Abstract
Nuclear Inhibitor of PP1 (NIPP1) is a conserved regulatory subunit of protein phosphatase PP1. The selective deletion of NIPP1 in mouse liver parenchymal cells or skin epidermal cells culminates in a late-onset hyperproliferation of a subset of resident progenitor cells. Although a hyperplastic phenotype is usually tumor promoting, we show here that the absence of NIPP1 conferred a strong resistance to chemically induced hepatocellular or skin carcinoma. The ablation of NIPP1 did not affect the metabolism of the administered mutagens (diethylnitrosamine or 7,12-dimethylbenz[a]anthracene), but reduced the conversion of mutagen-induced covalent DNA modifications into cancer-initiating mutations. This reduced sensitivity to mutagens correlated with an enhanced DNA-damage response and an augmented expression of rate-limiting DNA-repair proteins (MGMT in liver, XPD and XPG in skin), hinting at an increased DNA-repair capacity. Our data identify NIPP1 as a repressor of DNA repair and as a promising target for novel cancer prevention and treatment therapies.
Subject terms: Cancer models, Cell biology
Introduction
NIPP1 (38 kDa), encoded by PPP1R8, is a ubiquitously expressed nuclear protein that targets phosphoproteins for regulated dephosphorylation by associated protein Ser/Thr phosphatase PP11–3. Phosphoproteins are recruited by the phosphate-binding loop of the forkhead-associated (FHA) domain of NIPP1 and include a transcription factor (protein methyltransferase EZH2), two pre-mRNA splicing factors (SF3B1/SAP155 and CDC5L), and a DNA-replication factor (protein kinase MELK)4–7. At first glance, the FHA ligands of NIPP1 are functionally disconnected. However, recent data suggest that they all contribute to the cellular response elicited by DNA damage. Indeed, CDC5L is an activator of the DNA-damage checkpoint kinase ATR8, SF3B1 and MELK promote the expression of DNA-repair factors9,10, and EZH2 contributes to the resolution of stalled replication forks11. Interestingly, NIPP1 itself may also have a role in the DNA-damage signaling as expression of a fusion of PP1 and NIPP1 induced DNA damage, including double-strand breaks3. PP1-NIPP1 fusions with inactivating mutations of the PP1 moiety or FHA-domain were less effective in eliciting DNA damage, suggesting that NIPP1 opposes DNA repair through its control on the PP1-mediated dephosphorylation of FHA ligands.
The global deletion of NIPP1 in mice was embryonic lethal at the gastrulation stage12. Likewise, the postnatal ablation of NIPP1 in testis resulted in a complete loss of germ cells13. In contrast, the selective deletion of NIPP1 in mouse skin epidermal cells or liver parenchymal cells was less detrimental, the major phenotype being a slow-onset hyperproliferation of specific epidermal or biliary progenitor cells, respectively14,15. This enabled us to use the skin knockout (SKO) and liver knockout (LKO) models to further delineate the contribution of NIPP1 to the cellular DNA-damage response. For these studies, we provoked DNA damage, and ultimately tumor formation, using well-characterized chemicals. Skin carcinogenesis was induced in two stages, using consecutively the tumor-initiating mutagen 7,12-dimethylbenz[a]anthracene (DMBA) and the tumor-promoting phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA)16. Hepatocellular carcinoma was provoked by a single injection of the DNA-alkylating agent diethylnitrosamine (DEN)17. Our data demonstrate that the NIPP1 SKOs and LKOs, despite the associated hyperproliferation phenotype, were strongly resistant to induced mutagenesis and carcinogenesis. This resistance correlated with an increased expression level of key DNA-repair factors. These findings further characterize NIPP1 as a repressor of the DNA-repair capacity and provide a rationale for the development of NIPP1 modulators in cancer prevention and treatment therapies.
