis workflow.
The schematic of (A) the overall workflow of MetaGalaxy analysis: proteins are organized into functional groups from known biology; analysis identifies patterns of protein expression for the functional groups (Functional Patterning); and finally, constellations, or patterns across functional groups are identified (Constellation Patterning). (B) steps in Functional Patterning include identifying the optimal groups for functional patterns via Progeny Clustering of the protein expression levels for the 209 AML patients; survival, relapse and other clinical covariate analyses; and signaling network analysis; and (C) steps in Constellation Patterning include co-clustering to identify which patients fall into each protein functional pathway (signatures) and how protein functional pathways are grouped (constellations); survival and other clinical analyses; drug target discovery for proteins that are significantly up- or down-regulated across patient groups; and decision tree modeling to identify the contribution of each protein constellation to patient outcomes (constellation analysis).
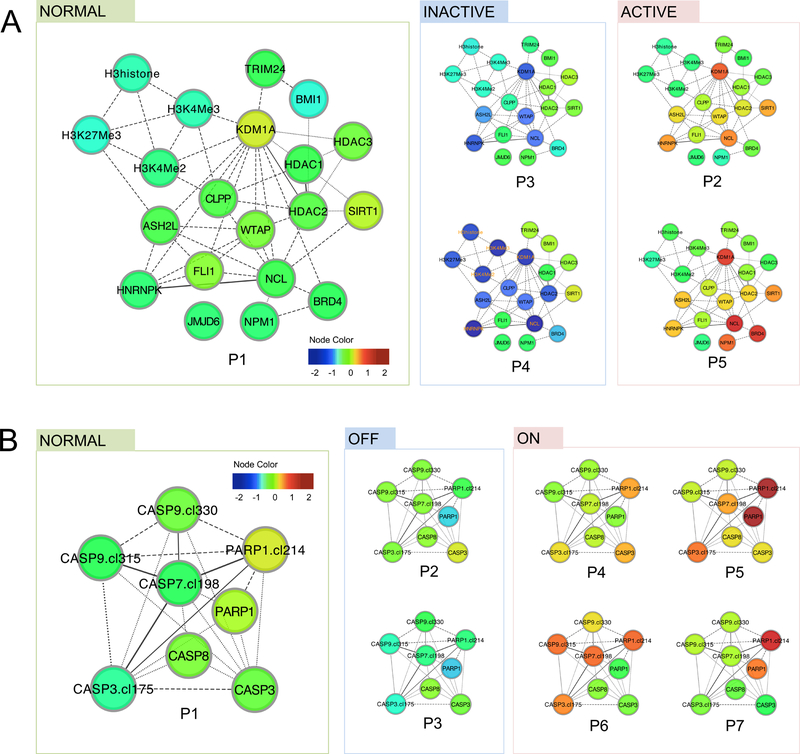
The variety of functional patterns in a functional group demonstrates the heterogeneity of mechanisms that AML cells can use to achieve a similar functional state. Here, a functional state refers to a qualitative state of the pathway activity, which is assessed based on key proteins in the functional group for interpretation purposes. In the Histone group for example, we found 5 patterns (P1 to P5) that can be grouped into 3 functional states based on the expression level of histone regulators (i.e. KDM1A, ASH2L and WTAP): a normal state (P1) that mimics patterns in control samples, an inactive state (P3 and P4) marked by reduced expressions of histone regulators, and an active state (P2 and P5) with higher expressions (Figure 2A). The observation that multiple distinct expression patterns underlie an active or inactive functional state indicates alternative regulation mechanisms of functional activities. A similar example can be found in the Apoptosis-Occurring group, where the apoptosis-on state arises from four distinct functional patterns (Figure 2B).
Figure 2. Functional patterns indicate varied functional states and alternative mechanisms.
Functional patterns were represented in protein networks, where the node color reflects the differential expression levels compared to the average expression levels in control samples. (A) In the Histone group, five functional patterns (P1-P5, ordered by similarity to patterns in control samples) were found. Specifically, P2 and P5 both result in an active state of histone modification (marked by high expression of histone regulators such as KDM1A and ASH2L), whereas P3 and P4 both result in an inactive state with reduced expression of these proteins. (B) In the Apoptosis-Occurring group, seven functional patterns were identified, where P4 to P7 all suggest heightened apoptosis activities (marked by higher levels of cleaved caspase 3, 7 and 9 as well as higher levels of cleaved PARP), despite their expression pattern differences.
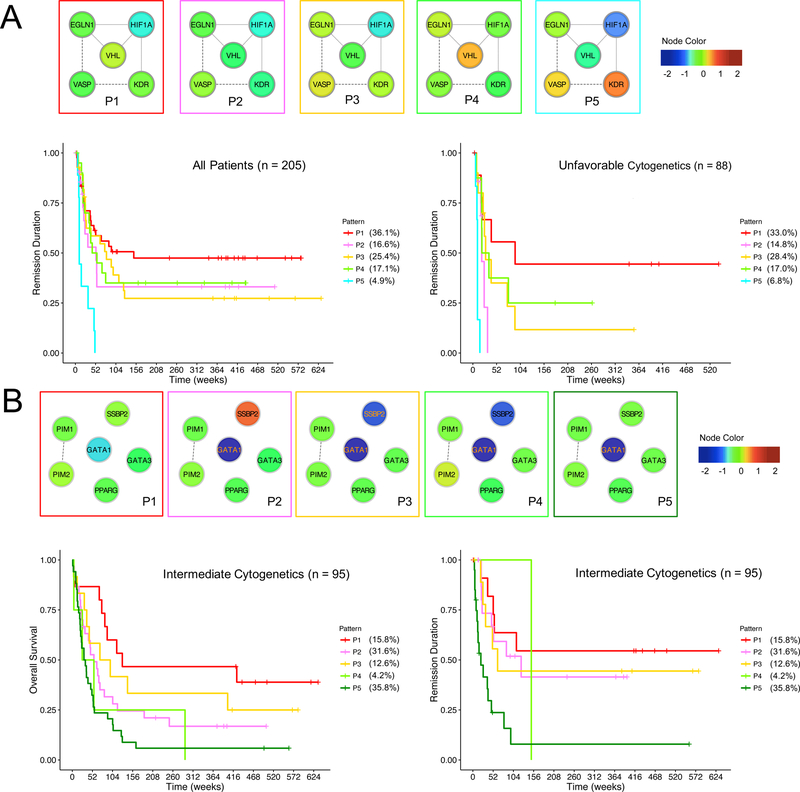
Characterizing functional patterns also reveals uncanonical protein expression relationships that could be AML specific. In the Hypoxia group for instance, we saw two main functional states among all AML patients based on the expression level of HIF1α: a hyperoxic state (P1, P2, P3 and P5) that features low HIF1α levels, and a normal state (P4) with comparable HIF1α level to that of control samples (Figure 3A). Contrary to the expectations that the hypoxic bone marrow environment harbors leukemic stem cells and thereby contains cells with high HIF1α levels (Benito et al., 2011), we didn’t observe any functional patterns that exhibit higher levels of HIF1α compared to that of control samples. Further, the canonical relationships among EGLN1, VHL and HIF1α were only seen in P1 to P3, but not in P4 and P5, indicating that P4 and P5 might be disease specific patterns. EGLN1 and VHL are known to regulate HIF1α by hydroxylation and subsequent degradation respectively, therefore higher levels of EGLN1 and VHL were expected to be associated with lower levels of HIF1α, which was seen in P1, P2 and P3. However, P4 and P5 do not agree with this conventional relationship, where the level of HIF1α is not reduced - despite the presence of high EGLN1 and VHL in P4, and it is abnormally lower without increases of EGLN1 and VHL levels in P5.
