Abstract
Tuberculosis remains one of the leading causes of death from a single pathogen globally. It is estimated that 1/4 of the world’s population harbors latent tuberculosis, but only a 5–10% of patients will develop active disease. During latent infection, Mycobacterium tuberculosis can persist unaffected by drugs for years in a non-replicating state with low metabolic activity. The rate of the successful tuberculosis treatment is curbed by the presence of these non-replicating bacilli that can resuscitate after decades and also by the spread of M. tuberculosis drug-resistant strains. International agencies, including the World Health Organization, urge the international community to combat this global health emergency. The thienopyrimidine TP053 is a promising new antitubercular lead compound highly active against both replicating and non-replicating M. tuberculosis cells, with an in vitro MIC of 0.125 μg/ml. TP053 is a prodrug activated by the reduced form of the mycothiol-dependent reductase Mrx2, encoded by Rv2466c gene. After its activation, TP053 releases nitric oxide and a highly reactive metabolite, explaining its activity also against M. tuberculosis non-replicating cells. In this work, a new mechanism of TP053 resistance was discovered. M. tuberculosis spontaneous mutants resistant to TP053 were isolated harboring the mutation L240V in Rv0579, a protein with unknown function, but without mutation in Rv2466c gene. Recombineering method demonstrated that this mutation is linked to TP053 resistance. To better characterize Rv0579, the protein was recombinantly produced in Escherichia coli and a direct interaction between the Mrx2 activated TP053 and Rv0579 was shown by an innovative target-fishing experiment based on click chemistry. Thanks to achieved results, a possible contribution of Rv0579 in M. tuberculosis RNA metabolism was hypothesized, linked to toxin anti-toxin system. Overall, these data confirm the role of Rv0579 in TP053 resistance and consequently in the metabolism of this prodrug.
Keywords: Rv0579, drug resistance, prodrug, tuberculosis, antitubercular drug
Introduction
Tuberculosis (TB), an old disease considered tied to the past, started again capturing the attention for different reasons, such as the spread of Mycobacterium tuberculosis drug-resistant strains (Koch et al., 2018). In the world, in 2018 about 500,000 people were infected by M. tuberculosis strains resistant to the most effective first-line drug rifampicin (RR-TB), and 78% of these isolates were multidrug-resistant (MDR-TB). Three countries accounted for almost half of the world’s cases of MDR-TB: India, China and the Russian Federation (World Health Organization [WHO], 2019). In order to fight drug-resistant TB, the treatment options have become limited, thus stressing the need for new therapies.
Another important threat is latent TB (LTB); according to WHO, LTB corresponds to a persistent immune response to stimulation by M. tuberculosis antigens without evidence of active TB (World Health Organization [WHO], 2018). The possible progression of LTB to active disease is a complex condition determined by bacterial, host, and environmental factors (Getahun et al., 2015).
About one-quarter of the world population develops LTB and it is considered as a persistent reservoir of active infection. Active TB will develop in 5–15% of individuals with LTB during their lifetimes. Several comorbidities could be related with increased risk of developing active TB, such as the HIV co-infection (Furin et al., 2019; Huaman and Sterling, 2019). Consequently, an effective treatment for LTB should be mandatory.
Current recommended LTB treatments include one of the following options: once-weekly isoniazid plus rifapentine for 3 months, daily rifampin for 4 months, daily isoniazid plus rifampin for 3–4 months, and daily isoniazid for 6–9 months (Huaman and Sterling, 2019). Treatments based on rifamycins seem to cause less adverse events and, consequently, to improve the therapy completion (Villa et al., 2019).
New and more effective drugs against LTB with fewer adverse events are needed in order to prevent active TB and to shorten treatment duration.
In this context, the thienopyrimidine (TP) TP053 is a new promising prodrug killing both replicating (MIC = 0.125 μg/ml) and non-replicating (MIC = 0.8 μg/ml) M. tuberculosis cells (Albesa-Jové et al., 2014). By microbiological, genetic, biochemical and crystallographic studies, its mechanism of activation was elucidated; TP053 is a prodrug activated by the reduced form of the DsbA-like mycoredoxin Mrx2 (encoded by Rv2466c gene), a mycothiol-dependent reductase with an unusual active-site motif CXXC (Albesa-Jové et al., 2014; Rosado et al., 2017). Mrx2 utilizes a chaperone-like mechanism of conformational changes, mainly in the CXXC active site motif, to recognize TP053 and promoting compound reduction (Albesa-Jové et al., 2015).