Results
Resistance of Ppp1r8−/− keratinocytes to DMBA/TPA-induced skin carcinogenesis
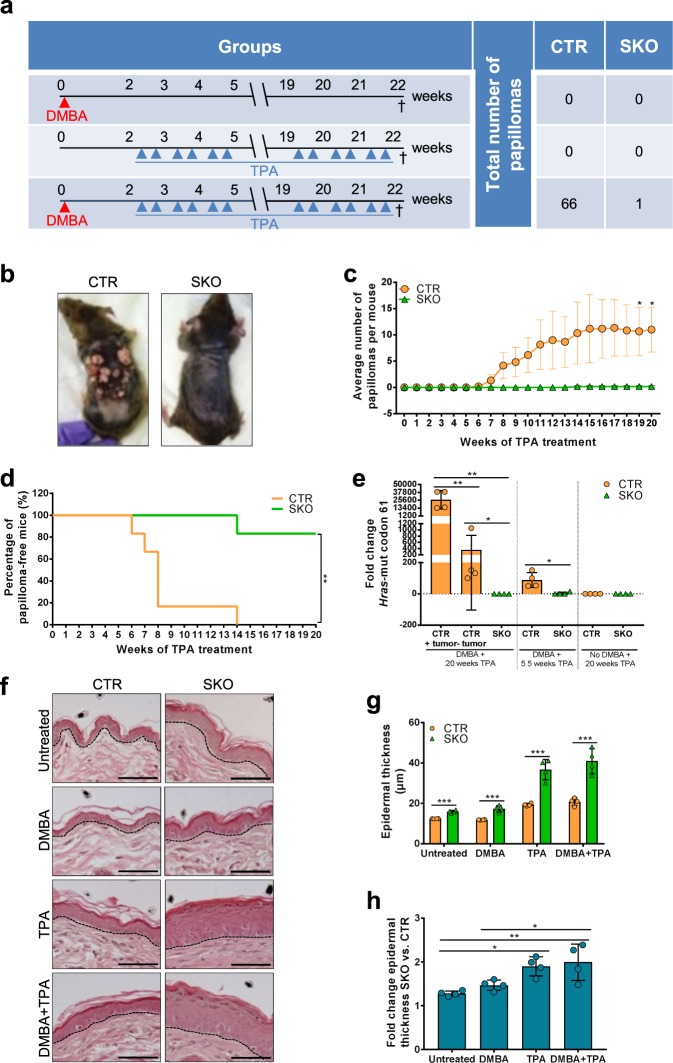
In the NIPP1 skin knockout mice (SKOs) that we generated one Ppp1r8 allele is disrupted in all somatic cells and the second, floxed Ppp1r8 allele, was selectively inactivated in keratinocytes by CRE recombinase under control of the Keratin-14 promoter15. Heterozygous Ppp1r8fl/+/Tg(Krt14-Cre) mice, which express (nearly) normal levels of NIPP1, were used as controls (CTRs). We compared the response of CTRs and SKOs with two-stage skin carcinogenesis, as induced by the topical administration of DMBA and/or TPA. Three different treatment regimens were adopted: (1) a single application of DMBA, (2) the administration of TPA twice a week for 20 weeks, and (3) a consecutive treatment with DMBA and TPA (Fig. 1a). No papillomas developed in the CTRs and SKOs after a treatment with DMBA or TPA alone (suppl. Fig. S1a, b), indicating that the deletion of NIPP1 does not initiate or promote tumor formation. With the combined treatment, the first papillomas in CTR mice appeared 6 weeks after the start of the TPA treatment (Fig. 1b–d; suppl. Fig. S1c, d). By 14 weeks, all CTR mice had developed 1–39 papillomas. In contrast, only one of six SKO mice had grown a (single) papilloma by this time, and this papilloma was small compared with most papillomas formed in CTR mice (suppl. Fig. S1e). These data demonstrated that NIPP1-deficient keratinocytes are strongly resistant to DMBA/TPA-induced skin carcinogenesis.
Fig. 1. Ppp1r8−/− keratinocytes are resistant to DMBA-induced mutagenesis.
a Procedure for the application of DMBA, TPA or DMBA + TPA on the back skin of CTR and SKO mice. Also shown are the total number of papillomas (6 mice in each condition, except for the TPA condition: 5 CTR and 7 SKO) that were macroscopically detected at the time of sacrifice (†). b Representative picture of a CTR and SKO mouse at the end of the combined DMBA + TPA treatment. All mice that received this treatment are pictured in suppl. Fig. 1c. c Average number of papillomas formed in CTRs and SKOs during the combined DMBA + TPA treatment. Results are means ± SEM (n = 6). d Percentage of papilloma-free CTR and SKO mice during the combined DMBA + TPA treatment. **p < 0.01 in Log-rank test (n = 6). e DMBA-induced mutations of Hras at codon 61, as detected by qPCR in genomic DNA isolated from the back skin of CTR and SKO mice treated with TPA, DMBA + TPA (5.5 weeks), or DMBA + TPA (20 weeks). For CTR mice, tumors and tumor-free tissue were analyzed separately. Actin was used as a housekeeping gene for normalization. *p < 0.05; **p < 0.01 in unpaired Student’s t-test. The results are expressed as means ± SD for 4 mice in each condition. f H&E staining of the back skin of (un)treated CTR and SKO mice. The treatment with TPA was for 20 weeks. Scale bars, 50 µm. g Quantification of epidermal thickness in (un)treated CTRs and SKOs. Data are represented as means (n = 4) ± SD. ***p < 0.001 in unpaired Student’s t-test. h Fold change of epidermal thickness in SKOs versus CTRs. Data are shown as means ± SD (n = 4). *p < 0.05; **p < 0.01 in two-way ANOVA with Tukey’s multiple comparisons test.
