Abstract
Objective
To perform a comprehensive multicompartment analysis of microRNA (miRNA) expression in multiple sclerosis (MS) linked to disease activity and compared with other neuroinflammatory diseases through a retrospective cross-sectional study.
Methods
One hundred twenty-seven miRNAs were measured by PCR arrays on pooled CSF, serum, and peripheral blood mononuclear cell (PBMC) samples of 10 patients with relapsing MS and 10 controls. Sixty-four miRNAs were then measured by quantitative PCR on individual CSF samples of patients with relapsing or remitting MS and controls (n = 68). Fifty-seven miRNAs were analyzed in the CSF from a second cohort (n = 75), including patients with MS, neuroinfectious, or neuroinflammatory diseases and controls. MiRNAs significantly dysregulated in the CSF were analyzed on individual serum/PBMC samples (n = 59/48) of patients with relapsing or remitting MS and controls. Post hoc analysis consisted of principal component analysis (PCA), gene set, and pathway enrichment analysis.
Results
Twenty-one miRNAs were differentially expressed, mainly upregulated in the CSF during MS relapses. Relapsing MS and neuroinfectious/inflammatory diseases exhibited a partially overlapping CSF miRNA expression profile. Besides confirming the association of miR-146a-5p/150-5p/155-5p with MS, 7 miRNAs uncharacterized for MS emerged (miR-15a-3p/124-5p/149-3p/29c-3p/33a-3p/34c-5p/297). PCA showed that distinct miRNA sets segregated MS from controls and relapse from remission. In silico analysis predicted the involvement of these miRNAs in cell cycle, immunoregulation, and neurogenesis, but also revealed that the signaling pathway pattern of remitting MS is more akin to controls rather than patients with relapsing MS.
Conclusions
This study highlights the CSF-predominant dysregulation of miRNAs in MS by identifying a signature of disease activity and intrathecal inflammation among neuroinflammatory disorders.
Multiple sclerosis (MS) is characterized by multifocal inflammatory lesions inducing myelin sheath damage and axonal degeneration. MS results from a complex interplay between genetic susceptibility and environmental and epigenetic factors, but its molecular determinants remain elusive. A predominance of patients present initially with a relapsing-remitting disease course.1
MicroRNAs (miRNAs) are single-stranded noncoding RNAs regulating posttranscriptionally the expression of a large spectrum of genes during various biological processes.2 Several miRNAs have been associated with MS, but only few were linked to disease activity.3,4 Most studies were performed in small cohorts, in single biological compartments, rarely compared with other neurologic disorders or were not replicated, hence large heterogeneity between study populations and analysis methods. Few studies have focused on CSF miRNAs,5 although these might be more relevant to understanding disease regulation.
We sought to characterize miRNA expression in 3 biological compartments, i.e., CSF, serum, and peripheral blood mononuclear cells (PBMCs) of patients with MS according to disease activity compared with that of controls and other neurologic disorders in the CSF. MiRNAs known for their involvement in immunity, inflammation, neurodegeneration, and lipid metabolism were measured by quantitative PCR (qPCR). Finally, a principal component analysis (PCA) and a pathway enrichment analysis were performed, altogether leading to a comprehensive multicompartment characterization of the miRNA expression profile in MS.
Methods
Patients
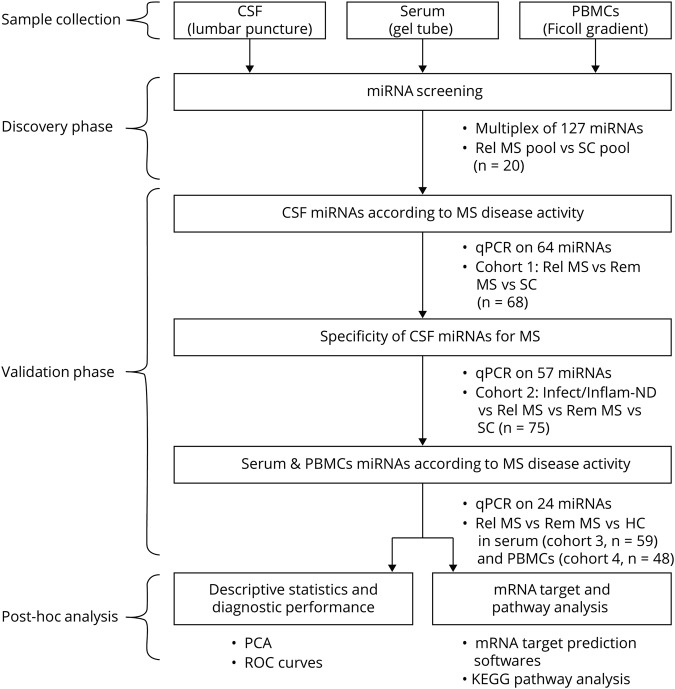
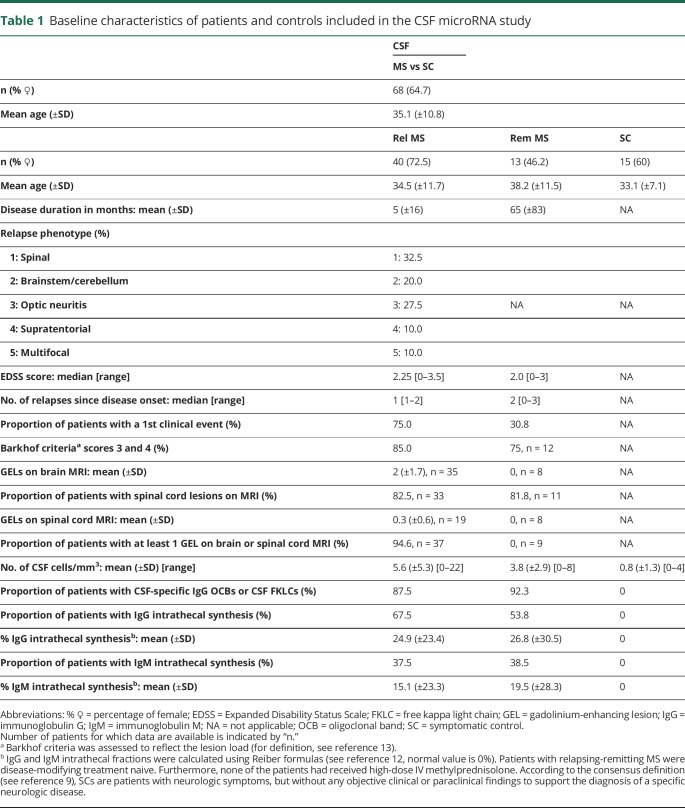
CSF and blood samples were collected at the Cliniques Universitaires Saint-Luc (Brussels) between March 2005 and March 2018, processed according to international guidelines within 1 hour before storage at −80°C until miRNA extraction.6,7 PBMCs paired with serum were processed as previously described.8 The study was conducted stepwise (figure 1). A group size of at least 9 patients was calculated using the CSF fold changes of relapsing MS vs controls in the 2 miRNA arrays, with alpha error at 0.05 and power of 0.80 (stat.ubc.ca/∼rollin/stats/ssize/n2.html). Patient cohorts were classified according to the BioMS-eu consortium definitions (table e-1, links.lww.com/NXI/A188).9 The 2017 McDonald criteria10 were used for MS diagnosis. Relapse was defined clinically and/or by the presence of gadolinium-enhancing lesions (GELs) on MRI for 93%–100% of patients, depending on the cohort.11 Remitting patients were clinically stable for 1 month up to 21 years. To reduce bias, patients were age and sex matched to controls, and none of the patients with MS were under any disease-modifying treatment (DMT). None of the relapsing patients had received high-dose IV methylprednisolone before sample collection. Clinical and paraclinical features of the patients are summarized in tables 1 and e-1.12,13 Diagnoses of other disease categories are listed in table e-2. Data were collected between February 2015 and July 2018.