Figure 3. Example prognostic functional patterns.
Functional patterns were represented in protein networks, where the node color reflects the relative expression levels compared to the average expression levels in control samples. (A) In the Hypoxia group, the functional patterns stratify the remission duration among all patients (left, log-rank test of p = 0.01, one-sided, BH corrected) and the remission duration among patients with unfavorable cytogenetics (right, log-rank test of p = 0.00004, one-sided, BH corrected). (B) In the Differentiation group, the functional patterns stratify the overall survival (left, log-rank test of p = 0.109, one-sided, BH corrected) and remission duration (right, log-rank test of p = 0.031, one-sided, BH corrected) among patients with intermediate cytogenetics.
Despite the fact that functional patterns were defined solely based on the proteomic data without taking any clinical information, some functional patterns are prognostic. For instance, the functional patterns of the Hypoxia group stratify remission duration in all patients (log-rank test of p = 0.010, BH corrected) and patients with unfavorable cytogenetics (log-rank test of p < 0.001, BH corrected) (Figure 3A). Notably, patients in P5 (which features high KDR, VASP and abnormal hypoxia regulation patterns) are associated with the most unfavorable outcome, whereas patients in P1 (which resembles the patterns in control samples) are subject to the most favorable outcome. The prognostic value of functional patterns was also seen in the Differentiation group (Figure 3B).
Most functional patterns were found not to associate with clinical factors (e.g. the French-American-British (FAB) class, cytogenetics) and commonly tested genetic mutations (e.g. FLT3, NPM1, TP53, RAS). This is consistent with the concept that functional patterns arise from a combination of multiple genetic, epigenetic, or environmental events. Only in some functional groups were proteomic patterns found to associate with specific genetic mutations. For example, the functional patterns of the TP53 group are associated with TP53 mutations, with 66.7% of TP53-P4 cases are TP53 mutated compared to a mutation rate of 14.8% across all patterns.
Integrating previously known protein interactions with the computationally derived interactions, we next built a protein network for each functional group consisting of the proteins in that functional group, their 1st-degree neighboring proteins in other functional groups, and their interactions (an example of SMAD-P5 shown in Figure S2). This functional-pattern-based network allowed us to quickly visualize and examine each functional pattern in the context of the proteins and functional groups in association, and extend our search for drug targets beyond the scope of individual functional groups.
Constellation patterning: characterizing global heterogeneities and hallmarks orchestrated by functional constellations
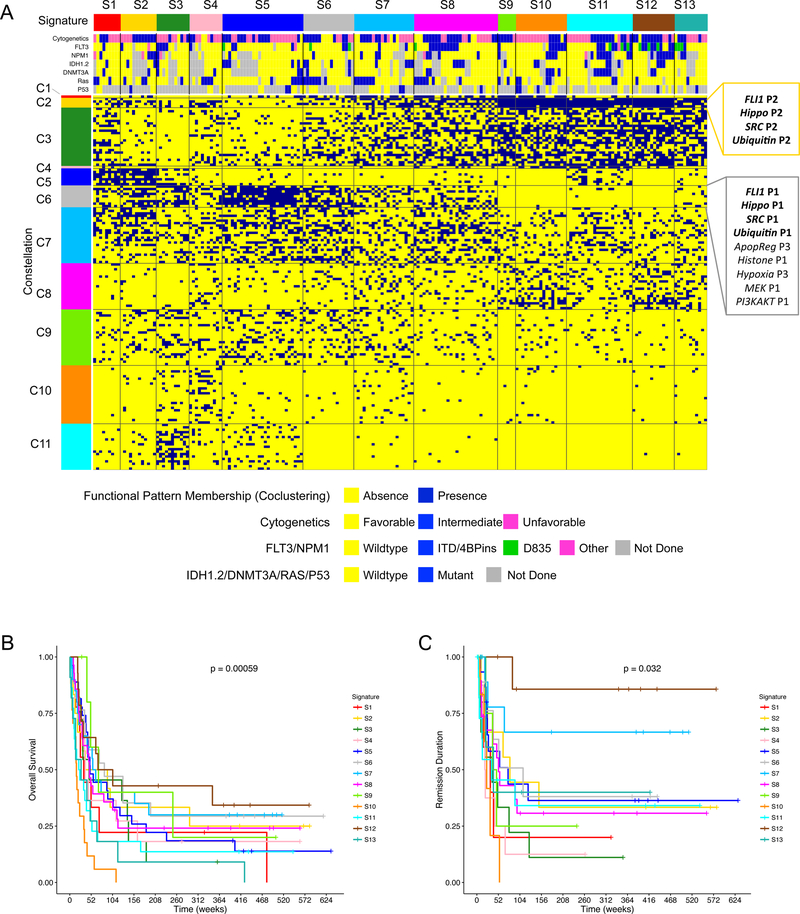
To recognize global proteomic heterogeneities across multiple functional groups, we co-clustered all 205 treated patients and the 154 functional patterns based on the binary functional pattern memberships assigned to each patient (i.e. 1 if the functional pattern is present in the patient, and 0 if it is absent). Based on the highest stability of clustering (assessed by Progeny Clustering23, a bootstrapping-based method, Table S8), we obtained 11 clusters of functional patterns and 13 clusters of patients (Figure 4A). Here, we define a cluster of functional patterns (which regularly co-occur in patient subpopulations) as a Constellation, and define a cluster of patients (with similar patterns of constellation membership) as a Signature.
Figure 4. The co-clustering of functional patterns generates biologically insightful constellations and prognostic signatures.
