ABSTRACT
High-grade neuroendocrine lung carcinomas (LCNEC, SCLC) are recalcitrant cancers for which no optimal management has been achieved. We have recently described two models of LCNEC and SCLC developed upon inactivation of 4 tumor suppressors genes (Rb1 (RB transcriptional corepressor 1), Rbl1 (RB transcriptional corepressor like 1), Pten (phosphatase and tensin homolog), Trp53 (transformation-related protein 53), which provide a suitable frame for preclinical intervention. A defined model for LCNEC had not been previously reported.
KEYWORDS: Lung cancer, LCNEC, SCLC, cell of origin, tumor suppressors, Rb1, Rbl1, Trp53, Pten, molecular imaging
Lung cancer is the leading cause of cancer-related deaths worldwide. Pulmonary neuroendocrine tumors are a subset of aggressive lung cancers, which are comprised of small-celllung carcinoma (SCLC), large-cell neuroendocrine carcinoma (LCNEC), atypical carcinoma (AC) and typical carcinoma (TC). SCLC and LCNEC are classified as high-grade neuroendocrine carcinomas and account for 15% and 3% of all cancer types, respectively.1 Furthermore, the relationship between them is a matter of debate. Critically, their survival rate is very poor and chemotherapy remains the first-line treatment option. The majority of cases are diagnosed at advanced stages, highlighting the urgent need to design better screening modalities and to understand the early events that underlie tumor progression. The paucity of human material from surgery makes mouse models of these diseases essential tools for basic research and preclinical intervention.
For these reasons, we have generated new mouse models of LCNEC and SCLC based on the loss of four tumor suppressor genes: Rb1, (RB transcriptional corepressor 1), Rbl1 (RB transcriptional corepressor like 1), Pten (phosphatase and tensin homolog), Trp53 (transformation-related protein 53). Mutations in TP53 and RB1 are a hallmark of human SCLC and LCNEC; PTEN alterations have also been identified in SCLC. SCLC models based on Rb1/Trp53 loss have been described (reviewed in Semenova et al.2), as well as models based on Rb1/Trp53/Pten loss rendering a mixture of SCLC, LCNEC, and NSCLC.3 We investigated the role of Rbl1 in the development of SCLC and LCNEC and developed both types of high-grade neuroendocrine carcinomas upon the inactivation of four tumor suppressors. Our study reports for the first time, a robust model of LCNEC, an under-studied type of lung cancer with no defined clinical management to date. The reduced latency, high incidence and uniform progression of both immunocompetent models provide unique tools that will facilitate the design and testing of novel tumor intervention strategies against these cancer types. In fact, their use in potential therapeutic studies is exemplified in the study with the use of novel molecular imaging methods based on 68Ga-DOTA peptides.4
Lung tumors were induced by intra-tracheal injection of Adeno-cre virus in an Rbl1-null background with Rb1, Pten and Trp53 floxed alleles (QKO, quadruple Knock-out mice). This procedure has proven to be a powerful method for modeling lung cancer in mice. We used two different approaches: either an Ad5-CMV-cre virus (which directs cre-mediated recombination in a wide variety of lung epithelial cells) or an Ad5-K5-cre virus (which targets cre-mediated recombination specifically to basal progenitor cells). Interestingly, the tumor spectrum was skewed depending on the type of cells switched (adeno-cre virus injected). Ad5-CMV-cre infection led to the development of LCNEC. However, the same gene modification (even in littermates) targeting a specific cell type residing in basal cells of tracheal epithelium or main bronchi and expressing keratin K5, yielded a different mouse model. Inactivation of the four tumor suppressors exclusively in basal K5 expressing cells, rendered a robust model of small-cell lung cancer. This constitutes the central finding of our work: delivery of cre recombinase under the control of promoters active in different cell types alters the distribution of the resulting lung cancers. It also implies that the two tumor classes generally arise from different cell types (Figure 1).
Figure 1.
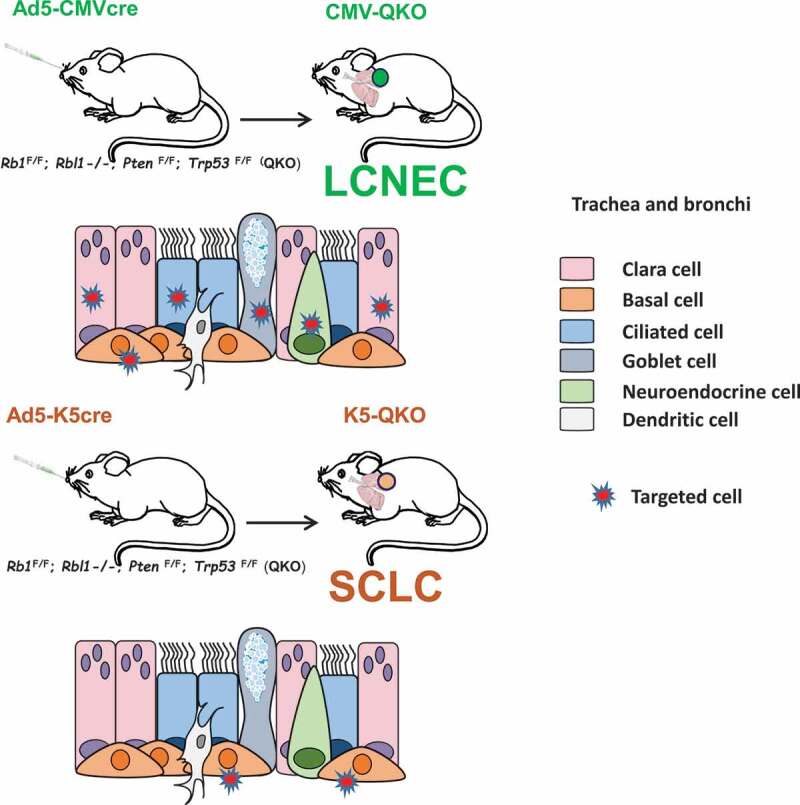
Genetic approach to generate new models of large-cell neuroendocrine carcinoma, LCNEC and small-cell lung carcinoma, SCLC. Upper panel: Rb1F/F (RB transcriptional corepressor 1); Rbl1-/- (RB transcriptional corepressor like 1); PtenF/F (phosphatase and tensin homolog); Trp53F/F (transformation-related protein 53) (quadruple mutant mice, QKO) mice were intratracheally infected with Ad5-CMVcre adenovirus (CMV-QKO), which targets all types of epithelial cells in the lungs, and large-cellneuroendocrine carcinomas were developed. Lower panel: QKO mice were infected with cell-type-restricted Ad5-K5cre adenovirus (K5-QKO), which targets specifically basal cells in trachea and bronchi, and small-cell lung carcinomas arose.