Figure 1. Workflow summary of different study steps.
AUC = area under the curve; HC = healthy control; Infect/Inflam-ND = patients with infectious or inflammatory neurologic disorders; KEGG = Kyoto Encyclopedia of Genes and Genomes; mRNA = messenger RNA; miRNA = microRNA; PBMC = peripheral blood mononuclear cell; PCA = principal component analysis; qPCR = quantitative PCR; Rel MS = relapsing MS; Rem MS = remitting MS; ROC = receiver operating characteristic; RRMS = relapsing-remitting MS; SC = symptomatic control.
Table 1.
Baseline characteristics of patients and controls included in the CSF microRNA study
MiRNA isolation
MiRNAs were isolated from CSF (400 μL) and serum (200 μL) using the miRNeasy Serum/Plasma Kit, and from 11-13.106 PBMCs using the miRNeasy Mini Kit, following the manufacturer's protocol (Qiagen, Hilden, Germany). Caenorhabditis elegans miR-39 mimic (Qiagen) was added to CSF and serum samples as spike-in control for relative quantification. MiRNA extraction, reverse transcription (RT), and PCR were performed sequentially.
MiRNA PCR array
MiRNA profiling was kindly performed by the manufacturer with “Inflammatory Response & Autoimmunity” and “T-cell & B-cell Activation” miScript miRNA PCR Arrays (Qiagen) on a pool of 10 patients with relapsing MS and 10 symptomatic controls (SCs, for CSF) or 10 healthy controls (HCs) (for serum and PBMCs).
RT, preamplification, and qPCR
RT was performed on 5 μL of miRNA using the miScript II RT Kit. One microliter of complementary DNA was preamplified using the miScript PreAmp PCR Kit following the manufacturer's recommendations (Qiagen). The spike-in control C. elegans miR-39 mimic was not preamplified. qPCR reactions were performed in duplicate, except for miR-24-3p in CSF, a putative internal reference gene,14 which was assayed in triplicate. Patients' subgroups and controls were equally distributed across each run to minimize interrun bias for a single miRNA target.
qPCR analysis
Sample selection was based on Ct variability within duplicates of a same sample and between samples, as well as melt curve morphology. Ct was set at 40 in case of inefficient amplification. The miRNA transcripts were normalized using references (exogenous miR-39 for CSF/serum or intracellular RNU6B [small nuclear ribonucleoprotein U6B, later referred to as RNU] for PBMCs). Relative expression was determined by the comparative Ct method to the mean of the SCs or HCs (2−ΔΔCt with ΔΔCt = ΔCt [target-reference]sample − ΔCt [target-reference]mean of controls).15 Extraction efficiency was considered by comparing the mean Ct of miR-39 or RNU amplification between subgroups and miRNA extraction series. When extraction efficiency appeared variable, results were expressed relative to the mean of controls extracted using the same RT enzyme lot.
Gene target prediction and pathway enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were investigated for miRNAs displaying a statistically significant relative expression difference above 1.5-fold between disease categories. MiRNAs were grouped in sets according to the disease subgroup and biological compartment. Two (table e-6, links.lww.com/NXI/A188) or three (figure 5) databases were combined comprising predicted and validated miRNA targets, namely mirPath v.3 by DIANA TOOLS based on TarBase v7.0 (filtered with Pathway Union and false discovery rate [FDR] correction; snf-515788.vm.okeanos.grnet.gr/), miRNet (filtered with a 2.0 cutoff degree and using a hypergeometric Fisher exact test algorithm; mirnet.ca/), and enrichment using the Cytoscape stringApp (with data set from the filtered miRNet analysis and a confidence score cutoff set at 0.6). The miRNA-gene network was generated from the miRNet database, filtered (2.0 cutoff degree of analysis and 40.0 betweenness) and sketched in the Cytoscape stringApp to show the most representative genes.
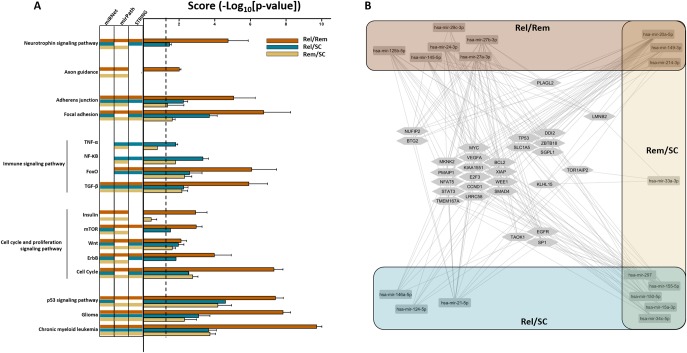
Figure 5. Predicted signaling pathways (SiPas) and genes targeted by dysregulated CSF miRNAs in MS.