(A) 11 constellations (rows) and 13 signatures (columns) were obtained from co-clustering the functional pattern memberships (rows) and patients (columns) with the cluster number determined by Progeny Clustering (stability scores available in Table S8). The functional members of Constellation 2 and 6 are shown at the right, with the association of functional groups FLI1-Hippo-SRC-Ubiquitin highlighted in bold. Patients’ cytogenetics and mutation information are included at the top. Sample size, n = 205 patients. The Kaplan-Meier curves for overall survival and remission duration based on 13 signatures are shown in (B) and (C) respectively. p-values shown are from the log-rank test, one-sided. n = 205 patients in both (B) and (C). See Table S4 for the detailed members of all constellations.
The constellations capture the dependent relations among functional groups and patterns. For example, four functional groups (Ubiquitin, SRC, Hippo and FLI1), are tightly associated, since the patterns from these groups appear together in both Constellation 2 and 6 (permutation test of p < 0.05). This association among the four functional groups is not entirely unexpected biologically, since previous studies have suggested that SRC proteins are subject to regulation by ubiquitination24, and that SRC is involved in regulating the hippo pathway25. Another coalition that we observed features functional groups of Transcription, TCell, Signal Transduction Pathway (STP), PKC, MEK and Differentiation, which co-occur in Constellation 3 and 5 (permutation test of p < 0.05).
The thirteen signatures characterized by the recurrent association of constellations are prognostic for both overall survival and remission duration based on Kaplan-Meier estimator (Figure 4B and 4C). Notably, these signatures defined by the MetaGalaxy analysis rendered more stratification of patients’ outcome compared to traditional approaches of clustering patients based on individual protein expression levels using k-means or hierarchical clustering (Figure S3). Furthermore, protein signature groups remain significant independent prognostic factors after adjusting for common prognostic factors (i.e. age, cytogenetics and white blood cell count) using Cox proportional hazards regression model (Table 1). In this case, three groups of protein signatures (favorable, intermediate and unfavorable) were formed similar to the common practice of forming risk groups from cytogenetics in AML, with each protein signature group consisting of signatures with similar outcome. The robustness of the prognostic implications of the protein signature groups were then validated in a training and testing subset of the data, which demonstrated high consistency (Figure S4). For each signature, proteins that were significantly up or down regulated compared to the normal range were identified as potential drug targets (Figure S5).
Table 1. Cox proportional hazard regression model results.
The model includes common prognostic factors including age at diagnostics (Age.at.Dx), white blood cell count (WBC) and cytogenetic categories (Cytogenetics-Intermediate, Cytogenetics-Unfavorable). The 13 protein signatures were grouped in three categories: Favorable (Signature 6,7,12), Intermediate (Signature 1,2,5,8,9,11,13), Unfavorable (Signature 3,4,10). SE: Standard Error. Sample size n = 205.
| Overall Survival | Remission Duration | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| beta | SE of beta | hazard ratio | z-score | p-value | beta | SE of beta | hazard ratio | z-score | p-value | |
| Age.at.Dx | 0.038 | 0.006 | 1.039 | 6.50 | 8.2e-11 | 0.015 | 0.008 | 1.016 | 1.98 | 0.048 |
| WBC | 0.006 | 0.001 | 1.006 | 4.33 | 1.5e-5 | 0.007 | 0.002 | 1.007 | 3.38 | 0.001 |
| Cytogenetics-Intermediate | 1.107 | 0.405 | 3.026 | 2.74 | 0.006 | 0.659 | 0.411 | 1.933 | 1.60 | 0.109 |
| Cytogenetics-Unfavorable | 1.488 | 0.404 | 4.428 | 3.69 | 2.3e-4 | 1.100 | 0.404 | 3.004 | 2.72 | 0.007 |
| Signature-Favorable | −0.950 | 0.252 | 0.387 | −3.77 | 1.7e-4 | −1.907 | 0.474 | 0.149 | −4.02 | 5.8e-5 |
| Signature-Intermediate | −0.737 | 0.207 | 0.478 | −3.57 | 3.6e-4 | −0.821 | 0.293 | 0.440 | −2.80 | 0.005 |
Signatures are also strongly associated with response to therapy. Primary resistance to induction therapy, observed in 30% of all patients, occurred in over 40% of patients in Signature 1, 5, 6, 7, 10 and 12, but occurred in fewer than 10% of patients in Signature 2, 3 and 4 (Pearson’s chi-square (PCS) of p = 0.004). Attainment of complete remission, seen in 58% of patients overall, occurred in over 80% of patients in Signature 2 and 3, but occurred in only 30% of patients in Signature 10 (PCS of p = 0.039). The occurrence of relapse was 59% of all cases, but was much higher in Signature 1, 3, 4, 9 and 10 (ranging from 75% to 100%) and lower in Signature 7 (27.3%) and 12 (12.5%) (PCS of p = 0.049). The signatures were found not to associate with most clinical factors and genetic mutations, but were associated with the FAB class (PCS of p < 0.001, BH corrected), the percent bone marrow or peripheral blasts, the percent bone marrow or peripheral monocytes (ANOVA of p < 0.001, BH corrected), and FLT3 mutations (PCS of p = 0.06, BH corrected).
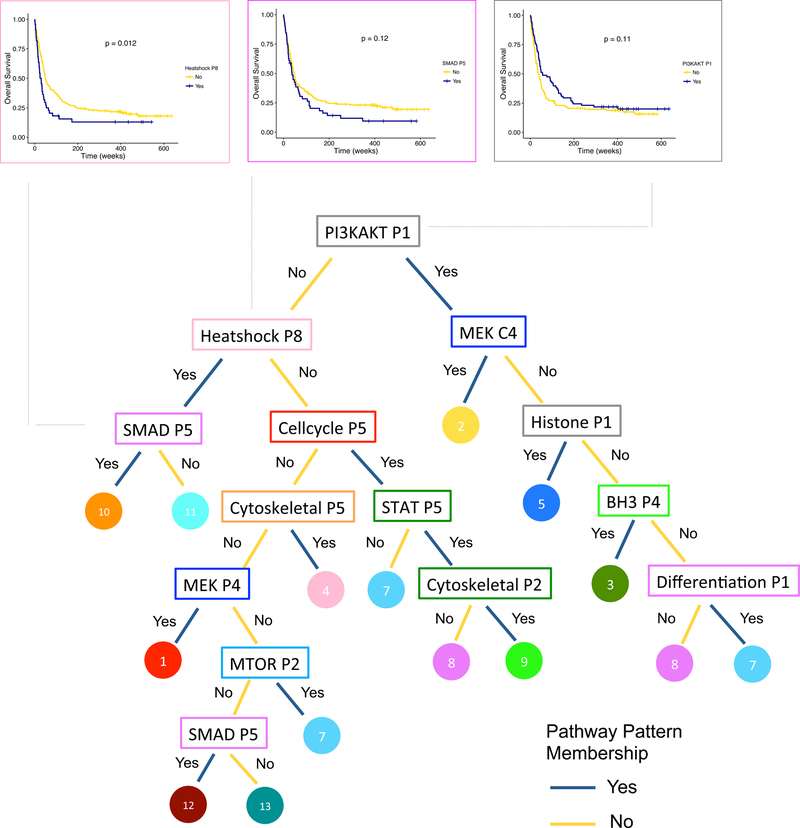
To understand the hallmarks of global heterogeneities in AML, we used a decision tree to identify key functional patterns that distinguish signatures (Figure 5). The 11 functional groups picked out by the tree demonstrate the breadth of the proteomic heterogeneity in AML, covering diverse cellular processes including various cell signaling cascades, cell fate decision, epigenetic regulation, cell structure and motion, and stress response. The hierarchical tree structure clearly illustrates the functional similarities and variations among signatures, as well as illustrates the multiplied prognostic value of functional patterns. For instance, Signature 10, the signature associated with the worst survival outcome, is characterized by a series of adverse functional patterns: not having an outcome-favorable pattern PI3KAKT-P1 and having two unfavorable patterns Heatshock-P8 and SMAD-P5 (Figure 5).