Recently, it has been demonstrated that Mrx2 works as a nitroreductase causing a release of nitric oxide (NO) from TP053 (Chiarelli et al., 2020). Thus, conceivably TP053 globally affects M. tuberculosis cell growth by NO release, which could be the cause of its activity against non-replicating bacilli, in a similar way to the NO releasing drug pretomanid, upon Ddn activation (Singh et al., 2008).
The study of the M. tuberculosis metabolic pathways affected by TP053 exposure, through transcriptional analysis, revealed features consistent with NO release, confirming the characteristic mechanism of action of TP053 against non-replicating bacilli (Chiarelli et al., 2020). Furthermore, it was identified a highly reactive metabolite, 2-(4-mercapto-6-(methylamino)-2-phenylpyrimidin-5-yl)ethan-1-ol, produced upon Mrx2 transformation of TP053, which could be co-responsible for the antimycobacterial effects on replicating and non-replicating M. tuberculosis cells (Chiarelli et al., 2020). However, no potential target of this active metabolite has been identified so far.
In this work, we demonstrated that the Rv0579 protein has a role in the mechanism of resistance of TP053, even if its physiological role in M. tuberculosis still remains unclear.
Materials and Methods
Bacterial Strains and Growth Conditions
Cloning steps were performed in Escherichia coli XL1-Blue, following standard methods (Sambrook and Russell, 2001) and using the oligonucleotides described in Supplementary Table S1. Protein expression was achieved in E. coli BL21(DE3).
Mycobacterium tuberculosis H37Rv and mutant strains were grown aerobically at 37°C either in Middlebrook 7H9 broth (Difco) or on Middlebrook 7H11 agar (Difco), both supplemented with 10% OADC Middlebrook Enrichment. When necessary, antibiotics were added at the following concentrations: ampicillin 100 μg/ml, hygromycin 200 μg/ml (20 μg/ml for M. tuberculosis), and kanamycin 50 μg/ml (20 μg/ml for M. tuberculosis).
All the experiments with M. tuberculosis were performed in Biosafety level 3 laboratory by authorized and trained researchers.
MIC Determination
MICs for the compounds were determined by means of the micro-broth dilution method. Dilutions of M. tuberculosis wild-type and mutant cultures (about 105–106 CFU/ml) were streaked onto 7H11 solid medium containing a range of drug concentrations. Plates were incubated at 37°C for about 21 days and the growth was visually evaluated. The lowest drug dilution at which visible growth failed to occur was taken as the MIC value. Results were expressed as the average of at least three independent replicates.
Isolation and Characterization of M. tuberculosis Mutants Resistant to TP053
The isolation of M. tuberculosis mutants was performed by plating ∼1010 cells from an exponential growth phase culture of M. tuberculosis H37Rv onto 7H11 medium containing different concentrations of TP053, ranging from 5- to 40-fold the MIC of the wild-type strain. Genomic DNA of M. tuberculosis resistant mutants was isolated and sequenced by using Illumina HiSeq2000 technology at IGA Technology Services S.R.L. (Udine, Italy). For the bioinformatic analysis of Illumina data, repetitive PE and PPE gene families were discarded as well as SNPs and Indels with less than 50% probability. The mutations identified were confirmed by Sanger sequencing (Eurofins Genomics), after PCR amplification using the primers described in Supplementary Table S1. PCR products were purified using the Wizard® SV Gel and PCR Clean-Up system (Promega).
Recombineering Method
Recombineering method utilizing mycobacteriophage-encoded functions was used in order to confirm the role of Rv0579 mutation in TP053 resistance, as previously described (van Kessel and Hatfull, 2008). Briefly, a M. tuberculosis strain that carries the recombineering plasmid pJV62 was induced by acetamide in mid-logarithmic growth and electrocompetent cells were prepared. Electroporation was performed with different concentrations (100 and 500 ng) of Rv0579rec oligonucleotide (Supplementary Table S1), which anneals to the template for discontinuous DNA synthesis (lagging strand), thus increasing recombination frequency. The introduced point mutation was co-selected by plating the transformations onto a 7H11 plate containing TP053 (1.25 and 5 μg/ml corresponding to 10× and 40× MIC, respectively) and kanamycin (20 μg/ml).