(A) Top enriched canonical pathways associated with the differentially expressed CSF miRNAs between each subgroup (Rel/Rem [orange bars], Rel/SC [blue bars], or Rem/SC [yellow bars]) are illustrated. Indicated pathways were found by performing Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis using the miRNet, mirPath, or Cytoscape stringApp databases. The columns on the left side of the graph indicate the number of databases from which a determined pathway was retrieved. The pathways retrieved by specific databases are indicated by the aforementioned color code (a void line indicates that a pathway was not found in the database from the corresponding column). On the right side, results are expressed as mean ± standard error of the enrichment score (−log10 of adjusted p value) obtained from the 3 databases, giving a score for the relative weight of a given SiPa between experimental subgroups; the dotted line designates the threshold of 1.3 (representing p value at 0.05). We identified SiPas consistent with MS pathogenesis such as cell cycle or apoptosis (Insulin, mTOR, Wnt, and ErbB SiPas with the following gene targets: MYC, CCND1, LMNB2, CDKN1A, MDM2, E2F3, TP53, NRAS, WEE1, MKNK2, BTG2, PMAIP1, TAOK1, DDI2, and NUFIP2), immunoregulation (TNF-α, NF-κB, TGF-β, FoxO SiPas, and gene targets: SMAD4, NFAT5, SP1, MYC, CCND1, SLC1A5, STAT3, BCL2, and XIAP), neuroprotective and neurodegenerative processes (axon guidance, ErbB, and neurotrophin SiPas with BCL2, TP53, VEGFA, EGFR, ZBTB18, PLAGL2, and TOR1AIP2 as targeted genes), and cancer-related SiPas (only glioma and myeloid leukemia were selected). (B) Cytoscape network representation of interactions between the 18 MS-associated miRNAs in the CSF and a panel of targeted genes extracted from the miRNet database. The resulting network comprises 47 nodes (the 18 dysregulated miRNAs with the 29 most represented genes) and 176 direct edges. The color code refers to the sets of miRNAs according to their differential expression across subgroups, as in A. miRNA = microRNA; NF-κB = nuclear factor kappa-light-chain-enhancer of activated B; Rel = relapsing MS; Rem = remitting MS; SC = symptomatic control TGF-β = transforming growth factor beta; TNF-α = tumor necrosis factor alpha.
Statistical analysis
For statistical analysis, a nonparametric 1-way analysis of variance Kruskal-Wallis test of all relative miRNA expression levels across subgroups was performed using GraphPad Prism 5.0, followed by a Dunn post-test for a 2-by-2 comparison of the subgroups. Dunn p values, with a significance threshold set at ≤0.05, were calculated and corrected by the Benjamini-Hochberg FDR method (astatsa.com/KruskalWallisTest/). Differential expression of miRNAs was considered only if statistically significant for both post-tests.
PCA of continuous variables obtained from the CSF miRNAs screening was performed using the R FactoMineR program. The presence/absence of oligoclonal bands (OCBs), MRI lesion load, presence/absence of GELs, and diagnostic category were analyzed as illustrative qualitative variables. The missing values were replaced by the mean value of the subgroup to which the sample belonged to.16 Results were plotted using Rcmdr (cran.r-project.org/web/packages/Rcmdr/index.html). For miR-146a-5p, -150-5p, and -155-5p in CSF, the receiver operating characteristic (ROC) curves were calculated providing a cutoff with an optimal compromise between sensitivity and specificity to discriminate MS from SC.
Standard protocol approvals, registrations, and patient consents
Patients sign a charter upon hospital admission stating that the residual CSF and serum samples, collected for routine diagnostic procedures or patient care, can be used for retrospective academic research, without an additional informed consent (ethical approval 2007/10SEP/233). PBMCs and paired serum were collected separately under a distinct research protocol upon signature of an informed consent (ethical approval 2007/31AOUT/222).
Data availability
Raw PCR files and detailed in silico and statistical outputs are not included in this article. Any unpublished and anonymized data will be shared upon request from a qualified investigator. Strengthening the Reporting of Observational Studies in Epidemiology guidelines have been respected.
Results
Multicompartment screening of immune-related miRNAs from a pool of patients with relapsing MS vs SCs/HCs
To investigate whether miRNA expression was modified in MS, particularly during relapses, we first performed a screening on a pool of CSF, serum, or PBMCs obtained from 10 patients with MS in relapse and 10 age- and sex-matched SCs or HCs (table e-1, links.lww.com/NXI/A188). The expression of 164 miRNAs was assessed using the “Human Inflammatory Response & Autoimmunity” and the “T-Cell & B-Cell Activation” miScript miRNA PCR Arrays. Thirty-seven miRNAs were assayed in both arrays. Absolute expression differences ranging between 3- (ΔCt = 1.59) and 148-fold (ΔCt = 7.21) were observed for 49 miRNAs, of which 35 originated from CSF, 9 from serum, and 8 from PBMCs. Of note, some miRNAs were found dysregulated in 2 compartments (miR-181c-5p and miR-210-3p in CSF and PBMCs; miR-34c-5p and miR-184 in PBMCs and serum). Most CSF miRNAs were overexpressed in the relapsing MS pool in comparison to the SC pool (ΔCt < 0), except miR-101-3p and miR-17-5p. Twenty-three miRNAs were discarded from subsequent analysis because of low expression levels (Ct ≥ 30) (figure e-1).
Quantification of miRNAs in the CSF of patients with relapsing and remitting MS vs SCs
As most miRNAs were found dysregulated in the CSF, we aimed to confirm these trends in individual CSF samples of patients with MS, according to disease activity (n = 40 in relapse and n = 13 in remission) and compared with age- and sex-matched SCs (n = 15) (table 1). In total, 64 miRNAs were amplified from individual CSF samples (table e-3, links.lww.com/NXI/A188). Those were either selected from the differentially expressed targets identified in the screening arrays, from previously published studies in MS or for their involvement in immunity, inflammation, lipid metabolism, or neurodegeneration.
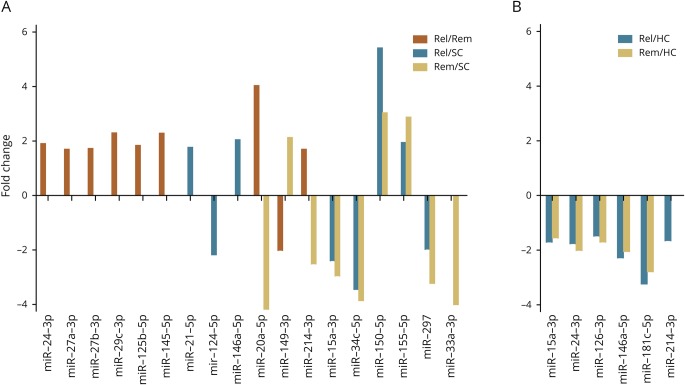
Eighteen miRNAs were differentially expressed in at least 1 subgroup, with statistically significant (p ≤ 0.05) median fold changes ranging from −4.17 to 5.41 (figure 2A, figure e-2 and table e-4A, links.lww.com/NXI/A188). MiR-150-5p and miR-155-5p were upregulated, whereas miR-15a-3p, -34c-5p, and -297 were downregulated in patients with MS, irrespective of disease activity in comparison to SCs. Most miRNAs (n = 8) were upregulated in patients with relapsing MS compared with patients with remitting MS or SCs, except miR-124-5p, which was downregulated compared with SCs. Three miRNAs (miR-20a-5p, -33a-3p, and -214-3p) were downregulated in patients with remitting MS compared with patients with relapsing MS and/or SCs, whereas miR-149-3p was upregulated. In addition, miR-24-3p was differentially regulated in patients with MS and could therefore not be used as an internal reference gene in this setting.
Figure 2. miRNA expression fold change in MS according to disease activity.
