Abstract
Dengue virus is a major pathogen and severe infections can lead to life threatening dengue hemorrhagic fever (DHF). Dengue exists as four serotypes and DHF is often associated with secondary heterologous infections. Antibody dependent enhancement (ADE) may drive higher virus loads in these secondary infections, and is purported to result from antibodies that recognize dengue but fail to fully neutralize. We have characterized two antibodies, 2C8 and 3H5, which bind to the envelope protein. 3H5 is highly unusual as it is both potently neutralizing, but promotes little if any ADE, whereas 2C8 has strong capacity to promote ADE. We show that 3H5 shows resilient binding in endosomal pH conditions and neutralizes at low occupancy. Immune complexes of 3H5 and dengue virus do not efficiently interact with Fcγ receptors, which we propose is due to the binding mode of 3H5 and which constitutes the primary mechanism of how ADE is avoided.
Introduction
Dengue is the most significant viral illness transmitted between humans and mosquitoes. The geographic spread of the Aedes aegypti mosquito vector is steadily expanding and disease incidence has increased considerably with an estimated 390 million infections occurring each year, a quarter of which are symptomatic1. Frequent outbreaks of dengue in Southeast Asia, South America and the West Pacific regions are a major burden on the healthcare systems and economies of affected countries. Dengue fever is a self-limiting febrile illness caused by four frequently co-circulating virus serotypes, DENV-1 to DENV-4 which differ in amino acid sequence by up to 30%. Infection with one serotype usually results in long-lasting immunity to this serotype. In a secondary infection with a different serotype, however, antibodies elicited during the primary infection may instead enhance dengue through a mechanism termed antibody dependent enhancement (ADE)2. ADE occurs when insufficiently neutralizing pre-existing antibodies opsonize virus and the infectious virus-antibody complexes are internalized by Fcγ receptor bearing cells such as monocytes or macrophages3. Almost all neutralizing antibodies promote ADE at sub-neutralizing concentrations and it is generally believed that ADE drives disease severity in secondary DENV infections4–6. Dengue viruses frequently co-circulate with the related Zika virus (ZIKV) and mutual ADE between them has been demonstrated7,8. Phase III trials of the most advanced dengue vaccine, CYD-TDV, a tetravalent vaccine directed against all four DENV serotypes, has shown higher hospitalization rates than controls in vaccinated seronegative children under 9 years of age, possibly due to ADE9,10. To circumvent exacerbation of dengue infections, vaccines and potential therapeutic antibodies should minimize the facilitation of ADE.
DENV is a member of the Flaviviridae family of small enveloped viruses with a single-stranded, positive-sense RNA genome of around 11kb length. Besides DENV, the Flaviviridae family contains other important arthropod-borne pathogens including West Nile virus (WNV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), tick-borne encephalitis virus (TBEV), and ZIKV. The DENV genome encodes seven non-structural proteins NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5, as well as the three structural proteins capsid (C), precursor membrane (prM), and envelope (E) protein. The E protein on the viral surface is the major target of neutralizing antibodies and consists of three distinct domains termed EDI, EDII, and EDIII. Of these, EDIII mediates attachment to target cells, prior to internalization into host endosomes11. EDII harbours the fusion loop, which becomes exposed through conformational changes triggered by the low pH environment of the endosome and is inserted into the host endosomal membrane in the course of the fusion process12.
The mature form of the viral particle has been studied at high resolution via cryo-EM and displays a smooth quasi-icosahedral surface of 90 E dimers lying flat on the viral membrane in a “herringbone” arrangement13. Averaging imposed in cryo-EM image reconstruction, however, does not fully capture that there is considerable dynamic motion in the E protein shell. These dynamics allow numerous neutralizing antibodies to bind “cryptic” epitopes which seem sterically inaccessible on the virion based on high resolution reconstructions5,14–16. The phenomenon, termed viral breathing, may also be the underlying cause for the irreversible transition to an expanded “bumpy” virus morphology and loss of icosahedral order which has been reported for some DENV-2 strains incubated at elevated temperatures17,18.
Human immune sera in a primary DENV infection are dominated by antibodies which target the conserved fusion loop of EDII and which are cross-reactive between DENV serotypes, cause ADE, and are poorly neutralizing19,20. In contrast to this poorly-neutralizing response, the neutralizing human humoral response was shown to be mostly serotype-specific and targets complex quaternary epitopes which are present only on intact virions21–23. In addition, broadly neutralizing antibodies that bind dimeric E by recognizing the interface between two E monomers, have been reported24,25. Finally, antibodies against EDIII represent a small fraction and play a minor role in the neutralizing response, but can be among the most potent21,26,27. The non-immunodominant EDIII has been recently suggested as an important epitope in immunotherapy for flaviviral infections14,28.
We describe the structural and in vitro characterization of two serotype-specific murine antibodies, 2C8 and 3H5, directed against EDIII of DENV-2. Both antibodies are potently neutralizing, yet possess greatly differing ADE properties. Contrary to 2C8, 3H5 displays dramatically reduced infection enhancement. An analysis of the binding affinities showed significant pH-dependent differences, with binding of 3H5 being more robust at endosomal pH. Crystal structures of 2C8 and 3H5 Fabs in complex with EDIII define the binding footprints of the antibodies. Compared to 2C8, 3H5 targets residues buried between E dimers and located close to the viral membrane. Importantly, we show that, in contrast to 2C8, 3H5-DENV2 immunecomplexes show either no or weak interaction with Fcγ receptors, providing a mechanistic explanation of the low ADE activity displayed by 3H5. Our structural data along with the binding data demonstrate how binding of few molecules of the potent 3H5 can exert neutralization while avoiding ADE, whereas 2C8 causes classical ADE.
Results
Neutralization and enhancement properties of 2C8 and 3H5
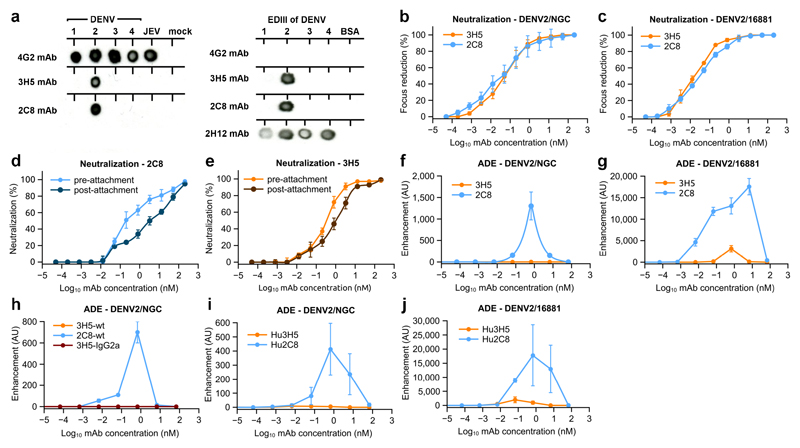
3H5 is a IgG1 monoclonal antibody (mAb) raised by immunizing mice with DENV2 strain New Guinea C (NGC)29. 2C8 is an IgG2a mAb raised in mice immunized with DENV2 16681 in our laboratories that has not been previously reported. Both are serotype specific to DENV2, demonstrated by dot blot and react to recombinant EDIII (Fig. 1a). Focus reduction neutralization assays (Fig. 1b and c) showed highly potent neutralization of DENV2 NGC and 16681 for both antibodies with 50% neutralization titers in the sub-nanomolar range (50% FRNT at 0.044 nM and 0.025 nM for 3H5 and 2C8, respectively). To test if the antibodies block binding to the cell or entry of the virus, we performed pre- and post-attachment neutralization assays with both antibodies (Fig. 1d and e). 2C8 and 3H5 were still capable of significantly inhibiting infection after the virus had bound to cells.
Figure 1. Neutralization and enhancement characteristics of the anti-DENV2 antibodies 3H5 and 2C8.
a, Dot blot showing specificity of 3H5 and 2C8. Virus or recombinant EDIII of the 4 DENV serotypes were probed with the indicated antibodies. The dot blot was repeated three times independently with similar results. Antibodies 4G2 and 2H12 were included as controls. 4G2 is a cross-reactive anti-dengue and anti-Japanese encephalitis virus (JEV) mAb directed against EDII. 2H12 is a cross-reactive, EDIII-specific mAb. b and c, Neutralization activities of 3H5 (orange) and 2C8 (blue) with DENV2 strains NGC and 16681 as indicated. d and e, Pre- and post-attachment neutralization of DENV2 by 2C8 and 3H5 as indicated. Pre-attachment neutralization was determined with virus preincubated with the corresponding antibody prior to transfer to Vero cells. For post-attachment neutralization virus was incubated with the Vero cells before addition of antibody. f-j, Enhancement properties of 3H5, 2C8, 3H5 switched to IgG2a isotype (3H5-IgG2a), and humanized 3H5 and 2C8 (Hu3H5 and Hu2C8) as indicated. For all neutralization assays and antibody-dependent enhancement assays three technical replicates were carried out and the data are shown as mean ± s.e.m.