Figure 5. A tree of proteomic hallmarks in AML and its clinical relevance.
The tree illustrates the key functional patterns (in rectangles, color corresponds to constellation color in Figure 3A) that distinguish signatures (in circles, color corresponds to signature color in Figure 3A). The Kaplan-Meier curves for overall survival are shown for PI3KAKT-P1 (right), Heatshock-P8 (top left) and SMAD-P5 (second left), in which the blue curve represents patients displaying the functional pattern, and the yellow curve represents patients not displaying the pattern. PI3KAKT-P1 sample size n = 205 (Yes = 78, No = 127). Heatshock-P8 sample size n = 205 (Yes = 44, No = 161). SMAD-P5 sample size n = 205 (Yes = 51, No = 154). p-values shown for log-rank test, one-sided.
Proteomic matching of leukemic cell lines to AML patient samples
Leukemic cell lines are extensively utilized in the investigation of leukaemia including drug screening, but the conditions that lead to immortalization and stable cell line formation likely induce changes in the biology of these cells that may be divergent from that of native leukaemia blasts. We next generated an RPPA with 111 commonly used leukemic cell line samples (Table S5), and investigated whether these leukemic cell lines fully or partially recapitulate the proteomic profiles in AML patient.
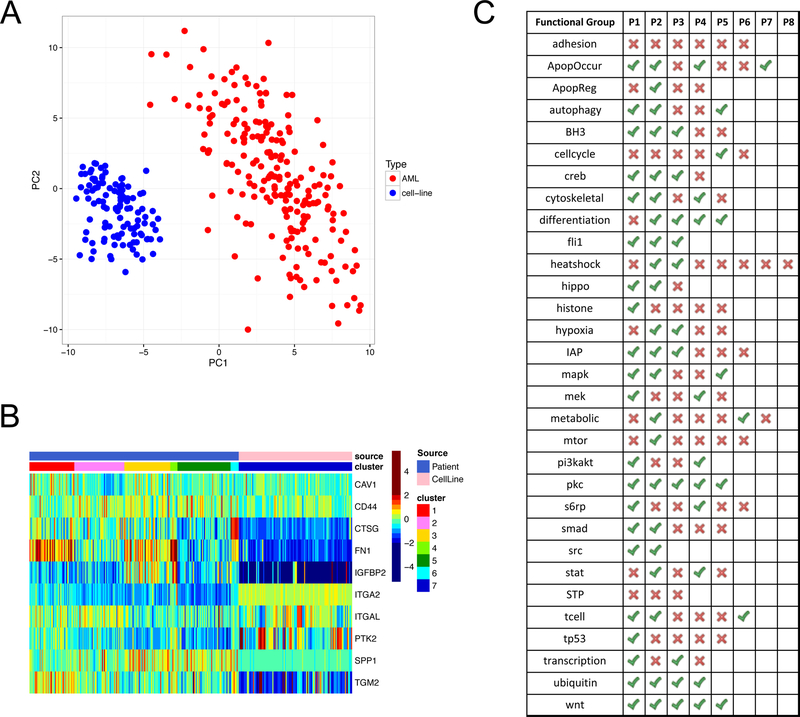
Overall, the proteomic profiles of cell lines are distinct from that of AML patient samples, as seen in both Principal Component Analysis (Figure 6A) and cluster analysis (Figure S6). However, a varying degree of proteomic similarity between cell lines and patient samples were found at the functional levels. In total, 73 (47.4%) of the 154 patient functional patterns have cell line analogues that mimic the expression patterns. Notably, none of the functional patterns in the Adhesion group and the STP group were seen in cell lines, revealing dramatic functional differences between cell lines and patient samples in these two groups. Meanwhile, 5 functional groups (i.e. FLI1, PKC, SRC, Ubiquitin and WNT) have cell line analogues for all of their functional patterns, indicating that cell lines could be a faithful replicate of AML patients for these groups.
Figure 6. Proteomic comparison between AML patients and leukaemia cell lines.
(A) The first and second principal components of the protein expression levels show a complete separation between AML patients and cell lines. Patient sample size (n = 205), cell line sample size (n = 111). (B) The heatmap illustrates the expression patterns of both patients and cell lines in the Adhesion group. None of the cell lines (pink in top row) mimicked any of the 6 functional patterns identified in patients (blue in top row) and were therefore grouped into a new cluster (P7). (C) The table summarizes whether a functional pattern (identified in patients) can be found in any cell line tested in this study. Green ticks indicate that there is at least one cell line that has a similar expression pattern as the functional pattern, and red crosses indicate that no cell lines are found to mimic the functional pattern. Also see Table S5 for detailed matching between all cell lines and functional patterns.
Next, we investigated whether any cell lines mimic the constellations observed from AML patient samples. Since only 47.4% of the functional patterns exist in the cell lines we tested, the chances of finding cell lines that match each constellation were greatly undermined. For constellations made up of a single functional pattern (i.e. Constellation 1 and 4), no cell lines display these patterns since the functional pattern Cellcycle-P6 (Constellation 1) and Heatshock-P8 (Constellation 4) are not captured by any cell lines. Though most constellations do not exist in the cell lines tested in this study, we did observe a weak presence (i.e. presence of at least half of the functional patterns in a constellation) of Constellation 2 and 6 in a few cell lines. For instance, cell lines BV173 and Molm13-P53, mimic 5 out of 9 functional patterns in Constellation 6, which is a combination of Apoptosis-regulating-P3, FLI1-P1, Hippo-P1, Histone-P1, Hypoxia-P3, MEK-P1, PI3KAKT-P1, SRC-P1, and Ubiquitin-P1. In particular, cell line BV173 displays functional patterns of FLI1-P1, Histone-P1, SRC-P1, and Ubiquitin-P1, representing the FLI1-Histone-SRC-Ubiquitin functional group association that we found in AML patient samples. However, none of the 13 signatures are replicated in any of the cell lines.