Decreased mutagenic response to DMBA in SKOs
Metabolically activated DMBA forms covalent DNA adducts with adenine (A) that base pairs with A during DNA replication18. During the subsequent round of replication, the mutation is fixed as thymine (T) and is incorporated opposite to A. To examine whether the absence of NIPP1 hampers tumor initiation by DMBA, we quantified oncogenic A → T transversions in codon 61 of Hras19, which account for a major fraction of oncogenic mutations in DMBA-induced skin carcinoma20. For this purpose, genomic DNA was isolated from the skin of mice treated with TPA for 20 weeks or with DMBA (single application) plus TPA for either 5.5 weeks or 20 weeks. The oncogenic A → T transversion in codon 61 of Hras (HRASQ61L) was massively enriched in tumors of CTR mice (Fig. 1e; suppl. Fig. S1f-h). To a lesser extent, this mutation was also present in tumor-free skin of CTR mice that had been treated with DMBA and TPA for either 5.5 weeks (no papillomas formed yet) or 20 weeks (papillomas formed). In contrast, the examined Hras mutation was barely detectable in the skin of CTRs treated with TPA alone and in SKOs treated with DMBA + TPA for 5.5/20 weeks. These data suggested that the deletion of NIPP1 severely impeded the mutagenic response to DMBA.
Next, we investigated whether the resistance of SKOs to papilloma formation was due to a skewed metabolism of DMBA. DMBA binds to the aryl hydrocarbon receptor (AHR), resulting in the upregulation of the Cytochrome P450 enzymes CYP1A1 and CYP1B121. Together with epoxide hydrolase 1 (EPHX1), these P450 enzymes convert DMBA into a thiol epoxide that forms covalent DNA adducts22. We observed no differences in the (altered) expression of Ahr, Cyp1a1, Cyp1b1, Ephx1, and the AHR nuclear translocator (Arnt) between CTR and SKO mice following a treatment with DMBA for 0–24 h (suppl. Fig. S1i), hinting at a similar DMBA-biotransformation potential. Application of DMBA also provoked a similar apoptotic response in CTR and SKO mice, as indicated by similar expression levels of pro- and anti-apoptotic genes, and by similar numbers of cleaved Caspase-3-positive cells in CTR and SKO mice (suppl. Fig. S1j-l).
In the DMBA/TPA-induced skin carcinogenesis model, TPA induces inflammation and epidermal hyperplasia, providing a beneficial microenvironment for the survival and expansion of keratinocytes with oncogenic Hras mutations16,23. This prompted us to examine whether the absence of NIPP1 limits papilloma formation by reducing the hyperplastic response to TPA. However, the hyperproliferation phenotype of SKO epidermis was unaffected by a treatment with DMBA alone while a treatment with TPA increased epidermal thickness in both CTR and SKO mice (Fig. 1f–h). Strikingly, epidermal thickness in TPA-treated mice was 1.90 ± 0.22-fold more increased in the SKOs than in the CTRs (Fig. 1h), indicating that the ablation of NIPP1 even amplified the hyperplastic response to TPA. Overall, our findings suggested that the reduced sensitivity of the SKOs to induced skin carcinogenesis stemmed from a decreased conversion of DMBA-induced DNA adducts to overt mutations, despite an increased hyperplastic response to TPA.
