ABSTRACT
A significant association between high blood-based tumor mutational burden (bTMB) and improved progression-free survival (PFS) was observed in advanced non-small cell lung cancer (NSCLC) receiving atezolizumab. However, this result was unrepeatable in a recent prospective study. We hypothesized that there might be a non-linear association between bTMB and survival. This study used the clinical and genetic data from POPLAR (n = 105, training set) and OAK (n = 324, validation set) trials. The non-linear association between bTMB and survival was assessed using restricted cubic spline (RCS). The cutoff values for bTMB were calculated via X-tile software. Non-linear relationships were observed between bTMB and PFS and overall survival (OS) in RCS plots (both P non-linearity < 0.001). The optimal cutoff values of bTMB for predicting PFS and OS were 7 and 14 mutations/Mb, respectively. The median PFS and OS of patients with low and high bTMB were significantly longer than those of patients with medium bTMB in the training, validation, and combined sets. Low and high bTMB were also associated with longer PFS and OS in high-programmed death-ligand 1 (PD-L1) expression population. In conclusion, there was a positive non-linear association between bTMB and survival in NSCLC patients receiving atezolizumab. Patients with low bTMB could also derive benefit from immunotherapy.
KEYWORDS: Non-small cell lung cancer, blood tumor mutation burden, atezolizumab, prognosis
Introduction
Immune checkpoint inhibitors (ICIs), including anti–programmed cell death-1 (anti-PD-1) and anti–programmed death-ligand 1 (anti-PD-L1) blockade, are successful in achieving long-term overall survival (OS) for advanced non-small cell lung cancer (NSCLC) patients.1−6 However, only a subset of patients responded to single-agent ICIs.7,8 PD-L1 expression and tumor mutational burden (TMB) serve as biomarkers for selecting the candidates who might benefit from ICIs.9,10 TMB analysis using circulating tumor DNA (ctDNA), also known as blood-based TMB (bTMB), has become an attractive method considering that not all NSCLC patients could provide adequate tumor tissue for biomarker analysis. However, whether bTMB could predict survival for patients receiving immunotherapy has been debated. Gandara et al. demonstrated that high bTMB (≥16 mutations/Mb) was associated with significant improvement in progression-free survival (PFS) from atezolizumab in NSCLC.11 However, a prospective, phase II, B-F1RST study only showed a trend toward a PFS benefit (hazard ratio (HR) = 0.66; 90% confidence interval (CI): 0.42–1.02; P = .12) and an OS benefit (HR = 0.77; 90% CI: 0.41–1.43; P = .48).12
Cancer is a genetic disease, and multiple genetic mutations may lead to resistance to therapy, including immunotherapy.13 Recent studies revealed that bTMB reflected overall tumor burden.11,14 Therefore, we hypothesized that 1) there might be a non-linear association between bTMB and survival in patients receiving ICIs, 2) patients with low bTMB, just like patients with high bTMB, may also have longer PFS and OS than those with medium bTMB.
Results
Clinical characteristics of the patient population
A total of 144 patients and 425 patients were randomly assigned to receive atezolizumab in POPLAR and OAK trials, respectively. Among these patients, 29 patients and 101 patients were ineligible for bTMB analysis. Finally, 105 patients and 324 patients were enrolled in the training set and the validation set, respectively (Figure 1). Baseline demographic and clinical characteristics were similar in the substudies and the whole POPLAR and OAK trials (eTables 1 and 2). Except for age and line of therapy, no significant difference was observed between the training and validation sets, including median PFS, OS, objective response rate (ORR), disease control rate (DCR), and durable clinical benefit (DCB) (eTable 3). The median follow-up was 14.8 months for the training set and 21 months for the validation set.
Figure 1.
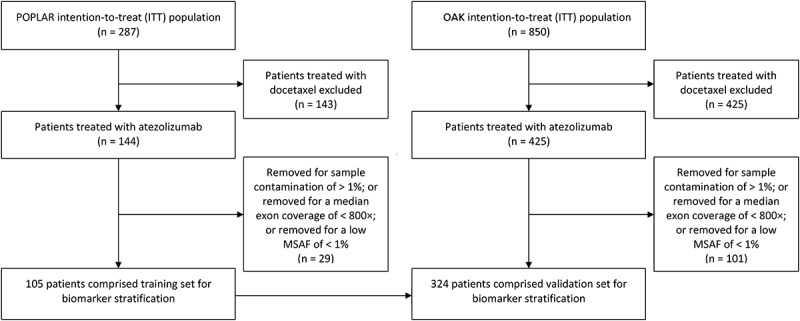
Study flowchart. 105 patients and 324 patients were enrolled in the training set and the validation set. MSAF, maximum somatic allele frequency.