AML proteome atlas
We have built a web portal (https://www.leukemiaatlas.org/) to make the full analysis results from this study accessible to researchers worldwide. The portal enables researchers to investigate protein expression patterns within AML cells in the context of patients’ clinical and genetic features as well as protein association networks – offering a user-friendly resource to inspire and facilitate new leukaemia studies.
Discussion
We characterized the heterogeneity of protein expression patterns in AML from specific functional groups to integrated functional constellations, which offer us a deeper and richer understanding of the diverse cellular and signaling activities in AML. Despite the great genetic heterogeneities that underlie AML, proteins do form a limited number of recurrent expression patterns in AML, and these patterns are prognostic of outcome. In particular, we found that the association between proteomic patterns and clinical indicators (e.g. cytogenetics, genetic mutations) is limited, and showed, by multivariate analysis, that the proteomic-defined signatures are independent prognostic factors able to further stratify patient subcategories defined by existing clinical practices – both facts support the potential of using proteomics to complement existing patient stratification practices in the clinic. Since many genetic mutations cannot be targeted, the proteomic profiling and target identification from this study provide a means to recognize multiple downstream therapeutic targets and to design combination therapies for individual patients based on their proteomic signatures. For future clinical applications, diagnostic kits can be developed using a reduced set of antibodies for rapid profiling of patients, and simple classifiers such as decision trees can be built to quickly classify patients into protein expression signatures.
Two types of resources are provided by this study to the research community. First, the multi-level proteomic profiles uncovered from this analysis can directly inspire new hypotheses of disease mechanisms, drug development and clinical trials for the AML community and beyond. The AML Proteome Atlas portal we developed can serve as a database for researchers to quickly look up the expression patterns of a functional group or an individual protein of interest and investigate its clinical significance in AML. For researchers specialized in certain protein or pathway studies outside the field of AML, the proteomic profile (the drug target list and cell line matching list in particular) allows them to explore opportunities of repurposing an existing drug for AML.
The second resource is the MetaGalaxy analysis, which can be applied broadly to quantify multi-level expression hallmarks in other cancers and diseases. In contrast to traditional analyses that typically cluster proteins and patients directly (e.g. using hierarchical clustering), the MetaGalaxy analysis uses a two-step approach to first identify patterns in functional groups consisting of functionally related proteins and then combines these patterns to discover protein expression signatures globally. As an analogy, if proteins were cities that people visited over the year, the traditional approach groups small cities together and big cities together, whereas the MetaGalaxy approach provides a map of how small cities are connected to large cities and then uses this map to identify frequent traveling routes both regionally and globally. In taking this relationship-based approach, MetaGalaxy analysis examines protein expressions within the context of other functionally related proteins, and obtains more clinically interesting patient groupings compared to traditional approaches.
Beyond the scope of AML, this study also raises three broader issues that are worth noting for future studies. First, the degree of post-translational modification seen for a given protein does not correlate with the total protein expression levels for most proteins (39 of 47 were independent), indicating that mRNA expression levels may not serve as faithful surrogates for some signaling events. Though gene set enrichment analysis of transcriptome-wide RNA-seq could potentially reach similar pathway-level conclusions drawn from RPPA due to the wide coverage of genes, more studies are merited to compare, contrast and match findings between these two.
Second, the proteomic resemblance between patients and disease-derived cell lines are limited. Cell lines were unable to recapitulate any of the 13 protein signatures seen in AML patients, and they can only recapitulate less than half of all functional patterns. For functional patterns that were recapitulated, these cell lines may serve as faithful analogues for patient samples for experiments designed to test within that functional group. However, cell lines cannot faithfully serve to test hypotheses that involve multiple constellations or signatures. Therefore, cell lines should be selected with caution for future clinical studies so that they are representative of patients’ proteomic patterns. In addition, it would be interesting to investigate whether a similar phenomenon is observed in other cancers.
Third, the proteomic profiles of cryopreserved samples and fresh samples differ dramatically in single protein expression as well as functional pattern utilizations, showing that the freeze-thaw procedure either induces changes in protein expression or acts as a selection mechanism for cryopreservation-tolerant cells which have different starting profiles from cryopreservation-intolerant cells. This observation is consistent with prior research by others. The effects of sample handling, including temperature and cryopreservation on sample integrity and results, are well described throughout the literature in leukaemic cells26,27. Significant changes can occur in mRNA-based gene expression profiling if samples are not kept refrigerated promptly for as little as 4 hours, and similar changes were seen after cryopreservation28. In a proteomic study using assessment by IMAC and SILAC, cryopreservation of three AML cell lines and one patient sample did not lead to “major global proteome and phosphoproteome changes” but did lead to significant changes in many apoptosis, signal transduction and mitochondrial respiratory chain proteins29. Similarly, cryopreservation was demonstrated to alter the expression of CD34 class 1 epitopes in AML progenitor cells30, and in another study led to the loss of phospho-STAT signals in some cases31.
To our knowledge, this study is unique in its evaluation of the differences between fresh and cryopreserved proteomes in a large cohort of human clinical samples. Although differences were noted for some individual proteins, the clear relationship of proteomic patterns to cryopreservation was apparent only upon clustering of each protein functional group, where clusters populated solely by cryopreserved samples were observed. This finding has important implications for cooperative group trials relying on samples shipped under variable conditions and processed after variable time delays, or after cryopreservation, when the analyte is labile, such as protein, mRNA, miRNA or metabolites. Careful consideration must be given to these preanalytic effects. It was for this reason that we opted to only consider the freshly prepared samples for inclusion in this report. While a full analysis of the changes associated with cryopreservation is beyond the scope of this manuscript, future studies are merited to further investigate the effects of cryopreservation and to establish consensus on how discoveries from cryopreserved samples can be translated to valid conclusions for fresh samples.
The difference between fresh and cryopreserved samples also raises an important concern about the normal CD34+ controls utilized in this study as they utilized cryopreserved specimens. When this array was constructed, the normal CD34+ supplier only provided cryopreserved material. We arranged to perform another RPPA that compared expression in fresh samples (which were stored on ice, shipped and processed within 24 hours) to the identical samples cryopreserved for a month and prepared in a similar manner to the cryopreserved specimens used in this study. While the unbiased hierarchical clustering separated fresh from cryopreserved normal CD34+ cells, about half of the proteins were unaffected, and of those that were altered 85% still had expression within the normal range. For proteins that were altered in fresh vs. cryopreserved normal CD34+ cells, the manner in which protein expression changed was not uniform across samples and hence no direct normalization correction was feasible. Notably, in this study, the expression patterns were solely dependent on the data of the AML samples, whereas the normal CD34+ cells were used only as a reference for what levels of expression in the AML samples were outside the normal range. Therefore, little would be anticipated to change in this analysis had fresh normal CD34+ comparators been available.