We analyzed ADE using U937 cells, a human Fcγ-receptor (FcγR) expressing monocytic cell line which is resistant to dengue infection in the absence of antibody (Fig. 1f and g). 2C8 demonstrated typical infection enhancement with peak titers of over 1000-fold enhancement over background and a wide range of concentrations at which enhancement occurred. In contrast, 3H5 showed no enhancing capacity for DENV2 NGC and dramatically reduced enhancement of DENV2 16681 at a very narrow concentration range. Similarly, 3H5 showed much reduced enhancement compared to 2C8 in K562 cells (Supplementary Fig. 1a and b). As 2C8 and 3H5 belong to different IgG subtypes (IgG2a and IgG1, respectively) it was necessary to rule this difference out as a cause of varying enhancement. We cloned cDNA encoding the heavy chain variable region of 3H5 onto the constant region of IgG2a and co-expressed it with the 3H5 light chain. 3H5-IgG2a showed no enhancement of infection for DENV2 NGC (Fig. 1h) while still potently neutralizing infection. Some ADE has been previously reported for 3H5 using P388D1 – a mouse macrophage-like cell line30. To control for incompatibility of murine Ig-Fc with human Fc receptors, we repeated the assays with humanized 2C8 and 3H5 (Hu2C8 and Hu3H5 on a human IgG1 background). Hu3H5 showed little or no enhancement for DENV2/16681 and DENV2/NGC compared to robust enhancement for Hu2C8 (Fig.1i and j).
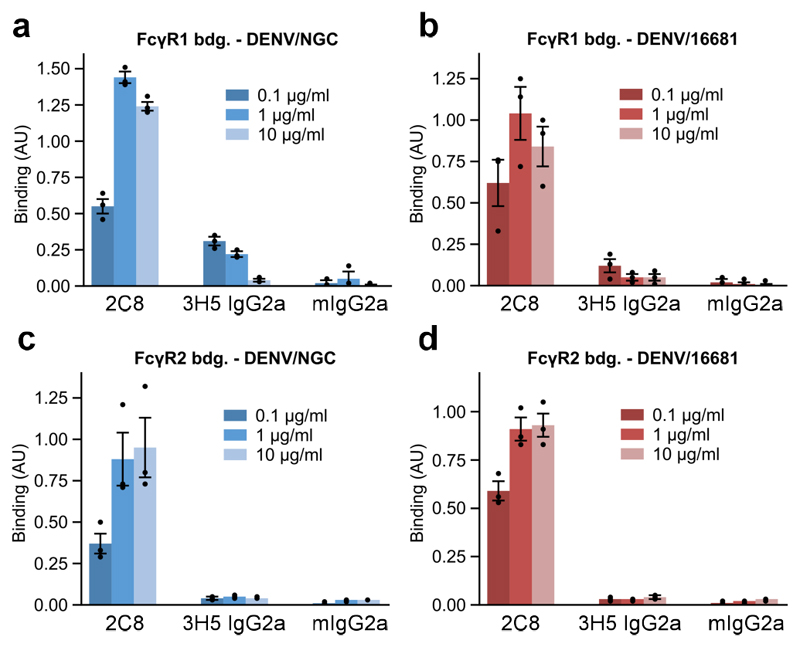
FcγR-mediated enhancement occurs mainly via FcγR1 in U937 cells and mainly via FcγR2a in K562 cells as evidenced by antibody blocking of Fc receptors (Supplementary Fig. 1c and d). To assess the interaction between virus-attached antibodies and Fcγ receptors, we measured binding of preformed antibody/virus immune complexes to immobilized Fcγ1 and Fcγ2a receptors for both DENV2/NGC and DENV2/16681 (Fig. 2a-d). In contrast to 2C8 complexes, which efficiently bound to Fcγ receptors, 3H5/DENV2 complexes showed close to no interaction with FcγR2a and greatly reduced interaction with FcγR1. As a control, to confirm that FcγR-mediated enhancement is indeed occurring with 2C8, we disrupted FcγR interaction by introducing LALA mutations into the Fc region of Hu2C8 and compared neutralization and enhancement (Supplementary Fig. 2a-d). As expected, enhancement was abolished in the Hu2C8-LALA antibodies. Our neutralization, enhancement, and FcγR binding experiments demonstrate that both 2C8 and 3H5 are potent neutralizers of dengue, however, in stark contrast to 2C8, 3H5 shows dramatically reduced ADE and fails to efficiently interact with Fc receptors.
Figure 2. Binding of DENV immunecomplexes to Fcγ receptors.
a-d, ELISA binding assays of 2C8-DENV2 or 3H5-DENV2 immune complexes to Fcγ-receptor 1 (FcγR1) or Fcγ-receptor 2a (FcγR2a). Data is shown for DENV2 strains NGC and 16681 as indicated. Antibody/virus complexes were pre-incubated and added to plates coated with FcγR1 or FcγR2a. Mouse IgG2a (mIgG2a) was included as a negative control. Three technical replicates of all ELISA experiments were performed and data are shown as mean ± s.e.m.
2C8 and 3H5 show varying binding in low pH conditions
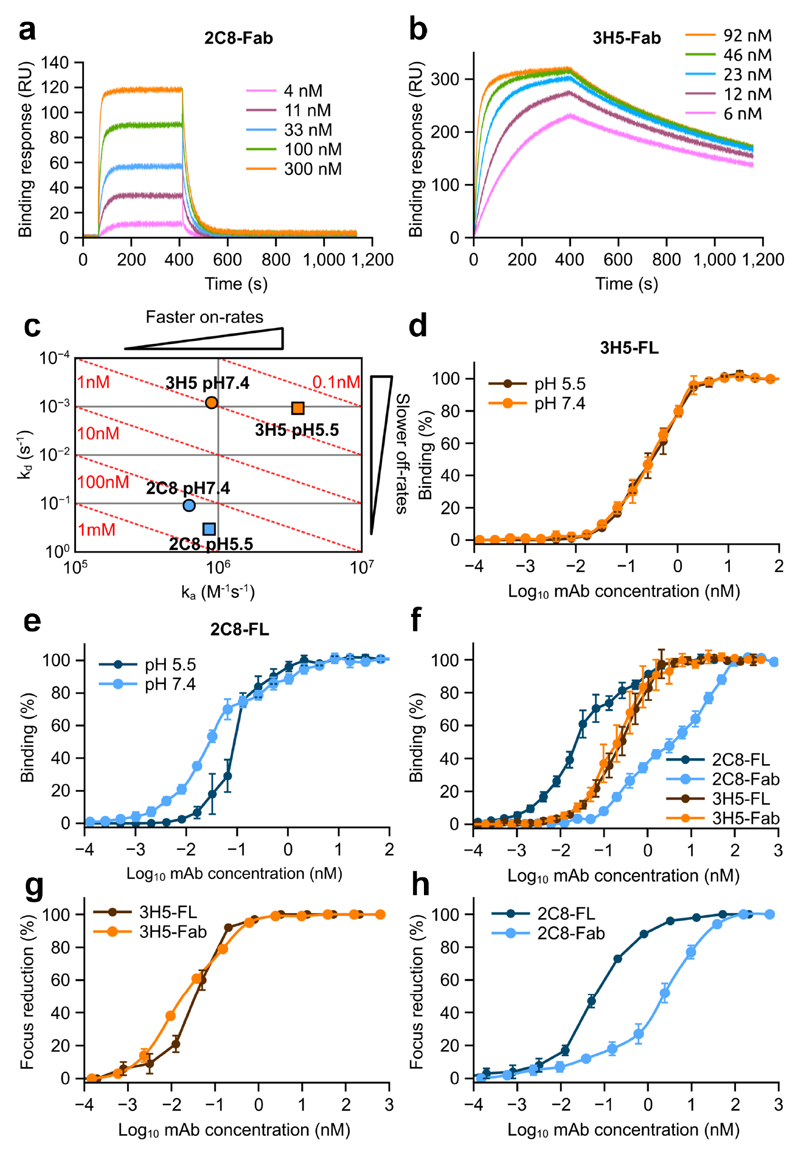
To further investigate the interaction of 2C8 and 3H5 with dengue we performed a series of surface Plasmon resonance (SPR) binding experiments to recombinant EDIII. Although 3H5 and 2C8 have similar neutralization potency, their affinities to EDIII revealed significant differences. 3H5 Fab showed nearly 200-fold higher affinity to the antigen than 2C8 Fab (1.0 nM vs. 176.7 nM, respectively) (Supplementary Table 1a). The discrepancy is an effect of varying binding kinetics. SPR sensograms (Fig. 3a and 3b) show that 3H5 Fab binds EDIII rapidly (ka ~ 9.0x105 M-1s-1) and dissociates slowly (kd ~ 8.3x10-4 s-1), while 2C8 associates comparatively slowly (ka ~ 6.2x105 M-1s-1) and dissociates quickly (kd ~ 0.1 s-1) (Supplementary Table 1a). We also assessed the robustness of binding at acidic pH (5.5), mimicking the conditions in the late endosomal compartment during viral fusion12,31. 3H5 Fab and 2C8 Fab reacted differently to the acidic environment; the affinity of 3H5 increased 3-fold (~1.0 nM at neutral pH to ~0.3 nM at low pH) while that of 2C8 decreased 2-fold (~176.7.0 nM at neutral pH to ~362.5 nM at low pH) (Fig. 3c and Supplementary Table 1a). Of note, at low pH the dissociation rate of 2C8 Fab was ~300-fold higher than that of 3H5 Fab.