Mouse models have contributed to the identification of the cells at the origin of distinct tumor types. Studies using cell-type specific-cre recombinase conditional knockout mice have identified neuroendocrine cells as cells of origin of SCLC. Sutherland et al.5 showed that the CGRP (calcitonin related polypeptide alpha) promoter which is active specifically in neuroendocrine cells gave rise to SCLC. In that study, the same genetic disruption of Rb1 and Trp53 in alveolar type II cells lead, to a minor extent, to the development of SCLC, thus defining the lung lineages from which SCLC arises.2 Our study provides evidence that keratin 5 expressing basal cells act as a cell of origin of SCLC. Unfortunately, Ad5-CMVcre targeting of lung epithelial cells does not allow the identification of a specific cell type, if any, acting as a cell of origin of LCNEC. In addition, different cell lineages are at the origin of the adenocarcinoma6,7 and lung squamous cell carcinoma tumor types,8 which highlights the plasticity of different putative cancer stem cell compartments in the lung.
We went on to characterize the molecular basis of mouse LCNEC and SCLC tumors and the relationships between them, as well as with other mouse SCLC described models and with their human counterparts. The data revealed significant differences at the gene expression level between both mouse tumor types, and a significant similarity between our SCLC model and a well-established Trp53/Rb1 mouse model.9 A particularly significant correlation was observed with human LCNEC and SCLC gene expression profiles using a human LCNEC/SCLC classifier.10 Mutation burden is much lower than that reported in human tumors.10 It seems that the loss of the four tumor suppressors and the targeted initiating cells are the drivers that determine the type of tumor to be developed over mutation burden, which poses an interesting question about their impact on the development of these tumors, at least in mice.
Finally, we investigated the possibility of early detection by means of a noninvasive molecular imaging modality. Taking advantage of the expression of the somatostatin receptors (SSTR) in neuroendocrine tumors and their ability to bind 68Ga-DOTA-peptides, we showed that mouse SCLC and LCNEC clearly had a remarkably higher avidity for 68Ga-DOTATOC than for the most widely used radiotracer 18F-FDG) (18F-fluorodeoxyglucose). This finding holds the potential to detect and stratify patients harboring neuroendocrine lung cancers.
The significance of our work lies in the novelty of the first described mouse model for LCNEC, the differentiation of its origins from the origins of SCLC, the identification of a new cell acting as a cell of origin of SCLC, and the description of a potentially useful imaging tool for such tumors. Our study is the beginning of a journey toward the development and testing of (novel) therapeutic interventions for LCNEC and SCLC.
Funding Statement
This work was funded by projects PI12/01959, PI15/00993, PI18/000263, and CIBERONC CB/16/00228 from the Instituto de Salud Carlos III (Ministry of Science, Innovation and Universities) and cofunded by the European Regional Development Fund.
Acknowledgments
A special thanks goes to the researches and assistants involved along the development of the work, Dr. Corina Lorz for critical reading of the manuscript and Norman Feltz for copyreading the manuscript.
Disclosure of potential conflicts of interest
The author reports no conflict of interest.
References
- 1.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1–3. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 2.Semenova EA, Nagel R, Berns A.. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, MacPherson D, McFadden DG, Farago A, Jacks T, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol. 2015;10:553–564. doi: 10.1097/JTO.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazaro S, Perez-Crespo M, Lorz C, Bernardini A, Oteo M, Enguita AB, Romero E, Hernandez P, Tomas L, Morcillo MA, et al. Differential development of large-cell neuroendocrine or small-cell lung carcinoma upon inactivation of 4 tumor suppressor genes. Proc Natl Acad Sci USA. 2019;116:22300–22306. doi: 10.1073/pnas.1821745116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111:4952–4957. doi: 10.1073/pnas.1319963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mainardi S, Mijimolle N, Francoz S, Vicente-Duenas C, Sanchez-Garcia I, Barbacid M. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci USA. 2014;111:255–260. doi: 10.1073/pnas.1320383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Danes A, Blanpain C. Deciphering the cells of origin of squamous cell carcinomas. Nat Rev. 2018;18:549–561. doi: 10.1038/s41568-018-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, Sweet-Cordero EA, Sage J. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 2010;70:3877–3883. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George J, Walter V, Peifer M, Alexandrov LB, Seidel D, Leenders F, Maas L, Muller C, Dahmen I, Delhomme TM, et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat Commun. 2018;9:1048. doi: 10.1038/s41467-018-03099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]