Expression and Purification of M. tuberculosis Rv0579
Mycobacterium tuberculosis H37Rv Rv0579 gene from genomic DNA was amplified by standard PCR, using the primers reported in Supplementary Table S1, and PCR fragments were cloned in the pET-SUMO vector (Invitrogen), to give pET-SUMO/Rv0579 recombinant plasmid. Rv0579 enzyme was produced in E. coli BL21(DE3), by overnight induction with 0.5 mM isopropyl-β-thiogalactopyranoside (IPTG) at 25°C. Thereafter, cells were harvested, re-suspended in 150 ml of buffer A (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 0.5 mM dithiothreitol (DTT), 5% glycerol, 2 mM MgCl2) containing 1 mM PMSF, disrupted by sonication and centrifuged for 30 min at 50,000 × g. The supernatant was loaded on HisTrap Crude column (1 ml, GE Healthcare), washed with 50 ml of buffer A containing 50 mM of imidazole, then Rv0579 elution was performed with 250 mM of imidazole in buffer A. The eluted protein was dialyzed using a HiPreP Desalting 26/10 column equilibrated in 50 mM Tris–HCl pH 7.5, 50 mM imidazole, 100 mM NaCl, 0.5 mM DTT, 2 MgCl2, 5% glycerol, incubated at 4°C in the presence of 15 μl SUMO-protease and then re-purified on HisTrap Crude column. The purified protein was checked by SDS-PAGE, concentrated to 5 mg/ml, and stored at −80°C. The protein concentration was determined by absorbance at 280 nm (ε: 21,680 M–1 cm–1).
Target Fishing
To demonstrate a direct interaction between Rv0579 protein and TP053, a TP053-azide-PEG3-biotin conjugate complex was used.
The 11526119 derivative of TP053, prepared as described in Supplementary Material, was firstly characterized to verify that it showed the same features as TP053. For this reason, Mrx2 enzymatic activity using 11526119 as substrate was assayed as previously described (Albesa-Jové et al., 2014), to confirm that this compound is activated like TP053.
Then, 11526119 was used to perform azide-alkyne cycloaddiction reaction with Azide-PEG3-biotin conjugate, to form an adduct which can be immobilized on a streptavidin agarose resin (Supplementary Figure S1). To this purpose, in a final volume of 500 μl, 11526119 (200 μM) was incubated with 2-fold excess of Azide-PEG3-biotin conjugate, in the presence of 250 μM of CuSO4 and 5 mM Na-ascorbate, overnight at room temperature. The reaction was checked by thin layer chromatography, then incubated 1 h with Streptavidine Agarose resin, and washed with phosphate saline buffer (PBS) to remove any unbound material.
The complex was incubated with Rv0579 (0.5 mg/ml) in a final volume of 200 μl, in the presence of Mrx2 (2 mg/ml, prepared according to Albesa-Jové et al., 2014), DTT (1 mM) and Mycobacterium smegmatis methanolic extract (2%) at 37°C for 30 min, then incubated 1 h with Streptavidine Agarose resin. After the incubation, the resin was washed five times with PBS to remove unbound proteins and then analyzed in SDS-PAGE. As negative control, the same incubation was performed with the only streptavidine-agarose resin, or in absence of M. smegmatis methanolic extract, to avoid Mrx2 activation.
Quantitative Real-Time PCR
Quantitative Real-Time PCR was performed on RNA extracted from M. tuberculosis H37Rv and Rv0579 mutant strains, both treated with TP053 (0.06 μg/ml; 0.5× MIC) or untreated using the Direct-zolTM RNA MiniPrep Kit (Zymo Research). The cDNA was obtained from about 1 μg of total RNA by the Quantitec reverse transcription kit (Qiagen). The RT-PCR experiments were performed using the QuantiTect SYBR Green PCR Master Mix kit (Qiagen) and the “Rotor Gene 6000” thermocycler (Corbett Life Science).
The reaction was carried out in a final volume of 15 μl and contained: 7.5 μl of 2× SYBR Green Buffer (supplied by the kit), cDNA, primers (7.5 pmol) and RNase-free water. All reactions were repeated in triplicate and the mean value was considered. The primers used to assess the transcriptional analysis of sigA and Rv0579 genes are present in Supplementary Table S1.