Figure 3. Binding properties of 2C8 and 3H5.
a and b, Representative SPR profiles of 2C8 and 3H5 Fabs, respectively, binding to recombinant EDIII. Varying concentrations of Fab were flowed over immobilized EDIII of DENV2. c, On/off rate map showing binding affinities and rate constants for 2C8 (blue) and 3H5 (orange) at pH 7.4 (circles) and pH 5.5 (squares). Log-scale plot of kd (y-axis) against ka (x-axis). In this plot, affinity (kd/ka) increases from bottom left to top right, resulting in iso-affinity diagonals (red dotted lines). d and e, ELISA binding assays of full-length 2C8 and full-length 3H5 (2C8-FL and 3H5-FL) measured at pH 7.4 and pH 5.5 as indicated. (f) ELISA binding assay of full-length 3H5 (brown) and 2C8 (light blue) and corresponding Fabs (orange and dark blue, respectively) to DENV2 virions. (g and h) Neutralization activities of Fabs and full-length antibodies (FL) of 2C8 (g) and 3H5 (h). Three technical replicates of all neutralization and ELISA assays were carried out and the data are shown as mean ± s.e.m.
Next, we measured binding avidities of Fabs and full-length antibodies for whole virus at both neutral and endosomal pH via ELISA (Supplementary Table 1b). For full-length dimeric 2C8 IgG we observed that the shift to lower pH was accompanied by a decrease in avidity (0.03 nM at neutral pH to 0.1 nM at low pH), consistent with the effect observed for 2C8 Fab, while there was no difference in the case of 3H5 IgG (0.31 nM at neutral pH to 0.35 nM at low pH) (Fig. 3d and e). Finally, comparison of Kd values obtained for full-length 3H5 and 3H5 Fab (~0.3 nM and 0.2 nM respectively), suggests a monovalent mode of antibody binding to the virus, while the large difference in Kd for 2C8 Fab and full-length 2C8 (4.76 nM vs. 0.03 nM respectively) may indicate a bi-valent mode of interaction (Fig. 3f and Supplementary Table 1b). In line with the notion of bivalent attachment of 2C8, 2C8 Fabs showed greatly reduced neutralization potency vs full-length 2C8, while there was little difference in the case of 3H5 (Fig. 3g and h).
3H5 neutralizes at low occupancy
For neutralization to occur a sufficient number of antibodies need to engage their epitopes on the virion, i.e. an occupancy threshold must be exceeded. For example, in the case of West-Nile virus (WNV), the antibody E16 needs to bind 30% of all available sites on the virion in order to neutralize 50% of the virus. We analyzed the occupancy thresholds of 2C8 and 3H5 with the binding data described above. Using the binding constant and concentration of half-maximal neutralization5 we calculated the fraction of accessible epitopes on the virion that are bound by antibody when 50% of virus is neutralized. These data indicate that 2C8 needs to engage 45% of all available epitopes in order to neutralize 50% of the virus. However, the non-enhancing antibody 3H5 can achieve 50% neutralization when only 14% of epitopes are engaged.
Crystal structures of 3H5 Fab and 2C8 Fab in complex with EDIII
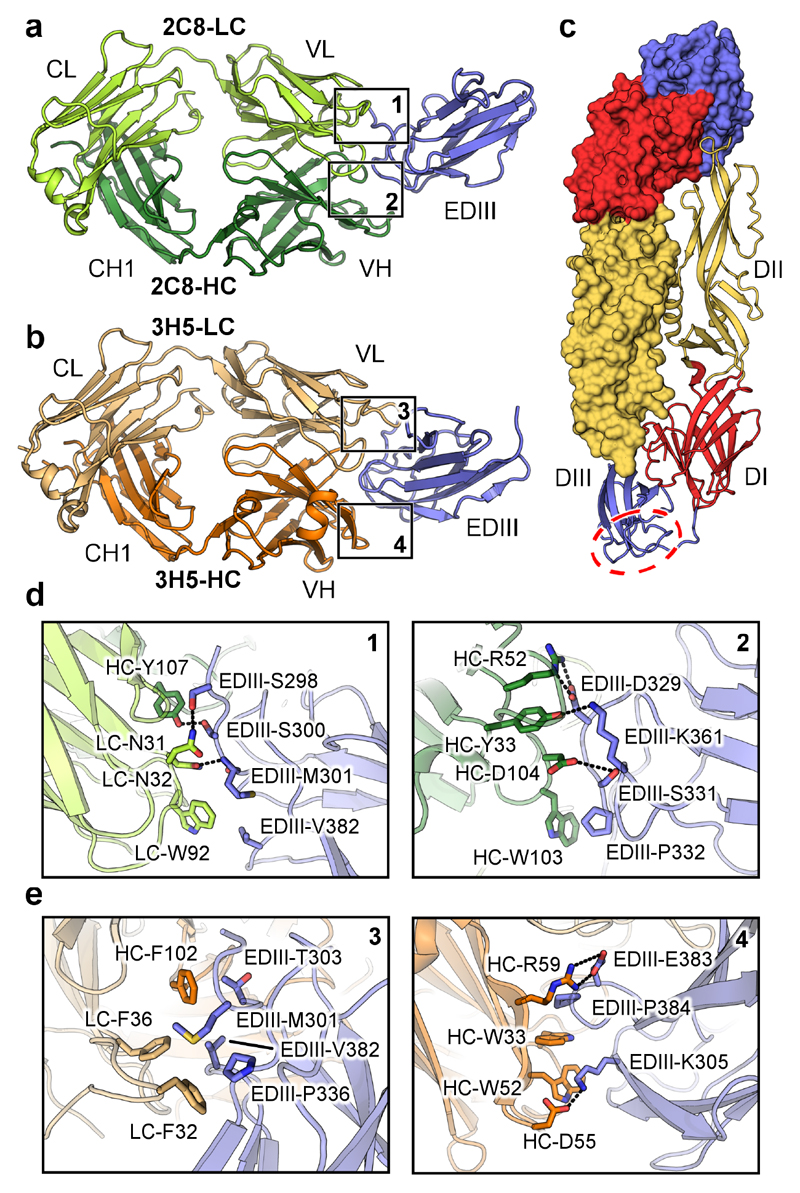
We determined the structures of 2C8 Fab and 3H5 Fab bound to refolded EDIII of DENV2 NGC (Fig. 4a, 4b and Table 1). The structure of 3H5-EDIII was determined for two crystal forms. Crystal form 1 diffracted to 3.0 Å and contained two 3H5-EDIII complexes per asymmetric unit (ASU), crystal form 2 diffracted to 2.2 Å and contained one complex per ASU. The elbow angles (the angle between the constant and variable domains of the Fab) in the 2 crystal forms (134° and 137° in crystal form 1 and 146° in crystal form 2) differed slightly. However, there is good agreement between the structures as evidenced by the low r.m.s.d. between equivalent Cα atoms of the constant domains of the Fab (0.29 Å), the variable domains of the Fab (0.24 Å) and the EDIII (0.30 Å). As there are no differences in the complex interfaces the higher resolution data obtained for crystal form 2 were used for structural analysis (representative electron density shown in Supplementary Fig. 3). The 2C8-EDIII crystals diffracted to 2.0 Å and contained two complexes per ASU. The Fabs in the ASU differed in elbow angles by 30° (187.2° and 157.2°) consistent with previous observations that the hinge region is flexible32. The r.m.s.d. values between equivalent Cα atoms of the constant domains of the Fab, the variable domains of the Fab and the EDIII in the two molecules were 0.53 Å, 0.31 Å and 0.39 Å respectively.
