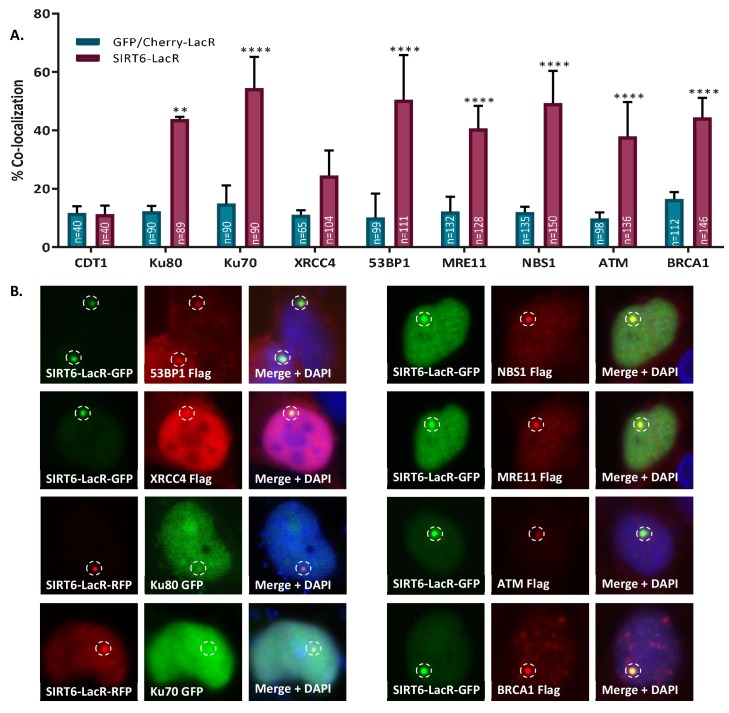
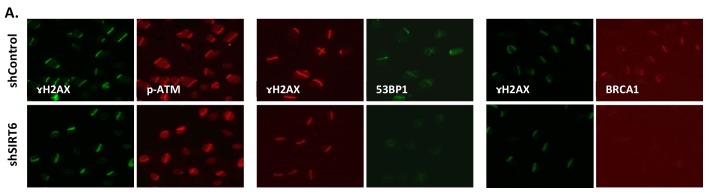
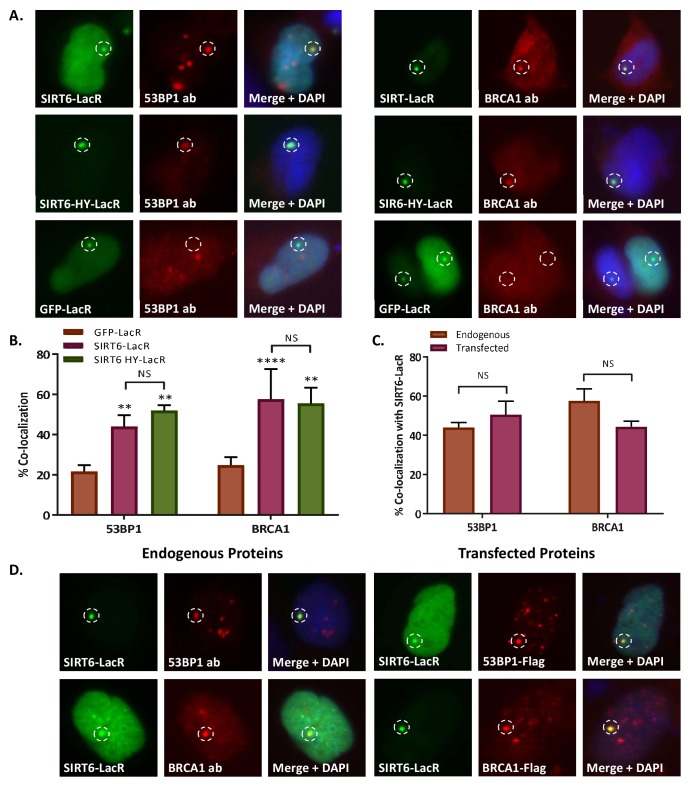
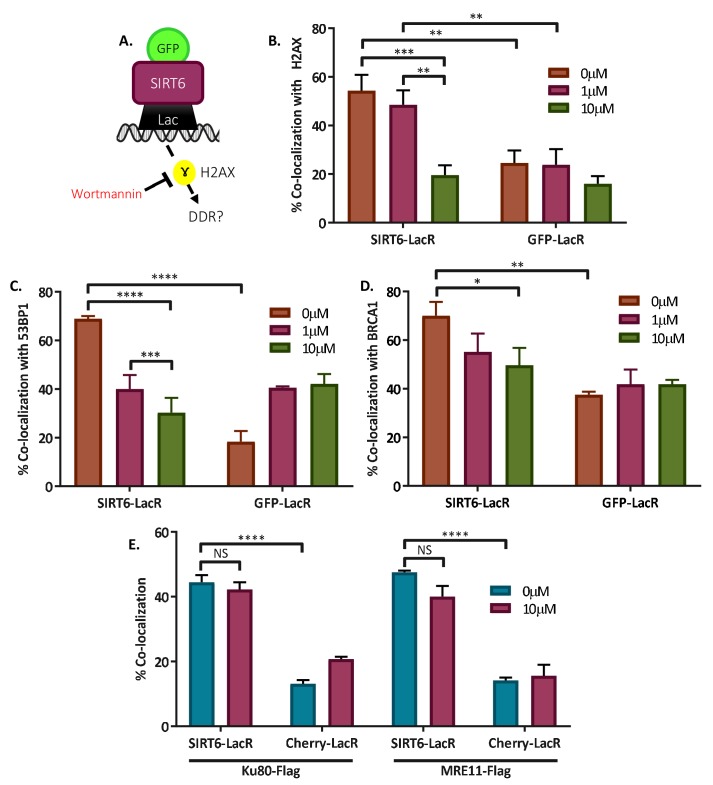
(A) Schematic representation of the Tethering assay with inhibition of signaling using Wortmannin. (B–D) Co-localization of γH2AX (B), 53BP1 (C), and BRAC1 (D) with SIRT6-LacR-GFP or GFP/Cherry-LacR after 24 hr with or without Wortmannin. Data are averages for three experiments (error bars are SEMs). (B) Co-localization with γH2AX (0 μM: n[SIRT6] = 43, n[GFP] = 41; 1 μM: (n[SIRT6] = 42, n[GFP] = 42; 10 μM: (n[SIRT6] = 42, n[GFP]=41). (C) Co-localization with 53BP1 (0 μM: n[SIRT6] = 32, n[GFP] = 32; 1 μM: n[SIRT6] = 30, n[GFP] = 32; 10 μM: n[SIRT6] = 33, n[GFP] = 31). (D) Co-localization with BRCA1 (0 μM: n[SIRT6] = 30, n[GFP] = 32; 1 μM: n[SIRT6] = 31, n[GFP] = 31; 10 μM: n[SIRT6] = 32, n[GFP] = 31). (E) Co-localization of Ku80-Flag or MRE11-Flag with SIRT6-LacR-GFP or GFP/Cherry-LacR after 24 hr with or without Wortmannin. Data are averages for three experiments (error bars are SEMs). For Ku80-Flag — 0 μM: n[SIRT6] = 45, n[Cherry] = 53; 10 μM: n[SIRT6] = 45, n[Cherry] = 58. For MRE11-Flag — 0 μM: n[SIRT6] = 50, n[Cherry] = 56; 10 μM: n[SIRT6] = 60, n[Cherry] = 51. *, p<0.05; **, p<0.005; ***, p<0.0005; ****, p<0.00005).