Results
The Mutation in Rv0579 Gene Is Responsible for TP053 Resistance
To identify the TP053 cellular target(s), two M. tuberculosis mutants resistant to TP053 (MIC = 2.5 μg/ml, 20× MIC of wild type strain) have been isolated using a M. tuberculosis culture overexpressing mrx2 gene, coding for the activator (M. tuberculosis/pSODIT-mrx2) (Albesa-Jové et al., 2014). In this way, the selection of mutation(s) in mrx2 could be avoided. After the confirmation of the TP053 resistance, to find the mutation(s) responsible for TP053 resistance, whole genome sequencing (WGS) of the new isolated resistant mutants was performed using Illumina method (Table 1). Interestingly, M. tuberculosis 2466.3 and 2466.4 mutants revealed the same mutation in Rv0579 gene (C718G → L240V), coding for a conserved hypothetical protein (Table 1). The presence of the mutation in the Rv0579 gene was confirmed by Sanger sequencing. No mutation was detected in mrx2 gene on either the chromosome or the plasmid.
TABLE 1.
Characteristics of the M. tuberculosis spontaneous TP053 resistant mutants.
| M. tuberculosis strains | Concentration of isolation of TP053 resistant colonies (μg/ml) |
Mutation in mrx2 |
Mutation in Rv0579 (aa change) | MIC to TP53 (μg/ml) | |
| On genome | On plasmid | ||||
| M. tuberculosis/pSODIT/mrx2 | – | – | – | – | 0.125 |
| 2466.3 mutant | 0.5 (4× MIC) | No | No | C718G (L240V) | 2.5 (20× MIC) |
| 2466.4 mutant | 1 (8× MIC) | No | No | C718G (L240V) | 2.5 (20× MIC) |
The specific contribution of the mutation in Rv0579 to TP053 resistance was confirmed by recombineering method (van Kessel and Hatfull, 2008) using different TP053 concentrations (10× and 40× MIC of the wild type strain), thus allowing the generation of the C718G substitution in chromosomal Rv0579 of wild-type H37Rv (Supplementary Table S2). The generated TP053 resistant colonies were screened for the introduction of the Rv0579 C718G substitution by Sanger sequencing and their TP053 resistance profile confirmed by MIC determination. None of the recombineering-generated mutants harboured mutations in mrx2. In every tested mutant, the only presence of Rv0579 mutation (C718G) conferred TP053 resistance (MIC = 2.5 μg/ml, 20× MIC of wild type strain). These results confirmed the role of Rv0579 in the mechanism of resistance to TP053.
Thus, the possibility that the resistance to TP053 might be due to the overexpression of Rv0579 gene was excluded by quantitative PCR of Rv0579 transcripts from both M. tuberculosis wild-type and TP053 resistant (Rec5 mutant obtained by recombineering) strains, in presence/absence of prodrug; in fact, no significant difference in the expression of Rv0579 was observed (Supplementary Table S3).
Rv0579 Protein Is Able to Directly Interact With the Activated TP053
In order to further investigate the role of Rv0579 in the TP053 resistance, the Rv0579 protein was recombinantly produced in E. coli and purified to homogeneity (Supplementary Figure S2). The produced protein was found to be bound to nucleic acids. Subsequent DNase and RNase digestion revealed RNA to be the main contaminant (Supplementary Figure S3). However, an efficient digestion of RNA was achieved only after partial denaturation of the protein, indicating a strong interaction. For this reason, DNase and RNase were added in the lysis buffer (100 μl, 2 mg/ml) for additional purification steps. Despite several attempts using different chromatographical approaches, the nucleic acids were not completely removed in any case (data not shown).