Methods
Patient demographics and treatment
A summary of the patient demographics is shown in Table S6. Among the 205 AML patients with available fresh samples and that were treated at MDACC and evaluable for outcome, 155 patients received high dose ara-C (HDAC) (109 with an anthracycline, 31 with fludarabine (FLAG, FA), 11 with clofarabine, and 4 with other agents); 12 patients received standard dose ara-C (10 with clofarabine and 2 with zarnestra); one patient received a low dose ara-C based regimen. Idarubicin and troxacitabine was used in 2 cases and VNP40101M was used for 6 cases. Demethylating or histone deacetylating agents were used alone or in combination for 18 patients. Targeted agents were utilized in 11 cases, among which 5 received Gemtuzumab Ozogamicin (GO) in combination with Interleukin-11, and 6 received phase 1 agents. Of all 205 treated cases, 118 (57.6%) achieved complete remission (CR), among which 69 (58.5%) relapsed and 39 are alive as of July 2015. Thirty-three underwent allogeneic stem cell transplantation from a related (n=18) syngeneic (n=1) or unrelated (n=14) donor, after primary resistance (n=8), in first CR (n=8) or after relapsing (n=15).
Patient sample collection and preparation
Samples were acquired during routine diagnostic assessments in accordance with the regulations and protocols (Lab 01–473) approved by the Investigational Review Board (IRB) of MDACC. Informed consent was obtained in accordance with Declaration of Helsinki. Samples were analyzed under an IRB-approved laboratory protocol (Lab 05–0654, MD Anderson Cancer Center IRB Board). Samples were enriched for leukaemic cells by performing Ficoll separation to yield a mononuclear fraction followed by CD3/CD19 depletion to remove contaminating T and B cells, if they were calculated to be > 5% based on the post Ficoll differential. The samples were normalized to a concentration of 1×104 cells/μL and a whole cell lysate was prepared as described in publications11,21. In addition to AML patient samples, each array includes the same ten cryopreserved normal bone marrow CD34+ samples (NLBM CD34+) from healthy subjects (AllCells, Alameda, CA).
Mutation Analysis
All mutation analysis for RAS, and mutation analysis for FLT3 and NPM1 in patients accrued after the recognition of these mutations in AML, were performed as part of routine diagnostic studies in a CLIA certified lab. For cases with available DNA, mutation analysis for IDH1, IDH2, TP53 and DNMT3a, as well as analysis for FLT3 and NPM1 for older cases, not routinely studied, was performed by PCR amplification of known hot spot regions followed by routine sequencing (S.M.K. Lab). Additionally, 47 cases were also analyzed by the Foundation of Medicine Heme panel covering 405 genes and 265 translocations. In total, mutation data was available for FLT3-ITD (n = 197), FLT3-D835Y (n =196), RAS (n = 178), NPM1 4 basepair insertion (n = 165), TP53 (n = 55), IDH1 or 2 (n = 145), and DNMT3a (n = 133). Details on the proteomic patterns related to mutational data is provided (Figure 4, Supplemental Table S6).
RPPA methodology
The methodology and validation of the technique are fully described in publications11,21. Briefly, patient samples were printed in 5 serial dilutions onto slides along with normalization and expression controls. Slides were probed with 230 strictly validated primary antibodies and a secondary antibody to amplify the signal, and finally a stable dye is precipitated32. Two antibodies were excluded due to poor array quality, resulting in a proteomic profile of 228 antibodies. This included antibodies against 169 different proteins along with 52 antibodies targeting phosphorylation sites, 6 targeting Caspase and Parp cleavage forms and 3 targeting histone methylation sites. The manufacturer and the antibody name, along with the primary and secondary antibody concentrations utilized, are listed in Table S1. The stained slides were analyzed using Microvigene® software (Vigene Tech, Carlisle, MA) to produce quantified data.
Antibody Nomenclature
Since neither HUGO33, HUPO34 or MiMI35 account for post-translational modifications, we developed a nomenclature in which the HUGO name is followed by a period, then the type of post-translational modification, “p” for phosphorylated, “cl” for cleaved or “Me” for methylation, followed by the letter code for the affected amino acid and its sequence position. For example, AKT1.pT308 is AKT1 phosphorylated on Threonine at position 308. Placing the post-translational modifications after the protein name enables alphabetical sorting and inclusion of the affected site, which is impossible if it comes before the protein name.
Cell line selection, array preparation and processing
The cell line array included 111 commonly used leukaemic cell line samples from AML (e.g. U937, HL-60, KG-1, ML-1, OCIAML3), APL (e.g. MR2), ALL (e.g. Jurkat), CML (e.g. KBM5) and Lymphoma (e.g. Raji). (e.g. U937, HL-60, KG-1, ML-1, OCIAML3), APL (e.g. MR2), ALL (e.g. Jurkat), CML (e.g. KBM5) and Lymphoma (e.g. Raji). To cover a wide range of cell lines that are used in various laboratories worldwide, we particularly included cell line variants that contain different molecular modifications. Additionally, mycoplasma infection is a common problem in cell culture, but its effect on cell biology is not well defined. We therefore included mycoplasma infected cell lines along with a post-treatment mycoplasma-free version on the array. The array was probed with 181 antibodies that were in overlap with the AML patient array. The raw data was first processed using the same computational procedures as used for the AML patient array. Similar to the normalization procedure applied for the AML patient array, we then mean normalized the expression levels of each protein using the mean expression level of the cryopreserved normal bone marrow CD34+ samples included on the array, which enabled us to compare the cell line array to the AML patient array. Since the overall expression patterns of cell lines and primary samples were found to be completely separated (Figure 6A) and since cell lines are assigned to a functional pattern independently, the inclusion of non-AML cell lines and mycoplasma infected cells in this array does not affect our conclusions drawn on AML cell lines.
Data processing, normalization and source comparison
The raw data from the array was first processed by multiple computational steps to ensure proper slide alignment36, background noise control37 and sample loading control38. Supercurve algorithms were used to generate a single value from the 5 serial dilutions39. Loading control38 and topographical normalization37 procedures were performed to account for protein concentration and background staining variations on each array. Since most samples have replicates printed on the same slide, the mean average expression level of all replicates was used as a single expression level for each sample. Replicates-based Normalization (RBN)36 was used to align samples from two different slides. The concordance correlation between sample replicates on each slide was assessed to ensure single slide quality, and the correlation between duplicated samples on different slides was checked to ensure slide alignment quality. In general, each protein array demonstrated high concordance between sample replicates printed on two separate slides with a median concordance correlation coefficient (CCC) of 0.91. Two antibodies, GRP78 and CASP9, were excluded from this study due to poor concordances (GRP78 CCC = 0.23, CASP9 CCC = 0.31).