Differences in CSF (A) and serum (B) miRNA expression levels. The fold change is calculated as the ratio between the median of the relative miRNA expression level of the respective subgroups. HC = healthy control; miRNA = microRNA; Rel = relapsing MS; Rem = remitting MS; SC = symptomatic control.
Specificity of the differentially expressed CSF miRNAs for MS
qPCR was performed for 57 miRNAs (table e-3, links.lww.com/NXI/A188) on individual CSF samples of 14 patients having infectious neurologic disorders (Infect-NDs), 12 patients having inflammatory neurologic disorders (Inflam-NDs), 15 patients with relapsing MS, 9 patients with remitting MS, and 25 SCs.
Twenty-four miRNAs were differentially expressed in at least 1 subgroup reaching a statistically significant threshold of 0.05 following Benjamini-Hochberg correction (table e-4B, links.lww.com/NXI/A188). These were all upregulated in Infect-ND compared with SC except miR-22-3p.
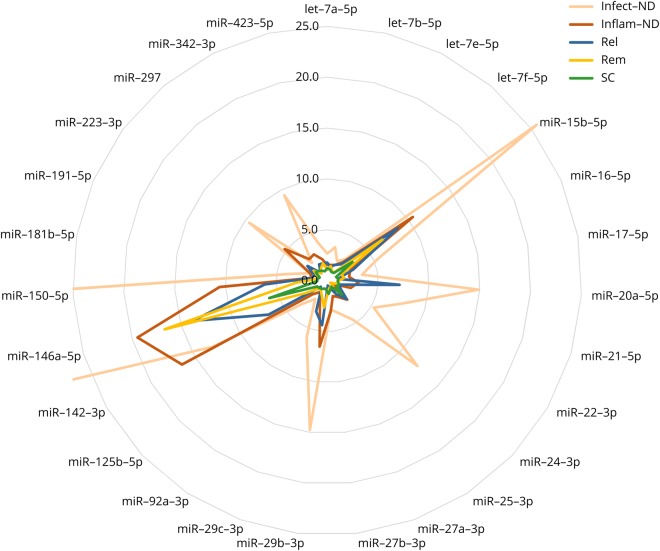
The levels of miR-21-5p, -142-3p, -223-3p, -342-3p, -423-5p, and let-7f-5p were increased in the CSF of both Infect- and Inflam-ND vs SC, whereas miR-15b-5p, -29b-3p, and -29c-3p were upregulated in Infect-ND, Inflam-ND, and relapsing MS compared with SC. miR-150-5p was the only miRNA upregulated in all disease subgroups in comparison to SC. The profile of overexpressed miRNAs in these disease categories was strikingly similar except for the expression levels that were 2- to 6-fold higher in the Infect-ND subgroup and even 14-fold higher for miR-150-5p compared with relapsing MS (figure 3).
Figure 3. Comparative CSF miRNA expression profile of MS vs neuroinfectious/neuroinflammatory diseases and controls.
Relative CSF miRNA expression levels in the different subgroups. Of note, values exceeding the graph scale were of 25.66, 61.48, and 85.94 for miR-15b-5p, -146a-5p, and -150-5p, respectively. Infect-ND = infectious neurologic disorder; Inflam-ND = inflammatory neurologic disorder; miRNA = microRNA; Rel = relapsing MS; Rem = remitting MS; SC = symptomatic control.
In addition, 46 of these miRNAs were investigated on CSF samples of 16 patients having neurodegenerative disorders and 17 patients with peripheral nervous system disorders, but no differences in expression levels were found, except compared with Infect-ND (data not shown).
Quantification of miRNAs in the serum of relapsing and remitting MS vs HCs
To determine whether the dysregulated miRNAs identified in the CSF were also differentially expressed in peripheral compartments, qPCR was performed for 24 miRNAs (table e-3, links.lww.com/NXI/A188) on individual serum samples of 20 patients with relapsing MS, 19 patients with remitting MS, and 20 HCs.
Relative quantification of 6 miRNAs reached statistical significance, with fold change in the median miRNA expression level ranging between −3.23 and −1.47 (figure 2B and table e-4A, links.lww.com/NXI/A188). Five miRNAs (miR-15a-3p, -24-3p, -126-3p, -146a-5p, and -181c-5p) were downregulated in both relapsing and remitting MS compared with HC, whereas miR-214-3p was downregulated in relapsing MS compared with HC only.
Quantification of miRNAs in PBMCs of patients with relapsing and remitting MS vs HCs
qPCR was performed for 24 miRNAs (table e-3, links.lww.com/NXI/A188) on individual PBMC samples of 18 patients with relapsing MS, 16 patients with remitting MS, and 14 HCs. Only 1 miRNA, miR-34a-5p, was significantly downregulated in relapsing MS compared with remitting MS with a fold change of −1.89 (table e-4A).
PCA and correlation with CSF parameters
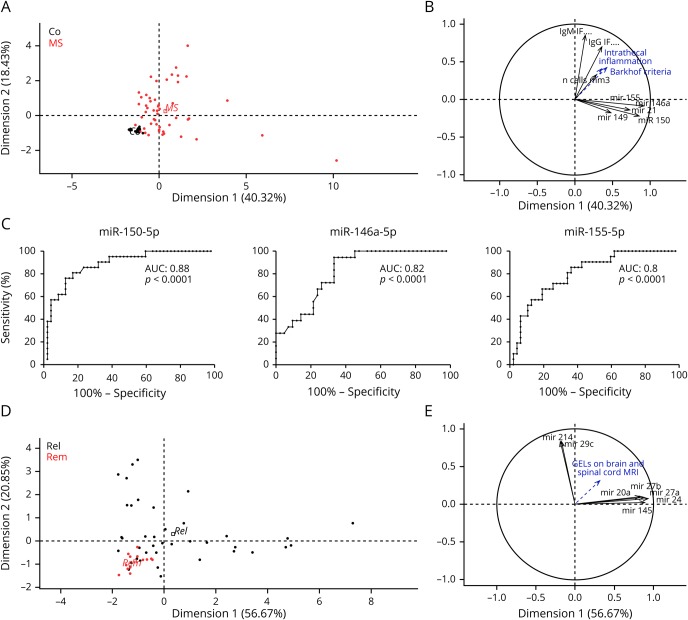
To identify the combination of variables that best explain the differences between patients with MS and SCs, we performed a PCA on upregulated CSF miRNAs that showed more than 1.5-fold variation between these groups. Significantly upregulated miRNAs in patients with MS (miR-150-5p, -155-5p, -146a-5p, -21-5p, and -149-3p) accounted for 40.32% of the sample population variance, whereas other numerical variables such as the immunoglobulin G (IgG) or immunoglobulin M intrathecal fractions and CSF pleocytosis accounted for 18.43% of the variance, allowing an excellent discrimination between patients with MS and SCs (figure 4, A and B). The relative contribution of selected miRNAs to the variability of the population was significant as indicated by their cos2 value, with the most significant miRNAs being miR-146a-5p and miR-150-5p (table e-5A, links.lww.com/NXI/A188). Conversely, the analysis performed with the miRNAs that were significantly downregulated in MS vs SC (miR-15a-3p, -20a-5p, -33a-3p, -34c-5p, -124-5p, -214-3p, and -297) accounted only for 31.95% of the sample population variance (data not shown). ROC curves were plotted for miR-150-5p, -146a-5p, and -155-5p, with an area under the curve of 0.88, 0.82, and 0.8, respectively (p < 0.0001). Relative cutoff value for miR-150-5p was 1.9, with a sensitivity of 80% and a specificity of 81% for MS diagnosis (figure 4C). In addition, miR-150-5p expression levels were moderately correlated with CSF pleocytosis, but not with other CSF parameters, both in the MS vs control cohort and in the cohort of different Infect-NDs/Inflam-NDs (Spearman r = 0.47 and r = 0.58, respectively, p < 0.0001). miR-150-5p levels were also significantly upregulated in patients with CSF-specific IgG OCBs (data not shown).