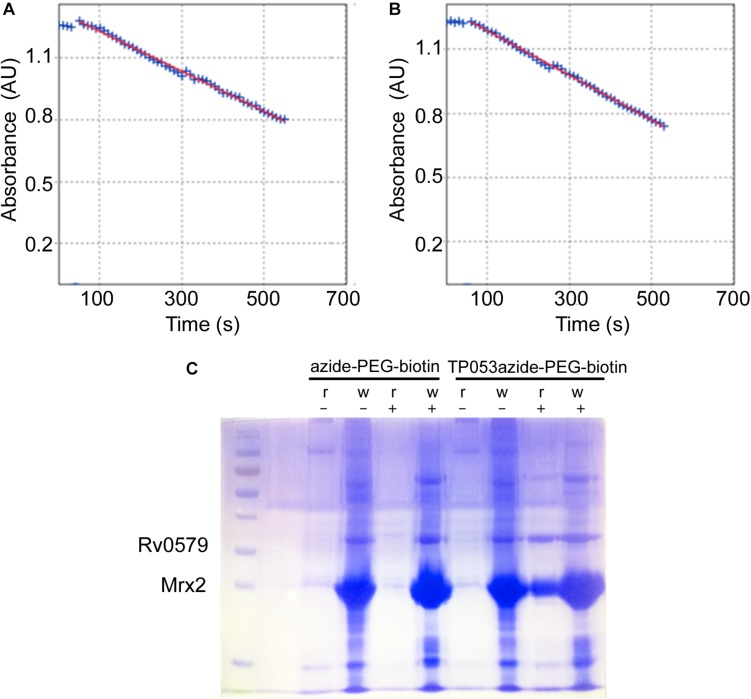
The achieved recombinant protein was then exploited to demonstrate a possible direct interaction between Rv0579 and the activated TP053 through a target-fishing experiment based on click chemistry (Figure 1). For this experiment, a TP053 derivative (11526119; MIC = 0.5 μg/ml) (Supplementary Material) was used, which was shown to retain TP053 ability to be activated by Mrx2 by an in vitro enzymatic assay (Figures 1A,B). Consequently, the Rv0579 protein was incubated with a TP053azide-PEG3-biotin conjugate complex in presence of the activator Mrx2, then bound on Streptavidin Agarose resin. After several washing steps, the resin was analyzed by SDS-PAGE (Figure 1C). As control, the same reaction was performed also in the absence of mycothiols, to avoid Mrx2 activation, or using the only azide-PEG3-biotin.
FIGURE 1.
Target fishing experiments. (A,B) The enzymatic activity assay of Mrx2 using 11526119 as a substrate (A), was compared with the assay using TP053 (B) demonstrating that the compound retaining the same capability to be metabolized by the enzyme. Then, it was suitable for target fishing experiments. (C) SDS-PAGE analysis of the target fishing experiments. The streptavidine-agarose resin was incubated 1 h with the TP53-azide-PEG-biotin, or with azide-PEG-biotin complex, reacted with Rv0579. After the incubation the samples were washed with buffer, and both resin and wash fractions analyzed by SDS-PAGE. r, resin; w, wash; +, incubation with Rv0579 in the presence of mycothiols, to allow Mrx2 activation; –, incubation with Rv0579 without mycothiols as blank control. Rv0579 did not show any reactivity with the streptavidine-agarose resin alone, since the protein was found only in wash fractions (lanes 5 and 6). By contrast, Rv0579 was found bound the resin-TP053 complex, but only after Mrx2 activation (lane 9). This experiment demonstrates a direct interaction between the protein and the activated compound, which confirms a role of Rv0579 in resistance to TP053.
The obtained data demonstrated that the activated prodrug binds not only to Mrx2, as expected, but also to Rv0579 (Figure 1C), indicating a direct interaction between the activated TP053 and Rv0579.
These results confirm the role of Rv0579 in the mechanism of TP053 resistance.
Understanding the Role of Rv0579 in TP053 Resistance
Previous RNA-seq experiments demonstrated that the exposure to TP053 (6 μg/ml; ∼50× MIC) of M. tuberculosis cells caused an over-expression of some toxin-antitoxin (TA) systems (e.g.: HigAB, VapBC16) (Chiarelli et al., 2020).
Interestingly, on M. tuberculosis genome, Rv0579 is located near to Rv0581 (coding for the antitoxin VapB26) and Rv0582 (coding for the toxin VapC26) (Kang et al., 2017). Taking advantage of the availability of our Rv0579 mutant (Rec5 mutant obtained by recombineering), we performed Real-Time PCR using both M. tuberculosis wild type and Rv0579 mutant cultures treated with TP053 (0.06 μg/ml; 0.5× MIC) (Supplementary Table S3) to investigate possible changes in the expression levels of the genes encoding VapBC26 TA system. Untreated cultures were used as control.
Surprisingly, only in M. tuberculosis wild type strain an up-regulation of the gene encoding the VapC26 toxin was found; in the Rv0579 mutant the Rv0582 over-expression was not detected (Supplementary Table S3). These results suggest a possible link between Rv0579 and VapBC26 system.