For each protein, the expression levels for all samples were subtracted by the mean of the normal bone marrow CD34+ samples printed on the same array. Protein expression differences between sources (i.e. bone marrow vs. blood, fresh vs. cryopreserved) were assessed using two-sided t-test, and the p-values were corrected for multiple testing to control the FDR at 5% using Benjamini-Hochberg Procedure22.
MetaGalaxy analysis
The MetaGalaxy analysis is a computational framework (Figure 1), consisting of a suite of statistical and machine learning methods, to characterize expression patterns and patient heterogeneities. As a pre-processing step, a set of functional groups were formed by including proteins with similar functionalities and pathway associations based on the KEGG database40 as well as by including proteins with strong correlation with each other within the dataset. Most functional groups correspond to conventional pathways. Since the apoptosis pathway contained too many protein members with patterns in one set of members potentially obscuring patterns in other members, we divided these proteins into multiple smaller functional groups related to sub-components of the pathway (i.e. ApopOccur, ApopReg, BH3 and IAP). As a result of this expert-driven procedure, 31 functional groups were created from the 228 proteins covered by the RPPA (see Table S3 for memberships of each group).
The MetaGalaxy analysis first focuses on each functional group to identify functional expression patterns regionally, a step termed Functional Patterning. In the Functional Patterning step, the cluster analysis was first performed for all samples (including both fresh and cryopreserved) using a combination of k-means41 (for generating cluster memberships) and Progeny Clustering23 (a bootstrapping and stability based method for selecting cluster number), from which the clustering results of fresh samples were extracted. The functional patterns (i.e. clusters) were ordered based on their similarities (i.e. Euclidean distance between the cluster centers) to the normal CD34+ samples, with Functional Pattern 1 (P1) being the most similar to normal CD34+ samples. Survival curves were generated using the Kaplan-Meier method, and the p-values were corrected for multiple testing (across the total number of functional groups) to control FDR at 5% using BH procedure22. The associations between cluster memberships and categorical clinical variables were assessed by Pearson’s Chi-squared test. The associations between cluster memberships and continuous clinical variables were assessed by one-way ANOVA. The protein network was built by combining literature-based protein interactions queried from the database STRING42 and proteomics-based protein interactions inferred from the RPPA data using graphical lasso43 and StARS44 (for model selection based on stability). Graphical lasso was chosen as the reverse network inference algorithm of chose as it is able to handle the static form of RPPA data present in this dataset, and it has been extensively vetted43,45,46. We have applied other protein network building algorithms in the past to RPPA datasets, and alternative methods could be chosen based on a user’s preference47–49. The edges of the network help provide a way to visualize the signaling pathways involved, while the edge associations do not contribute to the remainder of the MetaGalaxy analysis. Since the STRING database does not consider post-translational modifications, the protein names were used to query literature-based interactions for post-translational modification sites.
Based on these functional patterns, the analysis then pieces functional patterns together to discover global expression patterns, a step termed Constellation Patterning. In the Constellation Patterning step, binary block clustering50 was used to co-cluster functional patterns and patients. The optimal cluster numbers were determined using Progeny Clustering23, which uses bootstrapping to assess clustering solution stability. The survival analysis and clinical association analysis were conducted using the same methods as in Functional Patterning. The decision tree was built using CART51 (Classification and Regression Trees) with Gini index. The permutation test was performed by repeatedly randomizing the functional pattern assignments and co-clustering the randomized data with the same number of clusters for one hundred iterations. The likelihood of observing certain functional pattern coalition is the number of occurrences divided by the total number of repeats. Note that all clustering algorithms used in this study are unsupervised. The implementation and parameter specification of MetaGalaxy analysis is summarized in Table S7. A tutorial and demo files for the online tool implementing MetaGalaxy are provided in the Supplement and also available online at https://www.leukemiaatlas.org/code.
Supplementary Material
Acknowledgments
This research was funded in part by a translational research grant from the Leukemia and Lymphoma Society to S.M.K., NSF CAREER 1150645, NSF NCS 1533708, and NIH R01 GM106027 grants to A.A.Q., and a HHMI Med-into-Grad fellowship to C.W.H.
Footnotes
Code availability: Access to all code used in the MetaGalaxy analysis is listed in Supplemental Table S7. In particular, Progeny Clustering code is available on the R repository: https://cran.r-project.org/web/packages/progenyClust/index.html, and the MetaGalaxy pipeline is available in an R Shiny portal: https://meta-pathway-analysis.shinyapps.io/mpa_r/, with a corresponding tutorial and demo files (main.csv and pathway.csv) available in the Supplement and online at the Leukemia Proteome Atlas (https://www.leukemiaatlas.org/code).
Data availability: The datasets generated and analysed during the study are available on the Leukaemia Proteome Atlas: LeukemiaAtlas.org (direct dataset-download at https://www.leukemiaatlas.org/adultaml). Source data for the figures in this study are also provided in .xlsx and .csv formats in figshare at https://figshare.com/s/5ec75fb971747383c0be52 and are accessible directly from the Atlas (https://www.leukemiaatlas.org/adultaml).
Competing interests
The authors declare no competing interests.