Increased DNA-damage response and DNA-repair capacity in SKOs
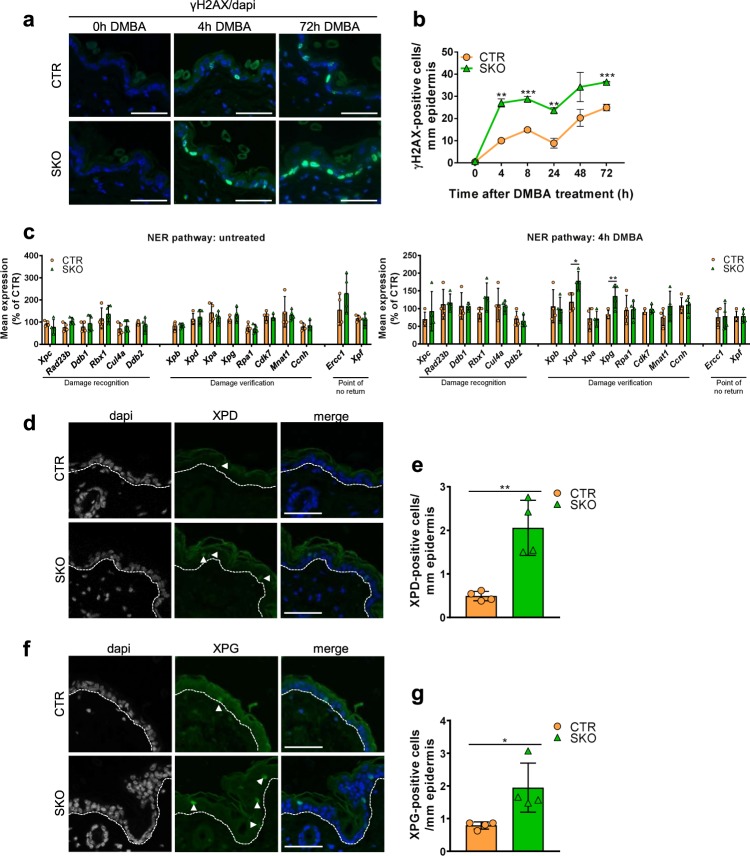
To obtain more insights in the reduced mutagenic response of the SKOs to DMBA, we examined the DNA-damage response at various time points after the administration of DMBA. The histone H2A variant H2AX is rapidly phosphorylated at S139 (γH2AX) when DNA is damaged, e.g., by DMBA administration19,24. Strikingly, the accumulation of γH2AX in DMBA-treated skin was about 2-fold more pronounced in the SKOs, as compared with that in the CTRs, hinting at an increased DNA-damage response (Fig. 2a, b). DMBA-induced DNA adducts are mainly detected and repaired through the nucleotide-excision-repair (NER) pathway, but can also be converted to DNA double-strand breaks25. Quantitative qRT-PCR analysis of all key regulators of the NER pathway did not reveal differences in transcript levels between non-treated CTRs and SKOs. However, following the administration of DMBA, the transcripts encoding the XPD helicase and the XPG endonuclease, which are required for DNA melting around the lesion and lesion excision, respectively, were increased in SKO mice (Fig. 2c). Immunostaining confirmed an increased number of XPD- and XPG-positive cells in the SKOs following DMBA treatment (Fig. 2d–g). Together, these data indicated that the accumulation of DMBA-induced mutations was reduced in Ppp1r8−/− keratinocytes by an increased DNA-damage response as well as an upregulation of rate-limiting DNA-repair factors.
Fig. 2. Enhanced DNA-damage response and repair in SKOs.
a Immunostaining for γH2AX at the indicated time points after DMBA administration. Dapi was used for nuclear staining. Scale bars, 50 µm. b Quantification of γH2AX, as shown in panel a. The data are expressed as means ± SEM (n = 4). c qRT-PCR analysis of NER factors in the back skin of mice not treated (left panel) or treated for 4 h with DMBA (right panel). Hprt was used as a housekeeping gene for normalization. The data are expressed as means ± SD (n = 4). d Immunostaining for XPD in the back skin of mice treated for 24 h with DMBA. Arrows indicate positive stained cells. Dapi was used as a nuclear counterstain. Scale bars, 50 µm. e Quantification of the data in panel (d). The data are expressed as means ± SD (n = 4). f Immunostaining for XPG in the back skin mice treated for 24 h with DMBA. Arrows indicate positive stained cells. Dapi was used as a nuclear counterstain. Scale bars, 50 µm. g Quantification of the data in panel (f). The data are expressed as means ± SD (n = 4). *p < 0.05; **p < 0.01; ***p < 0.001 in unpaired Student’s t-test.