Figure 4. Crystal structures of 2C8 and 3H5 EDIII complexes.
a, 2C8 Fab (heavy chain in dark green, light chain in light green) in complex with EDIII (blue). Light chain (LC) and heavy chain (HC) as well as constant light (CL), variable light (VL), constant heavy (CH1), and variable heavy (VH) domains are labelled. b, 3H5 Fab (heavy chain in orange, light chain in light orange) in complex with EDIII (blue). c, Overview of the E-protein architecture. A head-to-tail dimer is shown with one monomer rendered as surface and colored by domain (DI in red, DII in yellow and DIII in blue). The lateral ridge region of DIII is indicated by a red dotted circle. d and e, Key contacts between heavy chain (HC) and light chain (LC) residues of 2C8 and 3H5 with EDIII. The locations of the highlighted interaction surfaces on the full structures is indicated by numbered boxes in a and b. Polar contacts are indicated by dotted lines. Domains are colored as above.
Table 1. Data collection and refinement statistics.
| 3H5-EIIID2 crystal form 1 | 3H5-EIIID2 crystal form 2 | 2C8-EIIID2 | |
|---|---|---|---|
| Data collection | |||
| Space group | C 2 2 21 | P212121 | P1 |
| Cell dimensions | |||
| a, b, c (Å) | 40.93, 264.28, 272.47 | 38.49, 131.57, 139.44 | 42.56, 61.42, 104.59 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 77.54, 81.11, 85.53 |
| Resolution (Å) | 40.0-2.9 | 21.3-2.2 | 33.7-2.0 |
| Rsym or Rmerge | 0.125 (0.738) | 0.089 (0.547) | 0.087(0.67) |
| I / σI | 20.5(2.9) | 14.8(2.8) | 15.0(1.5) |
| Completeness (%) | 100(100) | 100 (100) | 97(96) |
| Redundancy | 9.7(9.1) | 6.6 (6.5) | 3.9(4.0) |
| Refinement | |||
| Resolution (Å) | 40.0-2.9 | 21.3-2.2 | 33.7-2.0 |
| No. reflections | 33359 | 37183 | 66849 |
| Rwork / Rfree | 20.3/22.1 | 17.6/21.8 | 18.0/21.6 |
| No. atoms | |||
| Protein | 8129 | 4056 | 8143 |
| Non-protein | 14 | 444 | 624 |
| B-factors | |||
| Protein | 67.6 | 43.5 | 36.5 |
| Non-protein | 63.1 | 49.9 | 41.6 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.010 | 0.010 | 0.008 |
| Bond angles (°) | 1.3 | 1.2 | 1.1 |
Both 3H5 and 2C8 recognize the lateral ridge region of EDIII (indicated in Fig. 4c). The total occluded surface areas of the interfaces are similar for the 3H5-EDIII (792 Å2 for EDIII and 730 Å2 for Fab) and 2C8-EDIII complexes (730 Å2 for EDIII and 664 Å2 for Fab). The 2C8-EDIII interface is dominated by an extensive network of hydrogen bonding and polar contacts (Supplementary Table 2), with a prominent salt bridge forming between HC-R52 and EDIII-D329 (Fig. 4d). In the case of 3H5, there is a comparatively large proportion of involved hydrophobic interactions, as well as two salt bridges between HC-R59 and EDIII-E383 and HC-D55 and EDIII-K305 (Fig. 4e and Supplementary Table 3). EDIII residues M301, K305, E383 and P384, located at the binding site (Fig. 4e), have been shown previously by mutation to affect 3H5 affinity33. The binding surface of 3H5 involves five out of six complementarity determining regions (CDRs) (heavy chain: H31-35, H47-59, H99-102, light chain: L27-36, and L95-100); while for the 2C8 complex all six CDRs contribute to antigen binding (H33, H50-56, H102-107, L30-32, L49-53, and L91-96) (Supplementary Fig. 4, Supplementary Tables 2 and 3). CDR-L1 of 3H5 contributes an especially prominent hydrophobic interaction (LC-F32) which is inserted into the surface of EDIII (Supplementary Fig. 4c).
3H5 targets additional C-strand and CD loop residues on EDIII
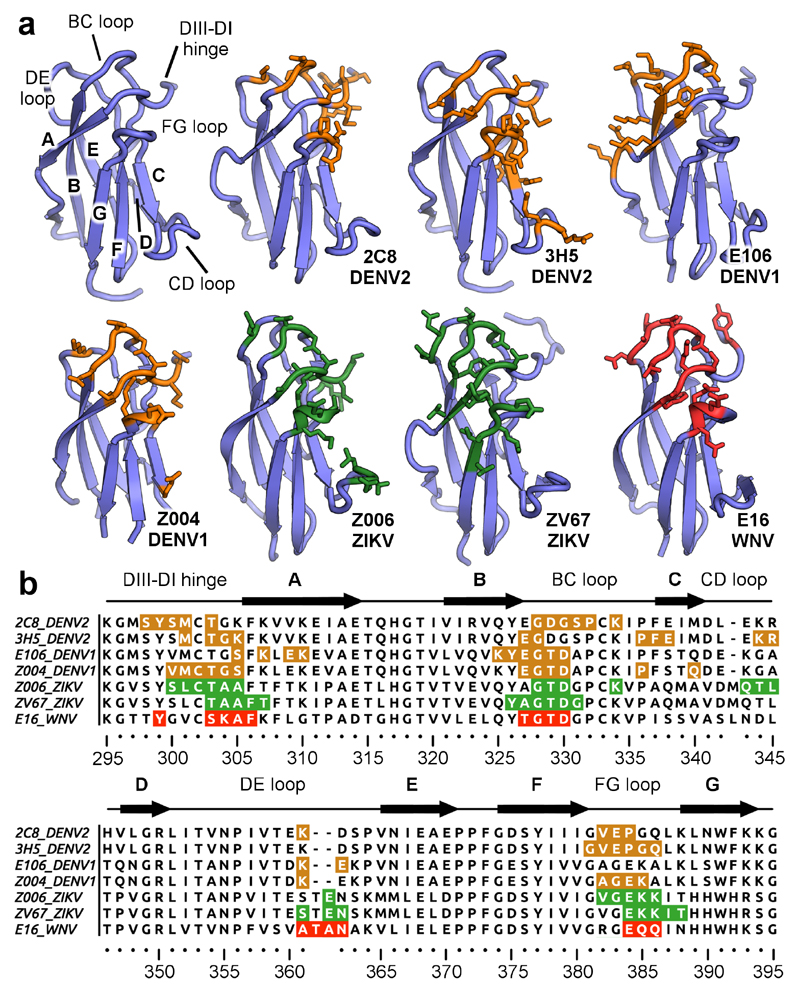
We determined the epitopes of 3H5 and 2C8 on EDIII with our crystal structures and compared these to published data on antibodies targeting the lateral ridge in flaviviruses (Fig. 5a and 5b)14,34–36. The lateral ridge region is composed of the DIII-DI hinge, A-strand, BC-loop, DE-loop, and FG-loop of EDIII (Fig. 5a, top left). 2C8 predominantly recognizes the N-terminal region of EDIII (DIII-DI hinge residues 298-303) and the BC loop (residues 328-334), but residues K361, V282, E383 and P384 also contact the Fab. The 2C8 Fab binding footprint on EDIII is not dissimilar to that of other lateral ridge antibodies and shows overlap with WNV-specific E16 or ZIKV-specific ZV67. In comparison, the DENV1-specific E106 slightly differs from other lateral ridge antibodies in that it does not bind the FG loop.
Figure 5. Epitope recognition by 3H5 and 2C8.
a, EDIII epitopes recognized by lateral ridge antibodies directed against different flaviviruses. The antibody designations and targeted viruses are indicated. The general architecture of EDIII is shown in the top left and important regions are labelled. Residues engaged by antibodies on dengue virus EDIIIs are colored in orange, ZIKV EDIIIs in green and WNV EDIIIs in red. All contacted residues are rendered as sticks. b, Sequence alignment of DENV, ZIKV, and WNV EDIIIs with highlighted antibody epitopes (coloring as above, antibodies and viruses indicated on the left of sequences). EDIII regions are labelled above the sequences.