Since Rv0579 protein possesses the two domains Ub-Mut7C and Mut7-C, usually related to RNase activity (Marchler-Bauer et al., 2017), we hypothesised that the link with VapBC26 could involve this possible activity. Unfortunately, we were not able to determine it, because of the nucleic acid contamination of the purified protein (data not shown). Further experiments to support this hypothesis currently were limited until this issue is solved.
Discussion
TP053 is a very promising compound active against replicating and non-replicating M. tuberculosis cells with a peculiar mechanism of action (Albesa-Jové et al., 2014; Chiarelli et al., 2020). In fact, this prodrug is activated by Mrx2 producing an active metabolite by NO release. Furthermore, TP053 inhibits M. tuberculosis cell growth by influencing several metabolic pathways such as RNA metabolism (Chiarelli et al., 2020).
In this work, a new mechanism of resistance to TP053 was discovered. M. tuberculosis spontaneous mutants resistant to TP053 were isolated harboring a mutation in Rv0579 gene. The contribution of this mutation to TP053 resistance was confirmed by recombineering method. Moreover, target fishing experiment demonstrated that Rv0579 is able to bind TP053, but only upon Mrx2 activation of the compound.
Rv0579 is a non-essential gene (Griffin et al., 2011), encoding a conserved hypothetical protein with unknown function. The sequence of Rv0579 protein shows two important domains, usually linked to RNase activity: Ub-Mut7C and Mut7-C (Marchler-Bauer et al., 2017) (Supplementary Table S4).
Mut7-C is a RNAse domain of the PIN fold with an inserted zinc ribbon located at the C terminus of the protein (Anantharaman et al., 2002); this domain is typical of VapC toxin (toxin-antitoxin system). The mutation found in TP053 resistant M. tuberculosis mutants is located at the end of this domain. Ub-Mut7C domain is occasionally present at the N-terminus of the protein with the Mut7-C domain, suggesting an RNA-binding role (Iyer et al., 2006) (Supplementary Table S4). In our case, this hypothesis is also corroborated by the finding that the recombinant protein tightly binds RNA (Supplementary Figure S3).
Iyer et al. (2006) identified a group of proteins containing the two domains, Ub-Mut7C and Mut7-C, fused; Rv0579 belongs to this group containing several proteins with unknown function belonging to different bacterial species (Supplementary Figure S4 and Supplementary Table S4). Interestingly, the alignment of these proteins shows that the mutation in Rv0579 responsible for TP053 resistance is present at the end of Mut7-C domain and it corresponds to a change of an amino acid conserved in some bacterial species (L240; Streptomyces coelicolor, Nostoc punctiforme, Nocardia farcinica, Thermobifida fusca) (Supplementary Figure S4). In other tested species, this amino acid changes in Met.
It is noteworthy that TP053 releases NO, consequently affecting RNA metabolism and causing inhibition of respiration, oxidative stress, and DNA damage (Chiarelli et al., 2020). This again suggests that Rv0579 has a role in RNA metabolism, one of the pathways mainly affected by NO release (Darwin et al., 2003; Voskuil et al., 2003; Manjunatha et al., 2009).
From the achieved results, and thanks to a deep analysis of the Rv0579 sequence, a role of Rv0579 in RNA metabolism could be hypothesized, linked to toxin anti-toxin systems (e.g. VapBC26). Indeed, TP053 treatment in wild-type M. tuberculosis caused an up-regulation of the Rv0582 gene encoding the VapC26 toxin, while no effect has been seen in M. tuberculosis resistant mutant carrying mutation in Rv0579. These evidences thus suggest a role of this protein in the regulation of this toxin-antitoxin system.
Even if the physiological role of Rv0579 is still under investigation, this work unveils some important aspects of the protein, paving the way for further evidences that will allow to clearly elucidate its role in M. tuberculosis metabolism and in TP053 resistance.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
GM, BO, LC, and MP designed the study. LC, VM, GR, and MP wrote the manuscript and interpreted the data. GM, BO, LC, GD, OR, and JS performed the experiments. All authors approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. G. F. Hatfull for kindly giving us pJV62 plasmid to perform recombineering method.