Resistance of Ppp1r8−/− livers to DEN-induced hepatocellular carcinoma
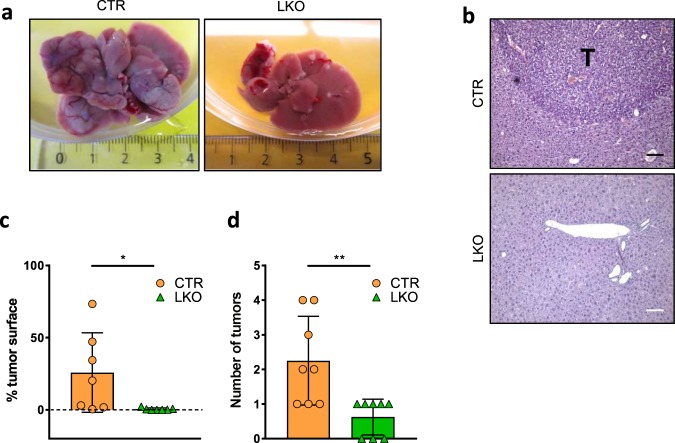
Next, we asked whether the resistance of Ppp1r8−/− cells to mutagenesis also applies to an unrelated mouse model of induced tumorigenesis. For that reason, we compared the response of liver control mice (CTRs; Ppp1r8fl/+/Tg(Alfp-Cre)) and NIPP1 liver knockout mice (LKOs; Ppp1r8fl/−/Tg(Alfp-Cre)) to chemically induced hepatocellular carcinoma (HCC). In the LKOs, the second floxed Ppp1r8 allele was selectively disrupted in hepatoblast-derived epithelial cells, i.e., hepatocytes, cholangiocytes and their bipotential liver progenitor cells, by a Cre recombinase under control of the albumin promoter with enhancer elements from both the albumin and α-fetoprotein genes14. HCC was induced with a single intraperitoneal injection of the alkylating agent diethylnitrosamine (DEN). In accordance with published data26–28, numerous tumor nodules were macroscopically detected after 10 months in all CTR livers (Fig. 3a). H&E stainings showed that ≈25% of the liver sections of CTR mice consisted of tumor tissue (Fig. 3b, c). In contrast, few nodules appeared in the LKOs and these were generally much smaller (Fig. 3b, d).
Fig. 3. Ppp1r8−/− livers are less sensitive to DEN-induced hepatocellular carcinoma.
a Representative picture of CTR and LKO livers 10 months after a single intraperitoneal injection of DEN (25 mg/kg) in male mice of 14 days. b H&E stainings of the DEN-treated livers. Scale bar, 100 µm; T, tumor tissue. c The tumor surface area in DEN-treated livers was quantified on H&E-stained sections. The data are expressed as means ± SD (n = 7). d Number of tumors, as detected in on H&E sections (n = 8). All Data are expressed as means ± SD. *p < 0.05; **p < 0.01 (unpaired Student’s t-test).
Increased expression of the DNA-alkylation repair enzyme MGMT in Ppp1r8−/− cells
A metabolite of DEN causes HCC because it ethylates guanine (G) at O6 or N7 and adenine (A) at O327. The predominant mutagenic lesion caused by DEN is an ethylation of guanine at O6. O6-ethylguanine base pairs with thymine (T). Hence, if the alkylation of a guanine remains unrepaired it generates a G → A transition during the subsequent round of DNA replication. Using an antibody that is specific for alkylated O6-guanine, we found that LKO mice were protected from acute DEN-mediated guanine alkylation (Fig. 4a, b), consistent with their much reduced tumor burden 10 months later (Fig. 3). A reduced alkylation can be due to a deficient conversion of DEN into its alkylating metabolite and/or more efficient alkylation repair. Biotransformation of DEN involves its cytochrome P450-mediated hydroxylation, resulting in the release of acetaldehyde and a reactive ethyldiazonium ion27. The expression of the P450-encoding Cyp genes was not significantly affected by the deletion of NIPP1, except for a small increase in the expression of Cyp1a2 (suppl. Fig. S2a), hinting at a normal biotransformation potential. Therefore, we explored whether the LKOs showed a more efficient ethylation repair. Alkylated O6-guanines are mainly repaired by the suicide enzyme O6-methylguanine-DNA methyltransferase (MGMT)29. qRT-PCR analysis showed that the Mgmt transcript level was increased by 50–100% in both untreated (Fig. 4c) and DEN-treated LKOs (Fig. 4d). The increased Mgmt transcript in LKOs was validated in the previously published RNA sequencing data14. The catalytic activity of MGMT was also significantly increased following the depletion of NIPP1 (Fig. 4e). The increased ethylation-repair potential of the LKOs was confirmed by sequencing of randomly selected gene fragments from three genes, which identified a 75% reduction in DEN-induced G → A conversions in the LKOs (Fig. 4f). Importantly, the number of another type of DEN-induced mutation (A → T transversions), which is not repaired by MGMT, was similar in CTRs and LKOs. This represents further evidence for a normal biotransformation of DEN in the LKOs. Collectively, these data indicated that DEN was equally efficient in the ethylation of DNA in CTR and LKO mice, but that O6-ethylated guanines were more efficiently repaired in the LKOs because of an increased expression of MGMT.