The epitope of 3H5 overlaps with that of 2C8, yet is nevertheless distinct. It shares residues in the lateral ridge including T303, G304, E383 and P384, however, has a reduced footprint in the BC loop as compared to the panel of antibodies (Fig. 5b). 3H5 has a further area of recognition within the C-strand and also additional contacts in the CD loop, which are not found in the classical lateral ridge epitope and are not bound by 2C8, E16, E106, or ZV67. The lateral ridge epitope targeted by antibodies such as 3H5 and 2C8 is distinct from recently characterized antibodies with different quaternary epitopes, e.g. 2D22 or C8-EDE which bind roughly on the opposite face of EDIII (Supplementary Fig. 5)24,25,37,38. Taken together, our comparison shows that while 2C8 targets the classical lateral ridge epitope, the 3H5 binding footprint extends to additional residues outside of this region.
Binding of 3H5 distorts the virion
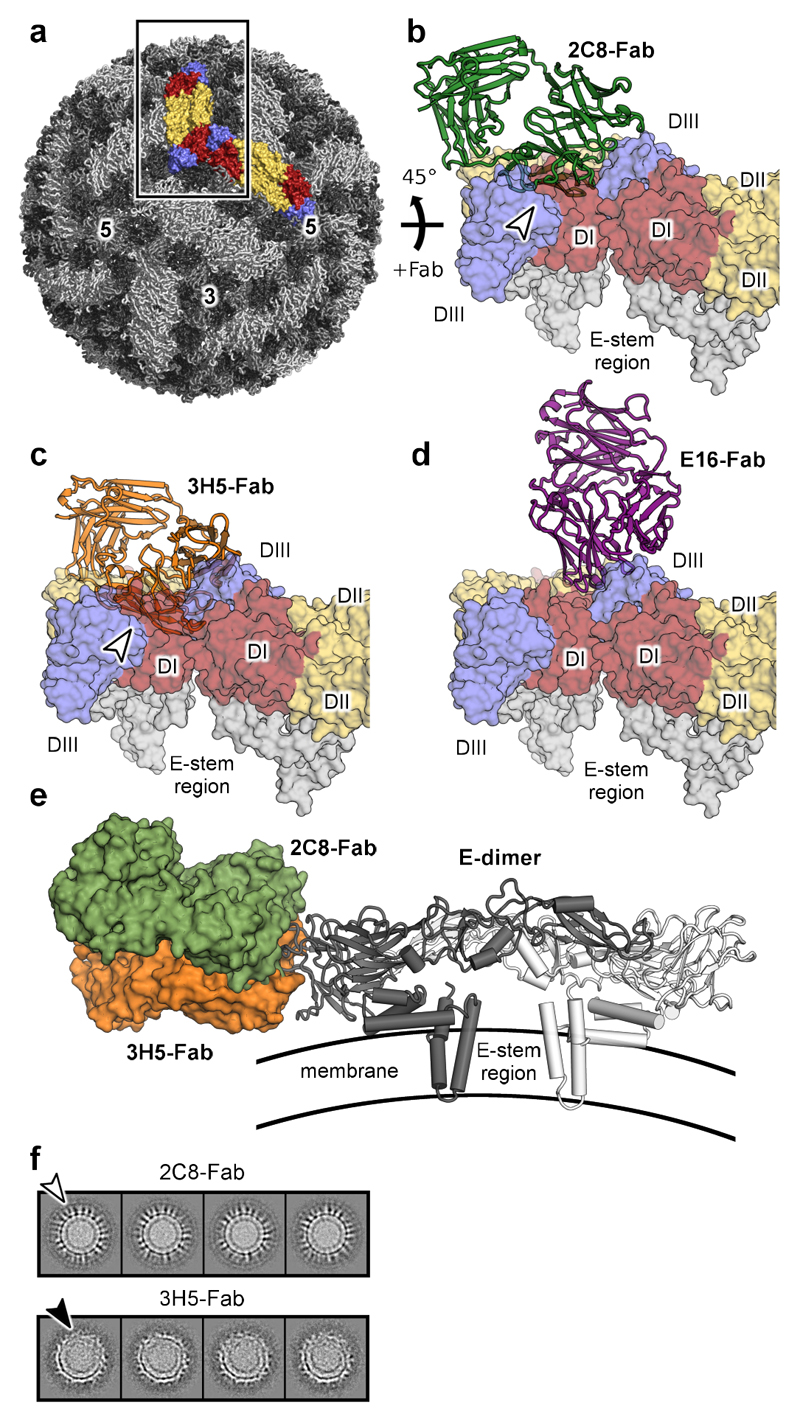
To explore the differences in binding of 2C8 and 3H5 in the context of the whole virus, the Fab-EDIII complexes were docked onto the 3.5 Å resolution cryo-EM reconstruction of DENV2 (Fig. 6a)13. Docking was carried out by aligning the EDIII portion of our structures onto the cryo-EM structure of the virion (pdbID 3J27). Although the epitope of 2C8 shares similarities to the classic accessible epitope of E16 on the lateral ridge, the two antibodies nevertheless display varying binding modes. While E16 can be docked without clashes onto the virion, 2C8 binding introduces minor steric clashes between the 2C8 variable light domain and neighboring E-dimers (compare Figs. 6b and 6d). However, because the lateral ridge areas recognized by 2C8 (primarily the BC loop, the DIII-DI hinge, and the FG loop) are exposed at the viral surface it is feasible that small-scale rearrangements of these loops could accommodate an upright and clash-free binding of 2C8. For 3H5 the observed clashes are substantially more severe than is the case for 2C8 (Fig. 6c) due to 3H5 contacting additional C-strand and CD loop residues (specifically residues K344 and R345) which lie buried deep between E-dimers. This would result in 3H5 binding closer to the viral membrane (Fig. 6e). Thus, in the case of 3H5, large-scale conformational changes would be necessary to accommodate clash-free binding.
Figure 6. Fab binding in the context of the mature virion.
a, Structure of mature DENV2 at 3.5 Å resolution (pdbID: 3J27). Two neighboring E-protein dimers on the viral surface are highlighted and rendered as surfaces (DI colored in red, DII in yellow, and DIII in blue). b-d, Docking of 2C8, 3H5, and E16 Fabs onto the virion. Two neighboring E dimers (as indicated by the box in A and rotated by 45°) on the mature virus are shown as surfaces with domains labelled. Clashes with antibodies are indicated with white arrows. e, Comparison of 2C8-Fab and 3H5-Fab docked onto a E dimer. 2C8 (green) and 3H5 (orange) Fabs were docked onto pdbID 3J27 by aligning the EDIII potion of the structures. The Fabs are shown as surfaces and the E-dimer is displayed in cartoon representation. A side view is of the E dimer on the viral surface is shown. The approximate location of the viral membrane is shown schematically. f,2D-class averages from cryo-EM of DENV-2 bound to the indicated Fabs. The white arrow indicates density corresponding to 2C8-Fabs protruding from the viral surface. The black arrow indicates deformed regions of the virion with disordered appearance.
Next we collected cryo-EM images of virus-2C8 and virus-3H5 Fab complexes. As we observed only incomplete engagement with Fab at room-temperature (Supplementary Fig. 6), consistent with the partial accessibility of the epitopes, we pre-incubated virus with Fabs at 37°C to facilitate temperature-induced viral breathing as described15,16,39,40. Virus-Fab particles were highly heterogenous for both 2C8 and 3H5, precluding reliable icosahedral reconstruction. Class averages derived from 2C8 data showed distinguishable lobed density for Fabs protruding perpendicularly from the virus surface (white arrow, Fig. 6f). In contrast, the 3H5 complexes lacked similar upright Fab density, suggesting that 3H5 Fabs attach to virus at a flatter angle. 3H5 complexes also showed a differing morphology, with noticeably distorted class averages often displaying a locally disordered hemisphere with discontinuous E protein layer and membrane (black arrow, Fig. 6f). These analyses show that while 2C8 can be sterically accommodated on the mature virus with minimal loop rearrangements, 3H5 binding coincides with severe clashes, promoting deformation of the virion.
Discussion
It is generally assumed that the vast majority of antibodies directed against flaviviruses, can cause ADE at sub-neutralizing concentrations40. For instance, in a previous study ADE properties of 145 antibodies isolated from hospitalized patients targeting a variety of epitopes, including quaternary epitopes, were characterized revealing robust enhancement for all samples24. Here, we have identified the minimally-enhancing and non-Fcγ receptor binding properties of the murine anti-EDIII Ab 3H5 and have carried out a side-by-side structural and functional analysis with Ab 2C8, which is similar in neutralization properties and recognized epitope but shows classic ADE over a wide range of antibody concentrations.