Prognostic value of bTMB in the training set
The restricted cubic spline (RCS) models showed non-linear associations between the level of bTMB and HR for PFS and OS (P non-linearity < 0.001; eFigure 1). X-tile was used to determine the optimal cutoff values of bTMB. The optimal cutoff values were 7 mutations/Mb and 14 mutations/Mb which showed the most significant prognostic effects in predicting PFS (eFigure 2) and OS (eFigure 3). The clinical characteristics for the low (≤7 mutations/Mb), medium (8–13 mutations/Mb), and high bTMB (≥14 mutations/Mb) subgroups were shown in eTable 4. Low and high bTMB were associated with longer PFS and OS (Figure 2). The results remained significant in multivariate analyses, after adjusting for clinical and pathologic factors (PFS: low vs. medium: HR 0.367; 95% CI 0.212–0.637; P < .001; high vs. medium: HR 0.207; 95% CI 0.105–0.410; P < .001; OS: low vs. medium: HR 0.441; 95% CI 0.239–0.814; P = .009; high vs. medium: HR 0.336; 95% CI 0.161–0.704; P = .004) (eTable 5). Both low and high bTMB were significantly associated with higher DCR and DCB (eFigure 4).
Figure 2.

Overall survival (OS) and progression-free survival (PFS) in training set.
(a) Kaplan–Meier estimates of PFS in low, medium, and high bTMB subgroups in training set. (b) Kaplan–Meier estimates of OS in low, medium, and high bTMB subgroups in training set.
Prognostic value of bTMB in the validation set
The RCS analysis revealed a non-linear relationship between bTMB and HR for PFS and OS (eFigure 5). The non-linear relationship was statistically significant (P non-linearity < 0.001). The cutoff values of bTMB were applied to the validation set. The demographic and disease characteristics at baseline were shown in eTable 6. For patients receiving atezolizumab, both PFS and OS were longer among patients with low and high bTMB than those with medium bTMB (Figure 3). In the multivariate Cox proportional hazard regression model, the association between bTMB and survival remained significant (eTable 7). Although DCB was similar among bTMB subgroups, both low and high bTMB were significantly associated with higher DCR (eFigure 6).
Figure 3.

Overall survival (OS) and progression-free survival (PFS) in validation set.
(a) Kaplan–Meier estimates of PFS in low, medium, and high bTMB subgroups in validation set. (b) Kaplan–Meier estimates of OS in low, medium, and high bTMB subgroups in validation set
Prognostic value of bTMB in the combined set
In the combined set, we found that both low and high bTMB significantly predicted longer PFS and OS in multivariate Cox analysis (eFigure 7 and eTable 8). Again, we observed that both low and high bTMB were associated with higher DCR and DCB (eFigure 8). Additionally, patients with low bTMB had the highest proportion of stable disease (low vs. medium vs. high: 44.5% vs. 21.6%: 32.5%; eFigure 8). The proportion of patients with an objective response was similar between low and medium bTMB subgroups (12.1% vs. 12.6%). However, median duration of response in low bTMB subgroup was notably longer than that in medium bTMB subgroup (not reached vs. 10.38 months, HR 0.356; 95% CI 0.140–0.906; P = .030; eFigure 9). In the multivariate logistic regression analysis, we found that low and high bTMB status were significantly associated with DCB (eTable 9).
Since EGFR mutated and ALK rearrangement patients may have lower bTMB and better outcome, we performed the analysis again after excluding these patients. PFS and OS were significantly longer in the low and high bTMB groups than in the medium bTMB group (data not shown).
Collectively, these results suggested that NSCLC patients with low and high bTMB had better prognosis compared to those with medium bTMB.
Mutations in specific genes associated with response and prognosis to atezolizumab
Genetic variants and clinical characteristics differed between low, medium, and high bTMB groups (eFigure 10). We first evaluated if gene mutations were associated with response and clinical benefit to atezolizumab in low and medium bTMB subgroups. Mutations of TP53 and KEAP1 were seen predominantly in patients with shorter PFS, shorter OS, and NDB (all FDR q < 0.05; eFigure 11). STK11 mutations were enriched in patients with shorter PFS (FDR q = 0.040; eFigure 11).