Fig. 4. Increased expression of MGMT in liver cells upon depletion of NIPP1.
a O6-alkyl guanine levels were measured by dot-blot assays on genomic DNA isolated from livers of male 14-days-old CTR and LKO mice that had been injected intraperitoneally with 25 mg/kg DEN 48 h before sacrifice (n = 4). Genomic liver DNA was loaded onto a nitrocellulose blotting membrane and probed with an antibody against O6-alkyl guanine (Squarix, EM2-3). b Quantification of the O6-alkyl guanine levels. Hprt, as determined by qPCR, was used for normalization. The data are expressed as means ± SD (n = 6). c, d Mgmt transcript levels were measured by qRT-PCR in the livers of untreated CTR and LKO mice (n ≥ 3) of two weeks (c) and 48 h after a DEN injection (25 mg/kg) in 14-days-old CTR and LKO mice (d; n ≥ 6). Hprt was used for normalization. The data are expressed as means ± SD. Total RNA was isolated from snap-frozen mouse livers using the GenElute®Mammalian Total RNA Miniprep kit (Sigma). e MGMT activity in liver extracts from untreated CTR and LKO mice of two weeks old. MGMT activity was measured according to manufacturer guidelines (MD0100, Sigma-Aldrich). The data are expressed as means ± SD (n ≥ 3). Immunoblotting with Tata-binding protein (TBP; Abcam, ab51841) was used for normalization. f Graphic display of mutations in the indicated gene fragments, 1 month after a single injection of DEN (25 mg/kg) in male CTR and LKO mice of 14 days. Proofreading PCR for Hnf4a, Ctnnb1, and Hras was performed on genomic liver DNA from 4 CTR and 4 LKO mice. PCR products were cloned into the pGEM®-T easy Vector System (Promega) and processed for sequencing. All observed mutations are indicated in the figure. g Negative correlation between PPP1R8 and MGMT mRNA expression levels in human liver samples with hepatocellular carcinoma. The graph was derived from data generated by the TCGA Research Network (http://cancergenome.nih.gov/ and www.cbioportal.org). h, i HepG2 cells were treated during 72 h with control (Ctr) siRNA or two independent NIPP1 siRNAs (NIPP1 KD, siRNA#1 or siRNA#2). Knockdown efficiency and MGMT expression were analyzed by qRT-PCR. HPRT was used for normalization. All data are means ± SD (n ≥ 4). *p < 0.05; **p < 0.01; ***p < 0.001 (unpaired Student’s t-test).
Interestingly, database mining also revealed a negative correlation between the transcript levels of PPP1R8 and MGMT in human HCC (Fig. 4g), indicating that NIPP1 also opposes the expression of MGMT in human liver. Consistent with this notion, the knockdown of PPP1R8 in the human HCC cell line HepG2 with two different siRNAs was associated with a significantly increased level of the MGMT transcript, albeit to a somewhat different extent (Fig. 4h, i). Immunoblotting analysis after the siRNA-mediated knockdown of NIPP1 in human HAP1 cells also revealed an increased MGMT level (Fig. S2b, c). Hence, the regulation of MGMT by NIPP1 is conserved in man.
Discussion
Here we have demonstrated that the deletion of NIPP1 in epithelial cells of mouse liver or skin confers resistance to chemically induced mutagenesis and carcinogenesis. This finding was unexpected as the SKO and LKO models develop a spontaneous hyperproliferation phenotype, which is usually associated with an increased sensitivity to tumorigenesis30,31. Our data identify NIPP1 as a repressor of DNA repair, which is consistent with our previous finding that overexpression of NIPP1, as a fusion with PP1, causes a spontaneous accumulation of DNA damage3. Different types of DNA lesions evoke distinct DNA-repair mechanisms25. In the present study, we focused on DNA damage repaired through the MGMT or NER pathways. However, PP1-NIPP1 is also implicated in the repair of DNA double-strand breaks3, indicating that NIPP1 is a more general repressor of DNA repair. The resistance of the NIPP1 SKOs and LKOs to genotoxic stress correlated with an enlarged activation of the DNA-damage response as well as an increased expression of DNA-repair factors, two key determinants of the DNA-repair capacity.
The mechanism underlying the increased DNA-repair capacity of the NIPP1 SKOs and LKOs is currently unclear. We speculate that the loss of NIPP1 results in the hyperphosphorylation and activation of FHA ligands known to promote DNA-damage signaling, expression of DNA-repair proteins or DNA repair itself (see Introduction for references). However, since NIPP1 acts as both an inhibitor and activator of associated PP132,33, it cannot be excluded that the deletion of NIPP1 actually reduces the phosphorylation of (some) FHA ligands. In addition, hitherto unidentified FHA ligands may be involved in the NIPP1-mediated response to DNA damage. Finally, we cannot rule out a more indirect mechanism. For example, the hair-follicle stem cells in the SKOs are less abundant and show a reduced stemness15. Since hair-follicle stem cells are the primary target for the induction of papillomas by DMBA/TPA34, it seems possible that the SKOs do not provide the stem-like state that is required to induce and propagate the oncogenic Hras mutation in codon 61.