Two exceptions to this have been reported, the cross-reactive antibodies 2H12 and 3E31 which target the AB-loop epitope of EDIII, away from the lateral ridge, and show little or no enhancement. However, 3H5 is substantially more potent, with 2H12 and 3E31 possessing only moderate neutralization potency41,42. The binding modes of 2H12 and 3E31 are such that attachment to EDIII would not only cause clashes with neighboring E proteins, but also within a single E monomer itself and E monomers would need to partially unfold for binding to occur42. The associated energetic barrier perhaps is what might limit the potency for this epitope by impeding binding to native E on the virion. This is illustrated by the fact that binding of 2H12 is dramatically reduced when measured to full virions vs. isolated EDIII41. Because EDIII needs to first be pulled apart from the other domains of an E monomer for binding of 2H12 or 3E31 to occur, these Abs may well engage their epitope at a flat angle, potentially restricting FcγR interaction, similar to that seen with 3H5.
A critical step in the flavivirus life cycle is the fusion with host membranes which predominantly occurs in late endosomes at ~pH 5.511. The significant structural rearrangements from pre-fusion dimeric E to postfusion trimeric E include a 70° rotation of EDIII12. Our post-attachment neutralization assays indicated that both 2C8 and 3H5 are still able to neutralize infection, even after virus had bound to cells, suggesting that a post-attachment step, perhaps fusion, may be inhibited. Blocking of the conformational changes of E by locking the virus into a non-fusogenic state has been previously put forward as a mechanism for similar lateral-ridge antibodies34,43,44. We examined binding properties of 2C8 and 3H5 at endosomal and neutral pH and observed distinct discrepancies. The dissociation rate of 2C8-Fab from its antigen is ~130-fold more rapid than that of 3H5-Fab, and this difference further increases to ~300-fold at endosomal pH. Furthermore, binding of 2C8 IgGs to virions was 3-fold less avid at low pH than at neutral pH, while 3H5 IgGs were unaffected. The greater pH-dependence of 2C8 might be a result of the presence of pH-sensitivehistidine residues near the 2C8 interface (HC-His106 and LC-His93), whilst there are none for 3H5. It has been suggested that in many cases antibodies may fail to maintain stable binding in the endosomal environment after FcγR-mediated internalization, leading to sub-neutralizing concentrations of bound virus and, consequently, ADE conditions45. Our data indicate that, in contrast to 2C8, 3H5 can remain securely attached to its antigen in the endosomal environment.
The crystal structures of 2C8 and 3H5 Fabs in complex with EDIII defined the binding footprints of the antibodies. Both recognize the lateral ridge epitope of EDIII which is associated with potent neutralization27. Docking of our structures onto the high-resolution EM reconstruction of DENV2 revealed minor clashes in the case of 2C8, whilst 3H5 introduced substantially more severe collisions with neighboring E proteins mainly due to the recognition of additional residues in the buried CD loop of DIII. It is perhaps the high kinetic on-rate of 3H5 which assists the antibody in rapidly attaching to this transiently exposed epitope via conformational selection. Cryo-EM images of virions decorated with 2C8 Fabs suggest binding in an upright orientation. In contrast, virions decorated with 3H5 Fabs lacked similar upright density and, instead, showed pronounced local distortions of the viral surface, leading to an asymmetric morphology.
We favor a model in which the distortions induced by 3H5 are capable of hindering the conformational acrobatics of E required for viral fusion at low occupancies. It is tempting to speculate that this unusual binding of 3H5 occurs at a flat angle to the viral surface, leaving the Fc region poorly accessible, in line with our observation that 3H5-virus complexes do not bind Fcγ receptors. In contrast, the enhancing antibody 2C8 likely uses a different strategy, perhaps one similar to what has been described for Ab E10634. 2C8 and E106 both recognize lateral ridge residues and there is evidence that 2C8 attaches to virions bivalently, similar to E106. E106 has thus been suggested to arrest virus in a pre-fusion conformation by cross-linking E monomers via its two Fabs34. We propose that 2C8 displays typical ADE properties because 2C8-virus complexes efficiently bind to Fcγ receptors, possess a substantially higher occupancy requirement for neutralization, and quickly dissociate in an endosomal pH environment which restores fusion competence of the virus.
Despite substantial efforts no antivirals against acute dengue are currently available and the sole licensed tetravalent vaccine (CYD-TDV) is not completely protective9. In addition, clinical trials showed worrying outcomes in sero-negative children under 9, possibly related to enhancement of infection. We propose that the properties of the potent anti-EDIII Ab 3H5 may have implications for the development of non-enhancing, therapeutically viable Abs without the need to modify Fc regions. Antibodies recognizing the lateral ridge epitope of EDIII have been previously shown to be protective in vivo against different flaviviruses14,44,46,47. Furthermore, the EDIII is not the major target of the human humoral response. Targeting of non-immunodominant epitopes in passive immunotherapy avoids interfering with the active humoral response48,49. A recent study demonstrated how structural methods can be used to successfully engineer a therapeutic antibody with altered serotype-specificity28. A similar approach may be applicable utilizing 3H5 as a template to engineer non-enhancing antibodies against other dengue serotypes and related flaviviruses.
Methods
Neutralization and enhancement assays
Serially diluted Ab was mixed with virus and incubated for 1 hr at 37°C, transferred to Vero cell (Armed Forces Research Institute of Medical Sciences, AFRIMS) monolayers and incubated for 2-3 days. The focus forming assay was performed using anti-E Ab (4G2), gift from AFRIMS, followed by polyclonal goat anti-mouse IgG (P0447; Dako), conjugated with HRP at 1:1000 dilution and visualized by the addition of DAB substrate. 50% FRNT were determined from graphs of percent reduction versus concentration of Abs. For the ADE assay, serially diluted Ab was pre-incubated with virus for 1 hr 37°C, then transferred to U937 cells (human monocyte, histiocytic lymphoma, Fc receptor-bearing cells, gift from Prida Malasit, previously described in50) or K562 cells (human myelogenous leukemia, described in51) and incubated for 4 days. Supernatants were harvested and titrated on Vero cells by a focus forming assay as described above. All cell lines were tested and found free from mycoplasma contamination.
Fcγ receptor blocking experiments
K562 or U937 cells were pre-incubated with polyclonal goat anti-FcγR1 (AF1257; R&D) or mouse monoclonal anti-FcγR2a, clone IV.3 (60012; STEMCELL Technologies) antibodies at a concentration of 10 μg/ml for 1 hour. After a washing step, the cells were incubated with 2C8 or 3H5 immunecomplexes with the appropriate virus, in presence of anti-FcγR antibodies for 4 days. Finally, virus supernatants were harvested and titrated on Vero cells for a focus forming assay, as described above.
ELISA binding assay of antibody and virus complexes on FcγR2a and FcγR1
DENV2/NGC, DENV2/16681 and mock-infected culture supernatant were pre-incubated with serial dilutions of 2C8, 3H5 IgG2a and mouse IgG2a (negative control) at 37°C for 1 hour. Virus/antibody complexes were added to ELISA plates coated with 0.5 μg of Fcγ receptor protein (Sino Biological Inc) and incubated for 2 hours at 37°C. The binding of Antibody/virus complexes was then determined by incubation with human anti-FLE mAbs (0.1 μg/ml), followed by alkaline phosphatase-conjugated goat anti-human IgG (Fc-specific) at 1:10000 dilution (A9544 Sigma). The reaction was developed with PNPP substrate and stopped with NaOH. The absorbance was read at 405 nm and subtracted with mock.
Pre- and post-attachment neutralization assays
The pre-attachment neutralization assay was carried out by incubating the virus with serially diluted Ab at 4 °C for 1 hour prior to transfer to Vero cell monolayers. After incubation for a further hour at 4 °C, cells were washed three times with cold MEM media, and then incubated at 37°C for 3 days before performing the focus forming assay. For post-attachment neutralization assays, virus was added to cells and incubated for 1 hour at 4 °C. The virus-bound cells were then washed three times with cold MEM to remove unbound virus. Serially diluted antibody was then added to cells and incubated for 1 hour at 4 °C. The cells were washed once with cold MEM and incubated for another 3 days at 37 °C. Finally, the cells were stained for DENV foci.