References
- 1.Hanahan D & Weinberg RA The hallmarks of cancer. Cell 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674, doi: 10.1016/j.cell.2011.02.013 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Society, A. C. Cancer Facts & Figures 2018 (Atlanta: American Cancer Society; ). (2018). [Google Scholar]
- 4.Byrd JC et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 100, 4325–4336, doi: 10.1182/blood-2002-03-0772 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Grimwade D et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood 92, 2322–2333 (1998). [PubMed] [Google Scholar]
- 6.Slovak ML et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood 96, 4075–4083 (2000). [PubMed] [Google Scholar]
- 7.Mardis ER et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361, 1058–1066, doi: 10.1056/NEJMoa0903840 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research, N. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368, 2059–2074, doi: 10.1056/NEJMoa1301689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valk PJ et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 350, 1617–1628, doi: 10.1056/NEJMoa040465 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Paweletz CP et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 20, 1981–1989, doi: 10.1038/sj.onc.1204265 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Tibes R et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther 5, 2512–2521, doi: 10.1158/1535-7163.MCT-06-0334 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Masuda M & Yamada T Signaling pathway profiling by reverse-phase protein array for personalized cancer medicine. Biochim Biophys Acta 1854, 651–657, doi: 10.1016/j.bbapap.2014.10.014 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Byers LA et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2, 798–811, doi: 10.1158/2159-8290.CD-12-0112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey MS et al. Functional proteomic analysis of advanced serous ovarian cancer using reverse phase protein array: TGF-beta pathway signaling indicates response to primary chemotherapy. Clin Cancer Res 16, 2852–2860, doi: 10.1158/1078-0432.CCR-09-2502 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubb RL et al. Signal pathway profiling of prostate cancer using reverse phase protein arrays. Proteomics 3, 2142–2146, doi: 10.1002/pmic.200300598 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Nishizuka S et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A 100, 14229–14234, doi: 10.1073/pnas.2331323100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Angulo AM et al. Open-label randomized clinical trial of standard neoadjuvant chemotherapy with paclitaxel followed by FEC versus the combination of paclitaxel and everolimus followed by FEC in women with triple receptor-negative breast cancerdagger. Ann Oncol 25, 1122–1127, doi: 10.1093/annonc/mdu124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierobon M et al. Pilot phase I/II personalized therapy trial for metastatic colorectal cancer: evaluating the feasibility of protein pathway activation mapping for stratifying patients to therapy with imatinib and panitumumab. J Proteome Res 13, 2846–2855, doi: 10.1021/pr401267m (2014). [DOI] [PubMed] [Google Scholar]
- 19.Posadas EM et al. A phase II and pharmacodynamic study of gefitinib in patients with refractory or recurrent epithelial ovarian cancer. Cancer 109, 1323–1330, doi: 10.1002/cncr.22545 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornblau SM et al. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin Cancer Res 16, 1865–1874, doi: 10.1158/1078-0432.CCR-09-2551 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornblau SM et al. Functional proteomic profiling of AML predicts response and survival. Blood 113, 154–164, doi: 10.1182/blood-2007-10-119438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol, 289–300 (1995).
- 23.Hu CW, Kornblau SM, Slater JH & Qutub AA Progeny Clustering: A Method to Identify Biological Phenotypes. Sci Rep 5, 12894, doi: 10.1038/srep12894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris KF et al. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc Natl Acad Sci U S A 96, 13738–13743 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim NG & Gumbiner BM Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J Cell Biol 210, 503–515, doi: 10.1083/jcb.201501025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gjertsen BT et al. Analysis of acute myelogenous leukemia: preparation of samples for genomic and proteomic analyses. J Hematother Stem Cell Res 11, 469–481, doi: 10.1089/15258160260090933 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Rai AJ et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics 5, 3262–3277, doi: 10.1002/pmic.200401245 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Dvinge H et al. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc Natl Acad Sci U S A 111, 16802–16807, doi: 10.1073/pnas.1413374111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aasebo E et al. Freezing effects on the acute myeloid leukemia cell proteome and phosphoproteome revealed using optimal quantitative workflows. J Proteomics 145, 214–225, doi: 10.1016/j.jprot.2016.03.049 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Lanza F et al. Assessment of distribution of CD34 epitope classes in fresh and cryopreserved peripheral blood progenitor cells and acute myeloid leukemic blasts. Haematologica 84, 969–977 (1999). [PubMed] [Google Scholar]
- 31.Xia Z, Baer MR, Block AW, Baumann H & Wetzler M Expression of signal transducers and activators of transcription proteins in acute myeloid leukemia blasts. Cancer Res 58, 3173–3180 (1998). [PubMed] [Google Scholar]
- 32.Hunyady B, Krempels K, Harta G & Mezey E Immunohistochemical signal amplification by catalyzed reporter deposition and its application in double immunostaining. J Histochem Cytochem 44, 1353–1362 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Eyre TA et al. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res 34, D319–321, doi: 10.1093/nar/gkj147 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hermjakob H et al. The HUPO PSI’s molecular interaction format--a community standard for the representation of protein interaction data. Nat Biotechnol 22, 177–183, doi: 10.1038/nbt926 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Jayapandian M et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res 35, D566–571, doi: 10.1093/nar/gkl859 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbani R et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun 5, 3887, doi: 10.1038/ncomms4887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neeley ES, Baggerly KA & Kornblau SM Surface Adjustment of Reverse Phase Protein Arrays using Positive Control Spots. Cancer Inform 11, 77–86, doi: 10.4137/CIN.S9055 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neeley ES, Kornblau SM, Coombes KR & Baggerly KA Variable slope normalization of reverse phase protein arrays. Bioinformatics 25, 1384–1389, doi: 10.1093/bioinformatics/btp174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J et al. Non-parametric quantification of protein lysate arrays. Bioinformatics 23, 1986–1994, doi: 10.1093/bioinformatics/btm283 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartigan JA & Wong MA Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Soc. Ser. C Appl. Stat, 100–108 (1979).
- 42.Franceschini A et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research 41, D808–D815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman J, Hastie T & Tibshirani R Sparse inverse covariance estimation with the graphical lasso. Biostatistics 9, 432–441, doi: 10.1093/biostatistics/kxm045 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Roeer K & Wasserman L Stability approach to regularization selection (stars) for high dimensional graphical models. Advances in Neural Information Processing Systems, 1432–1440 (2010). [PMC free article] [PubMed]
- 45.Zuo Y, Cui Y, Yu G, Li R & Ressom HW Incorporating prior biological knowledge for network-based differential gene expression analysis using differentially weighted graphical LASSO. BMC Bioinformatics 18, 99, doi: 10.1186/s12859-017-1515-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sulaimanov N & Koeppl H Graph reconstruction using covariance-based methods. EURASIP J Bioinform Syst Biol 2016, 19, doi: 10.1186/s13637-016-0052-y (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill SM et al. Inferring causal molecular networks: empirical assessment through a community-based effort. Nat Methods 13, 310–318, doi: 10.1038/nmeth.3773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.York H, Kornblau SM & Qutub AA Network analysis of reverse phase protein expression data: characterizing protein signatures in acute myeloid leukemia cytogenetic categories t(8;21) and inv(16). Proteomics 12, 2084–2093, doi: 10.1002/pmic.201100491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kornblau SM et al. Proteomic profiling identifies distinct protein patterns in acute myelogenous leukemia CD34+CD38- stem-like cells. PLoS One 8, e78453, doi: 10.1371/journal.pone.0078453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Govaert G & Nadif M Clustering with block mixture models. Pattern Recognition 36, 463–473 (2003). [Google Scholar]
- 51.Therneau T, Atkinson B & Ripley B rpart: Recursive Partitioning and Regression Trees. R package version 41–10 (2015). [Google Scholar]
- 52.Hu CW et al. Dataset for “A quantitative analysis of heterogeneities and hallmarks in acute myelogenous leukaemia.”. figshare https://figshare.com/s/5ec75fb971747383c0be doi: https://figshare.com/s/5ec75fb971747383c0be (2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.