Figure 4. Patient stratification according to diagnosis or disease activity by PCA.
PCA was applied using the significantly upregulated CSF miRNAs in patients with MS and other CSF parameters (IgG or IgM IF, CSF pleocytosis). PCA can distinctly separate MS from SC (A) in a 2-dimensional representation comprising in total 58.75% of sample population variability. The upregulated miRNA panel (i.e., miR-21-5p, -146a-5p, -149-3p, -150-5p, and -155-5p) segregates from other diagnostic criteria (Barkhof imaging criteria, IgM and IgG IF). Cos2 values for each variable are plotted in abscissa and ordinate (B). They represent the strength of the individual variables in indicating population variability according to each dimension (significant above 0.5). ROC curves are plotted (C) for miR-150-5p, -146a-5p, and -155-5p as diagnostic biomarkers for MS with respective areas under the curve of 0.87, 0.82, and 0.8, respectively (p < 0.0001). PCA can also partially separate patients with Rel MS from patients with Rem MS (D) in a 2-dimensional representation comprising in total 77.52% of sample population variability. MiR-20a-5p, -24-3p, -27a-3p, -27b-3p, and -145-5p segregate from miR-29c-3p and miR-214-3p (E). AUC = area under curve; GEL = gadolinium-enhancing lesion; IF = intrathecal fraction; IgG = immunoglobulin G; IgM = immunoglobulin M; miRNA = microRNA; PCA = principal component analysis; Rel = relapsing MS; Rem = remitting MS; ROC = receiver operating characteristic; SC = symptomatic control.
PCA on miRNAs whose expression was modified in patients with relapsing vs remitting MS (miR-20a-5p, -145-5p, -214-3p, -27a-3p, -27b-3p, -24-3p, and -29c-3p), using the presence/absence of GELs as illustrative variable for disease activity, showed a good separation between patients with relapsing and remitting MS. This miRNA combination accounted for 77.52% of the population variance (figure 4, D and E), with the most representative miRNAs being miR-24-3p, -27a-3p, and -145-5p (table e-5B, links.lww.com/NXI/A188). Adding miR-149-3p, which is decreased 2-fold in relapsing vs remitting MS, did not improve disease activity discrimination (data not shown).
Biological significance and targeted genes of the differentially expressed miRNAs in MS
To obtain insight regarding the functions of the dysregulated miRNAs, we performed pathway enrichment analysis for the differentially expressed miRNA sets (more than 1.5-fold difference with corrected p value ≤ 0.05) between subgroups, as defined below and in figure 5. Some pathways were affected, independently from disease activity, such as cancer (p53 signaling pathway [SiPa], glioma, or chronic myeloid leukemia), cell cycle and proliferation (Wnt), immune (transforming growth factor beta and FoxO), cellular adhesion (focal adhesion and adherens junction) SiPas. We nevertheless observed that the enrichment score (defined as the -log10[p value])17 was always stronger for the relapsing vs remitting (Rel/Rem) subgroup compared with the 2 other subgroups. More strikingly, some pathways were found specifically in relapsing patients (neurotrophin, mTOR, and ErbB) compared with the 2 other groups, suggesting some relevance of these pathways in modulating disease activity. On the other hand, nuclear factor kappa-light-chain-enhancer of activated B cells was shown as general signaling for relapsing-remitting MS (RRMS). Some pathways were only affected in the Rel/Rem subgroup (axon guidance and insulin) or specifically in the Rel/SC subgroup (tumor necrosis factor alpha) (figure 5A). The comparison between up- and downregulated miRNAs in the different compartments showed a parallelism between pathways targeted by miRNAs upregulated in the CSF of relapsing MS and downregulated in the CSF of remitting MS (table e-6, links.lww.com/NXI/A188). In addition, the most representative genes retrieved from the analysis of all differentially expressed miRNAs in the CSF were introduced in a miRNA-gene interaction network. Several genes (MYC, VEGFA, MKN2, BCL2, PMAIP1, KIAA1551, XIAP, NFAT5, E2F3, WEE1, STAT3, CCND1, SMAD4, TMEM167A, and LRRC58) were targeted by at least 4 miRNAs species from the 3 different sets, revealing possibly important miRNA-related genes affected in MS (figure 5B).
Discussion
CSF-predominant miRNA dysregulation and miRNA species newly associated with MS
This study sought to perform a comprehensive analysis of the differential expression of carefully preselected miRNAs in patients with RRMS, according to disease activity, vs controls in 3 different biological compartments. Patients were free of DMT and devoid of high-dose steroids. A second CSF cohort was analyzed to compare the MS miRNA profile with that of other neurologic diseases. Strikingly, we observed 18 significantly dysregulated miRNAs in the CSF vs 6 in the serum and only 1 in PBMCs, pointing toward specific intrathecal processes ongoing in MS and confirming the results of the preliminary miRNA array analysis performed on pooled samples. We have identified 7 miRNAs, namely miR-15a-3p, -124-5p, -149-3p, -29c-3p, -33a-3p, -34c-5p, and -297 that have not yet been described in MS to the best of our knowledge, but rather found in cancer either as tumor suppressors or as oncogenes.18 Other miRNA species (miR-21-5p, -27b-3p, -145-5p, -146a-5p, -150-5p, -155-5p, -20a-5p, -214-3p, -24-3p, -27a-3p, and -125b-5p) were previously described as differentially expressed in RRMS in at least 1 of the following sample types: PBMCs or immune cell subpopulations, serum, whole blood, or active brain lesions.3 Moreover, this study suggests that a specific set of miRNAs could serve as potential biomarkers for MS disease activity.