Antibody sequencing and preparation of 3H5 and 2C8 Fab fragments
mRNA was directly purified from hybridoma cells using the RNeasy and Oligotex mRNA Mini Kit (Qiagen). cDNA of heavy and light chains of all antibodies were prepared for sequencing with GeneRacer Kit from Invitrogen. Recombinant 3H5 Fab was expressed in HEK 293T cells. In brief, 3H5 mRNA was amplified by One-step RT-PCR kit (Qiagen) with the following primers for heavy and light chains: 3H5_heavy_FWD: TGGGTTGCGTAGCTCAGGTTCAGCTCCAGCAGTCTGG, 3H5_heavy_REV: GGGTGTCGTTTTGGCTGAGGAGACGGTGACTGAGGTTCC, 3h5_light_FWD: TGGGTTGCGTAGCTAACATTGTAATGACCCAATCTCCC, 3h5_light_REV: TGAAGCATCAGCCCGTTTTATTTCCAGCTTGGTCCC. The gene products were cloned into pOPINVH and pOPONVL vectors52 using the In-Fusion cloning technique (Clontech). The recombinant Fab, of which the heavy chain was tagged with 6-histidines at C-terminus, was purified using nickel affinity chromatography and gel filtration on a Superdex 75 16/60 column (GE Healthcare) equilibrated in 150mM NaCl, 20mM Tris, pH 7.5. 2C8 Fab was generated by digestion of purified IgG with preactivated papain agarose (1-10 mg/ml) (Sigma) for 4 h at 37 °C in PBS buffer containing 10 mM EDTA and 10 mM cysteine. Supernatant containing digested antibody was buffer exchanged into 50mM Tris pH 8.0. Fab fragments were purified by cation exchange chromatography at pH 8.0 (MonoQ HR 5/5, GE Healthcare) and gel filtration over a Superdex 75 16/60 column (GE Healthcare) in 100mM NaCl, 25mM Tris, pH 7.5.
Recombinant EIIID2 expression
The DNA sequence coding for residues 295-401 of E of DENV2 serotype NGC was PCR amplified and cloned into the pET3c vector (Novagen). No tags were used. EDIII was also cloned into pET3c-AviTag, adding the peptide DPLHHILDAQKMVWNHRD to the C-terminus of the protein which is recognized by biotin ligase (BirA). The protein was expressed and refolded as previously described 53.
Surface Plasmon Resonance
Recombinant EIIID2 was biotinylated using BirA (Avidity, Denver, CO) and immobilized on a streptavidin–coated CM5 chip at 3 different densities (25, 80 and 180 RU). The Fab binding experiments were performed at 37°C using a flow-rate of 50μl/min. Binding at neutral pH (7.4) was tested in HBS-EP running buffer (150 mM NaCl, 10 mM Hepes, pH 7.4, 3.4 mM EDTA and 0.005 % Tween 20), that at endosomal pH (5.5) in MBS-EP (150 mM NaCl, 10 mM MES, pH 5.5, 3.4mM EDTA and 0.005% Tween 20). The kinetics of binding were measured in multi-cycle mode. Analyte was injected for 400 s and allowed to dissociate for 700 s. At the end of each run the antigen surface was regenerated with 10mM hydrochloric acid for 30 s and washed with buffer for 200s. Analyte binding was tested in duplicate at 5 different concentrations prepared by two or three fold dilutions.
ELISA and occupancy calculation
To determine the binding avidity and affinity of Ab and Fab, mock or DENV from infected culture media was captured onto plates pre-coated with purified immunoglobulin from pooled dengue convalescent serum and then incubated with serial dilutions of either full length IgG or Fab fragment followed by alkaline phosphatase-conjugated polyclonal goat anti-mouse IgG (Fc-specific (A2429) or Fab-specific (A1293), respectively) at 1:2000 dilution, both from Sigma. Mock–infected culture supernatant was used as a negative control. The reaction was developed by the addition of PNPP substrate and stopped with NaOH. The absorbance was read at 405 nm. Results are expressed as the percentage of total binding, with 100% binding corresponding to the Ab concentration at maximum absorbance. GraphPad PRISM was used to perform nonlinear regression curve-fitting analyses to estimate dissociation constants (Kd). Percent occupancy at 50% FRNT was determined by using the following formula: Percent occupancy = [Ab]/(Kd+[Ab]), where [Ab] is the concentration of Ab required to reach 50% FRNT.
Crystallization of 3H5 Fab and 2C8 Fab complexed with E-DIII
Purified Fab was incubated with a molar excess of EIIID2 for 1 h prior to size exclusion chromatography. All crystals used for data collection grew at 21°C. The 3H5-EIIID2 complex yielded two different crystal forms. Crystal form 1 was obtained from 20% w/v PEG 3350, 0.2 M di-Sodium Tartrate at a protein concentration of 4.16 mg/ml. Crystal form 2 grew in 20% w/v PEG 3350, 0.2 M Ammonium di-Hydrogen Phosphate using a protein concentration of 6 mg/ml. The 2C8-EIIID2 complex crystallized at 13.4 mg/ml in 15% v/v glycerol, 25.5% w/v PEG 4000, 0.17M Sodium Acetate, 0.085 M Tris-HCl pH 8.5. Protein crystals were cryo-protected with 20% glycerol. X-ray data were collected at 100 K at the Diamond Light Source (beamline IO2 – 3H5-EIIID2 crystal form 1 at wavelength 0.951 Å) and European Synchrotron Radiation Source (beamlineID23-EH1 – 3H5-EIIID2 crystal form 2 at wavelength 0.933 Å, ID14-EH2 – 2C8-EIIID2 at wavelength 0.980 Å). Bragg intensities were integrated and equivalent reflections were merged using the HKL2000 package54 or xia255.
Crystallographic structure determination
The structures of the complexes were solved by molecular replacement using Phaser56, with three search models: one for the Fab constant region (CH1 and CL), one for Fab variable region (VH and VL) and one for the antigen (EIIID2). The following models were used for the Fab: coordinates of pdb code 1A3R for 2C8 and pdb code 1ACY for 3H5. For EIIID2, domain III of pdb code 1OAN was used. Manual rebuilding was performed using COOT57 and structures were refined using autoBuster58. Non-crystallographic local structure similarity restraints were used during the refinement of all structures. Molecular graphics were prepared using PyMOL (DeLano Scientific) and sequence alignments with Jalview59.
Virus preparation and Fab complex formation
Dengue virus serotype 2 strains 16681 and New Guinea C (gifts from the Armed Forces Research Institute of Medical Sciences, AFRIMS) were grown in C6/36 cells (Armed Forces Research Institute of Medical Sciences, AFRIMS). Cell-free supernatants were collected and concentrated by Polyethylene Glycol (PEG) precipitation using PEG 8,000, to a final concentration of 8%. The supernatant/PEG mixture was stored overnight at 4°C before centrifugation at 3,200 x g for 90 minutes at 4°C. Pelleted viral particles were resuspended in cold NTE buffer (12 mM Tris, pH 8.0, 120 mM NaCl, 1mM EDTA), cleared by low speed centrifugation (3,200 x g, 5 min, 4°C), layered over a 22% sucrose cushion and centrifuged at 175,000 x g (Beckman Coulter Sw41) for 2 h at 4°C. The virus-containing fraction was resuspended in cold NTE buffer and kept overnight at 4°C and was subsequently applied to a 10-35% potassium tartrate step gradient was collected and the sample was buffer exchanged into NTE buffer by multiple rounds of dilution and centrifugation using a 100 kD cutoff centrifugal filter device (Amicon Ultra). The resulting virus preparation was utilized for cryo-EM. To form Fab-virus complexes purified virus was mixed with 3H5-Fab or 2C8-Fab at a molar ratio of roughly 5 Fabs per E protein and incubated either at room-temperature or at 37°C for 1 h.
Electron microscopy
Aliquots (3 µl) of virus and virus-Fab complexes incubated at either 37°C or room-temperature were applied onto glow-discharged holey carbon grids (Cflat; Protochips, Raleigh, NC), blotted with filter paper for 3s and vitrified in a ethane-propane mixture at liquid nitrogen temperature using a plunging device (CP3; Gatan, Pleasanton, CA). Data were acquired using a transmission electron microscope (F30 or F30 ‘Polara’; FEI, Hillsboro, OR) operated at 300kV and at liquid nitrogen temperature. Images were recorded on a 16-megapixel charge-coupled device camera (Ultrascan 4000; Gatan, Pleasanton, CA) at a calibrated magnification of 125,000x, yielding a pixel size of 2.4 Å/pixel. The defocus values ranged from -2.0 to -6.0 μm.
Additional data were acquired with a direct electron detector (K2 Summit; Gatan, Pleasanton, CA) at a calibrated magnification of 35,000x and a pixel size of 2.84 Å/pixel with defocus values ranging from -2.0 to -6.0 μm.
The contrast transfer function (CTF) parameters of each micrograph were determined using CTFFIND360 and CTF correction was performed with the Bsoft software package61. Particles were manually selected from micrographs with EMAN62. The final data sets comprised 2,456 particles of 3H5-Fab complexes at 37°C, 3,378 particles of 2C8-Fab complexes at 37°C, 1,182 particles of 2C8-Fab complexes at room-temperature and 1,200 particles of apo DENV-2 at room-temperature. The particles were iteratively centered until particle shifts converged and radial profiles were calculated from averages of all particles in each set using IMAGIC63. Subsequently, iterative rounds of multi-reference alignment (MRA), multivariate statistical analysis (MSA) and 2D-classification were carried out with IMAGIC.