Next, we analyzed gene mutations to nominate additional mediators of response or resistance in medium and high bTMB subgroups. Alterations in ATM were enriched in patients with complete or partial response, stable disease, DCB, longer PFS, and OS (eFigure 12). However, all these observations did not pass FDR correction. Among the high bTMB patients with tumor having an ATM mutation, PFS (HR 0.520; 95% CI 0.310–0.870; P = .013) and OS (HR 0.500; 95% CI 0.280–0.090; P = .019) strongly favored treatment with atezolizumab vs. those without an ATM mutation (eTable 10). The interaction P values for atezolizumab vs. docetaxel treatment were positive for PFS (P = .040) and marginally significant for OS (P = .050), indicating that the presence of an ATM mutation may predict better outcomes with atezolizumab in the high bTMB subgroup (eTable 10).
Impact of bTMB in high PD-L1 expression set
We next explored whether the PFS and OS with atezolizumab in high PD-L1 expression population differed in different bTMB subgroups. Because ICIs significantly improved PFS and OS of the patients harboring a high PD-L1 expression,4,5,9 X-tile was used to determine new optimal cutoff values in this population (n = 59). We found that 12 mutations/Mb and 19 mutations/Mb were the optimal cutoff values. Patients with low and high bTMB had longer PFS and OS compared with patients with medium bTMB (eTable 11).
Discussion
Our exploratory analysis observed PFS and OS on atezolizumab by bTMB status of advanced NSCLC. We found that bTMB was non-linearly related to PFS and OS among NSCLC patients receiving atezolizumab. The prognosis was better with atezolizumab in both low and high bTMB populations than that in medium bTMB population.
There were some potential explanations for the non-linear relationship between bTMB and survival. First, Cabel et al. suggested that baseline ctDNA was associated with tumor burden and could be a prognostic factor in patients with metastatic cancer receiving ICIs.15 We found a positive correlation between the longest baseline diameters and the bTMB score in this study (eFigure 13). Thus, bTMB may serve as a surrogate for tumor burden and low bTMB could be associated with better prognosis. Second, highly mutated tumors are more likely to produce neoantigens, which may be associated with increased tumor immunogenicity and response to ICIs.16 Third, we found that low bTMB patients had higher stable disease rate and longer duration of response than medium bTMB patients. Recent studies reported that KEAP1 and STK11 mutations were associated with low tumor inflammation and T cell-inflamed gene expression profile.17,18 In this study, medium bTMB patients had higher proportion of KEAP1 and STK11 mutations than those in low bTMB patients, which could be the reason why these patients were lack of benefit. However, if the apparent decrease of bTMB was due to including population of low tumor burden, this phenomenon may influence the detection of KEAP1 and STK11 mutations in low bTMB. Last, besides a lower number of mutations, low bTMB may correspond to low tumor shedding.
B-F1RST trial is conducted to answer whether the prespecified bTMB cutoff (16 mutations/Mb) can predict for the improved clinical outcome with atezolizumab. However, patients at this prespecified bTMB cutoff only had numerical benefit for PFS and OS.12 Two possible reasons might explain this result. First, tumor mutational burden is dynamic and could be changed after treatments. While OAK and POPLAR trials included patients with previously treated NSCLC, B-F1RST was designed for treatment-naive NSCLC. Therefore, bTMB cutoff derived from previously treated patients may not be applicable for patients in the first-line treatment. Second, we inferred that low bTMB patients could obtain much more clinical benefit from first-line atezolizumab treatment than medium bTMB patients because of lower tumor burden and lower frequency of KEAP1 and STK11 mutations. Thus, the treatment effect would be overestimated if medium bTMB patients were not distinguished from low bTMB ones.
Recently, Chea et al. indicated that higher ctDNA TMB was significantly correlated with shorter PFS and OS among patients treated with ICIs.14 Guardant360 (68-gene and 70-gene panels) NGS was used in their study. They suggested that Guardant360-based bTMB may only reflect tumor burden and more mutations should be included in the panels to reflect bTMB accurately.14 Wang et al. designed an NCC-GP150 (150-gene panel) NGS and found that higher bTMB estimated by NCC-GP150 was associated with superior PFS in patients with NSCLC treated with ICIs.19 However, Buchhalter et al. suggested that panels between 1.5 and 3 Mb were good to estimate tissue TMB (tTMB), whereas smaller panels tend to produce imprecise tTMB estimates for low to moderate tTMB.20 Thus, we inferred that NCC-GP150 might not be good at distinguishing low and medium bTMB patients. Furthermore, Budczies et al. found the inherent imprecision of panel-based tTMB estimates drastically increased for panel sizes <1 Mb and recommended a novel three-tier tTMB classification scheme to reduce the likelihood of misclassification.21 Therefore, large NGS panels (at least 1 Mb) with three-tier bTMB classification scheme might be considered in the future studies, although the influence of panel size in bTMB is still needed to be assessed in future.