The identification of NIPP1 as a general inhibitor of the DNA-repair capacity turns this protein into a potential target for therapeutic intervention. Patients with Xeroderma pigmentosum (XP) are extremely sensitive to UV-induced skin cancer, mostly because of an inactivating mutation of a key component of the NER pathway35. Our data suggest that the inhibition or removal of NIPP1 may offer resistance to UV-induced mutations for a subset of Xeroderma pigmentosum patients with hypomorphic mutations in NER pathway components. Conversely, induction of DNA damage through activation of NIPP1 is a possible new strategy for cancer therapy, as such or in combination with clinically used DNA-damaging treatments.
In conclusion, using two distinct mouse models for induced tumorigenesis, we have demonstrated that the loss of NIPP1 increases the DNA-repair capacity. This brings NIPP1 in the limelight as a novel target for cancer prevention or treatment therapies.
Materials and methods
Detailed information on the animal models, chemically induced carcinogenesis models, Quantitative reverse transcriptase PCR, quantification of the genomic mutations and histological, immunohistochemical and statistical analyses is available in the supplementary section. Primers for genotyping and for q(RT)-PCR and antibodies are listed in supplementary Tables 1–3.
Supplementary information
Acknowledgements
Fabienne Withof, Maud De Meyer, and Annemie Hoogmartens provided expert technical assistance. Myriam Baes is acknowledged for providing access to the microtome. Iris Verbinnen was a research fellow of the FWO-Flanders. This work was supported by the Belgian Foundation Against Cancer, FWO grant G078717N and a KU Leuven BOF-grant (GOA 10/18).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Iris Verbinnen, Shannah Boens
Contributor Information
Aleyde Van Eynde, Email: Aleyde.VanEynde@kuleuven.be.
Mathieu Bollen, Email: Mathieu.Bollen@kuleuven.be.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41389-020-0214-3).
References
- 1.Jagiello I, et al. Nuclear and subnuclear targeting sequences of the protein phosphatase-1 regulator NIPP1. J. Cell Sci. 2000;113:3761–3768. doi: 10.1242/jcs.113.21.3761. [DOI] [PubMed] [Google Scholar]
- 2.Van Eynde A, et al. Molecular cloning of NIPP-1, a nuclear inhibitor of protein phosphatase- 1, reveals homology with polypeptides involved in RNA processing. J. Biol. Chem. 1995;270:28068–28074. doi: 10.1074/jbc.270.47.28068. [DOI] [PubMed] [Google Scholar]
- 3.Winkler, C. et al. Overexpression of PP1-NIPP1 limits the repair capacity of DNA double-strand breaks. J. Cell Sci.131, 1–54 (2018). [DOI] [PubMed]
- 4.Boudrez A, et al. NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J. Biol. Chem. 2000;275:25411–25417. doi: 10.1074/jbc.M001676200. [DOI] [PubMed] [Google Scholar]
- 5.Boudrez A, Beullens M, Waelkens E, Stalmans W, Bollen M. Phosphorylation-dependent interaction between the splicing factors SAP155 and NIPP1. J. Biol. Chem. 2002;277:31834–31841. doi: 10.1074/jbc.M204427200. [DOI] [PubMed] [Google Scholar]
- 6.Vulsteke V, et al. Inhibition of spliceosome assembly by the cell cycle-regulated protein kinase MELK and involvement of splicing factor NIPP1. J. Biol. Chem. 2004;279:8642–8647. doi: 10.1074/jbc.M311466200. [DOI] [PubMed] [Google Scholar]
- 7.Nuytten M, et al. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene. 2008;27:1449–1460. doi: 10.1038/sj.onc.1210774. [DOI] [PubMed] [Google Scholar]
- 8.Maréchal A, et al. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell. 2014;53:235–246. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savage KI, et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell. 2014;54:445–459. doi: 10.1016/j.molcel.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beke L, et al. MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells. Biosci. Rep. 2015;35:e00267. doi: 10.1042/BSR20150194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rondinelli B, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 2017;19:1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- 12.Van Eynde A, et al. The nuclear scaffold protein NIPP1 is essential for early embryonic development and cell proliferation. Mol. Cell Biol. 2004;24:5863–5874. doi: 10.1128/MCB.24.13.5863-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira, M. et al. The protein phosphatase 1 regulator NIPP1 is essential for mammalian spermatogenesis. Sci. Rep.7, 1–13 (2017). [DOI] [PMC free article] [PubMed]