Statistical evaluation
Functional data are presented as mean ± s.e.m. with the number of technical replicates indicated in the corresponding figure legends.
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust, UK; the Newton-Medical Research Council, UK; the National Institute for Health Research Biomedical Research Centre funding scheme; the Thailand National Center for Genetic Engineering; and Biotechnology, the Academy of Finland; the Office of the Higher Education Commission and Mahidol University under the National Research Universities Initiative; the Research Chair Grant from the National Science and Technology Development Agency (NSTDA), Thailand; and Faculty of Medicine Siriraj Hospital, Mahidol University, Grant Number (IO)R015936005. The OPIC electron microscopy facility was founded by a Wellcome Trust JIF award (060208/Z/00/Z) and is supported by a WT equipment grant (093305/Z/10/Z). The Wellcome Trust is also acknowledged for providing administrative support (Grant 075491/Z/04). GRS is a Wellcome Trust senior investigator. MR and AF were supported by Wellcome Trust fellowships (204703/Z/16/Z and 080721/Z/06/Z). We thank Dr. A. Siebert for support with electron microscopy, T. Prommool and C. Tawilert for scale-up and purification of mAbs, P. Keelapang for good advice on NT and ADE assays and D. Bitto for assistance with cryo-EM sample preparation. We also thank the staff of the Diamond Light Source (proposal MX8423), and ESRF, K. Harlos and T. Walter for assistance with crystallization, and D. Stuart for scientific discussion.
Footnotes
Reporting Summary. Further information on experimental design is available in the accompanying Research Reporting Summary.
Data availability statement. The data supporting the findings of this study are available from the corresponding authors upon request. Structure factors and final refined coordinates of crystal structures have been deposited in the RCSB PDB (https://www.rcsb.org/pdb/) under the accession codes 6FLA (3H5 complex form 1), 6FLB (3H5 form2) and 6FLC (2C8).
Author Contributions
Experiments were conceived and designed by AF, MR, WD, JM, JTH, JMG and GRS. Experiments were performed by AF, MR, WD, PS, WW, KC, TD, AC and CMM. The data were analyzed by AF, MR, WD, CMM, JM, JTH, JMG, GRS. The paper was written by AF, MR, JM, JTH, JMG and GRS. CP, WK and PM provided antibodies.
Competing interests
The authors declare no competing interests.
References
- 1.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 3.Halstead SB, O’Rourke EJ. Antibody-enhanced dengue virus infection in primate leukocytes. Nature. 1977;265:739–741. doi: 10.1038/265739a0. [DOI] [PubMed] [Google Scholar]
- 4.Guzman MG, Alvarez M, Halstead SB. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch Virol. 2013;158:1445–1459. doi: 10.1007/s00705-013-1645-3. [DOI] [PubMed] [Google Scholar]
- 5.Pierson TC, et al. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe. 2007;1:135–145. doi: 10.1016/j.chom.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliphant T, et al. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dejnirattisai W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Priyamvada L, et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A. 2016;113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadinegoro SR, et al. Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N Engl J Med. 2015;373:1195–1206. doi: 10.1056/NEJMoa1506223. [DOI] [PubMed] [Google Scholar]
- 10.Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol. 2015;15:745–759. doi: 10.1038/nri3916. [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 12.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao H, et al. Structural Basis of Zika Virus-Specific Antibody Protection. Cell. 2016;166:1016–1027. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Austin SK, et al. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 2012;8:e1002930. doi: 10.1371/journal.ppat.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 2011;7:e1002111. doi: 10.1371/journal.ppat.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, et al. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci U S A. 2013;110:6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fibriansah G, et al. Structural changes in dengue virus when exposed to a temperature of 37°C. J Virol. 2013;87:7585–7592. doi: 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beltramello M, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai C-Y, et al. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol. 2008;82:6631–6643. doi: 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Alwis R, et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A. 2012;109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fibriansah G, et al. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun. 2015;6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fibriansah G, et al. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol Med. 2014;6:358–371. doi: 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dejnirattisai W, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouvinski A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 26.Dejnirattisai W, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lok S-M. The Interplay of Dengue Virus Morphological Diversity and Human Antibodies. Trends Microbiol. 2016;24:284–293. doi: 10.1016/j.tim.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Robinson LN, et al. Structure-Guided Design of an Anti-dengue Antibody Directed to a Non-immunodominant Epitope. Cell. 2015;162:493–504. doi: 10.1016/j.cell.2015.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentry MK, Henchal EA, McCown JM, Brandt WE, Dalrymple JM. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am J Trop Med Hyg. 1982;31:548–555. doi: 10.4269/ajtmh.1982.31.548. [DOI] [PubMed] [Google Scholar]
- 30.Morens DM, Halstead SB, Marchette NJ. Profiles of antibody-dependent enhancement of dengue virus type 2 infection. Microb Pathog. 1987;3:231–237. doi: 10.1016/0882-4010(87)90056-8. [DOI] [PubMed] [Google Scholar]
- 31.Stiasny K, Allison SL, Schalich J, Heinz FX. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J Virol. 2002;76:3784–3790. doi: 10.1128/JVI.76.8.3784-3790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Gromowski GD, Barrett ADT. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology. 2007;366:349–360. doi: 10.1016/j.virol.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 34.Edeling MA, et al. Potent dengue virus neutralization by a therapeutic antibody with low monovalent affinity requires bivalent engagement. PLoS Pathog. 2014;10:e1004072. doi: 10.1371/journal.ppat.1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nybakken GE, et al. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robbiani DF, et al. Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell. 2017;169:597–609.e11. doi: 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallichotte EN, et al. Human dengue virus serotype 2 neutralizing antibodies target two distinct quaternary epitopes. PLoS Pathog. 2018;14:e1006934. doi: 10.1371/journal.ppat.1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fibriansah G, et al. DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science. 2015;349:88–91. doi: 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lok S-M, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 40.Dowd KA, Mukherjee S, Kuhn RJ, Pierson TC. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J Virol. 2014;88:11726–11737. doi: 10.1128/JVI.01140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Midgley CM, et al. Structural analysis of a dengue cross-reactive antibody complexed with envelope domain III reveals the molecular basis of cross-reactivity. J Immunol Baltim Md 1950. 2012;188:4971–4979. doi: 10.4049/jimmunol.1200227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, et al. Structural and Functional Characterization of a Cross-Reactive Dengue Virus Neutralizing Antibody that Recognizes a Cryptic Epitope. Struct Lond Engl 1993. 2018;26:51–59.e4. doi: 10.1016/j.str.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, et al. A Human Bi-specific Antibody against Zika Virus with High Therapeutic Potential. Cell. 2017;171:229–241.e15. doi: 10.1016/j.cell.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliphant T, et al. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz FX, Stiasny K. The Antigenic Structure of Zika Virus and Its Relation to Other Flaviviruses: Implications for Infection and Immunoprophylaxis. Microbiol Mol Biol Rev MMBR. 2017;81 doi: 10.1128/MMBR.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sukupolvi-Petty S, et al. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol. 2010;84:9227–9239. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shrestha B, et al. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siegrist CA, et al. Determinants of infant responses to vaccines in presence of maternal antibodies. Vaccine. 1998;16:1409–1414. doi: 10.1016/s0264-410x(98)00100-5. [DOI] [PubMed] [Google Scholar]
- 49.Siber GR, et al. Interference of immune globulin with measles and rubella immunization. J Pediatr. 1993;122:204–211. doi: 10.1016/s0022-3476(06)80114-9. [DOI] [PubMed] [Google Scholar]
- 50.Sundström C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 51.Lozzio CB, Lozzio BB. Cytotoxicity of a factor isolated from human spleen. J Natl Cancer Inst. 1973;50:535–538. doi: 10.1093/jnci/50.2.535. [DOI] [PubMed] [Google Scholar]
- 52.Nettleship JE, et al. A pipeline for the production of antibody fragments for structural studies using transient expression in HEK 293T cells. Protein Expr Purif. 2008;62:83–89. doi: 10.1016/j.pep.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 53.Midgley CM, et al. An in-depth analysis of original antigenic sin in dengue virus infection. J Virol. 2011;85:410–421. doi: 10.1128/JVI.01826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Winter G, Lobley CMC, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Smart OS, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinforma Oxf Engl. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 61.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 63.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.