The optimal cutoff points for tTMB and bTMB may vary across tumor types, platforms, and ICIs. The median TMB ranged widely across cancer types.22 In addition, there is a lack of standardization for TMB calculation and different platforms may report different TMB scores. Furthermore, different ICIs have different treatment effects in different cancer types.
Some mutations such as KEAP1, STK11, and ATM mutations were associated with the effectiveness of ICIs.17,18,23 In this study, we also found KEAP1 and STK11 mutations were significantly associated with worse prognosis in low and medium bTMB groups. The medium and high bTMB group failed FDR correction although some genes mutations were enriched. Miao et al. suggested that detection of an ICI-related response mutation (approximately 1% frequency), even if highly specific, required thousands of patients.24 Therefore, larger scale cohorts are needed to find the response-associated mutations. Alterations in DNA damage response and repair (DDR) genes were associated with improved clinical outcomes in PD-1/PD-L1 blockade-treated metastatic urothelial carcinoma.23 In this study, we observed that ATM alterations might predict clinical benefit among high bTMB NSCLC patients treated with atezolizumab. This finding should be validated in other studies.
High PD-L1 expression was associated with high ORR and longer OS of immunotherapy.2,4,5 However, not all patients with high PD-L1 expression gained benefit from ICIs. We found that patients with high PD-L1 expression and medium bTMB had significantly shorter PFS and OS, suggesting that a combination of bTMB and PD-L1 expression may better predict prognosis to ICIs.
Recently, Bristol-Myers Squibb has withdrawn its marketing application for nivolumab plus low-dose ipilimumab for the first-line treatment of NSCLC patients with tTMB of at least 10 mutations/Mb. The data showed no difference in OS compared with chemotherapy between patients whose tumors had high or low levels of tTMB. Gandara et al. found a positive correlation between tTMB and bTMB.11 Thus, it is reasonable to see whether there is also a non-linear association between tTMB and OS in NSCLC patients with ICIs.
The agreement between the tTMB and the bTMB was an important issue. In Gandara et al. paper, they found that lower maximum somatic allele frequency (MSAF) was associated with higher discordance.11 Samples with lower MSAF tended to underestimate bTMB and MSAF ≥ 1% was necessary to accurately and reproducibly call bTMB scores. Thus, patients with a low MSAF of <1% were excluded from this study.
Previous OAK and POPLAR studies demonstrated an overall better outcome for patients receiving atezolizumab compared to docetaxel. The majority of evaluable patients (94%) had a bTMB interval of 0–30 mutations/Mb. Through assessing the relationship between bTMB and prognosis, we surprisingly observed a non-linear relationship. Patients with a bTMB interval of 8–14 mutations/Mb, accounting for approximately 20% of the overall population, were categorized into the median bTMB population base on the statistical analysis (X-tile software). We speculate that this result should be further validated with a larger sample size and more sophisticated methods.
There were four implications of this work for future research of bTMB. First, the findings of this work should be confirmed in B-F1RST study. Second, more studies are needed to explore if the non-linear association between bTMB and survival exists in other cancer types and ICIs. Third, the role of bTMB should be determined in new treatment patterns, such as combined chemo/immunotherapy. Last, it would be interesting to explore the non-linear association with other methodologies, such as whole-exome sequencing.
This study has several limitations. First, a moderate sample size limited the power to draw a definite conclusion. Second, the follow-up time was relatively short, which precluded the assessment of long-term survival analysis. Third, we could not assess the association between bTMB and survival in patients with liver or brain metastases due to the incomplete information about the metastasis site. Last, not all blood samples can pass the quality control and be used to calculate bTMB.
In summary, our study showed that bTMB was non-linearly associated with PFS and OS in NSCLC patient receiving atezolizumab. The findings indicated that both low and high bTMB were favorable prognostic factors of atezolizumab.