- 14.Boens S, et al. Brief Report: The deletion of the phosphatase regulator NIPP1 causes progenitor cell expansion in the adult liver. Stem Cells. 2016;34:2256–2262. doi: 10.1002/stem.2375. [DOI] [PubMed] [Google Scholar]
- 15.Verbinnen, I. et al. The phosphatase regulator NIPP1 restrains chemokine-driven skin inflammation. J. Invest. Dermatol. (2020). 10.1016/j.jid.2020.01.008 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 16.Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: Fundamentals and applications. Nat. Protoc. 2009;4:1350–1362. doi: 10.1038/nprot.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolba R, Kraus T, Liedtke C, Schwarz M, Weiskirchen R. Diethylnitrosamine (DEN)-induced carcinogenic liver injury in mice. Lab Anim. 2015;49:59–69. doi: 10.1177/0023677215570086. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz M, Münzel PA, Braeuning A. Non-melanoma skin cancer in mouse and man. Arch. Toxicol. 2013;87:783–798. doi: 10.1007/s00204-012-0998-9. [DOI] [PubMed] [Google Scholar]
- 19.Modi BG, et al. Langerhans cells facilitate epithelial DNA damage and squamous cell carcinoma. Science. 2012;335:104–108. doi: 10.1126/science.1211600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nassar D, Latil M, Boeckx B, Lambrechts D, Blanpain C. Genomic landscape of carcinogen-induced and genetically induced mouse skin squamous cell carcinoma. Nat. Med. 2015;21:946–954. doi: 10.1038/nm.3878. [DOI] [PubMed] [Google Scholar]
- 21.Miyata M, et al. Targeted disruption of the microsomal epoxide hydrolase gene. Microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J. Biol. Chem. 1999;274:23963–23968. doi: 10.1074/jbc.274.34.23963. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Lauer FT, Dunaway S, Burchiel SW. Cytochrome P450 1B1 is required for 7,12-dimethylbenz(a)-anthracene (DMBA) induced spleen cell immunotoxicity. Toxicol. Sci. 2005;86:68–74. doi: 10.1093/toxsci/kfi176. [DOI] [PubMed] [Google Scholar]
- 23.Rundhaug JE, Fischer SM. Molecular mechanisms of mouse skin tumor promotion. Cancers. 2010;2:436–482. doi: 10.3390/cancers2020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quesnot N, et al. Evaluation of genotoxicity using automated detection of γH2AX in metabolically competent HepaRG cells. Mutagenesis. 2016;31:43–50. doi: 10.1093/mutage/gev059. [DOI] [PubMed] [Google Scholar]
- 25.Marteijn Ja, Lans H, Vermeulen W, Hoeijmakers JHJ. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 26.Heindryckx F, Colle I, Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. Int. J. Exp. Pathol. 2009;90:367–386. doi: 10.1111/j.1365-2613.2009.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verna L, Whysner J, Williams GM. N-Nitrosodiethylamine mechanistic data and risk assessment: Bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol. Therapeutics. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 28.Lim IK. Spectrum of molecular changes during hepatocarcinogenesis induced by DEN and other chemicals in Fischer 344 male rats. Mech. Ageing Dev. 2002;123:1665–1680. doi: 10.1016/S0047-6374(02)00087-8. [DOI] [PubMed] [Google Scholar]
- 29.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong W, et al. Skin hyperproliferation and susceptibility to chemical carcinogenesis in transgenic mice expressing E6 and E7 of human papillomavirus type 38. J. Virol. 2005;79:14899–14908. doi: 10.1128/JVI.79.23.14899-14908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simanainen U, et al. Androgen resistance in female mice increases susceptibility to DMBA-induced mammary tumors. Horm. Cancer. 2012;3:113–124. doi: 10.1007/s12672-012-0107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minnebo N, et al. NIPP1 maintains EZH2 phosphorylation and promoter occupancy at proliferation-related target genes. Nucleic Acids Res. 2013;41:842–854. doi: 10.1093/nar/gks1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferreira M, Verbinnen I, Fardilha M, Van Eynde A, Bollen M. The deletion of the protein phosphatase 1 regulator NIPP1 in testis causes hyperphosphorylation and degradation of the histone methyltransferase EZH2. J. Biol. Chem. 2018;293:18031–18039. doi: 10.1074/jbc.AC118.005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sánchez-Danés A, Blanpain C. Deciphering the cells of origin of squamous cell carcinomas. Nat. Rev. Cancer. 2018;18:549–561. doi: 10.1038/s41568-018-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugasawa K. Xeroderma pigmentosum genes: functions inside and outside DNA repair. Carcinogenesis. 2008;29:455–465. doi: 10.1093/carcin/bgm282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.