The CSF miRNA profile parallels the extent of intrathecal inflammation, but is not specific to MS
Although studies on CSF miRNAs in MS are sparse, several miRNAs identified in this study were previously described by other groups either in the CSF or other sample types.4 Our work emphasizes, however, the predominance of CSF miRNA dysregulation in MS, in concordance with its pathogenesis. Furthermore, our results highlight the partially overlapping profile of CSF-upregulated miRNAs in infectious and inflammatory CNS disorders compared with relapsing MS. The CSF miRNA expression level was related to pleocytosis, as it was higher in Infect-NDs than in relapsing MS, especially regarding miR-150-5p expression, which was also associated with the presence of OCBs.
The upregulation of CSF miR-150-5p in MS is in agreement with the previously published studies by Quintana et al. and Bergman et al.19,20 The expression of this miRNA is also increased in serum and exosomes from activated lymphocytes and is therefore considered as a sensor for activation of adaptive immunity.21 In this study, however, miR-150-5p was not upregulated in peripheral compartments, again suggesting the predominance of intrathecally mediated immune processes. We illustrated by PCA that miR-150-5p, -146a-5p, and -155-5p could significantly segregate patients with RRMS from controls. ROC curve analysis was the strongest for miR-150-5p, with a sensitivity and specificity of 80% and 81%, respectively, an even stronger result than previously shown19 and comparable to ROC curves of kappa free light chains for MS diagnosis.7
The set of miRNAs (miR-20a-5p, -145-5p, -214-3p, -27a-3p, -27b-3p, -24-3p, and -29c-3p) whose expression was modified according to disease activity could be sufficient to segregate patients with relapsing MS from patients with remitting MS. Further studies, in larger independent cohorts, should investigate whether these miRNAs could serve as biomarkers of disease activity.
MiRNA profile in the serum and PBMCs of patients with MS
Five miRNAs were downregulated in the serum of both relapsing and remitting MS (miR-15a-3p, -24-3p, -126-3p, -146a-3p, and -181c-5p), whereas miR-214-3p was downregulated in the serum of relapsing MS only. Remarkably, only downregulation of miR-15a-3p was consistent in both serum and CSF, whereas miR-24-3p, -146a-5p, and -214-3p were found upregulated in the CSF. Comparably, miR-126-3p was downregulated in whole blood MS samples.22 Other miRNAs (miR-122-5p, -196b-5p, -301a-3p, and -532-5p) found in extracellular vesicles (EVs) were also decreased in the serum of patients with relapsing MS.23 This discrepancy between the CSF and peripheral compartments was also shown in Alzheimer disease and intracerebral hemorrhage, but is yet not understood.24,25 The expression patterns of miR-181c-5p remain controversial, as some have described its upregulation in the CSF and in active MS lesions.5,26 In our hands, the levels of miR-181c-5p were however too low to allow quantification in the CSF, but it was downregulated in the serum of patients with MS compared with controls. Finally, in agreement with the decrease of miR-34a-5p levels in CD4+ T cells of patients with MS, we found its expression significantly downregulated in the PBMCs of patients with relapsing MS.27
Target analysis of miRNAs dysregulated in MS
To generate hypothesis regarding the functional role of the identified miRNAs in CSF, we performed a pathway enrichment analysis on several KEGG pathway databases to reveal the most affected SiPas according to disease activity, but also in serum and PBMCs. As a single miRNA species is not sufficient to account solely for the disease, as discussed above, we grouped the different miRNAs in sets. For CSF miRNAs, we first observed the highest enrichment score for several cancer SiPas, but we only presented the results for glioma and chronic myeloid leukemia because (1) these pathways are relevant for the (neuro)immunopathologic aspect of MS disease and (2) most of the miRNAs were extensively investigated in cancer research, which naturally biases most analysis toward cancer-related pathways. Besides cancer-related SiPas, the score comparison of the different subgroups of patients (relapsing, remitting MS and SC vs each other) and the Cytoscape miRNA-messenger RNA (mRNA) network representation obtained from CSF showed pathways consistent with MS pathogenesis such as cell cycle or apoptosis, immunoregulation, neuroprotective, and neurodegenerative processes.
MiRNA molecular function prediction by gene ontology provided the highest score to several negative regulators of transcriptional activity through targeting of CCND1, STAT3, the E2Ffamily, MYC, SP1, ZBTB18, and PLAGL2 among others (findings obtained by miRNet; data not shown). This result confirms previous studies on the impact of miRNAs on the transcriptional network in MS pathogenesis.23,28–31
Our research demonstrates for the first time that the CSF profile of remitting MS (Rem) is more closely related to the profile of controls than relapsing MS (Rel) because (1) the SiPas target analysis scores were the lowest (if not absent) for the miRNAs differentially expressed between Rem vs SC (Rem/SC), whereas the scores were the highest for the ones between Rel/Rem and (2) that no common SiPas specifically stood out for the dysregulated miRNAs in Rel and Rem compared with controls. At the mRNA level, it is worth noting that, unlike most of the genes regulated by miRNAs from all groups, PLAGL2 and LMNB2 were specifically clustered to the Rel/Rem and Rem/SC subgroups. Of interest, LMNB2 was related to 2 miRNAs not previously associated with MS, namely miR-149-3p and miR-33a-3p. PLAGL2 is a transcription factor regulating Wnt signaling, and thus leading to the inhibition of neural stem cell differentiation.32 It could therefore be relevant for reparative processes during remission. LMNB2 codes for a member of the lamin family and was found 5-fold downregulated in CD4+ T cells of patients with remitting MS compared with HCs.33 Altogether, the pathway enrichment analysis with the miRNA sets identified in this study remarkably pinpoints to important pathways and miRNA-gene interactions pertinent to MS pathogenesis, as evidenced in earlier studies. The differentially expressed miRNAs have relevant mRNA interactions, especially in regard to the modulation of the immune response. For instance, miR-155-5p promotes Th17 and regulatory T-cell (Treg) differentiation, Th17 function, and microglia-mediated immune responses, through expression of interleukin (IL-) 6, tumor necrosis factor alpha, and iNOS, by targeting SOCS-1.34,35 MiR-155-5p also alters blood-brain barrier permeability and enhances leukocyte adhesion to endothelial cells, whereas miR-126-3p and miR-146a-5p counteract this by modulating the expression of brain endothelial ICAM1, VCAM1.36–38 Moreover, miR-146a-5p exerts its immunoregulatory effect by blocking Th17 differentiation through repression of TRAF6 and IRAK1, which are transducers of nuclear factor kappa-light-chain-enhancer of activated B cells and thus reduces IL-6 and IL-21 expression.39 MiR-27b-3p inhibits Treg differentiation but not their function, by downregulating SMAD4 and TGFBR1.40 MiR-21-5p represses lymphocyte migration through STAT3 regulation.41 The deletion of miR-150-5p in experimental autoimmune encephalomyelitis (EAE) mice affects CD3+, CD4+, and CD8+ cells and mRNA expression of proinflammatory cytokines compared with wild-type EAE, likely by regulating CMYB.42
Cellular source and carriers of CSF miRNAs
Although our study did not investigate the cellular source of the CSF-associated miRNAs, according to the cell-specific miRNA catalogue, nearly all immune cell subpopulations express the miRNAs found in our study.43 Yet, some miRNA species might be more specific of certain cell types, as EVs of Treg cells were specifically enriched with miR-21-5p, -146-5p, and -150-5p, whereas those of Th1 and Th17 cells preferentially expressed miR-155-5p.44
Whether miRNAs are carried exclusively by EVs such as exosomes in the CSF is still an open question. However, miRNAs were enriched in exosomal fractions of CSF.45 The number of CSF EVs increases during MS relapses and GELs on brain or spinal cord MRIs were associated with an increase in CSF EVs expressing markers of CD8+ memory T cells, Th2 and Th1 cells.46 The cellular source of miRNAs from the CSF and their carriers should be then further investigated in the setting of MS.