Patients and methods
Study design and patient population
POPLAR trial and OAK trial included patients with histologically confirmed squamous or non-squamous NSCLC with progression after at least one platinum-based chemotherapy. Eligibility criteria for POPLAR and OAK have been previously described.4,5 In brief, eligible patients were 18 years or older, had measurable disease per Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST v1.1), and had an Eastern Cooperative Oncology Group performance status (ECOG PS) of 0 or 1. Patients with EGFR mutations or an ALK fusion oncogene were required to have received previous tyrosine kinase inhibitor therapy. Atezolizumab was given as an intravenous 1200 mg fixed dose every 3 weeks; docetaxel was given intravenously at 75 mg/m2 every 3 weeks. Tumor assessments, according to RECIST v1.1, were performed at baseline, then every 6 weeks until week 36 and every 9 weeks thereafter, until disease progression. Our study was conducted without institutional review board approval, because all the clinical and genetic data were obtained from a recently published study by Gandara et al.11 This research complied with the Declaration of Helsinki.
Outcomes
PFS was defined as the time from randomization to disease progression or death from any cause. OS was defined as the time from randomization to death from any cause. DCB was defined as OS that last more than 12 months, and no durable benefit (NDB) was defined as OS that last less than 12 months.
Sample collection and genetic analysis
Patients included in POPLAR and OAK were needed to have archival blood samples available for exploratory assessment of tumor biomarkers. Thus, collection of blood samples was prearranged. Cell-free DNA was extracted from these blood samples. The blood genetic analysis has been previously described.11 FoundationOne next-generation sequencing (NGS) assay was conducted on hybridization-capture libraries for 1.1 Mb coding region of 315 cancer-related genes. The bTMB score was determined by identifying all base substitutions present at an allele frequency of ≥0.5% and filtering out germline mutations by comparing against the dbSNP and ExAC databases.11
PD-L1 testing
PD-L1 expression was evaluated with the VENTANA SP142 PD-L1 immunohistochemistry assay (Ventana Medical Systems, Inc., Tucson, AZ, USA). High PD-L1 expression was defined as PD-L1 expression on 50% or more of tumor cells and was defined as 10% or more of tumor-infiltrating immune cells.4
Statistical analysis
To examine the non-linear relationship between bTMB and survival, we used restricted cubic spline analysis in the Cox proportional hazard model,25 with predefined knots at the 5th, 35th, 65th, and 95th percentile of bTMB.26 Spline regression fits smooth polynomial functions between predefined knots on a graph and joins them in a piecewise manner. Splines could be used to produce a non-linear model between a continuous prognostic variable (e.g., bTMB) and an outcome (e.g., HR of PFS or OS). Optimal cutoff points for bTMB were determined using X-tile software version 3.6.1 (Yale University School of Medicine, New Haven, CT, USA). X-tile software is good for defining cut-points for U-shaped distributions (e.g., where both the high and low subpopulations do better or worse than the middle population).27 We used Mann–Whitney U test and Kruskal–Wallis test to compare the difference between two and three groups, respectively. The Fisher exact test was used to assess categorical variables. The OS and PFS were calculated using the Kaplan–Meier method. A Cox proportional hazard regression model was used to calculate HRs and 95% CIs. Assessment of enrichment of binary molecular features (e.g., wild-type or mutant gene) with response (e.g., complete or partial response versus progressive disease) and HR of survival (e.g., PFS and OS) was done with Fisher’s exact tests and Cox proportional hazard regression, respectively. Correction for multiple-hypothesis testing was done controlling for false discovery rate (FDR) by the Benjamini–Hochberg method.28 Logistic regression was performed to assess the correlations between clinical variables and DCB, with the results showed as odds ratios (ORs) and 95%CIs. All analyses were conducted with R (version 3.3.1). A two-sided P value of 0.05 was considered statistically significant.
Funding Statement
This study was sponsored by Shanghai Municipal Human Resource and Social Security Bureau Talent Project (No. 052), National Natural Science Foundation of China (No. 81601988, 81602078, 81502450, and 81472642), Science and Technology Commission of Shanghai Municipality, China (No. 18441904700), and Shanghai Chest Hospital Project of Collaborative Innovation (No. YJXT20190102).
Authors’ contributions
WN, JQ, MDX, and KG were involved in the literature search, figures, study design, data collection, data analysis, and writing. FFQ, JL, LG, XYZ, HMW, BY, BZ, SYW, FH, and CHL were involved in data collection. HZ and BHH were involved in the study design, writing, and article review.