Footnotes
Funding. This work was supported by the European Community’s Seventh Framework Program (Grant 260872) and the University of Pavia, Italy (“Universitiamo–Tubercolosi: un killer riemergente”). This work was also supported by the Italian Ministry of Education, University and Research (MIUR): Dipartimenti di Eccellenza Program (2018–2022) – Department of Biology and Biotechnology “L. Spallanzani,” University of Pavia.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00292/full#supplementary-material
References
- Albesa-Jové D., Chiarelli L. R., Makarov V., Pasca M. R., Urresti S., Mori G., et al. (2014). Rv2466c mediates the activation of TP053 to kill replicating and non-replicating Mycobacterium tuberculosis. ACS Chem. Biol. 9 1567–1575. 10.1021/cb500149m [DOI] [PubMed] [Google Scholar]
- Albesa-Jové D., Comino N., Tersa M., Mohorko E., Urresti S., Dainese E., et al. (2015). The redox state regulates the conformation of Rv2466c to activate the antitubercular prodrug TP053. J. Biol. Chem. 290 31077–31089. 10.1074/jbc.M115.677039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V., Koonin E. V., Aravind L. (2002). Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30 1427–1464. 10.1093/nar/30.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarelli L. R., Salina E. G., Mori G., Azhikina T., Riabova O., Lepioshkin A., et al. (2020). New insights into the mechanism of action of the thienopyrimidine antitubercular prodrug TP053. ACS Infect Dis. 6 313–323. 10.1021/acsinfecdis.9b00388 [DOI] [PubMed] [Google Scholar]
- Darwin K. H., Ehrt S., Gutierrez-Ramos J. C., Weich N., Nathan C. F. (2003). The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302 1963–1966. 10.1126/science.1091176 [DOI] [PubMed] [Google Scholar]
- Furin J., Cox H., Pai M. (2019). Tuberculosis. Lancet 393 1642–1656. 10.1016/S0140-6736(19)30308-30303 [DOI] [PubMed] [Google Scholar]
- Getahun H., Chaisson R. E., Raviglione M. (2015). Latent mycobacterium tuberculosis infection. N. Engl. J. Med. 373 1179–1180. 10.1056/NEJMc1508223 [DOI] [PubMed] [Google Scholar]
- Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011). High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251. 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaman M. A., Sterling T. R. (2019). Treatment of latent tuberculosis infection-an update. Clin. Chest. Med. 40 839–848. 10.1016/j.ccm.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L. M., Burroughs A. M., Aravind L. (2006). The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 7:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. M., Kim D. H., Lee K. Y., Park S. J., Yoon H. J., Lee S. J., et al. (2017). Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides. Nucleic Acids Res. 45 8564–8580. 10.1093/nar/gkx489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A., Cox H., Mizrahi V. (2018). Drug-resistant tuberculosis: challenges and opportunities for diagnosis and treatment. Curr. Opin. Pharmacol. 42 7–15. 10.1016/j.coph.2018.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunatha U., Boshoff H. I., Barry C. E., III (2009). The mechanism of action of PA-824: novel insights from transcriptional profiling. Commun. Integr. Biol. 2 215–218. 10.4161/cib.2.3.7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J., Lu S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45 D200–D203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado L. A., Wahni K., Degiacomi G., Pedre B., Young D., de la Rubia A. G. (2017). The antibacterial prodrug activator Rv2466c is a mycothiol-dependent reductase in the oxidative stress response of Mycobacterium tuberculosis. J. Biol. Chem. 292 13097–13110. 10.1074/jbc.M117.797837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Singh R., Manjunatha U., Boshoff H. I., Ha Y. H., Niyomrattanakit P., Ledwidge R., et al. (2008). PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322 1392–1395. 10.1126/science.1164571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel J. C., Hatfull G. F. (2008). Mycobacterial recombineering. Methods Mol. Biol. 435 203–215. 10.1007/978-1-59745-232-8_15 [DOI] [PubMed] [Google Scholar]
- Villa S., Ferrarese M., Sotgiu G., Castellotti P. F., Saderi L., Grecchi C., et al. (2019). Latent tuberculosis infection treatment completion while shifting prescription from isoniazid-only to rifampicin-containing regimens: a two-decade experience in Milan, Italy. J Clin Med. 9:E101. 10.3390/jcm9010101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil M. I., Schnappinger D., Visconti K. C., Harrell M. I., Dolganov G. M., Sherman D. R., et al. (2003). Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198 705–713. 10.1084/jem.20030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2018). Latent TB Infection: Updated and Consolidated Guidelines for Programmatic Management. Geneva: World Health Organization. [PubMed] [Google Scholar]
- World Health Organization [WHO] (2019). Global Tuberculosis Report 2018. Geneva: World Health Organization. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.