Study limitations
The strength of this study resides in the comprehensive stepwise strategy used, the large CSF sample size, the use of a confirmatory cohort for the CSF, the comparison between compartments, and finally the comparison with miRNA expression in other disease categories. Moreover, patients were systematically sex- and age-matched to controls. However, there is inevitable heterogeneity between reports on miRNA expression analysis in MS due to discrepancies in sample numbers, collection and storage procedures, patient characteristics, study design, molecular biology techniques (microarray or qPCR, with or without preamplification, TaqMan vs SYBR Green), data analysis (quantification method, choice of reference gene), and finally the type of statistical tests. These factors, altogether or separately, possibly affect results. In this study, qPCRs from CSF and serum were normalized with an external reference, as an internal reference could not be identified. Future studies could possibly combine a panel of nonsignificantly altered miRNAs as normalizers. Some selected miRNAs were below the PCR detection level, preventing further analysis. The targeted selection of miRNAs based on the literature and immune-related miRNA PCR arrays does not exclude that other miRNA species are also dysregulated in MS. Finally, the patient cohort from a single academic center may not be representative of a more global MS cohort. An international and multicentric study that includes miRNA sequencing could provide novel miRNA targets that would refine our current results.
In conclusion, we have demonstrated that 21 miRNAs were differentially expressed in RRMS, mostly in the CSF, which is at the core of the inflammation, 7 of which were never associated with MS so far. The miRNA expression profile varied according to disease activity. It was associated with the extent of intrathecal inflammation, notably CSF pleocytosis, not only in MS but also in neuroinfectious and other neuroinflammatory diseases. Considering this, a single miRNA is unlikely to be considered as a MS biomarker, but a set of miRNAs could rather be investigated for that purpose. The strength of this work resides in (1) the study of different disease categories, (2) the comprehensive miRNA analysis performed on 3 biological compartments in patients with MS, and (3) the validation of CSF results by our second cohort and some miRNAs already described in other studies. Our work emphasizes the potential of a miRNA set to reflect intrathecal inflammation and MS disease activity. The miRNA expression profile was in concordance with their known effects on immune and neuroinflammatory processes at play in MS. In silico analysis provided a view on affected SiPas, warranting further studies into their function, cellular source, and carriers, especially for the newly found miRNAs associated with MS. Ultimately, the present report will help fundamental research on MS by paving the way for the functional characterization of miRNAs and unraveling novel pathways for possible therapeutic intervention.
Acknowledgment
The authors thank Qiagen (USA) for kindly performing the miRNA PCR arrays.
Glossary
- DMT
disease-modifying treatment
- EAE
experimental autoimmune encephalomyelitis
- EV
extracellular vesicle
- FDR
false discovery rate
- GEL
gadolinium-enhancing lesion
- HC
healthy control
- IgG
immunoglobulin G
- Infect-ND
infectious neurologic disorder
- Inflam-ND
inflammatory neurologic disorder
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- miRNA
microRNA
- mRNA
messenger RNA
- MS
multiple sclerosis
- OCB
oligoclonal band
- PBMC
peripheral blood mononuclear cell
- PCA
principal component analysis
- qPCR
quantitative PCR
- ROC
receiver operating characteristic
- RRMS
relapsing-remitting MS
- RT
reverse transcription
- SC
symptomatic control
- SiPa
signaling pathway
- Treg
regulatory T-cell
Appendix. Authors
Study funding
This work was supported by the Université catholique de Louvain (Fonds Spécial de Recherche), the Charcot Foundation (fondation-charcot.org/en), Fondation Louvain (uclouvain.be/fr/chercher/fondation-louvain), and the Fonds National de la Recherche Scientifique (frs-fnrs.be/fr/). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Disclosure
O. Perdaens, H.A. Dang, and L. D'Auria report no disclosure. V. van Pesch received travel grants from Biogen, Sanofi, Merck, and Roche. His institution receives honoraria for consultancy and lectures from Biogen, Sanofi, Merck, Roche, Teva, and Novartis Pharma and research grants from Novartis Pharma, Sanofi, and Roche. Go to Neurology.org/NN for full disclosures.