Acknowledgments
The data of the OAK and POPLAR trials included in this analysis were provided online by F. Hoffmann-La Roche. We would like to thank Yingyi Qin from Department of Health Statistics, Second Military Medical University for his valuable comments on statistical analysis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Availability of supporting data
Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
Bao-Hui Han has consulted for AstraZeneca and Roche Pharmaceutical Company. He also received payment for speaking from AstraZeneca Pharmaceutical Company and Lily Pharmaceutical Company. All remaining authors have declared no conflicts of interest.
Supplementary material
Supplementary data for this article can be accessed on the publisher’s website.
References
- 1.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, et al. Pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol. 2019;37(7):537–7. doi: 10.1200/JCO.18.00149. [DOI] [PubMed] [Google Scholar]
- 2.Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, Castro G Jr, Srimuninnimit V, Laktionov KK, Bondarenko I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393(10183):1819–1830. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- 3.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F, et al. First-line nivolumab in stage iv or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376(25):2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols MC, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389(10066):255–265. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D, Artal-Cortes A, Lewanski C, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387(10030):1837–1846. doi: 10.1016/S0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 6.Gettinger S, Horn L, Jackman D, Spigel D, Antonia S, Hellmann M, Powderly J, Heist R, Sequist LV, Smith DC, et al. Five-year follow-up of nivolumab in previously treated advanced non-small-cell lung cancer: results from the CA209-003 study. J Clin Oncol. 2018;36(17):1675–1684. doi: 10.1200/JCO.2017.77.0412. [DOI] [PubMed] [Google Scholar]
- 7.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 9.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, et al. Pembrolizumab versus chemotherapy for PD-L1-Positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 10.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36(7):633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, Rittmeyer A, Fehrenbacher L, Otto G, Malboeuf C, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 12.Socinski M. Final efficacy results from B-F1RST, a prospective phase II trial evaluating blood-based tumour mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC). 2019 ESMO. LBA83.
- 13.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15(2):81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 14.Chae YK, Davis AA, Agte S, Pan A, Simon NI, Iams WT, Cruz MR, Tamragouri K, Rhee K, Mohindra N, et al. Clinical Implications of Circulating Tumor DNA Tumor Mutational Burden (ctDNA TMB) in non-small cell lung cancer. Oncologist. 2019;24(6):820–828. doi: 10.1634/theoncologist.2018-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabel L, Proudhon C, Romano E, Girard N, Lantz O, Stern MH, Pierga JY, Bidard FC. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat Rev Clin Oncol. 2018;15(10):639–650. doi: 10.1038/s41571-018-0074-3. [DOI] [PubMed] [Google Scholar]
- 16.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362(6411):pii: eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, Zhu B, Wang S, Zhuo M, Sun J, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol. 2019;5(5):696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchhalter I, Rempel E, Endris V, Allgäuer M, Neumann O, Volckmar AL, Kirchner M, Leichsenring J, Lier A, von Winterfeld M, et al. Size matters: dissecting key parameters for panel-based tumor mutational burden analysis. Int J Cancer. 2019;144(4):848–858. doi: 10.1002/ijc.31878. [DOI] [PubMed] [Google Scholar]
- 21.Budczies J, Allgäuer M, Litchfield K, Rempel E, Christopoulos P, Kazdal D, Endris V, Thomas M, Fröhling S, Peters S, et al. Optimizing panel-based tumor mutational burden (TMB) measurement. Ann Oncol. 2019;30(9):1496–1506. doi: 10.1093/annonc/mdz205. [DOI] [PubMed] [Google Scholar]
- 22.Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teo MY, Seier K, Ostrovnaya I, Regazzi AM, Kania BE, Moran MM, Cipolla CK, Bluth MJ, Chaim J, Al-Ahmadie H, et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol. 2018;36(17):1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM, Adeegbe D, Keliher D, Schilling B, Tracy A, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50(9):1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desquilbet L, Mariotti F. Dose–response analyses using restricted cubic spline functions in public health research. Stat Med. 2010;29(9):1037–1057. doi: 10.1002/sim.3841. [DOI] [PubMed] [Google Scholar]
- 26.Harrell FE Jr. Regression modeling strategies. New York (NY): Springer- Verlag; 2001. [Google Scholar]
- 27.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets analyzed during the current study are available from the corresponding author on reasonable request.