References
- 1.Reich DS, Lucchinetti CF, Calabresi PA. Multiple sclerosis. N Engl J Med 2018;378:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheleznyakova GY, Piket E, Marabita F, et al. . Epigenetic research in multiple sclerosis: progress, challenges, and opportunities. Physiol Genomics 2017;49:447–461. [DOI] [PubMed] [Google Scholar]
- 3.Martin NA, Illes Z. Differentially expressed microRNA in multiple sclerosis: a window into pathogenesis. Clin Exp Neuroimmunol 2014;5:149–161. [Google Scholar]
- 4.Dolati S, Marofi F, Babaloo Z, et al. . Dysregulated network of miRNAs involved in the pathogenesis of multiple sclerosis. Biomed Pharmacother 2018;104:280–290. [DOI] [PubMed] [Google Scholar]
- 5.Haghikia A, Haghikia A, Hellwig K, et al. . Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology 2012;79:2166–2170. [DOI] [PubMed] [Google Scholar]
- 6.Teunissen CE, Tumani H, Bennett JL, et al. . Consensus guidelines for CSF and blood biobanking for CNS biomarker studies. Mult Scler Int 2011;2011:246412. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Bayart JL, Muls N, van Pesch V. Free Kappa light chains in neuroinflammatory disorders: complement rather than substitute? Acta Neurol Scand 2018;138:352–358. [DOI] [PubMed] [Google Scholar]
- 8.Muls NG, Dang HA, Sindic CJ, van Pesch V. Regulation of Treg-associated CD39 in multiple sclerosis and effects of corticotherapy during relapse. Mult Scler 2015;21:1533–1545. [DOI] [PubMed] [Google Scholar]
- 9.Teunissen C, Menge T, Altintas A, et al. . Consensus definitions and application guidelines for control groups in cerebrospinal fluid biomarker studies in multiple sclerosis. Mult Scler 2013;19:1802–1809. [DOI] [PubMed] [Google Scholar]
- 10.Thompson AJ, Banwell BL, Barkhof F, et al. . Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 2018;17:162–173. [DOI] [PubMed] [Google Scholar]
- 11.Lublin FD, Reingold SC, Cohen JA, et al. . Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology 2014;83:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiber H. Flow rate of cerebrospinal fluid (CSF)—a concept common to normal blood-CSF barrier function and to dysfunction in neurological diseases. J Neurol Sci 1994;122:189–203. [DOI] [PubMed] [Google Scholar]
- 13.Barkhof F, Filippi M, Miller DH, et al. . Comparison of MRI criteria at first presentation to predict conversion to clinically definite multiple sclerosis. Brain 1997;120:2059–2069. [DOI] [PubMed] [Google Scholar]
- 14.Baraniskin A, Kuhnhenn J, Schlegel U, et al. . Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 2011;117:3140–3146. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 16.Lê S, Josse J, Husson F. FactoMineR: an R Package for multivariate analysis. J Stat Soft 2008;25:1–18. [Google Scholar]
- 17.Piket E, Zheleznyakova GY, Kular L, Jagodic M. Small non-coding RNAs as important players, biomarkers and therapeutic targets in multiple sclerosis: a comprehensive overview. J Autoimmun 2019;101:17–25. [DOI] [PubMed] [Google Scholar]
- 18.Ratovitski EA. Tumor protein p63/microRNA network in epithelial cancer cells. Curr Genomics 2013;14:441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quintana E, Ortega FJ, Robles-Cedeño R, et al. . miRNAs in cerebrospinal fluid identify patients with MS and specifically those with lipid-specific oligoclonal IgM bands. Mult Scler 2017;23:1716–1726. [DOI] [PubMed] [Google Scholar]
- 20.Bergman P, Piket E, Khademi M, et al. . Circulating miR-150 in CSF is a novel candidate biomarker for multiple sclerosis. Neurol Neuroimmunol Neuroinflamm 2016;3:e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Candia P, Torri A, Gorletta T, et al. . Intracellular modulation, extracellular disposal and serum increase of MiR-150 mark lymphocyte activation. PLoS One 2013;8:e75348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox MB, Cairns MJ, Gandhi KS, et al. . MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One 2010;5:e12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selmaj I, Cichalewska M, Namiecinska M, et al. . Global exosome transcriptome profiling reveals biomarkers for multiple sclerosis. Ann Neurol 2017;81:703–717. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Reddy PH. Are circulating microRNAs peripheral biomarkers for Alzheimer's disease? Biochim Biophys Acta 2016;1862:1617–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwuchukwu I, Nguyen D, Sulaiman W. MicroRNA profile in cerebrospinal fluid and plasma of patients with spontaneous intracerebral hemorrhage. CNS Neurosci Ther 2016;22:1015–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Junker A, Krumbholz M, Eisele S, et al. . MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009;132:3342–3352. [DOI] [PubMed] [Google Scholar]
- 27.Lindberg RL, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur J Immunol 2010;40:888–898. [DOI] [PubMed] [Google Scholar]
- 28.Baulina N, Kulakova O, Kiselev I, et al. . Immune-related miRNA expression patterns in peripheral blood mononuclear cells differ in multiple sclerosis relapse and remission. J Neuroimmunol 2018;317:67–76. [DOI] [PubMed] [Google Scholar]
- 29.Riveros C, Mellor D, Gandhi KS, et al. . A transcription factor map as revealed by a genome-wide gene expression analysis of whole-blood mRNA transcriptome in multiple sclerosis. PLoS One 2010;5:e14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liguori M, Nuzziello N, Introna A, et al. . Dysregulation of microRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Front Mol Neurosci 2018;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuzziello N, Vilardo L, Pelucchi P, et al. . Investigating the role of microRNA and transcription factor co-regulatory networks in multiple sclerosis pathogenesis. Int J Mol Sci 2018;19:E3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng H, Ying H, Wiedemeyer R, et al. . PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell 2010;17:497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berge T, Eriksson A, Brorson IS, et al. . Quantitative proteomic analyses of CD4(+) and CD8(+) T cells reveal differentially expressed proteins in multiple sclerosis patients and healthy controls. Clin Proteomics 2019;16:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao R, Ma YL, Liang W, et al. . MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One 2012;7:e46082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardoso AL, Guedes JR, Pereira de Almeida L, Pedroso de Lima MC. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology 2012;135:73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Ramirez MA, Wu D, Pryce G, et al. . MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J 2014;28:2551–2565. [DOI] [PubMed] [Google Scholar]
- 37.Wu D, Cerutti C, Lopez-Ramirez MA, et al. . Brain endothelial miR-146a negatively modulates T-cell adhesion through repressing multiple targets to inhibit NF-kappaB activation. J Cereb Blood Flow Metab 2015;35:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerutti C, Edwards LJ, de Vries HE, Sharrack B, Male DK, Romero IA. MiR-126 and miR-126* regulate shear-resistant firm leukocyte adhesion to human brain endothelium. Sci Rep 2017;7:45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Wang X, Choi IY, et al. . miR-146a modulates autoreactive Th17 cell differentiation and regulates organ-specific autoimmunity. J Clin Invest 2017;127:3702–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Severin ME, Lee PW, Liu Y, et al. . MicroRNAs targeting TGFbeta signalling underlie the regulatory T cell defect in multiple sclerosis. Brain 2016;139:1747–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Han J, Cui Y, Zhou X, Fan K. miRNA-21 inhibition enhances RANTES and IP-10 release in MCF-7 via PIAS3 and STAT3 signalling and causes increased lymphocyte migration. Biochem Biophys Res Commun 2013;439:384–389. [DOI] [PubMed] [Google Scholar]
- 42.Hu Z, Cui Y, Qiao X, et al. . Silencing miR-150 ameliorates experimental autoimmune encephalomyelitis. Front Neurosci 2018;12:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juzenas S, Venkatesh G, Hübenthal M, et al. . A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res 2017;45:9290–9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torri A, Carpi D, Bulgheroni E, et al. . Extracellular microRNA signature of human helper T cell subsets in health and autoimmunity. J Biol Chem 2017;292:2903–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yagi Y, Ohkubo T, Kawaji H, et al. . Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neurosci Lett 2017;636:48–57. [DOI] [PubMed] [Google Scholar]
- 46.Geraci F, Ragonese P, Barreca MM, et al. . Differences in intercellular communication during clinical relapse and gadolinium-enhanced MRI in patients with relapsing remitting multiple sclerosis: a study of the composition of extracellular vesicles in cerebrospinal fluid. Front Cell Neurosci 2018;12:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw PCR files and detailed in silico and statistical outputs are not included in this article. Any unpublished and anonymized data will be shared upon request from a qualified investigator. Strengthening the Reporting of Observational Studies in Epidemiology guidelines have been respected.